Abstract
Gamma-secretase, which is responsible for the intramembranous cleavage of Alzheimer's β-amyloid precursor protein (APP), the signaling receptor Notch, and many other substrates, is a multiprotein complex consisting of at least four components: presenilin (PS), nicastrin, APH-1, and PEN-2. Despite the fact that PEN-2 is known to mediate endoproteolytic cleavage of full-length PS and APH-1 and nicastrin are required for maintaining the stability of the complex, the detailed physiological function of each component remain elusive. Unlike that of PS, the transcriptional regulation of PEN-2, APH-1, and nicastrin has not been investigated. Here, we characterized the upstream regions of the human PEN-2 gene and identified a 238-bp fragment located 353 bp upstream of the translational start codon as the key region necessary for the promoter activity. Further analysis revealed a CREB binding site located in the 238-bp region that is essential for the transcriptional activity of the PEN-2 promoter. Mutation of the CREB site abolished the transcriptional activity of the PEN-2 promoter. Electrophoretic mobility shift assays and chromatin immunoprecipitation analysis showed the binding of CREB to the PEN-2 promoter region both in vitro and in vivo. Activation of the CREB transcriptional factor by forskolin dramatically promoted the expression of PEN-2 mRNA and protein, whereas the other components of the γ-secretase complex remained unaffected. Forskolin treatment slightly increases the secretion of soluble APPα and Aβ without affecting Notch cleavage. These results demonstrate that expression of PEN-2 is regulated by CREB and suggest that the specific control of PEN-2 expression may imply additional physiological functions uniquely assigned to PEN-2.
The γ-secretase complex is a high-molecular-weight complex that consists of at least four components: presenilins (PS) (including PS1 and PS2), nicastrin, APH-1 (anterior pharynx defective 1), and PEN-2 (presenilin enhancer 2) (9, 18, 20). The PS/γ-secretase complex controls proteolytic processing of a number of type 1 transmembrane proteins, including the amyloid precursor protein APP, which can be sequentially cleaved by β-secretase (BACE1) and the PS/γ-secretase complex to generate heterogeneous β-amyloid (Aβ) species. Most of the species are Aβ40 and the more deleterious Aβ42, which are generally believed to cause Alzheimer's disease (AD) (13, 16). The discovery of PS1 and PS2 as major pathogenic genes for familial AD, in addition to mutations in the APP gene (8, 38), demonstrates the importance of PS/γ-secretase in AD pathogenesis. Furthermore, the PS/γ-secretase complex also cleaves other functionally important proteins, such as Notch (10), E-cadherin (28), ErbB4 (31), CD44 (21), etc., suggesting the involvement of PS/γ-secretase in a broad range of biological activities. Interestingly, although knocking out PS (11), nicastrin (24, 25), or APH-1 (27) in mice results in lethality and abnormal embryonic phenotypes which resemble that of Notch null mice (6, 17, 41), the phenotypes among different gene knockouts are not identical, implying that each of these γ-secretase components may have its own unique physiological functions in addition to the γ-secretase activity.
Presenilins undergo endoproteolytic cleavage to generate an amino-terminal fragment (NTF) and a carboxyl-terminal fragment, which form functional PS heterodimers (42). The first identified protein cofactor of PS in the γ-secretase complex, nicastrin, is a type I transmembrane protein that can be highly glycosylated on its ectodomain (47, 48). Two additional PS/γ-secretase components, APH-1 and PEN-2, were identified through genetic screening of Caenorhabditis elegans (12, 15). There is only one PEN-2 homologue in mammals, whereas there are two APH-1 homologues in mammals, identified as APH-1a and APH-1b. Mammalian APH-1a has at least two splice variants: APH-1aL and APH-1aS. Recent studies have demonstrated that the four components of PS/γ-secretase regulate one another in a coordinate way; down-regulation/deficiency of one given component typically results in a decrease in the level of other components and/or a deficiency of their trafficking/maturation (9, 18). In the absence of PS1, PEN-2 is sequestered in the endoplasmic reticulum and not transported to post-endoplasmic reticulum components, where the mature γ-secretase complexes reside (44). PS deficiency also leads to destabilization of PEN-2 (26, 37), which is degraded via the proteasome-mediated pathway (2, 7). On the other hand, down-regulation of PEN-2 by small interfering RNA results in an accumulation of full-length PS1 and a reduction of PS1 fragments, suggesting that PEN-2 is involved in PS1 endoproteolysis (26). Intracellular trafficking and maturation of nicastrin are also PS dependent: in the absence of PS, nicastrin fails to reach the medial Golgi compartment and thus is incompletely glycosylated (23). Moreover, nicastrin deficiency reduces the levels of APH-1, PEN-2, and PS1 fragments (24, 25, 49), while in APH-1a knockout cells, the levels of nicastrin, PEN-2, and PS1 fragments are reduced (27).
Despite its broad biological importance, studies of transcription regulation of the PS/γ-secretase components are largely missing. Several studies have identified promoter regions of presenilins, showing that PS1 transcription may be regulated by the cyclic AMP-response element-binding protein (CREB) (30), Ets factors, Sp1 (33), and/or possibly HIF-1a (1) and that PS1 transcription regulation may be cell type specific (33). However, to our knowledge there is no such report on transcriptional regulation of PEN-2, APH-1, or nicastrin. In the current study, we identify and characterize the promoter region of PEN-2 and demonstrate that transcription of PEN-2 is regulated by CREB.
MATERIALS AND METHODS
Cell culture.
HeLa cells, SH-SY5Y cells, and HEK 293T cells were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. All cells were maintained at 37°C in an incubator containing 5% CO2.
Cloning of the PEN-2 promoter and construction of chimeric luciferase reporter plasmids.
PCR was performed to amplify the 5′-flanking regions of the PEN-2 gene by using human peripheral blood lymphocyte genomic DNA as the template. The primers used to generate different promoter deletion plasmids were as follows: forward, 5′-CGACGCGTTGCCCCCAGGCCTGGTCCCTTACTC-3′, 5′-CGACGCGTAGAGAGTGAGGCATGAAGGTTG-3′, 5′-CGACGCGTATAGGCGGGACATGGTGACGAC-3′, 5′-CGACGCGTGGCTGAATGTCGGCTTGTTGTG-3′, 5′-CGACGCGTAGGTGCAAAGACAAAGGTGAAC-3′, 5′-CGACGCGTTTCTCACTTGTCCTTCTCAGTC-3′, and 5′-CGACGCGTCTCCAGGTCTGGCTGCTCTTAC-3′; reverse, 5′-CCCTCGAGCGCGAGACGCCCCAGGTTTC-3′, 5′-CCCTCGAGATTGTAGTCTTCTTTGGGCGAG-3′, and 5′-CCCTCGAGATGCGCTGTTAAAAGAACTCCTACCG-3′.
Primers were designed to contain MluI or XhoI restriction sites so that the resulting PCR-amplified fragments could be cloned into the multicloning sites upstream of the luciferase reporter gene in the pGL3-enhancer expression vector (Promega). Site-directed mutations were integrated into the PEN-2 promoter fragments by overlapping PCR. The mutated fragments were then cloned into vector pGL3-basic (Promega). To generate these mutants, two end primers, 5′-CGACGCGTATAGGCGGGACATGGTGACGAC-3′ (forward) and 5′-CCCTCGAGCGCGAGACGCCCCAGGTTTC-3′ (reverse), were used in combination with various primers with mutations: AP-1mut1, 5′-CGACGCGTATAGGCGGGACATGGGTACGACGGC-3′; AP-1mut2, 5′-CTGCTCTTACGTACCTTCCGTTG-3′, and its reverse complementary sequence (RCS); HIF-1mut, 5′-CAGCGCATGTGTGCAGTGTTGC-3′, and its RCS; NF-ATmut1, 5′-CGCTCGGAATCGCCCCAGG-3′, and its RCS; NF-AT&c-relmut, 5′-GCTCGTGAACGCCCCAGG-3′, and its RCS; NF-ATmut2, 5′-CCCTCGAGCGCGAGACGCCCCAGGTTTAACTGGAGATTG 3′; CREBmut1, 5′-CGACGCGTATAGGCGGGACATGGTGACTACGGCCCCAG-3′; CREBmut2, 5′-CTGCTCTTATGTCACTTCCGTTG-3′, and its RCS; c-relmut1, 5′-CGACGCGTATAGGCGGGACATGGTGACGACGGCCCCAGAAGGAGAG-3′; c-relmut2, 5′-GCTCGGAAACGAACCAGGG-3′, and its RCS; and ATF-6mut, 5′-GAAACACCGCCCCAAAACCCAG-3′, and its RCS. Mutations are shown in boldface, underlined type. The PEN-2 promoter fragment from bp −889 to bp −290 was cut off from the 600-bp pGL3E plasmid by using MluI and XhoI and then linked into pGL3-basic to generate a wild-type plasmid for the positive control.
Transfection and luciferase assay.
HeLa cells were grown to approximately 90% confluence and transfected with 0.6 μg of plasmid DNA on 24-well plates with Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. In order to normalize the different transfection efficiencies of various luciferase constructs, the phRL-SV40 plasmid (where SV40 is simian virus 40) containing the Renilla luciferase gene was cotransfected into the cells in a molar ratio of 1:50 (phRL-SV40:pGL3). Cells were harvested 24 h (pGL3-enhancer chimeric plasmids) or 48 h (pGL3-basic chimeric plasmids) after transfection and lysed with passive lysis buffer (Promega). Firefly luciferase activities and Renilla luciferase activities were measured sequentially using a dual luciferase reporter assay system (Promega) and a luminometer (Beckman).
5′-RACE.
Total RNA was extracted from healthy human liver with Trizol reagent (Invitrogen). 5′ rapid amplification of cDNA ends (RACE) was performed using a BD Smart Race cDNA amplification kit (BD Bioscience). To amplify the 5′ untranslated region of PEN-2, a gene-specific primer located 77 bp upstream of ATG, 5′-GATCAGAACAACCACGCCCCAGGAGAGC-3′, was used. The PCR products were cloned into pGEM-T vector (Promega) and sequenced with primer SP6.
Electrophoretic mobility shift assay.
HeLa nuclear extracts were prepared using a nuclear extract kit (Activemotif). Oligonucleotides were synthesized and annealed with their respective reverse complements to generate double-stranded oligonucleotides, which include CREBwt (where wt is wild type), containing a putative CREB binding site corresponding to the PEN-2 promoter from bp −213 to bp −187 (5′-GCTGCTCTTACGTCACTTCCGTTGCTT-3′); CREBmut, with a C→T mutation at bp −203 (boldface and underlined), (5′-GCTGCTCTTATGTCACTTCCGTTGCTT-3′); CREB consensus sequence (Promega) (5′-AGAGATTGCCTGACGTCAGAGAGCTAG-3′); AP4, as described in reference 3 (5′-CACCCGGTCAGCTGGCCTACACC-3′); HIF-1, as described in reference 5, (5′-ACCGGCCCTACGTGCTGTCTCAC-3′); and SP1 (Promega) (5′-ATTCGATCGGGGCGGGGGAGC-3′). Double-stranded CREBwt and CREB consensus sequences were end labeled with [γ-32P]ATP by T4 polynucleotide kinase and separated from unincorporated nucleotides by Microspin G-25 columns (Amersham Biosciences). 32P-labeled CREBwt probes (35 fmol) were incubated with or without HeLa nuclear extracts (∼10 μg) in gel shift binding buffer containing 4% glycerol, 1 mM MgCl2, 0.5 mM EDTA, 0.5 mM dithiothreitol, 50 mM NaCl, 10 mM Tris-HCl (pH 7.5), and 50 μg/ml poly(dI-dC) · poly(dI-dC) at room temperature for 20 min. 32P-labeled CREB consensus sequences incubated with HeLa nuclear extracts were used for the positive control. For the competition assay, HeLa nuclear extracts were incubated with 1.75 pmol (50× excess) of unlabeled competition oligonucleotides for 10 min prior to addition of 35 fmol labeled probes. The samples were analyzed by 4% nondenaturing polyacrylamide gel electrophoresis (PAGE) and exposed to X-ray film at −70°C.
ChIP analysis.
Chromatin immunoprecipitation (ChIP) assays were performed using SH-SY5Y cells, as previously described (39, 40). The following primers were used: for human PEN-2 promoter, 5′-TTTTACCCAAGCCCTCCAGG-3′ (forward) and 5′-CTCCTACCGGGGAGTCTATG-3′ (reverse); and for human GAPDH (gene encoding glyceraldehyde-3-phosphate dehydrogenase) exon 8 amplification, 5′-ATCACTGCCACCCAGAAGACTGTGGA-3′ (forward) and 5′-TCATACCAGGAAATGAGCTTGACAAA-3′ (reverse).
Forskolin treatments.
HEK 293T cells were treated with forskolin (Sigma) dissolved in dimethyl sulfoxide at different dosages for the indicated times.
Northern blot hybridization.
Total RNA was extracted from HEK 293T cells after forskolin treatments by use of Trizol reagent (Invitrogen). Equal amounts of RNA from differentially treated cells were run on formaldehyde gels and then transferred to nylon membranes. For Northern blot hybridization, a PEN-2 fragment excised from a human PEN-2 construct (26) by PstI and BglII, which contains the first 236 bp of the PEN-2 protein encoding sequence, 12 bp of the pRK5 vector sequence, and a hemagglutinin tag encoding sequence, was used as the probe. The probe for GAPDH was prepared by PCR amplification using primers 5′-GTCAGTGGTGGACCTGACCT-3′ (forward) and 5′-TGCTGTAGCCAAATTCGTTG-3′(reverse) and using cDNA from HEK 293T cells as the template. Probes were 32P-labeled using a random primed DNA labeling kit (Roche Molecular Biochemicals) and hybridized to Northern blots.
Immunoblot and antibodies.
Cell lysates were resolved by 4 to 20% Tris-glycine sodium dodecyl sulfate (SDS)-PAGE, and proteins were detected by Western blotting using different antibodies as indicated. Rabbit anti-PEN-2 polyclonal antibody and anti-APH-1 polyclonal antibody were from Zymed. Rabbit anti-PS1 NTF antibody Ab14 and rabbit antinicastrin antibody 716 were developed in our lab (49). Mouse anti-phospho-CREB antibody was from Cell Signaling Technology. Mouse anti-α-tubulin antibody was from Sigma. Monoclonal antibodies 6E10 and 4G8 (Signet Laboratories), which recognize amino acids 1 to 17 and 17 to 24 of the human Aβ peptide, respectively, were used to detect soluble APPα (sAPPα) and Aβ.
sAPPα, Aβ, and Notch assays.
HEK 293T cells were transiently transfected with human APP or Notch ΔE-myc constructs. After 48 h, the cells were equally split into six plates and treated with or without 10 μM forskolin for 1, 2, or 4 h. The conditioned media were assayed for sAPPα by Western blotting with antibody 6E10. Aβ was immunoprecipitated from conditioned media using 4G8 and immunoblotted with antibody 6E10 as described in reference 45. Cell lysates were immunoblotted with anti-myc antibody 9E10 (to visualize Notch and its intracellular domain [NICD]). For assaying the nascent conversion of Notch to NICD, cells were pulse-labeled with 500 μCi/ml [35S]methionine for 15 min and then chased in the presence/absence of 10 μM forskolin for 1, 2, or 4 h. Cell lysates were immunoprecipitated with anti-myc antibody 9E10, followed by SDS-PAGE analysis and autoradiography.
RESULTS
Cloning of promoter of the human PEN-2 gene.
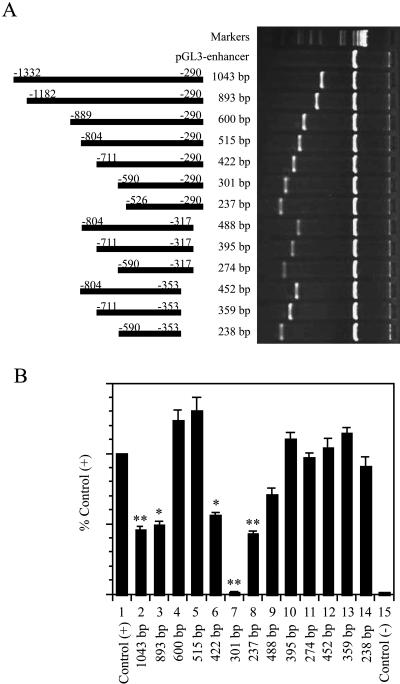
To study the transcriptional regulation of PEN-2, we analyzed the genomic DNA sequence of PEN-2 by using multiple-promoter prediction software. The prediction results suggested that the transcriptional initiation site is likely located at around 300 bp upstream of the translation start codon ATG. Two potential promoter regions together with their respective TATA boxes are located within a 600-bp region which is about 300 bp upstream of ATG. Therefore, we cloned and sequenced a 1,043-bp fragment of the 5′-flanking region. This 1,043-bp fragment includes the 600-bp region predicted as the potential promoter, which ranges from bp −889 to bp −290 upstream of PEN-2 translation start codon ATG (Fig. 1). A computer-based transcription factor binding site search showed several putative regulatory elements, including AP-1, CREB, c-rel, ATF-6, NF-AT, and HIF-1 binding sites, within the 600-bp region (Fig. 1).
FIG. 1.
Sequence features of the human PEN-2 gene promoter. The 600-bp fragment of the 5′-flanking region of the human PEN-2 gene is shown. The numbering was based on the translation start site, defined as +1. The putative transcription factor binding sites are underlined and in boldface. The major transcription initiation site is indicated by an arrow. m1 to m10 denote each putative transcription factor binding site that was mutated for the study shown in Fig. 3.
To determine the transcription initiation site of the human PEN-2 gene, a primer extension assay was carried out using an antisense primer located 77 bp upstream of the first ATG and using the cDNA reverse transcribed from liver RNA as a template. This reaction generated a PCR product of approximately 160 bp, which was cloned into pGEM-T vector and sequenced. The results indicated that the major transcription initiation site is located 302 bp upstream of the PEN-2 translation start codon (Fig. 1).
Identification of the minimal promoter and cis-acting regulatory region.
To determine whether the 600-bp fragment contains the promoter of the PEN-2 gene, we subcloned this fragment into a promoterless luciferase plasmid, pGL3-enhancer (Fig. 2A), and transfected it into HeLa cells. The luciferase activity was assayed using a dual luciferase system and normalized with cotransfected Renilla luciferase activity. As shown in Fig. 2B, the luciferase activity in cells transfected with the luciferase gene driven by an SV40 promoter (bar 1), which was used as a positive control and defined as 100% for normalization purposes, was approximately 120-fold higher than the negative control of cells transfected with empty pGL3-enhancer vector (bar 15). The level of luciferase gene expression driven by the 600-bp fragment was approximately 123.4% ± 7.3% of the positive control (Fig. 2B, bar 4), suggesting that the 600-bp fragment contains a functional promoter region of the human PEN-2 gene. However, extension of the 600-bp fragment's 5′ end to either bp −1332 or bp −1182 dramatically reduced its promoter activity (Fig. 2B, bars 2 and 3), implying a negative regulatory element located in the region between bp −1182 and bp −889.
FIG. 2.
Functional deletion analysis of the human PEN-2 gene promoter. (A) Schematic diagram of the PEN-2 promoter constructs consisting of a 5′-flanking region with serial deletions cloned into the promoterless vector pGL3-enhancer. The numbers represent the endpoints of each construct. The deletion plasmids were confirmed by sequencing and restriction enzyme digestion, and the digested samples were analyzed on a 1.5% agarose gel. The vector size is 5.06 kb, and the sizes of pen-2 promoter deletion constructs range from 0.24 to 1.04 kb. (B) The constructed plasmids were cotransfected into HeLa cells with phRL-SV40, and luciferase activity was measured after 24 h with a luminometer. Renilla luciferase activity expressed by phRL-SV40 was used to normalize the transfection efficiency. The values represent means ± standard errors (n = 4). *, P < 0.05 versus pGL3-control; **, P < 0.001 versus pGL3-control (both by analysis of variance with the post hoc Newman-Keuls test).
To further identify the minimal promoter region required for PEN-2 gene expression, a series of luciferase reporter gene plasmids with different deletions of the 600-bp fragments was constructed (Fig. 2A). Although a small truncation of 85 bp from the 5′ end showed no effect on luciferase expression (Fig. 2B, bar 5), a large truncation of 178 bp from the 5′ end significantly reduced luciferase expression (Fig. 2B, bar 6) and a larger truncation of 299 bp further reduced luciferase expression to the basal level (Fig. 2B, bar 7). However, a further truncation of 76 bp from the 5′ end of the 299-bp truncated fragment rescued luciferase expression to some extent (Fig. 2B, bar 8). These data suggest a positive regulatory cis-acting element existing between bp −804 and bp −590 and a negative regulatory element existing within the region of bp −590 to bp −526. On the other hand, for each 5′ deletion fragment with a reduced luciferase expression level (422 bp and 301 bp), an additional truncation of 27 bp or 63 bp from the 3′ end rescued luciferase expression to a level similar to that of the positive control (Fig. 2B, bars 10 and 13 versus bar 6; bars 11 and 14 versus bar 7), suggesting a negative regulatory element located between bp −290 and bp −317. Interestingly, although the fragment with an 85-bp truncation from the 5′ end had minimal effect on gene expression (Fig. 2B, bar 5), an additional truncation of 27 bp from its 3′ end greatly impaired luciferase expression (Fig. 2B, bar 9 versus bar 5), whereas a longer truncation of 63 bp from its 3′ end only marginally reduced luciferase expression (Fig. 2B, bar 12 versus bar 5). Together, analysis of the deletion plasmids demonstrated that a 238-bp fragment from bp −590 to bp −353 possesses minimal promoter properties and that the flanking regions contain several positive and negative regulatory elements.
The human PEN-2 gene promoter contains CREB binding sites.
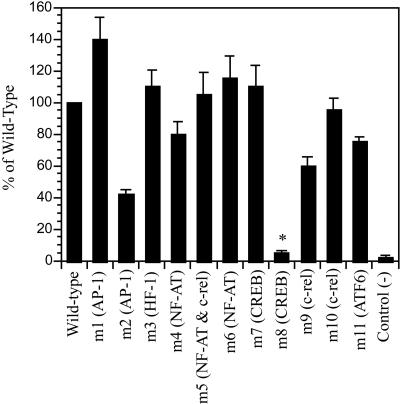
Sequence analysis revealed that the 600-bp fragment contains several potential transcriptional factor binding sites, including two AP-1 binding consensuses, two NF-AT binding consensuses, two c-rel binding consensuses, two CREB binding consensuses, one ATF-6 consensus, and one HIF-1 consensus (Fig. 1). To define the potential transcriptional factor for the PEN-2 promoter, we mutated these potential transcriptional factor binding sites individually or collectively. The mutated fragments were cloned into pGL3-enhancer luciferase reporter vector, followed by promoter activity analysis. The mutation of AP-1 located at bp −506 to bp −500 resulted in a 60% reduction of the promoter activity compared to that of the wild-type 600-bp promoter fragment (Fig. 3, m2). Interestingly, when a CREB binding site congregating to the AP-1 site located at bp −506 to bp −500 was mutated, the promoter activity of the 600-bp promoter was abolished (Fig. 3, m8). In contrast, mutation of other potential transcriptional binding sites had little effect on the promoter activity (Fig. 3). These results suggest that CREB is a potential transcriptional factor regulating PEN-2 transcription. Because the AP-1 and CREB sites gather together, mutation of the AP-1 site may indeed also impact CREB binding, thus resulting in the reduced promoter activity.
FIG. 3.
Mutation of the potential CREB binding site dramatically reduced PEN-2 promoter transcription activity. HeLa cells were transfected with wild-type plasmid (pGL3-basic inserted with the PEN-2 promoter 600-bp fragment), various mutant constructs, or pGL3-basic. Relative luciferase activity was determined in triplicate, and data were normalized to Renilla luciferase activity expressed by phRL-SV40. Data represent means ± standard errors. *, P < 0.05 versus the wild type by analysis of variance with the post-hoc Newman-Keuls test.
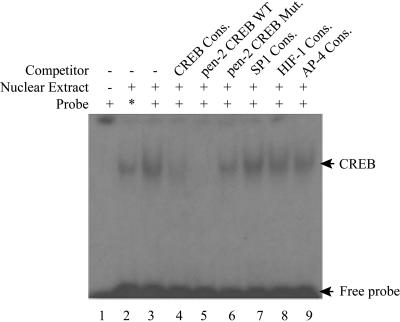
To further investigate the binding of CREB to the element located at bp −506 to bp −500, we performed gel shift assays. A 27-bp double-stranded oligonucleotide corresponding to the PEN-2 promoter region from bp −515 to bp −489 was synthesized and labeled as the probe. As shown in Fig. 4 (lane 3), a protein-DNA complex was detected after incubation of the probe with HeLa nuclear extracts. As a positive control, a classic CREB binding oligonucleotide probe (see Materials and Methods) was also shown to form a protein-DNA complex that migrated at the same position as the PEN-2 CREB probe-protein complex (Fig. 4, lane 2). Notably, the PEN-2 CREB probe was able to form the protein-DNA complex even more effectively than the classic CREB consensus probe. The complex formation between PEN-2 CREB probe-protein was dramatically inhibited and completely abolished by competitive binding of the CREB consensus (Fig. 4, lane 4) and the PEN-2 CREB probe (Fig. 4, lane 5), respectively. In contrast, the PEN-2 CREB probe-protein complex formation was not affected by competition from a PEN-2 CREB oligonucleotide with a mutated CREB binding site or the consensus probes of SP-1, HIF-1, and AP-4. The results indicate that the identified PEN-2 CREB binding element is able to form a complex with nuclear CREB protein in vitro.
FIG. 4.
Electrophoretic mobility shift assay for the PEN-2 gene promoter. The gel shift assay was performed as described in Materials and Methods. Lane 1, labeled CREBwt probe without nuclear extract. Incubation of 32P-labeled CREBwt with HeLa nuclear extracts retarded the migration rate of the labeled probe, which formed a new shifted DNA-protein complex band (lane 3) at the same position as the positive control (lane 2) formed by incubating the 32P-labeled CREB consensus (Cons.) oligonucleotide probe (*) with HeLa nuclear extract. Competition assays were performed by further adding a 50-fold molar excess of unlabeled competition oligonucleotides, consensus CREB (lane 4), CREBwt (lane 5), CREBmut (lane 6), consensus SP1 (lane 7), HIF-1 (lane 8), or AP4 (lane 9).
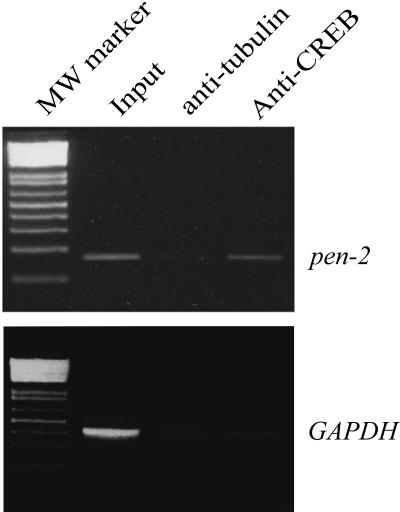
To confirm the PEN-2 CREB element binding to CREB transcriptional factor in vivo, we next performed chromatin immunoprecipitation analysis. Human neuroblastoma SH-SY5Y cells were cross-linked, followed by immunoprecipitation with an anti-CREB antibody or an antitubulin antibody. The precipitates were then PCR amplified with a pair of PEN-2 CREB-specific primers. The results showed that a specific fragment was amplified from the cell lysates and the anti-CREB antibody precipitates but not from the antitubulin precipitates (Fig. 5, upper panel). In contrast, a GAPDH DNA fragment was amplified only from the input lysates with a pair of specific GAPDH primers (Fig. 5, lower panel). These results suggest that the identified PEN-2 promoter is able to specifically bind CREB in vivo.
FIG. 5.
ChIP analysis of CREB binding to the PEN-2 promoter. Cross-linked chromatin extracts from SH-SY5Y cells were immunoprecipitated with either anti-CREB antibody or antitubulin antibody. The PEN-2 promoter fragment covering bp −539 to bp −370 and a DNA fragment from exon 8 of the human GAPDH gene were PCR amplified from the immunoprecipitated and the input chromatin, respectively. MW, molecular weight.
Activation of CREB up-regulates PEN-2 expression.
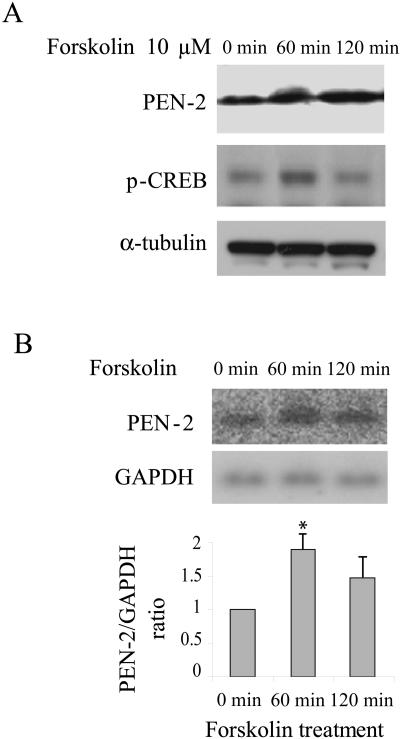
We next examined the role of CREB on transcription and protein expression of PEN-2 in vivo. It is well documented that forskolin treatment increases the phosphorylation of the active form of CREB (14, 22). In agreement with the previous reports, we showed that the treatment of HEK 293T cells with 10 μm forskolin resulted in increased phosphorylation of CREB (Fig. 6A). Accordingly, transcription of PEN-2 was increased about 1.8-fold after a 60-min treatment and 1.4-fold after a 120-min treatment compared to results for the control (Fig. 6B), suggesting that activation of CREB stimulates the transcription of PEN-2. The slight decrease of PEN-2 transcription after a 120-min forskolin treatment compared to that of a 60-min treatment was likely caused by reduced CREB sensitivity to forskolin. Treatment of cells with 10 μm forskolin for 60 or 120 min significantly increased the protein level of PEN-2 (Fig. 6A).
FIG. 6.
Activation of CREB by forskolin stimulated the gene transcription and protein expression of PEN-2. (A) HEK 293T cells were treated with 10 μM forskolin for 0, 60, or 120 min. Protein levels of PEN-2 and p-CREB were assayed by Western blot analysis. α-Tubulin was used as the loading control. (B) Total RNAs were extracted from forskolin-treated HEK 293T cells for Northern blot hybridization analysis. The mRNA levels of PEN-2 and GAPDH were quantified for comparison of the ratios of PEN-2 over GAPDH. Data represent averages of three separate experiments. *, P < 0.01 compared to 0-min treatment.
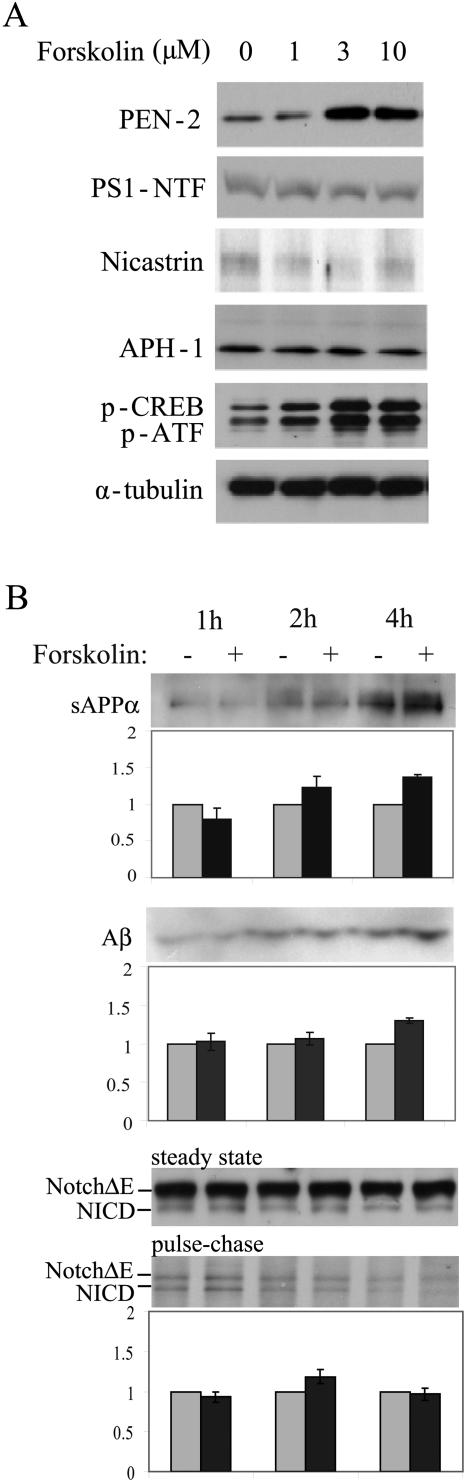
The stimulatory effect of forskolin on PEN-2 expression is concentration dependent: the effect was shown to be maximal at 3 μM and saturated at 10 μM, while 1 μM showed minimal effect. In contrast, the other three γ-secretase complex components, presenlin-1, nicastrin, and APH-1, were not changed by forskolin treatment at various concentrations (Fig. 7A). Thus, activation of CREB specifically stimulates PEN-2 expression. Consistent with previous observations (29, 46), we also demonstrated that treatment of cells with forskolin for 4 h slightly increased the secretion of Aβ (by 30%) and sAPPα (by 40%), a nonamyloidogenic cleavage product of APP by the α-secretase (Fig. 7B) (13, 16). However, forskolin treatment failed to affect the steady-state levels of Notch to NICD. We also found that forskolin treatment did not alter the de novo proteolytic conversion, by the γ-secretase activity, of Notch to NICD, as examined by a pulse-chase experiment paradigm (Fig. 7B).
FIG. 7.
Forskolin treatment increased PEN-2 protein expression and the secretion of sAPPα and Aβ without affecting protein expression of other γ-secretase components and Notch cleavage. (A) HEK 293T cells were treated with 10 μM forskolin for 60 min. The same amounts of cell lysates were analyzed and immunoblotted with antibodies against PEN-2, PS1 NTF, nicastrin, and APH-1. The levels of phospho-CREB (p-CREB) and α-tubulin were detected for the positive control and loading control, respectively. p-ATF, phospho-ATF. (B) HEK 293T cells were transiently transfected with human APP695 or Notch ΔE. After 48 h, the cells were equally split into six plates and treated with or without 10 μM forskolin for 1, 2, or 4 h. The conditioned media were assayed for sAPPα by immunoblotting with antibody 6E10 and for Aβ by immunoprecipitation using antibody 4G8 followed by Western blot analysis using 6E10. Cell lysates were immunoblotted with anti-myc antibody 9E10 to detect steady-state levels of Notch and NICD. Cells were also pulse-labeled with [35S]methionine for 15 min and then chased in the presence/absence of 10 μM forskolin for 1, 2, or 4 h. Cell lysates were immunoprecipitated with anti-myc antibody 9E10, followed by SDS-PAGE analysis and autoradiography. Data represent means ± standard deviations (n = 3). *, P < 0.04.
DISCUSSION
Aβ deposition is one of the most important pathological features in AD. Sequential proteolytic cleavages of APP by β-secretase and the PS/γ-secretase complex are essential for Aβ generation (13, 16). Dissecting β-secretase and γ-secretase complex components, including the mechanisms underlying their transcriptional regulation, will be greatly important for understanding AD pathogenesis. Furthermore, PS/γ-secretase is also involved in multiple physiological activities, such as the Notch signal pathway (10, 21, 28, 31, 36, 43), making it a target of extensive scrutiny from neurobiological to cancer and development fields. In the present study, we identified a 600-bp fragment upstream of the PEN-2 translational start codon as a critical PEN-2 promoter region responsible for its transcription. There are several potential transcriptional factor binding sites within this PEN-2 promoter region. However, only a CREB binding site is shown to be essential for PEN-2 transcription regulation: mutation of this CREB site resulted in abolishment of the 600-bp fragment's transcriptional activity. Furthermore, both electrophoretic mobility shift assays and ChIP analysis demonstrated the binding of CREB to the PEN-2 promoter both in vitro and in vivo. Finally, activation of CREB by forskolin significantly potentiates expression of PEN-2 at both mRNA and protein levels. Together, these results demonstrate that expression of PEN-2 is regulated by the CREB transcriptional factor.
It has previously been reported that the transcription of β-secretase (BACE1) is regulated by SP1 (4). Among the four γ-secretase components, however, only the promoter of PS and its transcriptional regulation have been studied so far (1, 30, 33, 34, 35). Pastorcic and Das (33) showed that Ets factors were involved in regulation of PS1 and that Ets factors were differentially utilized in human neuroblastoma SH-SY5Y and SK-N-SH cells. There are potential HIF binding sites on PS1/PS2 promoter regions. Hypoxia treatment potentiated PS1 gene expression in interleukin-1β- and Aβ42-treated human neural cells (1), implying that HIF-1a may be involved in stress-mediated PS transcriptional regulation. Interestingly, Mitsuda et al. (30) identified a CREB binding site on the PS1 promoter region and showed that, by using N-methyl-d-aspartate to activate CREB for 6 h, both PS1 transcription and translation were up-regulated in SK-N-SH cells. In our experiments, however, CREB activation by forskolin failed to stimulate protein expression of PS1, APH-1 and nicastrin in HEK 293T cells. This apparent discrepancy may be attributed to the use of the different cell types or the ways to activate CREB. Excitotoxic stimulants such as N-methyl-d-aspartate likely represent a stress which causes multiple cellular responses. In addition, another study showed that 12-O-tetradecanoylphorbol 13-acetate is able to activate transcription of the human presenilin 1 gene and that point mutations eliminating the −6 CREB/AP1 motif did not affect the stimulation by 12-O-tetradecanoylphorbol 13-acetate (34).
In addition to the amyloidogenic cleavage pathway by β- and γ-secretase, APP also undergoes a nonamyloidogenic cleavage by α-secretase activity to release sAPPα (13, 16). Consistent with previous reports (29, 46), we found that treatment of cells by forskolin for 4 h stimulated the secretion of both sAPPα and Aβ. However, forskolin did not affect the processing of Notch by γ-secretase (Fig. 7B). Our results have shown that CREB activation up-regulated only PEN-2 but not PS1, nicastrin, or APH-1, indicating that the γ-secretase activity, which requires precise stoichiometric interaction among all four components, is unlikely to be affected. Previous reports showed that activation of protein kinase A promotes trafficking of secretory vesicles, including APP-containing vesicles, from the trans-Golgi network to the plasma membrane (32, 46). We hence speculate that, rather than altering γ-secretase activity, forskolin treatment specifically increases the trafficking and release of APP- and Aβ-containing vesicles without affecting those vesicles carrying Notch molecules. In contrast to up-regulation of PEN-2 by CREB/protein kinase A activation, it is conceivable that inhibition of CREB activation may down-regulate PEN-2 expression and hence reduce γ-secretase activity and Aβ generation. Given the broad spectrum of γ-secretase substrates, however, whether down-regulation of PEN-2 by inhibiting CREB activation may specifically reduce Aβ processing without affecting other substrates (such as Notch) awaits further investigation.
In addition to the essential role of PS and its associated proteins in γ-secretase activity, numerous reports have assigned other physiological functions to PS and its partners, including roles in regulating protein trafficking, platelet-derived growth factor signaling, calcium homeostasis, skeletal development, neurite outgrowth, apoptosis, synaptic plasticity, and tumorigenesis (19, 23, 36, 43). Although mice lacking PS1 (11), nicastrin (24, 25), or APH-1 (27) individually are embryonically lethal and show defects resembling those found in Notch null embryos (6, 17, 41), these knockout models still exhibit patterning defects that are not identical to each other. In the present study, CREB activation stimulated by forskolin promoted only PEN-2 expression but not expression of other γ-secretase components, even though it has been reported that the PS1 promoter region possesses a CREB binding site (30). These results suggest that additional physiological functions may be uniquely assigned to PEN-2 through the specific control of PEN-2 expression.
Acknowledgments
This work was supported in part by National Institutes of Health grants (RO1 NS046673 to H.X., RO1 AG024895 to H.X., and RO1 DC006497 to Z.Z.); grants from the American Health Assistance Foundation (to H.X.) and the Alzheimer's Association (to H.X. and Z.Z.); and funds from the National Natural Science Foundation of China, no. 30370737, China's 863 project (2002BA711A07) and 973 project (2004CB518601, 2001CB510302). Y.-W.Z. is the recipient of National Institutes of Health training grant F32 AG024895.
REFERENCES
- 1.Bazan, N. G., and W. J. Lukiw. 2002. Cyclooxygenase-2 and presenilin-1 gene expression induced by interleukin-1beta and amyloid beta 42 peptide is potentiated by hypoxia in primary human neural cells. J. Biol. Chem. 277:30359-30367. [DOI] [PubMed] [Google Scholar]
- 2.Bergman, A., E. Hansson, S. E. Pursglove, M. R. Farmery, L. Lannfelt, U. Lendahl, J. Lundkvist, and J. Naslund. 2004. Pen-2 is sequestered in the endoplasmic reticulum and subjected to ubiquitylation and proteasome-mediated degradation in the absence of presenilin. J. Biol. Chem. 279:16744-16753. [DOI] [PubMed] [Google Scholar]
- 3.Cheng, C. K., R. L. C. Hoo, B. K. C. Chow, and P. C. K. Leung. 2003. Functional cooperation between multiple regulatory elements in the untranslated exon 1 stimulates the basal transcription of the human GnRH-II gene. Mol. Endocrinol. 17:1175-1191. [DOI] [PubMed] [Google Scholar]
- 4.Christensen, M. A., W. Zhou, H. Qing, A. Lehman, S. Philipsen, and W. Song. 2004. Transcriptional regulation of BACE1, the β-amyloid precursor protein β-secretase, by Sp1. Mol. Cell. Biol. 24:865-874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chun, Y. S., E. Choi, E. J. Yeo, J. H. Lee, M. S. Kim, and J. W. Park. 2001. A new HIF-1 alpha variant induced by zinc ion suppresses HIF-1-mediated hypoxic responses. J. Cell Sci. 114:4051-4061. [DOI] [PubMed] [Google Scholar]
- 6.Conlon, R. A., A. G. Reaume, and J. Rossant. 1995. Notch1 is required for the coordinate segmentation of somites. Development 121:1533-1545. [DOI] [PubMed] [Google Scholar]
- 7.Crystal, A. S., V. A. Morais, R. R. Fortna, D. Carlin, T. C. Pierson, C. A. Wilson, V. M. Y. Lee, and R. W. Doms. 2004. Presenilin modulates Pen-2 levels posttranslationally by protecting it from proteasomal degradation. Biochemistry 43:3555-3563. [DOI] [PubMed] [Google Scholar]
- 8.Delkoe, D. J. 1997. Alzheimer's disease: genotypes, phenotypes, and treatments. Science 275:630-631. [DOI] [PubMed] [Google Scholar]
- 9.De Strooper, B. 2003. Aph-1, Pen-2, and nicastrin with presenilin generate an active gamma-secretase complex. Neuron 38:9-12. [DOI] [PubMed] [Google Scholar]
- 10.De Strooper, B., W. Annaert, P. Cupers, P. Saftig, K. Craessaerts, J. S. Mumm, E. H. Schroeter, V. Schrijvers, M. S. Wolfe, W. J. Ray, A. Goate, and R. Kopan. 1999. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 398:518-522. [DOI] [PubMed] [Google Scholar]
- 11.Donoviel, D. B., A. K. Hadjantonakis, M. Ikeda, H. Zheng, P. St. George-Hyslop, and A. Bernstein. 1999. Mice lacking both presenilin genes exhibit early embryonic patterning defects. Genes Dev. 13:2801-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis, R., G. McGrath, J. Zhang, D. A. Ruddy, M. Sym, J. Apfeld, M. Nicoll, M. Maxwell, B. Hai, M. C. Ellis, A. L. Parks, W. Xu, J. Li, M. Gurney, R. L. Myers, C. S. Himes, R. Hiebsch, C. Ruble, J. S. Nye, and D. Curtis. 2002. aph-1 and pen-2 are required for Notch pathway signaling, gamma-secretase cleavage of betaAPP, and presenilin protein accumulation. Dev. Cell 3:85-97. [DOI] [PubMed] [Google Scholar]
- 13.Golde, T. E. 2005. The Abeta hypothesis: leading us to rationally-designed therapeutic strategies for the treatment or prevention of Alzheimer disease. Brain Pathol. 15:84-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gonzalez, G. A., and M. R. Montminy. 1989. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell 59:675-680. [DOI] [PubMed] [Google Scholar]
- 15.Goutte, C., M. Tsunozaki, V. A. Hale, and J. R. Priess. 2002. APH-1 is a multipass membrane protein essential for the Notch signaling pathway in Caenorhabditis elegans embryos. Proc. Natl. Acad. Sci. USA 99:775-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greenfield, J. P., R. S. Gross, G. K. Gouras, and H. Xu. 2000. Cellular and molecular basis of beta-amyloid precursor protein metabolism. Front. Biosci. 5:D72-D83. [DOI] [PubMed] [Google Scholar]
- 17.Huppert, S. S., A. Le, E. H. Schroeter, J. S. Mumm, M. T. Saxena, L. A. Milner, and R. Kopan. 2000. Embryonic lethality in mice homozygous for a processing-deficient allele of Notch1. Nature 405:966-970. [DOI] [PubMed] [Google Scholar]
- 18.Iwatsubo, T. 2004. The gamma-secretase complex: machinery for intramembrane proteolysis. Curr. Opin. Neurobiol. 14:379-383. [DOI] [PubMed] [Google Scholar]
- 19.Kang, D. E., I. Sang Yoon, E. Repetto, T. Busse, N. Yermian, L. Ie, and E. H. Koo. 2005. Presenilins mediate PI3K/Akt and ERK activation via select signaling receptors: selectivity of PS2 in PDGF signaling. J. Biol. Chem. 280:31537-31547. [DOI] [PubMed] [Google Scholar]
- 20.Kimberly, W. T., M. J. LaVoie, B. L. Ostaszewski, W. Ye, M. S. Wolfe, and D. J. Selkoe. 2003. γ-Secretase is a membrane protein complex comprised of presenilin, nicastrin, Aph-1, and Pen-2. Proc. Natl. Acad. Sci. USA 100:6382-6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lammich, S., M. Okochi, M. Takeda, C. Kaether, A. Capell, A. K. Zimmer, D. Edbauer, J. Walter, H. Steiner, and C. Haass. 2002. Presenilin-dependent intramembrane proteolysis of CD44 leads to the liberation of its intracellular domain and the secretion of an Abeta-like peptide. J. Biol. Chem. 277:44754-44759. [DOI] [PubMed] [Google Scholar]
- 22.Lamph, W. W., V. J. Dwarki, R. Ofir, M. Montminy, and I. M. Verma. 1990. Negative and positive regulation by transcription factor cAMP response element-binding protein is modulated by phosphorylation. Proc. Natl. Acad. Sci. USA 87:4320-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leem, J. Y., S. Vijayan, P. Han, D. Cai, M. Machura, K. O. Lopes, M. L. Veselits, H. Xu, and G. Thinakaran. 2002. Presenilin 1 is required for maturation and cell surface accumulation of nicastrin. J. Biol. Chem. 277:19236-19240. [DOI] [PubMed] [Google Scholar]
- 24.Li, J., G. J. Fici, C. A. Mao, R. L. Myers, R. Shuang, G. P. Donoho, A. M. Pauley, C. S. Himes, W. Qin, I. Kola, K. M. Merchant, and J. S. Nye. 2003. Positive and negative regulation of the gamma-secretase activity by nicastrin in a murine model. J. Biol. Chem. 278:33445-33449. [DOI] [PubMed] [Google Scholar]
- 25.Li, T., G. Ma, H. Cai, D. L. Price, and P. C. Wong. 2003. Nicastrin is required for assembly of presenilin/gamma-secretase complexes to mediate Notch signaling and for processing and trafficking of beta-amyloid precursor protein in mammals. J. Neurosci. 23:3272-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo, W. J., H. Wang, H. Li, B. S. Kim, S. Shah, H. J. Lee, G. Thinakaran, T. W. Kim, G. Yu, and H. Xu. 2003. PEN-2 and APH-1 coordinately regulate proteolytic processing of presenilin 1. J. Biol. Chem. 278:7850-7854. [DOI] [PubMed] [Google Scholar]
- 27.Ma, G., T. Li, D. L. Price, and P. C. Wong. 2005. APH-1a is the principal mammalian APH-1 isoform present in gamma-secretase complexes during embryonic development. J. Neurosci. 25:192-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marambaud, P., J. Shioi, G. Serban, A. Georgakopoulos, S. Sarner, V. Nagy, L. Baki, P. Wen, S. Efthimiopoulos, Z. Shao, T. Wisniewski, and N. K. Robakis. 2002. A presenilin-1/γ-secretase cleavage releases the E-cadherin intracellular domain and regulates disassembly of adherens junctions. EMBO J. 21:1948-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marambaud, P., N. Chevallier, K. Ancolio, and F. Checler. 1998. Post-transcriptional contribution of a cAMP-dependent pathway to the formation of alpha- and beta/gamma-secretases-derived products of beta APP maturation in human cells expressing wild-type and Swedish mutated beta APP. Mol. Med. 4:715-723. [PMC free article] [PubMed] [Google Scholar]
- 30.Mitsuda, N., N. Ohkubo, M. Tamatani, Y. D. Lee, M. Taniguchi, K. Namikawa, H. Kiyama, A. Yamaguchi, N. Sato, K. Sakata, T. Ogihara, M. P. Vitek, and M. Tohyama. 2001. Activated cAMP-response element-binding protein regulates neuronal expression of presenilin-1. J. Biol. Chem. 276:9688-9698. [DOI] [PubMed] [Google Scholar]
- 31.Ni, C. Y., M. P. Murphy, T. E. Golde, and G. Carpenter. 2001. gamma-Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science 294:2179-2181. [DOI] [PubMed] [Google Scholar]
- 32.Ohashi, M., and W. B. Huttner. 1994. An elevation of cytosolic protein phosphorylation modulates trimeric G-protein regulation of secretory vesicle formation from the trans-Golgi network. J. Biol. Chem. 269:24897-24905. [PubMed] [Google Scholar]
- 33.Pastorcic, M., and H. K. Das. 2004. Alternative initiation of transcription of the human presenilin 1 gene in SH-SY5Y and SK-N-SH cells. The role of Ets factors in the regulation of presenilin 1. Eur. J. Biochem. 271:4485-4494. [DOI] [PubMed] [Google Scholar]
- 34.Pastorcic, M., and H. K. Das. 2002. Activation of transcription of the human presenilin 1 gene by 12-O-tetradecanoylphorbol 13-acetate. Eur. J. Biochem. 269:5956-5962. [DOI] [PubMed] [Google Scholar]
- 35.Renbaum, P., R. Beeri, E. Gabai, M. Amiel, M. Gal, M. U. Ehrengruber, and E. Levy-Lahad. 2003. Egr-1 upregulates the Alzheimer's disease presenilin-2 gene in neuronal cells. Gene 318:113-124. [DOI] [PubMed] [Google Scholar]
- 36.Sisodia, S. S., S. H. Kim, and G. Thinakaran. 1999. Function and dysfunction of the presenilins. Am. J. Hum. Genet. 65:7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steiner, H., E. Winkler, D. Edbauer, S. Prokop, G. Basset, A. Yamasaki, M. Kostka, and C. Haass. 2002. PEN-2 is an integral component of the gamma-secretase complex required for coordinated expression of presenilin and nicastrin. J. Biol. Chem. 277:39062-39065. [DOI] [PubMed] [Google Scholar]
- 38.St. George-Hyslop, P. H. 2000. Molecular genetics of Alzheimer's disease. Biol. Psychiatry 47:183-199. [DOI] [PubMed] [Google Scholar]
- 39.Sun, P., and H. H. Loh. 2001. Transcriptional regulation of mouse delta-opioid receptor gene: role of Ets-1 in the transcriptional activation of mouse delta-opioid receptor gene. J. Biol. Chem. 276:45462-45469. [DOI] [PubMed] [Google Scholar]
- 40.Sun, P., and H. H. Loh. 2002. Transcriptional regulation of mouse delta-opioid receptor gene. Role of Ikaros in the stimulated transcription of mouse delta-opioid receptor gene in activated T cells. J. Biol. Chem. 277:12854-12860. [DOI] [PubMed] [Google Scholar]
- 41.Swiatek, P. J., C. E. Lindsell, F. F. del Amo, G. Weinmaster, and T. Gridley. 1994. Notch1 is essential for postimplantation development in mice. Genes Dev. 8:707-719. [DOI] [PubMed] [Google Scholar]
- 42.Thinakaran, G., D. R. Borchelt, M. K. Lee, H. H. Slunt, L. Spitzer, G. Kim, T. Ratovitsky, F. Davenport, C. Nordstedt, M. Seeger, J. Hardy, A. I. Levey, S. E. Gandy, N. A. Jenkins, N. G. Copeland, D. L. Price, and S. S. Sisodia. 1996. Endoproteolysis of presenilin 1 and accumulation of processed derivatives in vivo. Neuron 17:181-190. [DOI] [PubMed] [Google Scholar]
- 43.Thinakaran, G., and A. T. Parent. 2004. Identification of the role of presenilins beyond Alzheimer's disease. Pharm. Res. 50:411-418. [DOI] [PubMed] [Google Scholar]
- 44.Wang, H., W. J. Luo, Y. W. Zhang, Y. M. Li, G. Thinakaran, P. Greengard, and H. Xu. 2004. Presenilins and gamma-secretase inhibitors affect intracellular trafficking and cell surface localization of the gamma-secretase complex components. J. Biol. Chem. 279:40560-40566. [DOI] [PubMed] [Google Scholar]
- 45.Xu, H., D. Sweeney, R. Wang, G. Thinakaran, A. C. Lo, S. S. Sisodia, P. Greengard, and S. Gandy. 1997. Generation of Alzheimer beta-amyloid protein in the trans-Golgi network in the apparent absence of vesicle formation. Proc. Natl. Acad. Sci. USA 94:3748-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu, H., D. Sweeney, P. Greengard, and S. Gandy. 1996. Metabolism of Alzheimer beta-amyloid precursor protein: regulation by protein kinase A in intact cells and in a cell-free system. Proc. Natl. Acad. Sci. USA 93:4081-4084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, D. S., A. Tandon, F. Chen, G. Yu, H. Yu, S. Arawaka, H. Hasegawa, M. Duthie, S. D. Schmidt, T. V. Ramabhadran, R. A. Nixon, P. M. Mathews, S. E. Gandy, H. T. Mount, P. St. George-Hyslop, and P. E. Fraser. 2002. Mature glycosylation and trafficking of nicastrin modulate its binding to presenilins. J. Biol. Chem. 277:28135-28142. [DOI] [PubMed] [Google Scholar]
- 48.Yu, G., M. Nishimura, S. Arawaka, D. Levitan, L. Zhang, A. Tandon, Y.-Q. Song, E. Rogaeva, F. Chen, T. Kawarai, A. Supala, L. Levesque, H. Yu, D.-S. Yang, E. Holmes, P. Milman, Y. Liang, D. M. Zhang, D. H. Xu, C. Sato, E. Rogaev, M. Smith, C. Janus, Y. Zhang, R. Aebersold, L. Farrer, S. Sorbi, A. Bruni, P. Fraser, and P. St. George-Hyslop. 2000. Nicastrin modulates presenilin-mediated notch/glp-1 signal transduction and βAPP processing. Nature 407:48-54. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, Y. W., W. J. Luo, H. Wang, P. Lin, K. S. Vetrivel, F. Liao, F. Li, P. C. Wong, M. G. Farquhar, G. Thinakaran, and H. Xu. 2005. Nicastrin is critical for stability and trafficking but not association of other presenilin/gamma-secretase components. J. Biol. Chem. 280:17020-17026. [DOI] [PMC free article] [PubMed] [Google Scholar]