Abstract
Somatic cell nuclear cloning has repeatedly demonstrated striking reversibility of epigenetic regulation of cell differentiation. Upon injection into eggs, the donor nuclei exhibit global chromatin decondensation, which might contribute to reprogramming the nuclei by derepressing dormant genes. Decondensation of sperm chromatin in eggs is explained by the replacement of sperm-specific histone variants with egg-type histones by the egg protein nucleoplasmin (Npm). However, little is known about the mechanisms of chromatin decondensation in somatic nuclei that do not contain condensation-specific histone variants. Here we found that Npm could widely decondense chromatin in undifferentiated mouse cells without overt histone exchanges but with specific epigenetic modifications that are relevant to open chromatin structure. These modifications included nucleus-wide multiple histone H3 phosphorylation, acetylation of Lys 14 in histone H3, and release of heterochromatin proteins HP1β and TIF1β from the nuclei. The protein kinase inhibitor staurosporine inhibited chromatin decondensation and these epigenetic modifications with the exception of H3 acetylation, potentially linking these chromatin events. At the functional level, Npm pretreatment of mouse nuclei facilitated activation of four oocyte-specific genes from the nuclei injected into Xenopus laevis oocytes. Future molecular elucidation of chromatin decondensation by Npm will significantly contribute to our understanding of the plasticity of cell differentiation.
Epigenetic regulation of cell differentiation is surprisingly reversible, as demonstrated in many vertebrate species by somatic cell nuclear cloning, a procedure to create genetically identical animals by replacing egg nuclei with somatic cell nuclei (29, 38, 52). The most striking evidence of this reversibility is the establishment of fertile mouse clones by using nuclei isolated from terminally differentiated lymphocytes and olfactory sensory neurons (24, 33). Whereas the success rate of mouse cloning is less than 5% (72), some of the surviving mouse clones have unexpectedly normal gene expression profiles, as shown by proper expression of over 11,000 genes in the placentae and livers of newborn mouse clones (36, 69). Because no other experimental models with a comparable degree of genomic reversibility exist, with the exception of cell fusion between somatic cells and embryonic stem cells (20), nuclear cloning provides a valuable opportunity for us to investigate the mechanisms of genome-wide epigenetic reprogramming activities that are important for the future of regeneration medicine. One of the key questions in nuclear cloning is whether a few general reprogramming factors exist that can nonspecifically affect multiple genes in addition to the obviously necessary gene-specific activators and suppressors. Currently, there is no evidence to support the existence of such general reprogramming factors in egg cytoplasm.
Massive nuclear swelling accompanied by global chromatin decondensation is one of the hallmarks of nuclear reprogramming observed in Xenopus laevis cloning (29). When somatic nuclei are injected into Xenopus eggs (meiotic metaphase II), the nuclei swell up to 100-fold in volume within 1 hour, but they do not transcribe genes, reflecting physiological transcriptional silencing in eggs. When injected into oocytes (meiotic prophase), the nuclei swell more slowly, spending 3 days to accomplish the same 100-fold increase in volume (29), but they remain transcriptionally active during this period. The swollen nuclei in oocytes tend to show more active transcription than those that have not swollen, suggesting that the chromatin decondensation is not merely a morphological event but is also closely linked with an increase in overall nuclear activity. Given the significance of subnuclear compartmentalization and chromosomal domains as regulatory mechanisms for a number of genes (15), it is not surprising that the nuclear swelling and chromatin decondensation significantly impact the transcriptional status of the donor nuclei in oocyte cytoplasm. Nucleus-wide chromatin decondensation might facilitate reprogramming of the donor nuclei by derepressing condensed chromatin; however, there is a wide knowledge gap between chromatin decondensation at the microscopic level and derepression at the transcriptional level.
Nuclear swelling and chromatin decondensation in egg cytoplasm have mainly been studied in the more physiological context of sperm chromatin decondensation upon fertilization. Xenopus sperm decondensation is induced by the acidic nuclear protein nucleoplasmin (Npm), which is expressed in oocytes and early embryos (9). Npm was first purified from Xenopus eggs as a molecular chaperon which helps load histone onto DNA during nucleosome assembly in vitro (43). Through its histone-binding capacity, Npm also plays an important role in storing the maternally derived histone, especially histone H2A and H2B, to prepare for early development without zygotic transcription. Later, it was found that during sperm chromatin decondensation upon fertilization, Npm replaces sperm-specific basic proteins X and Y with egg histone H2A and H2B, resulting in assembly of somatic-type nucleosomes onto sperm DNA (59, 60). This phenomenon has been explained by interactions between the negatively charged Npm and the positively charged X and Y (32). Hyperphosphorylation of Npm, which occurs during maturation of oocytes into eggs, facilitates the histone replacement on sperm DNA and sperm decondensation (44), although exact phosphoamino acids have not been identified. Npm can also decondense erythrocyte nuclei by releasing the specialized linker histone H1o in addition to partial removal of H1 (22). Thus, chromatin decondensation by Npm has primarily been explained by histone exchanges through charge interactions between acidic Npm and histone or other basic proteins.
Xenopus Npm consists of 200 or 196 (four residues deleted) amino acids and forms a pentamer in vivo. The Npm monomer is composed of two domains, the N-terminal core domain containing 120 amino acids and the C-terminal tail domain. The core domain has a small acidic amino acid cluster (acidic tract A1) and an eight-stranded β-barrel that forms a wedge shape which provides an interface for pentamer assembly (23). The core domain binds to histone and is sufficient for in vitro nucleosomal assembly and sperm decondensation (3, 5). The tail domain contains two larger acidic tracts, A2 and A3, in addition to a bipartite nuclear localization signal. Homologous proteins have been isolated in Drosophila melanogaster (dNLP) (37), mice (mNpm2) (10), and humans (hNpm2) (10). mNpm2 is composed of 207 amino acids and shares 39.5% identity with Xenopus Npm at the amino acid level. Burns et al. found that sperm could be decondensed in mNpm2 knockout mouse oocytes, suggesting the existence of functionally redundant proteins in mouse oocytes (10). The primary defects in the mNpm2 null mice were dispersed nucleoli and female infertility.
We previously established an in vitro nuclear reprogramming assay by combining Xenopus egg extract and somatic cell nuclei to study the biochemistry of nuclear reprogramming in nuclear cloning (26, 40). In our current work we applied the assay to study widespread chromatin decondensation in somatic nuclei incubated in Xenopus egg extract, expecting that the decondensation factor might be one of the general reprogramming factors described above. In this assay we used decondensation of mouse cell centromeres as a convenient indicator to monitor the chromatin decondensation activity. Mouse centromeres are composed of two types of heterochromatin, centromeric heterochromatin and pericentric heterochromatin, each with distinct DNA components (46). Centromeric heterochromatin contains an approximately 300-kbp tandem repeat of the minor satellite sequence (120-bp repeating unit); pericentric heterochromatin contains an approximately 200- to 2,000-kbp repeat of the major satellite sequence (234-bp unit), depending on the chromosomes (39). These repeated sequences are also characterized by methylated DNA, histone H3 with trimethylated Lys 9 (tm-H3K9) (57, 62), and association with heterochromatin proteins HP1α and HP1β through binding to methylated H3K9 (4, 42, 50). Due to the highly condensed heterochromatin, mouse centromeres are clearly visible as DNA-dye-positive spots in interphase cells by using fluorescence microscopes. Such rich background information in centromeres makes them easy to detect and to characterize their decondensation in egg extract. For simplicity, we will refer to both centromeric and pericentric heterochromatin as centromeric heterochromatin in the following sections.
From our study based on the hypothesis that global decondensation of somatic cell chromatin in egg cytoplasm may facilitate reprogramming of the somatic cell genome, we found that Npm could decondense both euchromatin and centromeric heterochromatin, primarily in undifferentiated mouse nuclei. We show that this decondensation was not accompanied by histone release from DNA, unlike in sperm chromatin decondensation, but it was accompanied by a variety of epigenetic modifications. At the functional level, the chromatin decondensation facilitated new gene expression as shown by the nuclear transplantation into oocytes. This study provides new insight into the molecular and functional analyses of chromatin decondensation in the context of somatic cell nuclear cloning.
MATERIALS AND METHODS
Chromatin decondensation assay.
We prepared sperm nuclei and egg S-phase extract as described elsewhere (65). We treated F9 and other cells with 50 μg/ml digitonin to permeabilize the plasma membrane and nuclear envelope using the method described for lysolecithin (28). The standard chromatin decondensation reaction consisted of 1 × 105 nuclei, the energy-regenerating system (ERS; 1 mM ATP, 1 mM GTP, 20 mM phosphocreatine, and 100 μg/ml creatine phosphokinase), and egg extract or Npm at appropriate concentrations, with a total volume of 25 μl. Apyrase was used at 10 U/ml when needed. Buffer B (10 mM HEPES, pH 7.8, 110 mM NaCl, 4 mM MgCl2, 1 mM dithiothreitol, 1 μg/ml leupeptin, 2 μg/ml pepstatin A, 100 μM phenylmethylsulfonyl fluoride, 150 μM spermine, 500 μM spermidine, 10% glycerol, and 0.003% Triton X-100) was used as a control. After incubation of the reaction mix for 2 h at 25°C, the nuclei were fixed with 4% paraformaldehyde and spun onto coverslips by brief centrifugation for immunofluorescence microscopy. The nuclei were also collected by brief centrifugation after the reaction for immunoblotting. We added 10 μCi [γ-32P]ATP/reaction mixture to detect newly phosphorylated proteins by autoradiography. For the protein kinase A (PKA) reaction, 3 × 105 F9 nuclei were incubated with ERS and 60 μg/ml PKA (Sigma) for 2 h. Electron microscopy was performed as described previously (71).
Immunofluorescence microscopy and immunoblotting.
Sources of the primary antibodies were as follows: tm-H3K9, tm-H3K4, p-H3S10, p-H3S28, ac-H3K14, pan-acetylated histone H4, H1, and H1o/H5 (all from Upstate Biotechnology); p-H3T11 and p-H3T32 (Abcam); H2B, TIF1β, and ubiquitin (Chemicon); HP1β (Abcam and Santa Cruz); histone H3 and HDAC1 (EMD); poly(ADP)-ribose (BD Pharmingen); sumo (Zymed). DNA of F9 nuclei were counterstained with Topro 3 (Molecular Probes) and sperm DNA with Hoechst 33342 (Sigma). Fluorescence in situ hybridization (FISH) was performed as described elsewhere (67) using the major satellite probe of 597 bp that contained two 234-bp units. This probe was cloned from the F9 genomic DNA by PCR using the primers 5′-CGGGATCCTATGGCGAGGAAAACTGAAAAAGGTGG-3′ and 5′-CGGAATCCTTTCACGTCCTAAAGTGTGTATTTCTCATTTTCC-3′. Two hundred nuclei were observed in each experiment, and more than 80% of the nuclei displayed the results represented in the figures.
Purification of Npm from egg and oocyte extracts.
Xenopus egg extract and oocyte extract were applied to Q Sepharose, and bound protein was eluted by a linear gradient of NaCl from 75 mM to 1 M in buffer C (the same as buffer B except for the lack of spermine and spermidine). The fractions that showed chromatin decondensation, eluted by 200 to 350 mM NaCl, were pooled, dialyzed against buffer C, and applied to HiTrap heparin. Bound protein was eluted by an NaCl gradient in buffer C, and the activity was detected in the fractions containing 430 to 650 mM NaCl. In a similar way, the active fractions were sequentially applied to MonoS (350 to 460 mM NaCl eluted the activity) and hydroxyapatite HTP-10 (170 to 500 mM). All the columns, except for HTP-10 (Bio-Rad), were purchased from Amersham. Mass spectrometry of the purified protein was performed as previously described (26). Egg Npm (eNpm) was immunodepleted from egg extract using polyclonal anti-Npm antibody as described previously (22).
Preparation of rNpm.
cDNA corresponding to recombinant Npm (rNpm) wild type (WT), Npm146, and Npm120 were subcloned into the pET-21c(+) vector with six-His tag at the C terminus (Novagen). The recombinant proteins were expressed in Escherichia coli, extracted by sonication, and treated at 80°C for 20 min followed by centrifugation at 15,000 × g for 30 min. The proteins were purified from the supernatant with nickel resin (QIAGEN) and dialyzed against buffer C.
MNase digestion.
After incubation of F9 nuclei with the indicated reagents, the nuclei were isolated by brief centrifugation and digested with 3 U micrococcal nuclease (MNase) at 25°C for the appropriate time (14). The DNA was isolated with TRIzol (Invitrogen) and applied to Southern hybridization using the following probes: major satellite, the 69-bp fragment used previously (16); minor satellite, a 93-bp fragment taken from pMRI150 (41); histone H3, a 406-bp fragment prepared with KpnI and BamHI from the plasmid previously described (56); 18S rRNA, a 750-bp fragment prepared with BamHI and SphI from clone 63178 (ATCC).
Salt extraction of histone from F9 nuclei.
After incubation of F9 nuclei with 5 μM rNpm for 2 h, the reaction mix was centrifuged at 150 × g for 5 min to precipitate the nuclei. The supernatant was labeled as 0.11 M (see Fig. 3A, below). The pellet was resuspended with buffer B with 300 mM NaCl, incubated for 20 min at 25°C, and centrifuged to isolate extracted protein. We repeated the same procedure using buffer B with 600 mM NaCl and buffer B with 1 M NaCl sequentially to extract proteins from a single reaction mix. The supernatant and pellet were applied to two sodium dodecyl sulfate gels for staining with Coomassie brilliant blue R-250 and immunoblotting, respectively.
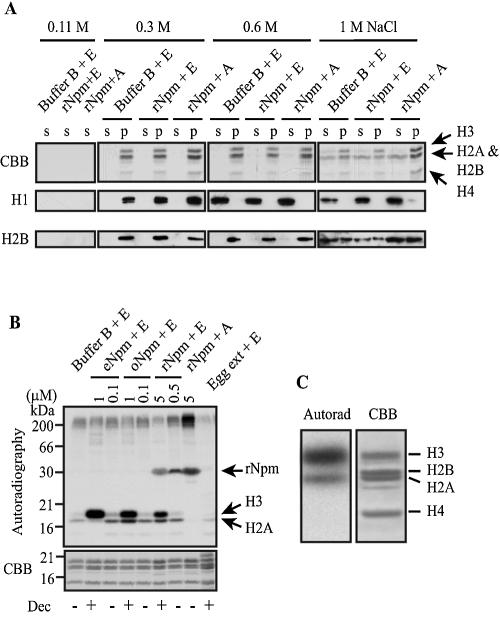
FIG. 3.
Histone was not released but phosphorylated by Npm. (A) Histone released from F9 nuclei into the supernatant (s) by various concentrations of NaCl and histone that remained in the nuclear pellet (p) were analyzed by Coomassie staining of the SDS gel (CBB) and immunoblotting with antibodies against histone H1 (H1) and H2B (H2B). The arrow indicates the height of each core histone confirmed by immunoblotting (not shown). (B) 32P was primarily incorporated into what appeared to be histone H3, H2A, and rNpm (reacted with anti-Npm antibody; not shown) when F9 nuclei were incubated with Npm. Autoradiography (top) and Coomassie staining (CBB) of the same gel are shown. Decondensation of centromeric heterochromatin was monitored by tm-H3K9 antibody and scored as + (with obvious decondensation) or − (without decondensation) at the bottom (Dec). (C) Higher magnification of the gel images to compare the sizes of the proteins incorporating 32P (Autorad) and core histones stained by the Coomassie dye (CBB). F9 nuclei were incubated with 5 μM rNpm and ERS. The gel used in panel C is different from that shown in panel B.
Nuclear transplantation into oocytes and PCR.
We defolliculated Xenopus oocytes, incubated at 18°C overnight in the MBS solution, and injected F9 nuclei into the surviving oocytes on the next day (11). We pretreated F9 nuclei with 1 μM eNpm, 3 μM polyglutamic acid (PGA), and buffer B separately for 2 h, injected 100 nuclei/oocyte into the germinal vesicles (GV) for each pretreatment, and cultured the oocytes in MBS with 50 μg/ml kanamycin at 18°C for up to 3 days. Total RNA was isolated from the pool of 10 oocytes at each day using TRIzol and treated with PCR-grade DNase I (Invitrogen). Each gene was amplified with 40 cycles of PCR using the MasterAmp reverse transcription-PCR (RT-PCR) kit (Epicentre). In each RT-PCR we compared gene activation in the three oocyte groups (eNpm, PGA, and buffer B) derived from a single frog to avoid variability among frogs. The primers for each gene were as follows: Msy2, 5′-TATGGTGGCAGAGGCTCCCTCAGGT-3′ and 5′-CCTTGGCTGGGCTTGGTCTCTCCAT-3′; H1foo, 5′-GGTGAGAAATCGAAGCCCTTGGCCAGC-3′ and 5′-GTCTTTGCCTTCCTGACCCTAGGTATGGGTTG-3′; cMos, 5′-CCCGAGGAAGGCAGGGAAGCTCTT-3′ and 5′-TCACTGATCAAAATGTTCGCTGGCTTCAGGTCC-3′; mNpm2, 5′-GCACCAGCAGCGTGACCGAAACC-3′ and 5′-GAGGCCTTCAGCGTAGCAATAGTGATTGGTGG-3′; Npm, 5′-AACCATGGTGGGCATTGAGCTGACC-3′ and 5′-TGGTGGCTGCAGGTCTCTTTACAGC-3′; major satellite, as described above. Identity of the PCR products was confirmed by DNA sequencing.
RESULTS
Xenopus Npm decondenses mouse centromeric heterochromatin.
Xenopus S-phase egg extract could induce massive swelling and decondensation of sperm chromatin within 30 min, as shown by DNA staining with Hoechst, which was consistent with previous reports (60) (Fig. 1A). The extract could also decondense centromeric heterochromatin in the mouse embryonal carcinoma F9 cells, as demonstrated by using anti-tm-H3K9 antibody and the DNA dye Topro 3. In contrast to the round centromeres observed in untreated F9 nuclei and the nuclei treated with buffer B for 2 h, the nuclei incubated with egg extract showed irregularly spread centromeres (Fig. 1A and B). Centromeric decondensation was also evident with FISH using the major satellite probe (Fig. 1A). The lack of obvious nuclear swelling in these experiments suggested that chromatin decondensation was not a secondary effect of nuclear swelling.
FIG. 1.
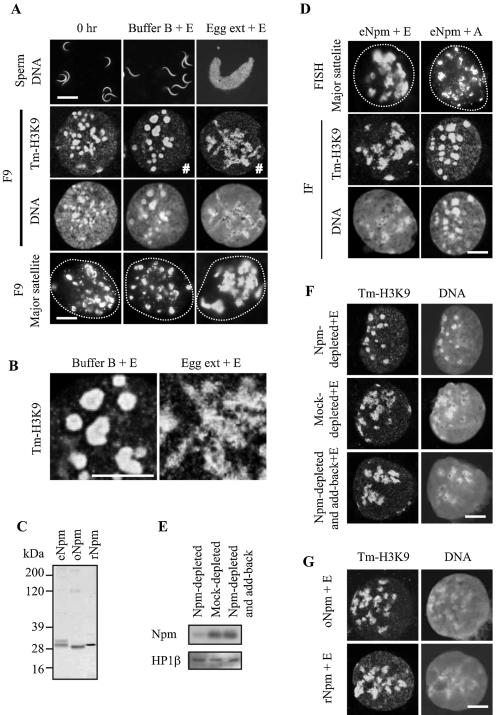
Decondensation of sperm and F9 chromatin by egg extract and Npm. (A) Decondensation of sperm chromatin and centromeric heterochromatin of F9 nuclei in egg extract. Sperm and F9 nuclei prior to incubation (0 h) and after incubation in buffer B and egg extract are shown. Incubation time was 2 h for F9 and 30 min for sperm, since swollen sperm nuclei became fragile after incubation for 30 min. ERS was included in the reactions indicated by +E. Sperm DNA was stained with Hoechst 33342, and F9 nuclei were stained with anti-tm-H3K9 antibody and Topro 3 (for DNA). F9 nuclei were also applied to FISH using the major satellite probe. Dotted lines in the FISH panels show nuclear contour. Bars, 10 μm for sperm and 3 μm for F9 in all images in the figure. (B) Central portions of the images marked by # in panel A were magnified threefold. (C) Silver-stained SDS-polyacrylamide gel electrophoresis gel loaded with Npm purified from eggs (eNpm) and from oocytes (oNpm) and rNpm. Hyperphosphorylation of eNpm and six-His tag on rNpm explain slower migration of these Npm compared with oNpm. (D) ERS-dependent centromeric heterochromatin decondensation in F9 nuclei by purified eNpm. F9 nuclei were incubated with 1 μM eNpm with ERS (+E) and apyrase (+A) for 2 h, followed by immunostaining and FISH. (E) Immunoblotting of egg extract immunodepleted with Npm antibody (Npm-depleted), with normal rabbit immunoglobulin G (Mock-depleted) and with anti-Npm antibody plus Npm add-back (Npm-depleted and add-back). The remaining Npm in these egg extracts was detected by anti-Npm antibody. HP1β was used as a loading control. (F) Immunofluorescence of F9 nuclei incubated in Npm-depleted egg extract followed by staining with tm-H3K9 antibody and Topro 3. (G) Immunofluorescence of F9 nuclei stained with tm-H3K9 antibody and Topro 3 after incubation with 1 μM oNpm and 5 μM rNpm in the presence of ERS for 2 h.
We purified the centromeric decondensation activity from egg extract by using dispersal of the tm-H3K9 signal as an indicator and found that eNpm was capable of decondensing centromeric heterochromatin (Fig. 1C and D). In addition, eNpm did not require any other factors supplied from egg extract for the heterochromatin decondensation, but it did require addition of ERS (Fig. 1D) (ERS regenerates ATP and GTP; apyrase hydrolyzes ATP and ADP). Although Npm, a thermostable protein, has traditionally been purified with heat treatment to precipitate and eliminate the majority of egg proteins (64), we did not use this method due to potential loss of other chromatin decondensation activities. During the purification process, the centromeric decondensation activity was always cofractionated with eNpm, as seen in immunoblotting using anti-Npm antibody (not shown). The purified active protein was confirmed to be Npm by mass spectrometry. By immunodepletion, we were able to confirm that eNpm was the protein responsible for the centromeric decondensation activity existing in egg extract. The extract depleted of eNpm lost the decondensation activity, unlike mock-depleted extract, and the activity was restored by adding back 1 μM eNpm (Fig. 1E and F). The eNpm pentamer concentration is estimated to be about 7 μM in egg extract (60), and the decondensation activity was still observed at up to a 10-fold dilution of the extract (not shown). Purified eNpm was effective at 1 μM or higher, roughly consistent with the results of diluted egg extract. Although oocyte extract could not decondense centromeric heterochromatin, Npm purified from oocytes (oNpm) could decondense centromeric heterochromatin as efficiently as eNpm, suggesting the existence of an Npm inhibitor(s) in oocytes (Fig. 1C and G). This is consistent with previous nuclear cloning studies that showed significantly slower swelling and chromatin decondensation in donor nuclei injected into oocytes compared with those injected into eggs (29). Bacterial rNpm could also decondense centromeres but required at least 5 μM for this purpose, presumably due to aggregation and/or lack of phosphorylation (Fig. 1G).
The centromeric heterochromatin decondensation by Npm and egg extract was also detectable in mouse embryonic stem (ES) cells, embryoid body cells, and embryonal carcinoma P19 and S2 cells. However, this decondensation was undetectable in mouse myoblasts C2C12, primary embryonic fibroblasts, established embryonic fibroblasts 10T1/2, and NIH 3T3 cells, melanoma B16 cells, and breast cancer 4T1 cells (Fig. 2A shows NIH 3T3 results). This tendency of undifferentiated cell-specific heterochromatin decondensation by Npm appears to be relevant to Gurdon's data, with nuclear swelling and chromatin decondensation upon injection into oocytes that were more rapid in embryonic nuclei than in differentiated cell nuclei (27).
FIG. 2.
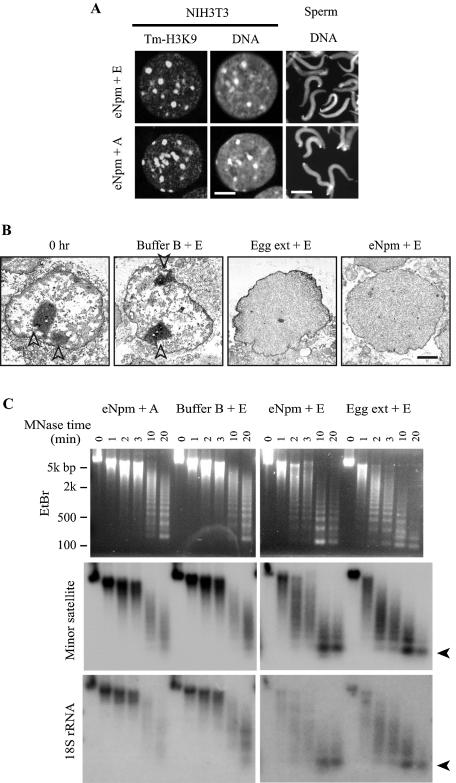
Decondensation of F9 chromatin by Npm and egg extract. (A) eNpm moderately decondensed sperm chromatin regardless of ERS but did not decondense centromeric heterochromatin in NIH 3T3 nuclei even with ERS. Both nuclei were incubated with 1 μM eNpm for 2 h and stained with Hoechst 33342 (sperm) and tm-H3K9 antibody plus Topro 3 (NIH 3T3). Bars, 3 μm for NIH 3T3 and 10 μm for sperm. (B) Electron microscopy of F9 nuclei after incubation in the described solutions. Bar, 1 μm. Arrowheads indicate nucleoli. (C) Npm facilitated sensitivity of F9 chromatin to the MNase digestion. F9 nuclei were incubated in each solution for 2 h and digested with MNase for the indicated time. Genomic DNA was isolated from the nuclei and applied to Southern hybridization. Ethidium bromide staining (top) and hybridization signal using the probes encoding minor satellite and 18S rRNA are shown. Arrowheads indicate mononucleosomes.
It has been well-established that Npm can decondense sperm chromatin regardless of the presence of ERS (Fig. 2A) (5, 60). However, this decondensation was less drastic than in egg extract due to the lack of additional components, such as histone H2A and H2B, which are necessary for full chromatin decondensation. In contrast, centromeric heterochromatin decondensation in F9 nuclei by Npm was strictly dependent on ERS, suggesting that the two types of chromatin decondensation were based on different mechanisms (discussed later). In subsequent assays we found that GTP was not essential for chromatin decondensation, but we continued to use both ATP and GTP so as not to miss subtle GTP-dependent decondensation if it existed. It is important to note that the F9 nuclei were deliberately permeabilized by the detergent digitonin, making nuclear import of Npm an unlikely mechanism for the ATP requirement in chromatin decondensation.
Npm widely relaxes chromatin structure.
Electron microscopy demonstrated a marked change in the subnuclear distribution of electron-dense material when F9 nuclei were treated with eNpm (Fig. 2B). Untreated F9 nucleoplasm contained many irregularly distributed electron-dense regions, especially under the nuclear envelope (perinuclear heterochromatin). The electron-dense material was more evenly distributed in the entire nucleoplasm in over 90% of 50 nuclear sections after incubation in egg extract or with eNpm in the presence of ERS, unlike in buffer B. The electron-dense material is usually interpreted to represent aggregation of protein, RNA, and DNA and does not necessarily correspond to transcriptionally inactive regions; however, this finding indicates that the influence of Npm was widespread, not limited to centromeres.
Npm increased the sensitivity of F9 chromatin to MNase, which preferentially digests linker DNA and produces a ladder of bands corresponding to the multiples of the nucleosomal core plus linker DNA (roughly 200 bp) (14). Increased MNase sensitivity is generally interpreted to reflect relaxed chromatin and is frequently associated with gene activation. Ethidium bromide staining of the agarose gels showed that the DNA ladder appeared after a 10-min treatment with MNase in the F9 nuclei that were incubated for 2 h with buffer B plus ERS, or with eNpm plus apyrase (Fig. 2C, EtBr). In contrast, the nuclei incubated with eNpm or egg extract, both with ERS, produced the ladder after only a 2-min treatment with MNase, which indicated that Npm significantly facilitated the MNase digestion of linker DNA.
Southern hybridization of the MNase-digested fragments with specific DNA probes showed chromatin decondensation more clearly. Four DNA probes, including major and minor satellites for heterochromatin and the genes encoding 18S rRNA and histone H3 for euchromatin, were used for easy detection, since they are all multicopy sequences. As shown in Fig. 2C, hybridization with the minor satellite and 18S rRNA probes showed that eNpm and egg extract clearly increased the sensitivity of F9 chromatin to MNase in the presence of ERS, compared with buffer B plus ERS and eNpm plus apyrase. The same results were obtained with the major satellite and histone H3 probes (not shown). These MNase data are evidence that both euchromatin and centromeric heterochromatin became decondensed at the nucleosomal level when F9 nuclei were incubated with egg extract and Npm with a strict requirement of ATP. In contrast, Npm could not increase MNase sensitivity in NIH 3T3 nuclei (not shown), consistent with the lack of centromeric heterochromatin decondensation in immunostaining. We also tested, by using DNA methylation-sensitive restriction enzymes, to see if DNA methylation in centromeres was diminished during decondensation in F9 nuclei, but no significant DNA demethylation was observed (not shown).
Npm induces multiple histone H3 phosphorylation in somatic nuclei.
Decondensation of sperm and erythrocyte chromatin by Npm is explained by the release of histone variants from DNA; however, we could not detect histone release from F9 nuclei after incubation with Npm (Fig. 3A, 0.11 M). In addition, step-wise washes of the nuclei with three different concentrations of NaCl showed that each histone subtype was extracted at the same concentration regardless of Npm and ERS, indicating that the binding strength of histone to DNA was not significantly altered by Npm (Fig. 3A, 0.3 M, 0.6 M, and 1 M NaCl). This lack of obvious histone release combined with the absolute dependence on ATP for the decondensation of F9 chromatin strongly argues that Npm decondenses chromatin in sperm and F9 nuclei by different mechanisms. Histone H1o, which could be released from erythrocyte nuclei by Npm, was undetectable in F9 nuclei by immunoblotting (not shown).
At least two possible mechanisms exist to explain the requirement of ATP during the decondensation of F9 chromatin by Npm. First, ATP may be necessary for phosphorylation of unidentified chromatin proteins during decondensation. Alternatively, ATP may be used as an energy source for chromatin remodeling proteins, such as SWI/SNF, that may be involved in relaxing nucleosomal structures. We have previously shown that one such member, ISWI, could release TATA-binding protein from somatic chromatin with the help of other unidentified molecules in egg extract (40), but ISWI alone could not decondense somatic chromatin (not shown). In the present work we focused on the first possibility by incubating F9 nuclei and Npm with [γ-32P]ATP for 2 h to study the global pattern of protein phosphorylation. With striking specificity, this experiment showed that a protein band, corresponding to the size of histone H3, was dominantly phosphorylated in the presence of Npm at concentrations that could induce chromatin decondensation (Fig. 3B and C, 1 μM eNpm and oNpm and 5 μM rNpm). In addition, a protein with the size of histone H2A was weakly phosphorylated, but its phosphorylation was undetectable when F9 nuclei were incubated with 1 μM eNpm, despite clear chromatin decondensation. This suggests that this phosphorylation may not be a prime candidate for the chromatin decondensation mechanism. It was perplexing at this stage that egg extract did not induce phosphorylation of what appeared to be histone H3 and H2A despite its capability to decondense chromatin, but later it was discovered that histone H3 was in fact phosphorylated and rapidly dephosphorylated in egg extract (see below). Because phosphorylation of what appeared to be histone H2A was not consistent with chromatin decondensation, we focused on the histone H3 phosphorylation in the following study.
Immunoblotting revealed that at least four amino acids, Ser 10, Ser 28, Thr 11, and Thr 32, in histone H3 were phosphorylated in F9 nuclei specifically in the presence of rNpm (Fig. 4A). Phosphorylation of Ser 10 (p-H3S10), observed in chromosome condensation in mitotic cells and gene activation (chromatin opening) in interphase cells, might be involved in chromatin relaxation to increase accessibility of chromatin to various factors (see Discussion). Physiological roles for p-H3S28 and other H3 phosphorylation sites in mitotic and interphase cells have not yet been elucidated (61).
FIG. 4.
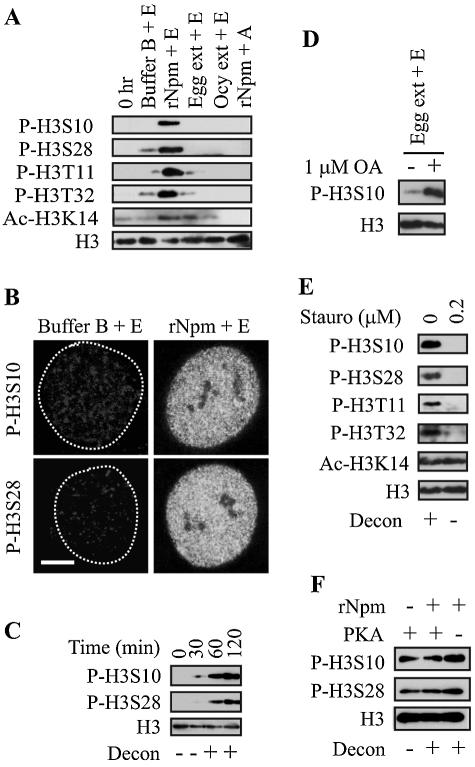
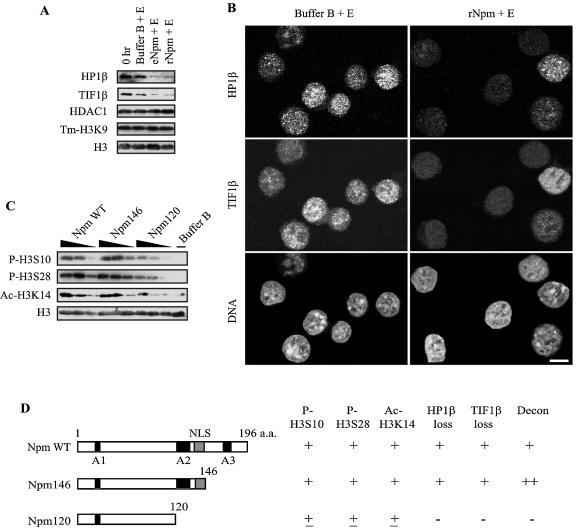
Histone H3 phosphorylation and acetylation by Npm. (A) Immunoblotting demonstrating histone H3 phosphorylation and acetylation in F9 nuclei by 5 μM rNpm. Total histone H3 was used as a loading control. The figures were prepared from more than one membrane in panels A and E because of the significantly diminished signal due to the repeated stripping of the membrane. (B) Immnofluorescence shows that H3S10 and H3S28 were globally phosphorylated when F9 nuclei were incubated with 5 μM rNpm. Bar, 3 μm. (C) Time course of histone H3 phosphorylation in F9 nuclei incubated with 5 μM rNpm. (D) Okadaic acid (OA) unmasked p-H3S10 in F9 nuclei incubated in egg extract. (E) Immunoblotting shows inhibition of histone H3 phosphorylation and chromatin decondensation by staurosporine. ac-H3K14 was not inhibited by staurosporine. (F) PKA phosphorylated histone H3 when incubated with F9 nuclei but could not induce centromeric heterochromatin decondensation without rNpm.
Several cytokines and hormones can induce both p-H3S10 and acetylation of Lys 14 in histone H3 (ac-H3K14), which work synergistically during gene induction by these external stimuli (17, 45). Consistent with these reports, ac-H3K14 was also induced by rNpm as well as by egg extract in F9 and NIH 3T3 nuclei (Fig. 4A for F9), but these histone modifications may not necessarily be mechanistically linked, as shown below. While p-H3S10 was observed in 100% of more than 50 experiments, ac-H3K14 was detected in approximately 50% of the cases, for unknown reasons.
Immunofluorescence microscopy demonstrated that p-H3S10 and p-H3S28 were distributed throughout the entire nuclei of F9 cells, revealing the nucleus-wide effect of Npm, which was distinct from p-H3S10 specific to a few activated genes in interphase cells mentioned briefly above. Phosphorylation of the four sites on histone H3 started to accumulate in F9 nuclei after incubation for 1 h with rNpm. It reached maximum intensity at 2 h and remained at the same level thereafter, which was consistent with the progress of chromatin decondensation in the nuclei (Fig. 4C for Ser 10 and 28). Phosphorylation of these amino acids was also observed in P19, ES, C2C12, B16, 10T1/2, and NIH 3T3 cells, but these nuclei, except for P19 and ES, did not show chromatin decondensation by Npm as monitored with tm-H3K9 antibody and the MNase digestion pattern (not shown). From this finding, H3 phosphorylation does not appear to be a consequence of chromatin decondensation (see Discussion).
As shown in Fig. 3B and 4A, histone H3 was not significantly phosphorylated in egg extract despite its strong chromatin decondensation capability, which potentially can be attributed to rapid dephosphorylation by unidentified phosphatases in egg extract. Through screening of phosphatase inhibitors, we found that phosphorylation of Ser 10 (Fig. 4D) and three other amino acids on histone H3 was unmasked in egg extract that was mixed with 1 μM or higher okadaic acid, a relatively specific inhibitor of protein phosphatases PP2A, PP4, and PP5 (31) (not shown). Okadaic acid treatment of intact HeLa cells at this concentration induces premature mitosis-like chromatin condensation (2), but in our case, egg extract treated with okadaic acid induced the same or even stronger chromatin decondensation than untreated egg extract (not shown). This finding leads us to speculate that the H3 phosphorylation in egg extract with okadaic acid was not due to premature entrance into mitosis. Similar rapid turnover of phosphorylation status has previously been reported in Drosophila polytene chromosomes, where heat-shock-induced p-H3S10 was masked by PP2A as revealed by using PP2A inhibitors (55).
After screening several protein kinase inhibitors, we found that staurosporine, a broad-spectrum inhibitor covering CaM kinase, myosine light chain kinase, and protein kinases A, C, and G, could, at 0.2 μM or higher concentrations, inhibit uptake of radioactive inorganic phosphate (not shown) as well as phosphorylation of the four amino acids on histone H3 (Fig. 4E). We then tested specific inhibitors to each of these protein kinases individually as well as in various combinations, but none of them could inhibit the H3 phosphorylation (not shown), which may indicate another unidentified kinase(s) was involved in the histone H3 phosphorylation. Importantly, 0.2 μM staurosporine also inhibited chromatin decondensation, potentially linking H3 phosphorylation and chromatin decondensation. However, we do not know at this stage if H3 phosphorylation was the cause or effect of chromatin decondensation, nor if staurosporine inhibited the H3 kinase directly or indirectly. Staurosporine did not inhibit ac-H3K14 induced by rNpm in F9 nuclei, suggesting that ac-H3K14 and histone H3 phosphorylation may not be mechanistically linked. To examine if H3 phosphorylation was sufficient to decondense chromatin, we incubated F9 nuclei with PKA, which is known to phosphorylate H3S10 and H3S28 (61). While PKA could indeed phosphorylate these two Ser residues, it could not decondense chromatin without Npm. This finding raises the possibility that p-H3S10 and p-H3S28 might be necessary but insufficient to induce chromatin decondensation. We also tested if other histone modifications were affected by Npm and egg extract but could not detect any significant changes in the level of other histone covalent modifications (not shown). These modifications included trimethylation of Lys 4 in histone H3 (enriched in transcriptionally active loci), acetylation of histone H4 (using pan-acetylated H4), ubiquitination, sumoylation, and poly(ADP-ribosyl)ation (involved in relaxation of Drosophila polytene chromosomes) (70). In summary, Npm specifically induces multiple H3 phosphorylation and ac-H3K14 during chromatin decondensation. Although H3 phosphorylation alone is not sufficient to induce chromatin decondensation, the staurosporine experiment shows the possible link between H3 phosphorylation and chromatin decondensation.
Npm releases heterochromatin proteins from F9 nuclei.
HP1α and HP1β are well-characterized heterochromatin proteins enriched in centromeres through binding to methylated H3K9 (4, 42, 50). TIF1β interacts with HP1 and functions as a corepressor for the KRAB domain-containing transcription factors (12). Both HP1β and TIF1β were substantially lost from F9 nuclei without significant loss of tm-H3K9 after incubation with Npm for 2 h as demonstrated by immunoblotting (Fig. 5A). Another centromeric component, histone deacetylase 1 (HDAC1) (21), was not decreased by Npm, suggesting selective release of heterochromatin components from centromeres. In immunofluorescence microscopy both HP1β and TIF1β, initially diffusely distributed throughout the nuclei, became almost undetectable in more than 80% of the nuclei after incubation with rNpm (Fig. 5B). HP1β and TIF1β were also lost from F9 nuclei incubated in egg extract (not shown). Unlike in F9 nuclei, these two proteins were highly localized in centromeres in more than 90% of NIH 3T3 cells and were also lost from the nuclei by Npm (not shown). Since NIH 3T3 nuclei did not show centromeric decondensation, the loss of these heterochromatin proteins was clearly insufficient for the decondensation. However, the loss of HP1β and TIF1β also required ATP and the loss was inhibited by 0.2 μM staurosporine (not shown), implying a mechanistic link between H3 phosphorylation (and potentially other proteins) and the release of the heterochromatin proteins.
FIG. 5.
Npm releases heterochromatin proteins from F9 nuclei. (A) HP1β and TIFβ were lost from F9 nuclei by Npm as shown by immunoblotting. (B) Immunofluorescence confirmed loss of HP1β and TIF1β from F9 nuclei by rNpm. Bar, 5 μm. (C) Dose-dependent histone H3 modifications by various forms of rNpm. Each rNpm was used at 5, 1.5, and 0.5 μM (black triangles). (D) Summary of epigenetic modification in F9 nuclei incubated with various forms of rNpm. The scores (− to ++) are intended to show overall tendency, not exactly quantitative values.
Acidic tract A2 of Npm is the key for chromatin decondensation, histone modifications, and release of heterochromatin proteins.
To identify the Npm domains necessary for chromatin decondensation, H3 phosphorylation and acetylation, and release of heterochromatin proteins, we prepared two deletion mutants and incubated them with F9 nuclei. Npm146, which lacked the C-terminal 50 amino acids, was slightly more potent in chromatin decondensation activity than the wild type, indicating that the third acidic tract A3 is not essential (Fig. 5D). In contrast, Npm120, which further lost the second acidic tract A2 and the nuclear localization signal, substantially lost activities of chromatin decondensation, H3 phosphorylation and acetylation, and release of heterochromatin proteins (Fig. 5C and D). It is likely that the result was due to the loss of A2 and not the nuclear localization signal, because digitonin had permeabilized the nuclear envelope of F9 cells. Polyanions, such as heparin and PGA, are known to induce chromatin decondensation, but the mechanisms have not been elucidated (6). In our in vitro assay 3 μM or a higher concentration of PGA (average, 75-kDa 510-mer) could indeed decondense centromeric heterochromatin, phosphorylate multiple H3 sites, acetylate H3K14, and release HP1β and TIF1β from F9 nuclei, all dependent on the presence of ATP, just like Npm (not shown). In addition, all these chromatin events were inhibited by 0.2 μM staurosporine, which is evidence that seemingly nonspecific chromatin decondensation by PGA was in fact based on specific mechanisms. Furthermore, these results indicate that such diverse epigenetic modifications could be accomplished by a simple acidic amino acid stretch without the specific three-dimensional protein structure observed in Npm.
Npm facilitates nuclear reprogramming in nuclear transplantation.
To understand functional consequences of chromatin decondensation by Npm, it would have been ideal if we could detect activation of new genes from the Npm-treated F9 nuclei in vitro, but F9 nuclei did not take up [32P]CTP in buffer B regardless of the Npm treatment (not shown). We also transfected F9 cells with Npm cDNA using a plasmid vector but could not obtain an expression level high enough to detect decondensation of centromeric heterochromatin (not shown). As an alternative approach we injected Npm-treated F9 nuclei into Xenopus oocytes and monitored activation of oocyte-specific genes from the F9 nuclei. We employed this strategy because of two advantages the oocytes provided for our purpose. First, the injected nuclei remain transcriptionally active over 7 days without DNA replication or mitosis, significantly simplifying interpretation of the results (30). Second, Xenopus oocytes can activate transcription from injected nuclei of other species, including mouse and human (11). It is important to note that although F9 cells are derived from undifferentiated embryonal carcinoma cells, they cannot differentiate into any tissues when injected subcutaneously into mice (nullipotent) (7), indicating that chromatin in F9 cells is not necessarily relaxed and ready to be activated prior to the Npm treatment.
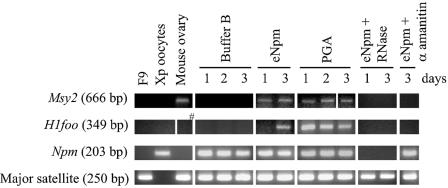
We pretreated F9 nuclei with eNpm, PGA, and buffer B for 2 h, injected these nuclei separately into oocyte nuclei (called GV), and incubated the oocytes for 3 days. We then monitored activation of four mouse oocyte-specific genes, Msy2 (encoding an mRNA-masking protein), H1foo (oocyte-specific histone H1 variant), c-Mos (arrests eggs at meiotic metaphase II), and mNpm2 (mouse nucleoplasmin), from the injected F9 nuclei by RT-PCR. Major satellite DNA (for the number of injected nuclei) and Xenopus Npm mRNA (for the amount of total RNA) were used for controls of PCR. The results showed that unspliced Msy2 (666 bp) was detectable from the eNpm- and PGA-pretreated F9 nuclei, but not from the buffer B-pretreated F9, between 1 and 3 days after oocyte injection in 9 out of 17 experiments (Fig. 6). When eNpm-pretreated F9 nuclei expressed Msy2, the PGA-pretreated F9 also expressed Msy2 and vice versa, as far as the oocytes used for RT-PCR were derived from a single frog. In addition, spliced Msy2 (239 bp) was detectable only from the eNpm- and PGA-pretreated F9 nuclei in two additional experiments (not shown). Likewise, unspliced H1foo (three out of six experiments) and mNpm2 and cMos (both in two experiments out of four) were expressed only from the eNpm- and PGA-pretreated nuclei injected into oocytes (Fig. 6 for H1foo). Spliced forms were undetectable for these three genes. In these experiments, there was a general tendency that the oocyte pools that expressed one of these genes expressed other genes as well, suggesting that the oocyte quality was the major limiting factor for activation of these dormant genes.
FIG. 6.
Expression of oocyte-specific genes from Npm and PGA-pretreated F9 nuclei injected into Xenopus oocytes. RT-PCR of Msy2 and H1foo (both unspliced forms) comparing intact F9 cells, Xenopus oocytes, and mouse ovary and Xenopus oocytes injected with buffer B-pretreated F9 nuclei (buffer B), injected with eNpm-pretreated F9 (eNpm), injected with PGA-pretreated F9 (PGA), or injected with Npm-pretreated F9 plus 30 μg/ml α-amanitin (eNpm+α amanitin). In eNpm+RNase, RNA isolated from Xenopus oocytes injected with Npm-pretreated F9 was treated with RNase prior to RT-PCR. Sizes of the amplified bands are shown in parentheses. Npm and major satellite were used as controls. The post-oocyte injection period is shown in days. Note that unspliced H1foo was undetectable in mouse ovary (#), the positive control. Only spliced H1foo was detectable in that sample (not shown).
It is unknown why mainly unspliced forms of these genes were detectable, but several reasons excluded the possibility that these amplified genes were in fact derived from genomic DNA. First, when we added DNase-free RNase into the RT-PCR tube, the PCR products became undetectable (Fig. 6). Second, all RNA was treated with DNase I prior to RT-PCR. Third, when we coinjected α-amanitin, an RNA polymerase II inhibitor, with F9 nuclei, these genes were not amplified (Fig. 6). Since the Epicentre RT-PCR kit used gene-specific primers for both reverse transcription and PCR to increase the sensitivity, there was no PCR without reverse transcription, the standard control reaction to test genomic DNA contamination. These findings indicate that Npm and PGA treatment increased the capability of F9 nuclei to transcribe new genes, most likely by increasing the accessibility of chromatin. We repeated the same experiments with B16 and NIH 3T3 cell nuclei, but they did not express new genes upon injection into oocytes (not shown).
DISCUSSION
In this work we found that Xenopus Npm could decondense centromeric heterochromatin as well as euchromatin primarily in undifferentiated mouse cell nuclei, rendering the nuclei competent to transcribe new genes upon injection into Xenopus oocytes. Immunodepletion experiments confirmed that Npm was essential for the chromatin decondensation activity existing in Xenopus egg extract. Although Npm's role in sperm chromatin decondensation is already known, the ATP requirement and lack of histone exchange indicate that the chromatin decondensation in F9 cells and sperm appear to be based on different mechanisms. The chromatin decondensation in F9 cells was accompanied by various epigenetic modifications which are associated with open chromatin structure: nucleus-wide multiple H3 phosphorylation, ac-H3K14, and release of heterochromatin proteins HP1β and TIF1β from the nuclei. Surprisingly, a simple stretch of Glu residues could trigger these specific and multiple epigenetic chromatin modifications as long as ATP was present. These findings identify Npm as one of the key reprogramming activities in eggs with wider effects on somatic nuclei than previously described.
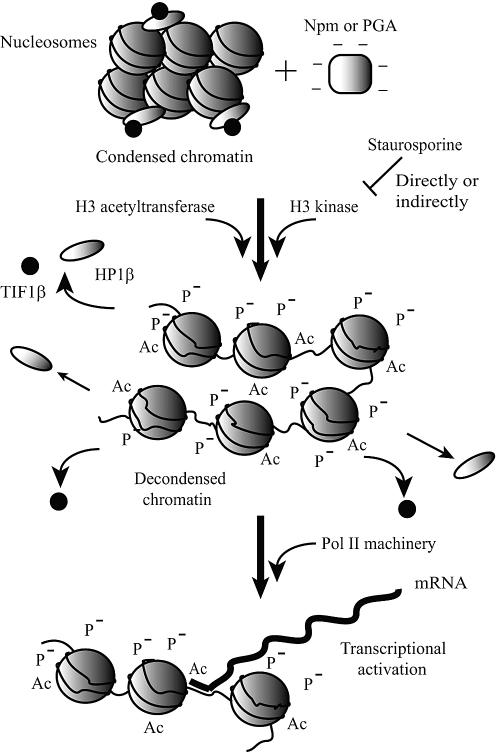
Although at this stage we do not know the precise order of these events or their causal relationships, the results of the A2 deletion mutant (diminished activity in all these chromatin events) and staurosporine (inhibited all except for ac-H3K14) potentially link these chromatin modifications. In addition, since Npm did not induce chromatin decondensation in NIH 3T3 cells despite H3 phosphorylation, ac-H3K14, and heterochromatin protein loss, chromatin decondensation appears to be downstream of these chromatin modifications. Based on these findings, we propose the following working hypothesis (Fig. 7). Npm and PGA activate or recruit a histone H3 kinase(s) and a histone acetyltransferase(s), which modify the surface charge and/or three-dimensional structure of chromatin, leading to release of heterochromatin proteins and potentially other histone binding proteins. As a cumulative effect of these multilayer epigenetic modifications, the chromatin structure relaxes and chromatin accessibility increases, which raises MNase sensitivity and creates the permissive status for activation of new genes when necessary transcription factors are provided externally. This hypothetical pathway does not explain why chromatin of NIH 3T3 and other differentiated cells could not be decondensed by Npm despite all these epigenetic modifications (see below for potential differences of chromatin structure between differentiated and undifferentiated cells). However, this hypothetical pathway will serve as a good starting point for the future investigation of the signaling cascade of chromatin decondensation. The literature states that the HP1 family proteins are constantly binding to and dissociating from heterochromatin without obvious loss of tm-H3K9 as shown by the fluorescence recovery after photobleaching experiments (18, 25). Also, the combination of p-H3S10 and ac-H3K14 facilitates dissociation of HP1 from methylated H3K9 (48). In addition to HP1, a number of other chromatin proteins are constantly associating with and dissociating from chromatin on the order of seconds (58). Therefore, it is possible that in our assay H3 phosphorylation and acetylation shifted the equilibrium between bound and unbound forms of heterochromatin proteins to the unbound side, creating a favorable environment for chromatin decondensation.
FIG. 7.
Model depicting the epigenetic modifications induced by Npm and gene activation. See Discussion for the details.
Histone H3 phosphorylation has been linked to two apparently contradictory chromatin statuses: mitotic chromosome condensation and chromatin relaxation during gene activation in interphase cells (54, 61). H3S10 and H3S28 are globally phosphorylated in the late G2-to-M phase by the Aurora B/Ipl1p kinase from yeast to human and by the NIMA kinase in Aspergillus nidulans (see references cited in reference 61). However, the functional meaning of this mitotic phosphorylation is not necessarily clear in some cases. For example, while p-H3S10 is essential for mitotic chromosome condensation in Tetrahymena thermophila (74), a yeast mutant lacking H3S10 and H3S28 shows proper mitotic progression (34). In addition, Drosophila cells show only a very weak correlation between chromosome condensation and p-H3S10 (1). In interphase cells there are a number of cases in which transcriptional activation is correlated with p-H3S10. Importantly, unlike global p-H3S10 in mitosis, interphase p-H3S10 is usually limited to the activated genes. For instance, p-H3S10 increases in transcriptionally active loci after heat shock in Drosophila polytene chromosomes (53). Various mitogens and stress can induce p-H3S10 in the proto-oncogenes c-fos and c-jun by recruiting the Msk1 and Msk2 kinases (19, 66). Based on these findings, it has been hypothesized that p-H3S10 does not define chromatin condensation or decondensation on its own; rather, it likely recruits or releases specific binding molecules, such as condensin for mitosis and SWI/SNF family proteins for gene activation, to alter chromatin structure in either way (68). p-H3S10 and p-H3S28 were clearly insufficient for chromatin decondensation, as shown by the PKA treatment (Fig. 4F) and by previous in vitro experiments in which p-H3S10 on isolated chromatin only slightly increased chromatin flexibility (49). From the result in egg extract, it does not seem to be necessary for H3 to be stably phosphorylated to support chromatin decondensation, which also argues that simple H3 phosphorylation is not the primary mechanism for the chromatin decondensation. Because there are 12 copies of the H3 gene in human and mouse (47), it is difficult to replace all the wild-type H3 with mutants to study if H3 phosphorylation is essential for chromatin condensation. Instead, we should probably identify the enzymes responsible for H3 phosphorylation and acetylation as well as interactions between these enzymes and Npm to connect more tightly the chromatin events we observed.
The chromatin decondensation in F9 nuclei is distinct in four aspects from the decondensation of sperm and erythrocyte chromatin, in which specialized histone variants play key roles in maintaining tightly packed and transcriptionally silent chromatin. First and most importantly, there is no known condensation-specific histone variant, such as H1o, or readily detectable histone release by Npm in F9 nuclei. Second, ATP was essential for decondensation of F9 chromatin while sperm chromatin could be decondensed moderately regardless of ATP. Third, oNpm, less phosphorylated than eNpm, is less potent in sperm decondensation, but both Npms were equally potent in F9 chromatin decondensation. Finally, whereas the A2 domain of Npm is dispensable for sperm chromatin decondensation, it is crucial for chromatin decondensation in F9 nuclei.
Currently, little is known about the nuclear structure specific to undifferentiated cells, such as embryonal carcinoma cells and ES cells, compared with more differentiated cells. Higher efficiency of postimplantation development of mouse clones derived from ES cell nuclei than from somatic cell nuclei (63) is presently explained by the possibility that already pluripotent ES cells (expressing Oct-4 and other pluripotent transcription factor genes) require less extensive nuclear reprogramming than regular somatic cell nuclei to support normal development of the clones. To this transcription factor-focused interpretation, our results of a more flexible chromatin structure at the nucleus-wide level, revealed only after the Npm and PGA treatment in embryonal carcinoma cell and ES cell nuclei, unravel another layer of difference betweens ES cells and somatic cells. There are some precedents that show differentiation-specific changes in centromeric heterochromatin components. For example, centromeres cluster to form large chromocenters during the progression of myogenic differentiation, depending on the methyl CpG-binding proteins MeCP2 and MBD2 (8). TIF1β, diffusely distributed in undifferentiated F9 cells, becomes highly localized to centromeric heterochromatin in differentiated F9 cells (13). Although biological meanings of these findings have not been elucidated, they clearly show that heterochromatin components can change, perhaps becoming more rigid during cell differentiation. An attractive hypothesis is that in general, undifferentiated cells contain more loosely packed chromatin than their differentiated counterparts to maintain many genes in a potentially open state to prepare them for future expression. This interpretation is further supported at the functional level by recent gene expression analyses in undifferentiated cells. Hematopoietic stem cells and progenitor cells promiscuously express seemingly unnecessary differentiated cell-specific genes of multiple lineages (35, 51). Some of the Xenopus early gastrula cells express marker genes for two or three germ layers (73). If we can identify the molecular mechanisms underlying the more flexible chromatin in undifferentiated cells by using Npm sensitivity as a novel indicator, we can potentially confer that mechanism on differentiated somatic cells so that they acquire the plasticity necessary to be smoothly reprogrammed into other lineages for therapeutic medical purposes. This will be an important step to realize the future of regeneration/transplantation medicine.
Acknowledgments
We thank K. Gonda for the preliminary work of this project, G. Ahlstrand for the electron microscopy, L. Higgins and T. Krick for the mass spectrometry, E. Coucouvanis for S2 cells, and M. Tatsuka for the histone H3 plasmid. We also thank J. B. Gurdon for teaching us the oocyte injection technique and M. A. Surani for comments on the work.
This work was supported by the Undergraduate Research Opportunity Program to D.N., a Grant-in-Aid for Scientific Research in Priority Areas (15080211) and Project for the Realization of Regenerative Medicine (the research field for the technical development of stem cell manipulation) to T.W., and the Minnesota Medical Foundation (J.W. and N.K.).
REFERENCES
- 1.Adams, R. R., H. Maiato, W. C. Earnshaw, and M. Carmena. 2001. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153:865-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ajiro, K., K. Yoda, K. Utsumi, and Y. Nishikawa. 1996. Alteration of cell cycle-dependent histone phosphorylations by okadaic acid. Induction of mitosis-specific H3 phosphorylation and chromatin condensation in mammalian interphase cells. J. Biol. Chem. 271:13197-13201. [DOI] [PubMed] [Google Scholar]
- 3.Arnan, C., N. Saperas, C. Prieto, M. Chiva, and J. Ausio. 2003. Interaction of nucleoplasmin with core histones. J. Biol. Chem. 278:31319-31324. [DOI] [PubMed] [Google Scholar]
- 4.Bannister, A. J., P. Zegerman, J. F. Partridge, E. A. Miska, J. O. Thomas, R. C. Allshire, and T. Kouzarides. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410:120-124. [DOI] [PubMed] [Google Scholar]
- 5.Banuelos, S., A. Hierro, J. M. Arizmendi, G. Montoya, A. Prado, and A. Muga. 2003. Activation mechanism of the nuclear chaperone nucleoplasmin: role of the core domain. J. Mol. Biol. 334:585-593. [DOI] [PubMed] [Google Scholar]
- 6.Barry, J. M., and R. W. Merriam. 1972. Swelling of hen erythrocyte nuclei in cytoplasm from Xenopus eggs. Exp. Cell Res. 71:90-96. [DOI] [PubMed] [Google Scholar]
- 7.Blelloch, R. H., K. Hochedlinger, Y. Yamada, C. Brennan, M. Kim, B. Mintz, L. Chin, and R. Jaenisch. 2004. Nuclear cloning of embryonal carcinoma cells. Proc. Natl. Acad. Sci. USA 101:13985-13990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brero, A., H. P. Easwaran, D. Nowak, I. Grunewald, T. Cremer, H. Leonhardt, and M. C. Cardoso. 2005. Methyl CpG-binding proteins induce large-scale chromatin reorganization during terminal differentiation. J. Cell Biol. 169:733-743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burglin, T. R., I. W. Mattaj, D. D. Newmeyer, R. Zeller, and E. M. De Robertis. 1987. Cloning of nucleoplasmin from Xenopus laevis oocytes and analysis of its developmental expression. Genes Dev. 1:97-107. [DOI] [PubMed] [Google Scholar]
- 10.Burns, K. H., M. M. Viveiros, Y. Ren, P. Wang, F. J. DeMayo, D. E. Frail, J. J. Eppig, and M. M. Matzuk. 2003. Roles of NPM2 in chromatin and nucleolar organization in oocytes and embryos. Science 300:633-636. [DOI] [PubMed] [Google Scholar]
- 11.Byrne, J. A., S. Simonsson, P. S. Western, and J. B. Gurdon. 2003. Nuclei of adult mammalian somatic cells are directly reprogrammed to oct-4 stem cell gene expression by amphibian oocytes. Curr. Biol. 13:1206-1213. [DOI] [PubMed] [Google Scholar]
- 12.Cammas, F., M. Herzog, T. Lerouge, P. Chambon, and R. Losson. 2004. Association of the transcriptional corepressor TIF1β with heterochromatin protein 1 (HP1): an essential role for progression through differentiation. Genes Dev. 18:2147-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cammas, F., M. Oulad-Abdelghani, J. L. Vonesch, Y. Huss-Garcia, P. Chambon, and R. Losson. 2002. Cell differentiation induces TIF1β association with centromeric heterochromatin via an HP1 interaction. J. Cell Sci. 115:3439-3448. [DOI] [PubMed] [Google Scholar]
- 14.Carey, M., and S. Smale. 1999. Transcriptional regulation in eukaryotes, p. 319-364. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Carmo-Fonseca, M. 2002. The contribution of nuclear compartmentalization to gene regulation. Cell 108:513-521. [DOI] [PubMed] [Google Scholar]
- 16.Chen, T., Y. Ueda, J. E. Dodge, Z. Wang, and E. Li. 2003. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 23:5594-5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheung, P., K. G. Tanner, W. L. Cheung, P. Sassone-Corsi, J. M. Denu, and C. D. Allis. 2000. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5:905-915. [DOI] [PubMed] [Google Scholar]
- 18.Cheutin, T., A. J. McNairn, T. Jenuwein, D. M. Gilbert, P. B. Singh, and T. Misteli. 2003. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299:721-725. [DOI] [PubMed] [Google Scholar]
- 19.Clayton, A. L., S. Rose, M. J. Barratt, and L. C. Mahadevan. 2000. Phosphoacetylation of histone H3 on c-fos- and c-jun-associated nucleosomes upon gene activation. EMBO J. 19:3714-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cowan, C. A., J. Atienza, D. A. Melton, and K. Eggan. 2005. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science 309:1369-1373. [DOI] [PubMed] [Google Scholar]
- 21.Craig, J. M., E. Earle, P. Canham, L. H. Wong, M. Anderson, and K. H. Choo. 2003. Analysis of mammalian proteins involved in chromatin modification reveals new metaphase centromeric proteins and distinct chromosomal distribution patterns. Hum. Mol. Genet. 12:3109-3121. [DOI] [PubMed] [Google Scholar]
- 22.Dimitrov, S., and A. P. Wolffe. 1996. Remodeling somatic nuclei in Xenopus laevis egg extracts: molecular mechanisms for the selective release of histones H1 and H10 from chromatin and the acquisition of transcriptional competence. EMBO J. 15:5897-5906. [PMC free article] [PubMed] [Google Scholar]
- 23.Dutta, S., I. V. Akey, C. Dingwall, K. L. Hartman, T. Laue, R. T. Nolte, J. F. Head, and C. W. Akey. 2001. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol. Cell 8:841-853. [DOI] [PubMed] [Google Scholar]
- 24.Eggan, K., K. Baldwin, M. Tackett, J. Osborne, J. Gogos, A. Chess, R. Axel, and R. Jaenisch. 2004. Mice cloned from olfactory sensory neurons. Nature 428:44-49. [DOI] [PubMed] [Google Scholar]
- 25.Festenstein, R., S. N. Pagakis, K. Hiragami, D. Lyon, A. Verreault, B. Sekkali, and D. Kioussis. 2003. Modulation of heterochromatin protein 1 dynamics in primary mammalian cells. Science 299:719-721. [DOI] [PubMed] [Google Scholar]
- 26.Gonda, K., J. Fowler, N. Katoku-Kikyo, J. Haroldson, J. Wudel, and N. Kikyo. 2003. Reversible disassembly of somatic nucleoli by the germ cell proteins FRGY2a and FRGY2b. Nat. Cell Biol. 5:205-210. [DOI] [PubMed] [Google Scholar]
- 27.Gurdon, J. B. 1968. Changes in somatic cell nuclei inserted into growing and maturing amphibian oocytes. J. Embryol. Exp. Morphol. 20:401-414. [PubMed] [Google Scholar]
- 28.Gurdon, J. B. 1976. Injected nuclei in frog oocytes: fate, enlargement, and chromatin dispersal. J. Embryol. Exp. Morphol. 36:523-540. [PubMed] [Google Scholar]
- 29.Gurdon, J. B., R. A. Laskey, E. M. De Robertis, and G. A. Partington. 1979. Reprogramming of transplanted nuclei in amphibia. Int. Rev. Cytol. Suppl. 9:161-178. [DOI] [PubMed] [Google Scholar]
- 30.Gurdon, J. B., G. A. Partington, and E. M. De Robertis. 1976. Injected nuclei in frog oocytes: RNA synthesis and protein exchange. J. Embryol. Exp. Morphol. 36:541-553. [PubMed] [Google Scholar]
- 31.Hardie, D. 1999. Analysis of signal transduction pathways using protein-serine/threonine phosphatse inhibitors, p. 53-67. In D. Hardie (ed.), Protein phosphorylation: a practical approach, 2nd ed. Oxford University Press, Oxford, England.
- 32.Hierro, A., J. M. Arizmendi, S. Banuelos, A. Prado, and A. Muga. 2002. Electrostatic interactions at the C-terminal domain of nucleoplasmin modulate its chromatin decondensation activity. Biochemistry 41:6408-6413. [DOI] [PubMed] [Google Scholar]
- 33.Hochedlinger, K., and R. Jaenisch. 2002. Monoclonal mice generated by nuclear transfer from mature B and T donor cells. Nature 415:1035-1038. [DOI] [PubMed] [Google Scholar]
- 34.Hsu, J. Y., Z. W. Sun, X. Li, M. Reuben, K. Tatchell, D. K. Bishop, J. M. Grushcow, C. J. Brame, J. A. Caldwell, D. F. Hunt, R. Lin, M. M. Smith, and C. D. Allis. 2000. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102:279-291. [DOI] [PubMed] [Google Scholar]
- 35.Hu, M., D. Krause, M. Greaves, S. Sharkis, M. Dexter, C. Heyworth, and T. Enver. 1997. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 11:774-785. [DOI] [PubMed] [Google Scholar]
- 36.Humpherys, D., K. Eggan, H. Akutsu, A. Friedman, K. Hochedlinger, R. Yanagimachi, E. S. Lander, T. R. Golub, and R. Jaenisch. 2002. Abnormal gene expression in cloned mice derived from embryonic stem cell and cumulus cell nuclei. Proc. Natl. Acad. Sci. USA 99:12889-12894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ito, T., J. K. Tyler, M. Bulger, R. Kobayashi, and J. T. Kadonaga. 1996. ATP-facilitated chromatin assembly with a nucleoplasmin-like protein from Drosophila melanogaster. J. Biol. Chem. 271:25041-25048. [DOI] [PubMed] [Google Scholar]
- 38.Jeanisch, R., K. Eggan, D. Humpherys, W. Rideout, and K. Hochedlinger. 2002. Nuclear cloning, stem cells, and genomic reprogramming. Cloning Stem Cells 4:389-396. [DOI] [PubMed] [Google Scholar]
- 39.Kalitsis, P., and K. H. A. Choo. 1997. Centromere DNA of higher eukaryotes, p. 97-142. In K. H. A. Choo (ed.), The centromere. Oxford University Press, Oxford, England.
- 40.Kikyo, N., P. A. Wade, D. Guschin, H. Ge, and A. P. Wolffe. 2000. Active remodeling of somatic nuclei in egg cytoplasm by the nucleosomal ATPase ISWI. Science 289:2360-2362. [DOI] [PubMed] [Google Scholar]
- 41.Kipling, D., H. E. Wilson, A. R. Mitchell, B. A. Taylor, and H. J. Cooke. 1994. Mouse centromere mapping using oligonucleotide probes that detect variants of the minor satellite. Chromosoma 103:46-55. [DOI] [PubMed] [Google Scholar]
- 42.Lachner, M., D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410:116-120. [DOI] [PubMed] [Google Scholar]
- 43.Laskey, R. A., B. M. Honda, A. D. Mills, and J. T. Finch. 1978. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature 275:416-420. [DOI] [PubMed] [Google Scholar]
- 44.Leno, G. H., A. D. Mills, A. Philpott, and R. A. Laskey. 1996. Hyperphosphorylation of nucleoplasmin facilitates Xenopus sperm decondensation at fertilization. J. Biol. Chem. 271:7253-7256. [DOI] [PubMed] [Google Scholar]
- 45.Lo, W. S., R. C. Trievel, J. R. Rojas, L. Duggan, J. Y. Hsu, C. D. Allis, R. Marmorstein, and S. L. Berger. 2000. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5:917-926. [DOI] [PubMed] [Google Scholar]
- 46.Martens, J. H., R. J. O'Sullivan, U. Braunschweig, S. Opravil, M. Radolf, P. Steinlein, and T. Jenuwein. 2005. The profile of repeat-associated histone lysine methylation states in the mouse epigenome. EMBO J. 24:800-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marzluff, W. F., P. Gongidi, K. R. Woods, J. Jin, and L. J. Maltais. 2002. The human and mouse replication-dependent histone genes. Genomics 80:487-498. [PubMed] [Google Scholar]
- 48.Mateescu, B., P. England, F. Halgand, M. Yaniv, and C. Muchardt. 2004. Tethering of HP1 proteins to chromatin is relieved by phosphoacetylation of histone H3. EMBO Rep. 5:490-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mazen, A., M. F. Hacques, and C. Marion. 1987. H3 phosphorylation-dependent structural changes in chromatin. Implications for the role of very lysine-rich histones. J. Mol. Biol. 194:741-745. [DOI] [PubMed] [Google Scholar]
- 50.Minc, E., Y. Allory, H. J. Worman, J. C. Courvalin, and B. Buendia. 1999. Localization and phosphorylation of HP1 proteins during the cell cycle in mammalian cells. Chromosoma 108:220-234. [DOI] [PubMed] [Google Scholar]
- 51.Miyamoto, T., H. Iwasaki, B. Reizis, M. Ye, T. Graf, I. L. Weissman, and K. Akashi. 2002. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev. Cell 3:137-147. [DOI] [PubMed] [Google Scholar]
- 52.Mullins, L. J., I. Wilmut, and J. J. Mullins. 2003. Nuclear transfer in rodents. J. Physiol. 554:4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nowak, S. J., and V. G. Corces. 2000. Phosphorylation of histone H3 correlates with transcriptionally active loci. Genes Dev. 14:3003-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nowak, S. J., and V. G. Corces. 2004. Phosphorylation of histone H3: a balancing act between chromosome condensation and transcriptional activation. Trends Genet. 20:214-220. [DOI] [PubMed] [Google Scholar]
- 55.Nowak, S. J., C. Y. Pai, and V. G. Corces. 2003. Protein phosphatase 2A activity affects histone H3 phosphorylation and transcription in Drosophila melanogaster. Mol. Cell. Biol. 23:6129-6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ota, T., S. Suto, H. Katayama, Z. B. Han, F. Suzuki, M. Maeda, M. Tanino, Y. Terada, and M. Tatsuka. 2002. Increased mitotic phosphorylation of histone H3 attributable to AIM-1/Aurora-B overexpression contributes to chromosome number instability. Cancer Res. 62:5168-5177. [PubMed] [Google Scholar]
- 57.Peters, A. H., S. Kubicek, K. Mechtler, R. J. O'Sullivan, A. A. Derijck, L. Perez-Burgos, A. Kohlmaier, S. Opravil, M. Tachibana, Y. Shinkai, J. H. Martens, and T. Jenuwein. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell 12:1577-1589. [DOI] [PubMed] [Google Scholar]
- 58.Phair, R. D., P. Scaffidi, C. Elbi, J. Vecerova, A. Dey, K. Ozato, D. T. Brown, G. Hager, M. Bustin, and T. Misteli. 2004. Global nature of dynamic protein-chromatin interactions in vivo: three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol. Cell. Biol. 24:6393-6402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Philpott, A., and G. H. Leno. 1992. Nucleoplasmin remodels sperm chromatin in Xenopus egg extracts. Cell 69:759-767. [DOI] [PubMed] [Google Scholar]
- 60.Philpott, A., G. H. Leno, and R. A. Laskey. 1991. Sperm decondensation in Xenopus egg cytoplasm is mediated by nucleoplasmin. Cell 65:569-578. [DOI] [PubMed] [Google Scholar]
- 61.Prigent, C., and S. Dimitrov. 2003. Phosphorylation of serine 10 in histone H3: what for? J. Cell Sci. 116:3677-3685. [DOI] [PubMed] [Google Scholar]
- 62.Rice, J. C., S. D. Briggs, B. Ueberheide, C. M. Barber, J. Shabanowitz, D. F. Hunt, Y. Shinkai, and C. D. Allis. 2003. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol. Cell 12:1591-1598. [DOI] [PubMed] [Google Scholar]
- 63.Rideout, W. M., III, T. Wakayama, A. Wutz, K. Eggan, L. Jackson-Grusby, J. Dausman, R. Yanagimachi, and R. Jaenisch. 2000. Generation of mice from wild-type and targeted ES cells by nuclear cloning. Nat. Genet. 24:109-110. [DOI] [PubMed] [Google Scholar]
- 64.Sealy, L., R. R. Burgess, M. Cotten, and R. Chalkley. 1989. Purification of Xenopus egg nucleoplasmin and its use in chromatin assembly in vitro. Methods Enzymol. 170:612-630. [DOI] [PubMed] [Google Scholar]
- 65.Smythe, C., and J. W. Newport. 1991. Systems for the study of nuclear assembly, DNA replication, and nuclear breakdown in Xenopus laevis egg extracts. Methods Cell Biol. 35:449-468. [DOI] [PubMed] [Google Scholar]
- 66.Soloaga, A., S. Thomson, G. R. Wiggin, N. Rampersaud, M. H. Dyson, C. A. Hazzalin, L. C. Mahadevan, and J. S. Arthur. 2003. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 22:2788-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spector, D. L., R. D. Goldman, and L. A Leinwand. 1997. Cells: a laboratory manual, p. 111.1-111.45. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 68.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 69.Suemizu, H., K. Aiba, T. Yoshikawa, A. A. Sharov, N. Shimozawa, N. Tamaoki, and M. S. Ko. 2003. Expression profiling of placentomegaly associated with nuclear transplantation of mouse ES cells. Dev. Biol. 253:36-53. [DOI] [PubMed] [Google Scholar]
- 70.Tulin, A., and A. Spradling. 2003. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science 299:560-562. [DOI] [PubMed] [Google Scholar]
- 71.Verheggen, C., S. Le Panse, G. Almouzni, and D. Hernandez-Verdun. 1998. Presence of pre-rRNAs before activation of polymerase I transcription in the building process of nucleoli during early development of Xenopus laevis. J. Cell Biol. 142:1167-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wakayama, T., A. C. F. Perry, M. Zucotti, K. R. Johnson, and R. Yanagimachi. 1998. Full term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature 394:369-374. [DOI] [PubMed] [Google Scholar]
- 73.Wardle, F. C., and J. C. Smith. 2004. Refinement of gene expression patterns in the early Xenopus embryo. Development 131:4687-4696. [DOI] [PubMed] [Google Scholar]
- 74.Wei, Y., L. Yu, J. Bowen, M. A. Gorovsky, and C. D. Allis. 1999. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell 97:99-109. [DOI] [PubMed] [Google Scholar]