Abstract
The genomic RNA of human T-cell leukemia virus type 1 encodes three polyproteins, Gag, Gag-Pro, and Gag-Pro-Pol, which are translated as a result of no, one, and two frameshifts, respectively. In this report we demonstrate that the 77 residues encoded at the C terminus of the Gag-Pro precursor can be collectively detected as an 8-kDa transframe protein (TFP) in virions. Mutant viruses with a C-terminally truncated TFP (19, 32, or 50 residues) had essentially a wild-type phenotype in vitro. However, a virus mutant that encoded only the Gag and Gag-Pro-Pol polyproteins due to a mutation in the second frameshift site, and hence did not produce TFP, was noninfectious. Mutation analysis of the proteolytic cleavage site between PR and TFP revealed the presence of an additional site and the existence of a p1 peptide separating protease and TFP. While removal of the cleavage site at the PR-p1 junction had a modest effect on virus replication, mutation of the p1-TFP cleavage site led to noninfectious virus and the loss of reverse transcriptase activity. Determination of the amino-terminal sequence of a hemagglutinin-tagged RT demonstrated that the same site is used in processing the Gag-Pro-Pol precursor and thus defines the start of mature RT. Neither mutation alone or in combination caused changes in the amounts or processing patterns of the Gag polyprotein, indicating that protease is active independent of its C terminus.
Human T-cell leukemia virus type 1 (HTLV-1) is the etiological agent of adult T-cell leukemia and HTLV-1-associated myelopathy and tropical spastic paraparesis (10, 14, 15, 18). The virus belongs to a small group of complex retroviruses consisting of HTLV-1 and HTLV-2, simian T-lymphotropic virus types 1 and 2 (STLV-1 and STLV-2), STLV-L, and bovine leukemia virus (BLV). One feature of this group of viruses is that they encode very compact Gag precursors consisting of only the canonical structural proteins MA, CA, and NC (Fig. 1A). Most other retroviruses encode additional peptides in their Gag precursors that often carry out important functions for virus maturation and replication. In HTLV-1, these functions either may be embedded in the Gag proteins or may reside in a peptide encoded by a different part of the genome. In addition to potentially coding for several proteins of an as yet poorly defined function at the 3′ end of the genome, HTLV-1 encodes an additional 77 residues beyond the cleavage site that defines the C terminus of PR at the end of the Gag-Pro precursor. Sequence comparison of all full-length HTLV-1 and STLV-1 isolates in GenBank shows that the reading frame is open in all but one and that the sequence is highly conserved (G. Heidecker and D. Derse, unpublished observation). The peptide has a predicted molecular weight of 8.5. The conservation of this sequence could be due to selective pressure on TFP itself or on RT, which shares the first 24 amino acids with TFP and is then encoded in a different reading frame. In contrast to HTLV-1, translation of the Gag-Pro precursor is terminated at the second frameshift site in BLV or 6 codons beyond it in HTLV-2/STLV-2 and STLV-L.
FIG. 1.
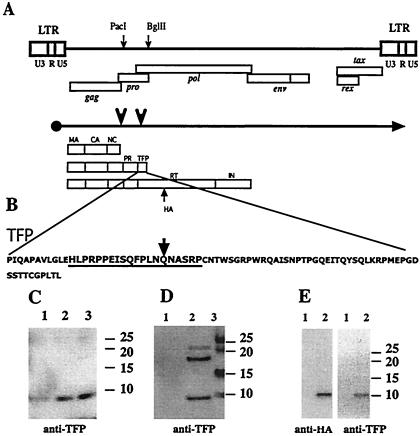
TFP is encoded between pro and pol and is a component of the HTLV-1 particle. (A) Major structural feature of the proviral sequences, the primary transcript, and translational products. The unique PacI and BglII sites used in the construction of mutant proviral clones are indicated by small arrows on the genome; the locations of the major genes are shown below the genome. Large arrows on the mRNA transcript mark the frameshift sites. The different precursor proteins are indicated below the mRNA transcript and reflect whether no frameshifts or one or two frameshifts occurred during translation. The mature processing products originating from the precursors are indicated. The position of the HA tag insertion into the pol reading frame is also shown. (B) The sequence of the synthetic peptide within TFP that was used to generate the rabbit antisera. The arrow marks the residue where the frameshift into RT would occur. (C) Immunoblot showing TFP reactive material from different amounts of banded virus from MT2 supernatants. Lanes 1, 2, and 3 contain 1, 2, and 5 μg of total viral protein, respectively. (D) TFP reactive material present in pelleted virus obtained from supernatants of 293 cells transfected with pX1MT (8). A total of 3 × 106 cells were transfected with 8 μg of plasmid DNA by using a calcium phosphate coprecipitation protocol (6). A forty-eight-hour posttransfection virus was pelleted from precleared supernatants for 90 min at 80,000 × g. Lane 1, mock transfection; lane 2, pX1MT; lane 3, marker. (E) Identical bands visible when supernatants from 293 cells transfected with the pCMV-gag-pro-TFPHA construct are probed with either anti-HA or with anti-TFP. Lane 1, mock transfection; lane 2, pCMV-gag-pro-TFPHA. All gels were 4 to 12% NUPAGE gels run in morpholineethanesulfonic acid buffer according to the manufacturer's conditions (Invitrogen, Carlsbad, Calif.). Proteins were transferred to an Immobilon (Amersham) membrane and probed with rabbit anti-TFP or monoclonal rat anti-HA (3F10). Secondary antibodies were coupled with horseradish peroxidase, and reactive material was visualized with Lumiglo (all from Cell Signaling). The positions of the prestained size markers are indicated for each gel.
No function has been attributed to TFP; indeed, TFP has not yet been described in the literature and is not shown on most maps of HTLV-1. As it is part of the Gag-Pro precursor, TFP should be incorporated into virus particles and appear after processing as an 8-kDa peptide. Rabbit antisera were raised against a synthetic peptide representing residues 12 to 31 following the C terminus of mature HTLV-1 protease (Fig. 1B). This region of TFP is also part of the Gag-Pro-Pol precursor and remains part of RT after processing, based on current predictions regarding RT maturation. The anti-TFP antibody detected a clear band migrating just ahead of the 10-kDa size marker on immunoblots of gels separating proteins of density-banded virus produced by MT2 cells (Fig. 1C). The antibody also detected TFP on immunoblots of gels separating proteins of virions pelleted from the supernatants of other chronically infected cell lines (data not shown) as well as from 293 cells transiently transfected with the full-length provirus clone, pX1MT (Fig. 1D). These results clearly demonstrated that TFP is incorporated into the virion. Additional higher-molecular-weight bands whose sizes are consistent with those of several Gag or RT precursors that would contain this TFP epitope were observed (data not shown). The blots also showed cross-reactive bands of about 18 and 22 kDa, which were often but not always observed and which appeared to be virus specific. Their origin is not clear at this point.
As an additional approach, we constructed an expression plasmid that encoded the Gag-Pro precursor with a hemagglutinin (HA) epitope tag at the carboxy terminus of TFP. Virus-like particles produced by cells transfected with the plasmid pCMV-gag-pro-TFPHA contained a protein which could be visualized with either anti-HA or anti-TFP antibodies (Fig. 1E). The HA-tagged TFP is predicted to be approximately 2 kDa larger than the wild-type protein, due to the additional 18 residues contributed by the HA tag and a Gly-Ala-Gly-Ala-Ser linker. These results again confirmed that TFP is part of the virion, that its carboxy terminus is indeed defined by the stop codon in the gag-pro reading frame, and that no additional proteolytic processing occurs close to this end of the protein.
To assess the importance of TFP for virus maturation and replication, we incorporated a series of stop codons into the TFP gene of the full-length proviral HTLV-1 clone, pX1MT. In general, mutations were made by overlapping PCR and transferred into pX1MT as a 576-bp PacI-to-BglII fragment. The sequence of the fragment was verified by DNA sequencing. The truncation clones were designated pX1MT-2600, pX1MT-2656, and pX1MT-2699 (Fig. 2A). The pX1MT-2600 mutation mimics the BLV configuration, introducing a termination codon right after the slippery stretch of the frameshift signal 25 codons following the C terminus of Pro. This mutation results in a change from a proline to a leucine residue in RT, whereas the other TFP mutations are silent in the pol frame and truncate TFP by 32 and 19 C-terminal residues, respectively. In addition to incorporating the stop codon mutations, we also eliminated TFP expression by inserting a nucleotide into the slippery stretch of the signal for the shift into the pol frame. Consequently, this construct (pX1MTpolfs) should express only Gag and Gag-Pro-Pol, not the Gag-Pro precursor.
FIG. 2.
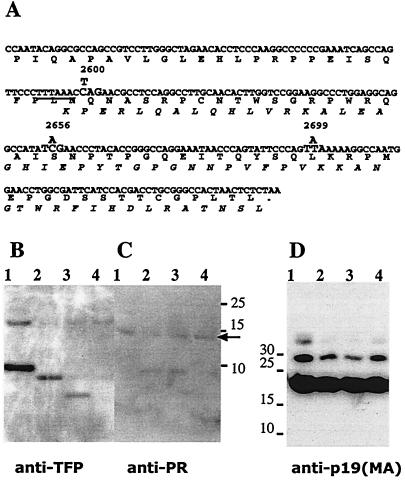
Immunoblots showing the effect of premature termination of TFP on Gag-Pro-derived peptide expression. (A) The coding and the predicted amino acid sequences of TFP and the amino terminus of RT after frameshifting at the slippery sequence (underlined). The nucleotide changes used to introduce stop codons into the TFP coding sequence are indicated above the sequence. (B) TFP reactive bands from representative immunoblot analysis of virus with no mutation (lane1), the pX1MT-2699 mutant (lane 2), the pX1MT-2656 mutant (lane 3), and the pX1MT-2600 mutant (lane 4). Samples were generated and processed as described in the legend for Fig. 1. (C) Parallel blot to that shown in panel B probed with anti-PR; the arrow indicates the PR band. The rabbit anti-PR antibody was raised against the synthetic peptide PAPRPLIKAQVDTQSHPKT. The bands seen at about 8 kDa are nonspecific contaminants from the media that are recognized by the secondary antibody. (D) The anti-p19 (MA) (mouse monoclonal antibody; ZeptoMetrix) blot of the same samples shown in panels B and C. All transfections were repeated a minimum of three times.
The truncated forms of TFP produced from the pX1MT-2656 and pX1MT-2699 mutants were detected in virus particles, while the truncated 25-residue-long peptide encoded by the pX1MT-2600 mutant could not be visualized by immunoblot procedures, regardless of gel system or transfer conditions (Fig. 2B). The most likely explanation is that the epitope was no longer recognized by the antiserum. The synthetic peptide, against which the antibodies were raised, extends another 6 amino acids beyond the end of the peptide encoded by pX1MT-2600. None of the mutations affected the size or level of PR in virus particles (Fig. 2C) or influenced the processing patterns of the Gag precursor (Fig. 2D). Similar results were found for pX1MTpolfs, where no TFP was observed but where PR and Gag peptides showed normal patterns (data not shown). These results, in conjunction with the infectivity studies discussed below, indicated that TFP itself does not play a significant role in virus maturation or infection. However, results with the deletion mutants did not address whether the proteolytic processing between PR and TFP is important in the regulation of PR activity.
Processing of the different precursor polyproteins must be tightly regulated and should occur only after assembly of the virus particle. The order of cleavage is mostly governed by the affinity of PR for its substrate sites, which is a function of primary sequence and protein conformation; the latter may change during the maturation process. The cleavage site at the end of PR was previously defined by amino acid sequencing (11, 12), and analyses by several groups using purified recombinant PR and short peptide substrates showed that the site was cleaved with the highest efficiency (5, 13). While it is not known whether this reflects the true order of cleavage in vivo, where at least initially this cleavage would have to be performed by the precursor, we reasoned that a mutant without the cleavage site might show impaired protease activity.
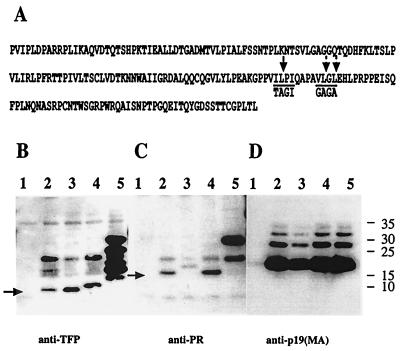
We mutated the known protease cleavage site, Val-Ile-Leu-Pro-Ile-Gln, to Val-Thr-Ala-Gly-Ile-Gln (Fig. 3A) in the proviral clone pX1MT-Psite1. Virus particles released from transfected 293 cells were analyzed by immunoblotting. The mutation resulted in an increase in the size of PR from 13 to 15 kDa without a compensatory change in size for TFP (Fig. 3B and C, lanes 3), suggesting the presence of an additional proteolytic cleavage site between PR and TFP. Inspection of the sequence offered only one possibility, Val-Leu-Gly-Leu, which was 8 residues distal of the known cleavage site. Given that the presence of a glycine in the P1′ position has been found to be detrimental (16), this site would be predicted to be cleaved very inefficiently (5, 9,16). We changed this sequence to Gly-Ala-Gly-Ala and analyzed virion proteins produced by the provirus mutant pX1MT-Psite2. As predicted, the PR from this mutant was indistinguishable from wild-type PR, while TFP increased in size (Fig. 3B and C, lanes 4). Virions derived from a proviral construct with mutations in both cleavage sites (pX1MT-Psite1/2) did not contain normal-size PR or TFP (Fig. 3B and C, lanes 5). Instead, the PR from this mutant appeared as a p24, of the size predicted for a Pro-TFP fusion; the anti-TFP antisera detected a heterogenous group of bands ranging in size from 11 to 24 kDa. Only the p24 and an additional band of about 18 kDa, which was present in all constructs, were shared between the anti-TFP and the anti-PR reactive material (see above). Taken together, the results indicate that both cleavage sites are necessary for the normal separation of PR and TFP, no other sites are normally utilized, and processing of the Gag-Pro precursor yields PR, p1, and p8 in addition to the Gag proteins MA, CA, and NC.
FIG. 3.
Effect of protease cleavage site mutations on proteolytic processing of precursor proteins. (A) Sequence of the PR-TFP part of the Gag-Pro precursor. The previously mapped cleavage site Psite1 (VIL/PIQ,) is indicated by a single downward arrow. Two arrows indicate the possible cleavage sites in Psite2. The sequences encoded by the mutants at the sites are indicated below the arrows. Samples were generated and processed as described in the legend for Fig. 1. Lane 1 contains the mock-transfected control. The provirus constructs used were pX1MT derived with either no mutation (lane 2), a Psite1 mutation (lane 3), a Psite2 mutation (lane 4), or both mutations (lane 5). (B to D) Immunoblots probed with anti-TFP (B), anti-PR (C), or anti-p19 (MA) (D). The arrows in panels B and C indicate the TFP and PR bands, respectively.
While the differences in relative amounts of TFP and PR in the single-mutation constructs were relatively small (Fig. 3B and C), there were always severalfold more anti-PR and considerably more anti-TFP reactive material in the pX1MT-Psite1/2 virions. Interestingly, none of the mutants showed a defect in Gag processing, as evidenced by the normal production of MA (Fig. 3D), indicating that neither the relative amounts of the protease nor the configuration at its C terminus had significant consequences for its activity.
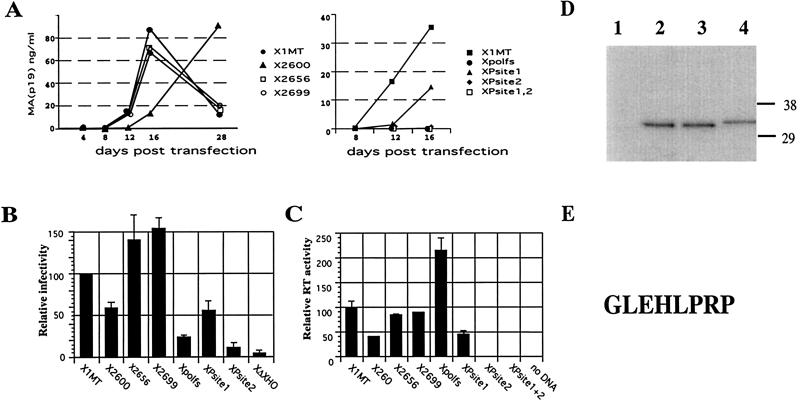
To assess the biological effects of the mutations, we performed spreading and single-cycle infections and measured reverse transcriptase activity. The ability of mutated viruses to sustain a spreading infection was tested in fetal rhesus lung cells (FRhL clone B5), which had previously been shown to support spreading infection with HTLV-1 (4). Infections were initiated by transfection of proviral DNA, and culture supernatants were tested for virus production by p19 (MA) antigen capture enzyme-linked immunosorbent assay (ELISA) at 4-day intervals over 14 to 16 days. The single-cycle assay monitors virus entry, reverse transcription, and integration by using recombinant virus particles generated by HTLV-1 vectors (6). Virus stocks were prepared by cotransfecting 293 cells with the wild-type or TFP mutant proviruses as packaging vectors in combination with an HTLV-1 transfer (reporter) vector. The reporter contains a cytomegalovirus promoter luciferase gene cassette, which replaces viral genes between the HTLV-1 long terminal repeats but retains the packaging signal. Filtered supernatants were used to infect 293 T cells, and extracts from these cells were prepared after 3 days and assayed for luciferase activity. To measure the reverse transcriptase activity in virus particles, we used filtered supernatants from 293 cells transfected with the provirus mutants. The reverse transcriptase activity of each sample is expressed as a percentage relative to that of the wild-type X1MT virus and was normalized with respect to the MA protein, whose concentration was determined by p19 (MA) antigen capture ELISA. All experiments were carried out several times, and representative results of these assays are shown in Fig. 4. Mutants with a C-terminal truncation of TFP behaved essentially like the wild type; only the pX1MT-2600 mutant showed a twofold drop in reverse transcriptase activity and in single-cycle infection, reflected in a delay of virus production in the spreading assay. The pX1MTpolfs mutant had twice the amount of polymerase activity but a fivefold drop in single-cycle infectivity and could not establish a spreading infection. Surprisingly, the pX1MT-Psite2 mutation abolished biological activity while the pX1MT-Psite1 mutation produced little effect. The pX1MT-Psite1 mutant showed only a twofold reduction in reverse transcriptase activity and single-cycle infectivity and a delayed onset of spreading infections. In contrast, the pX1MT-Psite2 and pX1MT-Psite1/2 mutants had activities comparable to that of the negative control in all assays.
FIG. 4.
Infectivity and reverse transcriptase activity of pX1MT derivatives with mutations in TFP. (A) Spreading infection of virus produced after transfection of the indicated constructs into B5 cells determined by measuring MA (p19) resulting from ELISA of supernatants. Results for stop codon mutants (left panel) and protease cleavage site mutants and pX1MTpolfs (right panel) are compared to those for pX1MT. The experiments were repeated three times with similar results. (B) Single-cycle infection assays. Full-length pX1MT and mutant derivatives were used to package a luciferase gene-carrying HTLV-1 retroviral vector following transfection of the two constructs into 293 cells. Two independent transfections were carried out for each construct in each experiment. Supernatants from the transfected cells were standardized relative to the p19 concentration and used to infect 293 T cells. Results for pX1MT were set as 100, and values obtained for the other constructs are shown relative to that value. X1MTΔXho is a negative control with a deletion in env. Experiments whose results are shown in panels A and B were carried out as previously described (6). (C) Reverse transcriptase activities associated with wild-type and mutant viruses. 293 cells were transfected with the indicated constructs, and reverse transcriptase activity was determined directly for 4- and 20-μl volumes of supernatant by using the HS sensitivity RT kit from Cavidi (Upsalla, Sweden) in accordance with the manufacturer's directions. The reverse transcriptase activity per nanogram of p19 in the supernatant, determined by p19 ELISA, was calculated, and the activity of pX1MT was set as 100. The reverse transcriptase activities of the other constructs are given relative to that value. Results are derived from at least three independent transfection experiments. (D) Immunoblot of viral proteins from pCMVHT (6)-derived constructs expressing a truncated HA-tagged RT (Fig. 1A) with either no mutation (lane 2), a Psite1 mutation (lane 3), or a Psite2 mutation (lane 4). Lane 1 was loaded with the pelleted supernatant from a mock transfection. (E) The sequence derived for the amino terminus of the HA-tagged RT peptide. Viral proteins produced by CMVHT-polfs-polHA, which has the U3 region of the 5′ LTR replaced by the CMV immediate-early promoter and an insertion of a nucleotide into the second frameshift site, were affinity purified on anti-HA-agarose (Roche). Bound proteins were separated on NUPAGE gels, followed by transfer to an Immobilon membrane and staining with Coomassie brilliant blue. N-terminal sequence determination by Edman degradation of protein in the candidate band was carried out in an Applied Biosystems model 494CLC Procise sequenator. Phenylthiohydantoin derivatives of amino acids were catalyzed online with an Applied Biosystems model 785A/140C/610A analyzer.
The logical interpretation of these data is that the additional cleavage site after PR is also used for the processing of RT and, thus, mutations affecting it may explain the RT-minus phenotype of the pX1MT-Psite2 mutant. To test this, we constructed plasmids that expressed a truncated RT with an HA tag at the end and contained either no mutation or the Psite1 or Psite2 mutation. As can be seen in Fig. 4D, the RT fragment produced by the construct with the Psite1 mutation is indistinguishable from that of the wild type, while changes at Psite2 resulted in a slightly larger protein. To definitively identify the cleavage site, we isolated the HA-tagged RT fragment by using a construct that overexpressed RT due to its pX1MTpolfs genotype. Amino-terminal sequencing performed on this protein verified that the cleavage site is indeed the one we identified by sequence inspection and determined the amino terminus of HTLV-1 RT to be GLEHLPR (Fig. 3E). Most earlier reports found that the start site of RT was defined by the cleavage site at the end of PR; however, these determinations had all been done in vitro with purified PR in trans (1, 17).
In summary, we found that the small transframe peptide from the end of the Gag-Pro precursor is retained in the HTLV-1 particle. However, only mutations affecting the first 24 residues that are shared with RT had significant consequences for virus replication in vitro, while deletion of the last 50 codons resulted only in a twofold drop in infectivity in our infectivity screen. This finding does not rule out the possibility that TFP is important for virus viability in vivo or in other cell types such as primary T cells or established T-cell lines. Deletion of the proximal half of the pX region of HTLV-1 did not have a deleterious effect on the virus in our experimental in vitro systems but was found to abolish infectivity in experimental animal model systems (2, 3, 7). Only two mutations affecting TFP expression prevented replication in vitro: one was a frameshift mutation resulting in loss of the Gag-Pro(-TFP) precursor expression. Although the virus with this mutation buds normally, and does not show any gross abnormalities on electron micrographs (data not shown), it seems likely that the additional amount of the bulky Gag-Pro-Pol precursor would impede virus assembly. The other mutation resulting in a negative phenotype affected the processing between PR and TFP. In the course of analyzing the effect of abolishing the cleavage site at the end of PR, we discovered an additional site 8 amino acids downstream. While changing the first site resulted in only a minor drop in replication, removal of the new site completely abolished infectivity, mainly due to the effects on RT. The N terminus of RT is defined by this newly discovered site, and RT is inactive with the 8-amino-acid extension at its amino terminus.
Acknowledgments
We thank S. Oroszlan for stimulating discussions, Young Kim for help with the protein sequencing, and Thomas Fanning for reading the manuscript.
This publication was funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. NO1-CO-12400.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does the mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
REFERENCES
- 1.Agbuya, P. G., N. E. Sherman, and L. K. Moen. 2001. Proteolytic processing of the human T-cell lymphotropic virus 1 reverse transcriptase: identification of the N-terminal cleavage site by mass spectrometry. Arch. Biochem. Biophys. 392:93-102. [DOI] [PubMed] [Google Scholar]
- 2.Bartoe, J. T., B. Albrecht, N. D. Collins, M. D. Robek, L. Ratner, P. L. Green, and M. D. Lairmore. 2000. Functional role of pX open reading frame II of human T-lymphotropic virus type 1 in maintenance of viral loads in vivo. J. Virol. 74:1094-1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collins, N. D., G. C. Newbound, B. Albrecht, J. L. Beard, L. Ratner, and M. D. Lairmore. 1998. Selective ablation of human T-cell lymphotropic virus type 1 p12I reduces viral infectivity in vivo. Blood 91:4701-4707. [PubMed] [Google Scholar]
- 4.Copeland, K. F., A. G. Haaksma, D. Derse, and J. L. Heeney. 1994. Detection of human T-cell leukaemia virus 1 permissive cells using cell lines producing selectable recombinant virions. J. Virol. Methods 50:219-225. [DOI] [PubMed] [Google Scholar]
- 5.Daenke, S., H. J. Schramm, and C. R. Bangham. 1994. Analysis of substrate cleavage by recombinant protease of human T cell leukaemia virus type 1 reveals preferences and specificity of binding. J. Gen. Virol. 75:2233-2239. [DOI] [PubMed] [Google Scholar]
- 6.Derse, D., S. A. Hill, P. A. Lloyd, H. K. Chung, and B. A. Morse. 2001. Examining human T-lymphotropic virus type 1 infection and replication by cell-free infection with recombinant virus vectors. J. Virol. Methods 75:8461-8468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Derse, D., J. Mikovits, and F. Ruscetti. 1997. X-I and X-II open reading frames of HTLV-I are not required for virus replication or for immortalization of primary T-cells in vitro. Virology 237:123-128. [DOI] [PubMed] [Google Scholar]
- 8.Derse, D., J. Mikovits, D. Waters, S. Brining, and F. Ruscetti. 1996. Examining the molecular genetics of HTLV-I with an infectious molecular clone of the virus and permissive cell culture systems. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 12:1-5. [DOI] [PubMed] [Google Scholar]
- 9.Ding, Y. S., D. H. Rich, and R. A. Ikeda. 1998. Substrates and inhibitors of human T-cell leukemia virus type I protease. Biochemistry 37:17514-17518. [DOI] [PubMed] [Google Scholar]
- 10.Gessain, A., F. Barin, J. C. Vernant, O. Gout, L. Maurs, A. Calender, and G. de The. 1985. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet ii:407-410. [DOI] [PubMed]
- 11.Hayakawa, T., Y. Misumi, M. Kobayashi, Y. Yamamoto, and Y. Fujisawa. 1992. Requirement of N- and C-terminal regions for enzymatic activity of human T-cell leukemia virus type I protease. Eur. J. Biochem. 206:919-925. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi, M., Y. Ohi, T. Asano, T. Hayakawa, K. Kato, A. Kakinuma, and M. Hatanaka. 1991. Purification and characterization of human T-cell leukemia virus type I protease produced in Escherichia coli. FEBS Lett. 293:106-110. [DOI] [PubMed] [Google Scholar]
- 13.Louis, J. M., S. Oroszlan, and J. Tozser. 1999. Stabilization from autoproteolysis and kinetic characterization of the human T-cell leukemia virus type 1 proteinase. J. Biol. Chem. 274:6660-6666. [DOI] [PubMed] [Google Scholar]
- 14.Osame, M., K. Usuku, S. Izumo, N. Ijichi, H. Amitani, A. Igata, M. Matsumoto, and M. Tara. 1986. HTLV-I associated myelopathy, a new clinical entity. Lancet i:1031-1032. [DOI] [PubMed]
- 15.Poiesz, B. J., F. W. Ruscetti, A. F. Gazdar, P. A. Bunn, J. D. Minna, and R. C. Gallo. 1980. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc. Natl. Acad. Sci. USA 77:7415-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tozser, J., G. Zahuczky, P. Bagossi, J. M. Louis, T. D. Copeland, S. Oroszlan, R. W. Harrison, and I. T. Weber. 2000. Comparison of the substrate specificity of the human T-cell leukemia virus and human immunodeficiency virus proteinases. Eur. J. Biochem. 267:6287-6295. [DOI] [PubMed] [Google Scholar]
- 17.Trentin, B., N. Rebeyrotte, and R. Z. Mamoun. 1998. Human T-cell leukemia virus type 1 reverse transcriptase (RT) originates from the pro and pol open reading frames and requires the presence of RT-RNase H (RH) and RT-RH-integrase proteins for its activity. J. Virol. 72:6504-6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshida, M., I. Miyoshi, and Y. Hinuma. 1982. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc. Natl. Acad. Sci. USA 79:2031-2035. [DOI] [PMC free article] [PubMed] [Google Scholar]