Abstract
Spt2/Sin1 is a DNA binding protein with HMG-like domains that has been suggested to play a role in chromatin-mediated transcription in Saccharomyces cerevisiae. Previous studies have suggested models in which Spt2 plays an inhibitory role in the initiation of transcription of certain genes. In this work, we have taken several approaches to study Spt2 in greater detail. Our results have identified previously unknown genetic interactions between spt2Δ and mutations in genes encoding transcription elongation factors, including members of the PAF and HIR/HPC complexes. In addition, genome-wide and gene-specific chromatin immunoprecipitation analyses suggest that Spt2 is primarily associated with coding regions in a transcription-dependent fashion. Furthermore, our results show that Spt2, like other elongation factors, is required for the repression of transcription from a cryptic promoter within a coding region and that Spt2 is also required for repression of recombination within transcribed regions. Finally, we provide evidence that Spt2 plays a role in regulating the levels of histone H3 over transcribed regions. Taken together, our results suggest a direct link for Spt2 with transcription elongation, chromatin dynamics, and genome stability.
The regulation of chromatin structure in eukaryotes is a fundamental aspect of all DNA-related processes in vivo, including transcription, replication, recombination, repair, and chromosome segregation. The basic unit of chromatin is the nucleosome, consisting of 146 bp of DNA wrapped around an octamer of histones (28). In addition to histones, several nonhistone proteins play important roles in chromatin structure and chromatin-related processes (67).
One such nonhistone component of chromatin in Saccharomyces cerevisiae is the HMG-like protein Spt2/Sin1. Spt2 was initially identified genetically, by mutations (called spt2) that suppress Ty and δ insertion mutations in the HIS4 promoter (69) and by mutations (called sin1) that suppress the loss of the Swi/Snf chromatin-remodeling complex for expression of an HO-lacZ fusion (58). Mutations in SPT2 have since been shown to be pleiotropic, suppressing initiation defects caused by mutations that abolish Swi/Snf (46) or the SAGA components Gcn5 and Ada3 (44, 46) and by deletion mutations in RPB1 that shorten the Rpb1 carboxy-terminal domain (45). In addition, spt2 mutations have been shown to allow increased expression of the SSA3 gene in an ssa1 ssa2 mutant background (4). While these strong mutant phenotypes suggested a negative role for Spt2 in transcription initiation, very little is actually understood concerning the function of Spt2 with respect to chromatin structure and transcription. Interestingly, while several studies show that an spt2Δ mutation affects expression of an HO-lacZ fusion (44, 46, 58), there is no effect of spt2Δ on transcription of HO itself (74). The finding that spt2 mutations impair chromosome segregation (23) also suggested a broad role in chromatin function.
Analysis of the Spt2 amino acid sequence has revealed several regions that are important for its function. First, Spt2 has two HMG-like domains, which are found in several proteins involved in chromatin structure and transcription (23, 25). HMG proteins can bind DNA and induce specific structural changes, thereby allowing the assembly of factors involved in processes such as transcription and recombination (for reviews, see references 63 and 64). Spt2 also contains a polar carboxy-terminal tail (amino acids 180 to 333) containing two subdomains (amino acids 226 to 249 and 277 to 303) that are very rich in acidic amino acids and important for Spt2 function (25). Recently, three domains of Spt2 were shown to bind four-way junction DNA (40), a property common for HMG-like proteins (76). The first domain (amino acids 1 to 96) overlaps with the first HMG-like box, while the other two (amino acids 224 to 304 and 303 to 333) are within the carboxy-terminal tail (40). Binding to four-way junction DNA often reflects the affinity of a factor for crossed DNA helices, a situation encountered at the entry-exit of a nucleosome (76).
To gain insight into the roles of Spt2, we have taken several approaches. First, by a combination of genetic methods, we have identified genetic interactions between Spt2 and two sets of transcription factors implicated in transcription elongation: the PAF complex and the HIR/HPC complex. Consistent with these results, genome-wide localization studies of Spt2 demonstrate physical association with the coding regions of actively transcribed genes. Standard chromatin immunoprecipitation (ChIP) analysis of Spt2 at specific genes supports these findings. Moreover, Spt2 collaborates with the PAF and the HIR/HPC complexes in inhibiting transcription initiation from a cryptic promoter within the FLO8 coding region, a role previously demonstrated for transcription elongation factors such as Spt6 and Spt16 (18, 35). In addition, we show that spt2Δ mutants have elevated recombination between inverted repeats in the genome. This phenotype was also previously shown for the spt4 and spt6 elongation factor mutants (29), and it was suggested that hyperrecombination in those mutants is probably caused by defects in chromatin structure. Finally, we find that loss of Spt2 results in a decrease of histone H3 associated with coding regions. This effect is dependent on active transcription and strongly suggests that Spt2 promotes genome stability through the maintenance of the chromatin structure of actively transcribed genes.
MATERIALS AND METHODS
S. cerevisiae strains, media, and genetic methods.
All S. cerevisiae strains (Table 1) are isogenic to a GAL2 derivative of S288C (70). Strains were constructed by standard methods, either by crosses or by transformation. The spt2Δ0::KANMX6, hir2Δ0::KANMX6, and hir3Δ0::KANMX6 alleles were constructed by replacing the open reading frames (ORFs) with the KANMX6 marker (3). The SPT2-13MYC, HIR1-13MYC, and HIR2-13MYC alleles, marked with KANMX6, were generated by integrating DNA encoding 13 copies of the Myc epitope at the 3′ end of the corresponding gene (27). The GAL1-FLO8-HIS3 reporter, to be described elsewhere, contains the HIS3 gene inserted out of frame into the 3′ coding region of FLO8 such that wild-type HIS3 product is expressed only when transcription initiates from a cryptic promoter within the FLO8 gene (18). In addition, the FLO8 promoter has been replaced by the GAL1 promoter. This construct is integrated in the genome, replacing FLO8. All strains were checked by PCR and Western blotting. The strains containing the alleles encoding the epitope-tagged proteins did not exhibit detectable mutant phenotypes compared to the untagged wild-type strain, with one exception: SPT2-13MYC suppresses the cold-sensitive growth of an rtf1Δ mutation (described in Results) and confers a weak Spt− phenotype when combined with hir1Δ or elf1Δ (data not shown). The strains bearing inverted repeats in the HIS3 locus were obtained by crosses with the previously described strains M137-11B, M236-12D, and ITE-1C (30). The plasmid pLL15 used for Fig. 1A was previously described (25). All oligonucleotide sequences used in strain constructions are listed in Table S1 at http://genetics.med.harvard.edu/%7Ewinston/Supp%20tables.html. The synthetic genetic array screen was done as previously described (65). Of the 14 candidates we obtained, we retested 5 by standard tetrad analysis. In each case, we found that the double mutants were viable, although they displayed synthetic phenotypes, including slow growth or thermosensitive phenotypes. For experiments involving GAL1 gene induction, cells were grown to an optical density of 600 nm of 0.5 in YP (1% yeast extract, 2% peptone) supplemented with 2% raffinose (YPraf). The cells were then centrifuged, resuspended in YP containing 2% galactose (YPgal), and grown for 2 hours before being harvested.
TABLE 1.
Saccharomyces cerevisiae strains
| Strain | Genotype | Source or reference |
|---|---|---|
| FY1235 | MATα ura3-52 his4-912δ leu2Δ1 lys2-128δ trp1Δ63 hir1Δ::LEU2 | F. Winston |
| FY1634 | MATα ura3-52 his4-912δ leu2Δ1 lys2-128δ spt5-276 | F. Winston |
| FY1856 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ | F. Winston |
| FY2116 | MATartf1Δ101::LEU2 his4-912δ lys2-128δ leu2Δ1 ura3-52 trp1Δ63 | F. Winston |
| FY2124 | MATα ura3-52 leu2Δ1 lys2-128δ his3Δ200 RPB3-HA::LEU2 SPT6-FLAG ctr9Δ::KANMX | F. Winston |
| FY2127 | MATα ura3-52 leu2Δ1 lys2-128δ his4-912δ RPB3-HA1::LEU2 SPT6-FLAG cdc73Δ::KANMX | F. Winston |
| FY2132 | MATα ura3-52 leu2Δ1 lys2-128δ his4-912δ RPB3-HA1::LEU2 SPT6-FLAG paf1Δ::KANMX | F. Winston |
| FY2427 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ SPT2-13MYC::KANMX6 | This work |
| FY2428 | MATaura3-52 his3Δ200 his4-912δ leu2Δ1 lys2-173 R2 | This work |
| FY2429 | MATahis4-912δ leu2Δ1 lys2-128δ snf2::LEU2 | This work |
| FY2430 | MATahis4-912δ leu2Δ1 lys2-173R2 snf2::LEU2 spt2Δ0::KANMX6 | This work |
| FY2431 | MATahis3Δ200 leu2Δ1 lys2-128δ spt2Δ0::KANMX6 | This work |
| FY2432 | MATα ura3-52 his3Δ200 leu2Δ1 lys2-128δ spt2Δ0::KANMX6 | This work |
| FY2433 | MATaura3-52 his3Δ200 leu2Δ1 lys2-128δ spt5-276 spt2Δ0::KANMX6 | This work |
| FY2434 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ ser33::KANMX srg1-1 SPT2-13MYC::KANMX6 | This work |
| FY2435 | MATaura3Δ0 his3Δ200 leu2Δ0 lys2-128δ ser33::KANMX SPT2-13MYC::KANMX6 | This work |
| FY2436 | MATα ura3Δ0 his3Δ200 leu2Δ1 lys2-128δ RPB3-HA1::LEU2 cdc73Δ::KANMX spt2Δ0::KANMX6 FLAG-SPT6 | This work |
| FY2437 | MATaura3-52 his3Δ200 his4-912δ leu2Δ1 lys2-128δ rtf1Δ101::LEU2 SPT2-13MYC::KANMX6 | This work |
| FY2438 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HIR1-13MYC::KANMX6 | This work |
| FY2439 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ HIR2-13MYC::KANMX6 | This work |
| FY2440 | MATahis3Δ200 leu2Δ1 lys2-128δ hir3Δ0::KANMX6 spt2Δ0::KANMX6 | This work |
| FY2441 | MATahis3Δ leu2Δ1 lys2-128δ hir3Δ0::KANMX6 | This work |
| FY2442 | MATaura3Δ0 his3Δ200 leu2Δ1 lys2-128δ | This work |
| FY2443 | MATahis3Δ200 leu2Δ0 lys2-128δ hir2Δ0::KANMX6 | This work |
| FY2444 | MATahis3Δ200 leu2Δ1 lys2-128δ hir2Δ0::KANMX6 spt2Δ0::KANMX6 | This work |
| FY2445 | MATaura3-52 his3Δ200 lys2-128δ RPB3-HA1::LEU2 spt2Δ0::KANMX6 KANMX6-PGAL1-FLO8-HIS3 SPT6-FLAG | This work |
| FY2446 | MATα ura3-52 his3Δ200 leu2Δ1 lys2Δ0 his4-912d RPB3-HA1::LEU2 paf1Δ::KANMX SPT2-13MYC::KANMX6 | This work |
| FY2447 | MATaura3-52 his3Δ200 leu2Δ0 lys2-128δ RPB3-HA1::LEU2 dst1Δ::KANMX SPT2-13MYC::KANMX6 | This work |
| FY2448 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ hir1Δ::LEU2 SPT2-13MYC::KANMX6 | This work |
| FY2449 | MATα ura3-52 leu2Δ1 spt4::URA3 SPT2-13MYC::KANMX6 | This work |
| FY2450 | MATα ura3Δ0 his3Δ200 leu2Δ0 lys2-128δ spt6-1004-FLAG SPT2-13MYC::KANMX6 | This work |
| FY2451 | MATα ura3-52 his3Δ200 his4-912δ leu2Δ1 lys2-128δ trp1Δ63 spt2Δ0::NAT hir1Δ::LEU2 | This work |
| FY2452 | MATaura3-52 his3Δ200 lys2-128δ RPB3-HA1::LEU2 KANMX6-PGAL1-FLO8-HIS3 | This work |
| FY2453 | MATaura3-52 his3Δ200 leu2Δ1 lys2-128δ RPB3-HA1::LEU2 KANMX6-PGAL1-FLO-8HIS3 SPT6-FLAG spt2Δ0::KANMX6 cdc73Δ::KANMX | This work |
| FY2454 | MATaura3-52 his3Δ200 lys2-128δ RPB3-HA1::LEU2 KANMX6-PGAL1-FLO8-HIS3 cdc73::KANMX | This work |
| FY2455 | MATaura3-52 his3Δ200 his4-912δ leu2Δ1 Lys2-128d spt2Δ0::NAT paf1Δ::KANMX SPT6-FLAG/pLL15 | This work |
| FY2456 | MATaura3-52 his3Δ200 leu2Δ1 lys2-128δ RPB3-HA1::LEU2 spt2Δ0::NAT ctr9::KANMX/pLL15 | This work |
| FY2503 | MATahis3Δ200 leu2Δ1 lys2-128δ trp1Δ63 hir1Δ::LEU2 KANMX6-PGAL1-FLO8-HIS3 | This work |
| FY2504 | MATaura3-52 his3Δ200 leu2Δ1 lys2-128δ trp1Δ63 spt2Δ0::KANMX6 hir1Δ::LEU2 KANMX6-PGAL1-FLO8-HIS3 | This work |
| leo1Δ | MATaura3Δ0 his3Δ1 leu2Δ0 met15Δ0 leo1Δ0::KANMX | Research Genetics |
| L1096 | MATaura3 his3p::INV::URA3::LEU2 leu2 lys2-128δ can1-100 spt2Δ0::KANMX6 | This work |
| L1097 | MATaura3-52 his3Δ200 leu2Δ1 lys2-128δ hpc2Δ0::KANMX | This work |
| L1098 | MATaura3-52 his3Δ1 leu2Δ1 lys2-128δ hpc2Δ0::KANMX spt2Δ0::KANMX6 | This work |
| L1099 | MATaura3 his3p::INV::URA3::LEU2 leu2 lys2-128δ can1-100 met15Δ0 rad51Δ0::KANMX | This work |
| L1100 | MATaura3 his3p::INV::URA3::LEU2 leu2 can1-100 met15Δ0 spt2Δ0::KANMX6 rad51::KANMX | This work |
| L1101 | MATα ura3 his3p::ITE::URA3::LEU2 leu2 lys2-128δ spt2Δ0::KANMX6 | This work |
| M137-11B | MATaura3 his3p::INV::URA3::LEU2 leu2 trp1 lys2-128δ can1-100 | 30 |
| M236-12D | MATaura3 his3p::INV::URA3::LEU2 leu2 lys2-128δ trp1 spt4-3 | 30 |
| ITE-1C | MATα ura3 his3p::ITE::URA3::LEU2 leu2 lys2-128δ trp1 can1-100 | 30 |
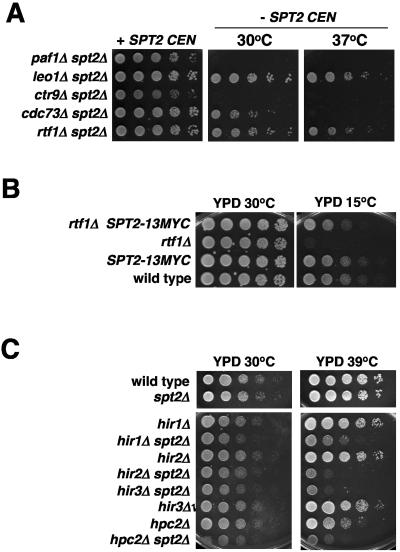
FIG. 1.
Genetic interactions of spt2Δ. (A) spt2Δ is synthetically lethal with paf1Δ and ctr9Δ. An spt2Δ mutant (FY2432) was transformed with pLL15, a CEN URA3 SPT2 plasmid (25). This strain was then mated with paf1Δ (FY2132), leo1Δ (Research Genetics), ctr9Δ (FY2124), cdc73Δ (FY2127), and rtf1Δ (FY2116) mutants. Diploids were sporulated and dissected, and representative progeny were spotted onto medium lacking uracil (+SPT2 CEN) or medium containing 5FOA (−SPT2 CEN) at 30°C and 37°C. (B) SPT2-13MYC suppresses the cold-sensitive phenotype of rtf1Δ. Serial dilutions of wild-type (FY2442), SPT2-Myc (FY2427), rtf1Δ (FY2116) and rtf1Δ SPT2-13Myc (FY2437) strains were grown in YPD for 2 days at 30°C or 7 days at 15°C. (C) spt2Δ interacts genetically with all the HIR/HPC complex members. Serial dilutions of cell cultures from wild-type (FY2442), spt2Δ (FY2431), hir1Δ (FY1235), hir1Δ spt2Δ (FY2451), hir2Δ (FY2443), hir2Δ spt2Δ (FY2444), hir3Δ spt2Δ (FY2440), hir3Δ (FY2441), hpc2Δ (L1097), and hpc2Δ spt2Δ (L1098) strains were grown on the indicated media for 2 or 3 days at the indicated temperature.
Chromatin immunoprecipitation experiments.
Chromatin immunoprecipitation experiments were performed as previously described (33). For the immunoprecipitation of Spt2-13Myc, Hir1-13Myc, and Hir2-13Myc, we used rabbit A14 anti-Myc serum (5 μl per immunoprecipitation; Santa Cruz Biotechnology). Immunoprecipitation of Rpb1 was performed using the 8WG16 anti-CTD antibody (2 μl per immunoprecipitation; Covance). The histone H3 immunoprecipitation was done using rabbit anti-H3 antibody (1 μl per immunoprecipitation; Abcam). The PCR amplification was performed using 1 to 2% of the precipitated material and 0.1 to 0.2% of the input DNA in a final volume of 15 μl. In the case of histone H3 immunoprecipitation, 0.2% to 0.4% of the immunoprecipitated material was used in the PCR analysis. When we analyzed histone H3 occupancy in the GAL1 coding region after galactose induction, 1 to 2% of the immunoprecipitated material was used. All oligonucleotide sequences used in ChIP experiments are listed in Table S1 at http://genetics.med.harvard.edu/%7Ewinston/Supp%20tables.html.
Genome-wide localization experiments.
Genome-wide localization experiments were performed using Spt2-13Myc (FY2427) and isogenic nontagged strains (FY1856) from cells grown in yeast extract-peptone-dextrose (YPD) to an optical density at 600 nm of 0.5 as described above. The immunoprecipitated DNA was amplified by ligation-mediated PCR and labeled with Cy5 (myc-Spt2) and Cy3 (untagged control), using an indirect incorporation method as described previously (10a ). The Cy5- and Cy3-labeled samples were mixed and hybridized to a microarray containing approximately 13,000 PCR products. The microarray contains one probe per open reading frame and one probe per intergenic region, except for long intergenic regions that are represented by two or three probes. Experiments were done in triplicate, and data were analyzed using a single-array error model and combined using a weighed average method as described previously (48a). Detailed protocols, details about the microarray as well as the complete data set can be found at http://www.ircm.qc.ca/microsites/francoisrobert/en/.
RNA analyses.
Total RNA was isolated using the hot-phenol method (2). Forty micrograms of RNA was separated on a 1% agarose formaldehyde-MOPS (morpholinepropanesulfonic acid) gel and transferred to a nylon membrane (60). For the analyses of the FLO8 transcripts, a probe located at the 3′ of the coding sequence (bp +1515 to + 2327) was amplified by PCR and radiolabeled by random priming. The probes used for SCR1 and PMA1 were also obtained by PCR amplification of DNA fragment corresponding to the coding regions of these genes. The sequences of the oligonucleotides used are listed in Table S1 at http://genetics.med.harvard.edu/%7Ewinston/Supp%20tables.html.
Determination of recombination frequencies.
The recombination frequencies presented are the averages and standard errors from three independent experiments. In each experiment, six independent colonies from the indicated strains were grown overnight in YPD and plated on either synthetic complete medium or medium lacking histidine. The recombination frequency for each independent culture is equal to the frequency of His+ colonies. The construction of strains with the inverted repeats was previously described (1, 30).
RESULTS
Genetic evidence that Spt2 genetically interacts with the PAF and HIR complexes.
To understand the cellular functions of Spt2, we conducted a synthetic genetic array screen (65) with an spt2Δ mutant as the query strain. By this procedure, we constructed and analyzed double mutants in which spt2Δ was combined with deletions of virtually all nonessential genes of S. cerevisiae. We found several candidate genes that are required for normal growth when combined with an spt2Δ mutation. Among these candidates, we have focused on five genes implicated in transcription elongation: CDC73, HIR1, HIR2, HIR3, and HPC2. In all five cases, tetrad analysis demonstrated that null mutations in these five genes confer growth defects and temperature-sensitive growth when combined with spt2Δ. CDC73 encodes one member of the PAF transcription elongation complex (57), composed of Paf1, Cdc73, Ctr9, Leo1, and Rtf1. While the exact role of the PAF complex is not known, its function is clearly associated with transcription elongation (10, 36, 50, 57). PAF has been shown to coordinate the recruitment of the Set1, Set2, and Dot1 histone H3 methyltransferases to transcribed regions of active genes (21, 22, 39, 71). We also uncovered the four members of the HIR/HPC complex (12a, 48), which is involved in several chromatin-related processes, including regulation of histone genes (43, 73), chromatin assembly (12a, 48, 53), kinetochore function (54), and transcription elongation (12). Interestingly, the HIR/HPC complex was recently purified and shown to contain four subunits (Hir1, Hir2, Hir3, and Hpc2) (12a).
To further test for genetic interactions between Spt2 and PAF components, we constructed pairwise double mutants between spt2Δ and a mutation in each of the PAF genes. The double mutants were constructed with a URA3 CEN plasmid containing the wild-type SPT2 gene. Double mutant phenotypes were assessed following growth on 5-fluoroorotic acid (5FOA) to select for loss of the SPT2-containing plasmid (Fig. 1A). We found that the spt2Δ paf1Δ and spt2Δ ctr9Δ double mutants were unable to grow on 5FOA medium, indicating a synthetic lethal phenotype. The spt2Δ cdc73Δ double mutant grew slowly at 30°C and was temperature sensitive for growth at 37°C. In contrast to these synthetic lethal phenotypes, there were no detectable growth defects for the spt2Δ rtf1Δ and spt2Δ leo1Δ double mutants. However, a genetic interaction was detected with respect to the cold-sensitive growth phenotype previously observed for rtf1Δ (5), as an SPT2-13MYC allele suppresses this cold sensitivity (Fig. 1B). Taken together, these double mutant phenotypes strongly suggest overlapping roles for Spt2 and PAF.
We also analyzed the genetic interactions between spt2Δ and hir/hpc mutations in greater detail. To do this, we constructed all the possible single and double mutants in an otherwise isogenic background and analyzed their growth on rich medium at both 30°C and 39°C (Fig. 1C). At 30°C, each of the four double mutants, i.e., the spt2Δ hir1Δ, spt2Δ hir2Δ, spt2Δ hir3Δ, and spt2Δ hpc2Δ mutants, grew more slowly than any of the single mutants, and at 39°C, all the double mutants grew very poorly. The temperature sensitivity of these null mutants may reflect a greater requirement for these factors at higher temperature, possibly due to altered physiology or the natural temperature sensitivity of a redundant factor. These synthetic phenotypes confirm that Spt2 genetically interacts with all members of the HIR/HPC complex. Previous evidence has implicated HIR/HPC in transcription elongation, as the HIR1 and HIR2 genes have been shown to interact with a number of genes encoding elongation factors, such as SPT16, PAF1, and CDC73 (12). In addition, a hir1 mutation was found to be synthetically lethal with mutations in the elongation factor genes, SPT4, SPT5, or SPT6 (A. Bortvin and F. Winston, unpublished results). The genetic interactions between Spt2 and the HIR/HPC and PAF complexes indicate a link between Spt2 and the transcription elongation process. This is further supported by our observation that spt2Δ genetically interacts with mutations in genes encoding several other elongation factors, including SPT5, SPT16, and ELF1 (data not shown).
Spt2 localizes primarily to the coding regions of active genes genome wide.
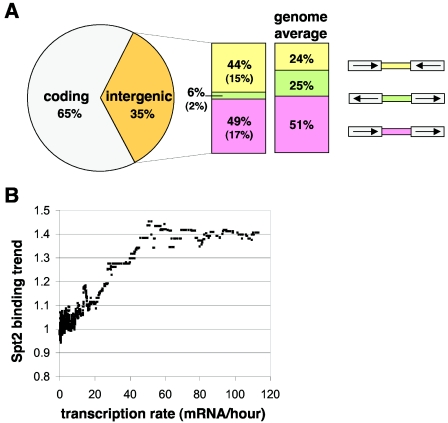
The results described above suggest that Spt2 is involved in transcription elongation. To test whether Spt2 is physically associated with transcribed regions, we performed genome-wide location analysis as described in Materials and Methods. In this experiment, 276 probes, mapping to 180 different chromosomal loci, were significantly enriched (P < 0.005) for Spt2. A list of these loci can be found in Table S2 at http://genetics.med.harvard.edu/%7Ewinston/Supp%20tables.html. At 65% of these loci, the coding region was preferentially enriched over flanking intergenic regions (Fig. 2A), supporting the idea that Spt2 is targeted mainly to transcribed regions. Among the loci where intergenic probes were more enriched than coding regions, 44% of the regions lacked a promoter (intergenic regions between two genes transcribed toward each other), compared to only 6% containing two promoters (regions between two divergently transcribed genes) (Fig. 2A). This represents approximately a twofold increase and a fourfold decrease for these respective intergenic regions relative to what is expected by chance (the genome average). This concomitant depletion for promoter regions and enrichment for terminator regions suggest that Spt2 binding to coding regions extends to the 3′ untranslated region (3′UTR). The remaining 49% of the intergenic probes contain both a promoter and a terminator and are neither enriched nor depleted relative to the genome average (Fig. 2A). Collectively, these data suggest that Spt2 localizes predominantly to transcribed regions, perhaps with a bias towards the 3′ end compared to the 5′ end. This was confirmed to be the case at the few genes where high-resolution ChIP assays were performed (described below).
FIG. 2.
Spt2 associates predominantly with actively transcribed regions. (A) A pie chart of the distribution of the Spt2-enriched loci (P < 0.005) between coding and intergenic regions. For the 180 loci enriched for Spt2, the most enriched array feature was classified as coding (gray) or noncoding (intergenic; orange). The intergenic features were further subdivided into those containing two terminators (yellow), two promoters (green), or one promoter and one terminator (pink). The distribution of these three classes of intergenic regions across the whole genome is also shown (genome average). (B) Spt2 association correlates with transcription rate genome wide. The Spt2 binding trend is plotted against the transcription rate for all yeast genes. The binding trend was determined as described previously by computing a sliding median of the Spt2 binding ratios across all genes ordered by transcription rate (24, 26, 38, 39, 49, 66). The genome-wide transcription rate was determined previously (14).
Manual inspection of the data suggested that Spt2 localizes predominantly to highly transcribed genes such as ribosomal protein genes, histone genes, genes coding for metabolic enzymes, and snoRNAs. This led us to hypothesize that Spt2 may be generally recruited to actively transcribed genes as opposed to genes involved in specific cellular functions. In order to test this idea, we looked at the correlation between Spt2 occupancy (as measured in our location analysis experiment) and the transcription rate for all yeast genes (determined as described in reference 14). Figure 2B shows that the association of Spt2 correlates with the transcription rate genome wide, supporting a general role for Spt2 in transcription elongation and suggesting that Spt2 is cotranscriptionally recruited to coding regions.
ChIP analysis of Spt2 at specific loci.
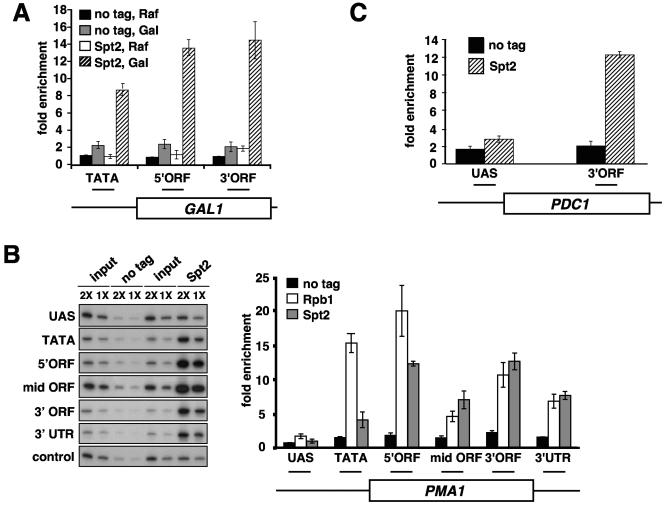
To analyze the localization of Spt2 in greater detail, we tested its physical association over four different loci by conventional ChIP. First, to test whether the association of Spt2 with coding regions is dependent upon transcription, we analyzed Spt2 association with the GAL1 gene under both noninducing and inducing conditions (Fig. 3A). Under noninducing conditions (medium containing raffinose as the carbon source), in which GAL1 is not transcribed, we did not observe significant association of Spt2 with any of the tested locations at GAL1. However, after induction in galactose, when GAL1 is transcribed at a high level, we found strong association of Spt2 over the GAL1 transcribed region. This result shows that the physical association of Spt2 to the transcribed regions of GAL1 is dependent upon active transcription, consistent with the genome-wide analysis of Spt2.
FIG. 3.
Spt2 is preferentially localized to transcribed regions of active genes. (A) Spt2-13Myc is recruited to the transcribed regions of GAL1 upon galactose induction. Yeast cells from the untagged strain (FY1856) or a strain expressing Spt2-13Myc (FY2427) were grown in YPraf medium. Cells were either formaldehyde fixed or shifted to YPgal medium for 2 hours prior to formaldehyde treatment. Chromatin immunoprecipitations were then performed using the A14 anti-Myc antibody. The horizontal bars in the diagram represent the regions assayed by PCR. The fold enrichment is calculated as the ratio of percent IP of the indicated region to percent IP of a nontranscribed control region. The values shown represent the averages and standard errors from three independent experiments. (B) Spt2 is associated with the transcribed region of PMA1. Yeast cells from an untagged strain (FY1856) or a strain expressing an Spt2-13Myc epitope-tagged protein (FY2427) were grown in YPD and cross-linked with 1% formaldehyde. Chromatin immunoprecipitations were performed using the A14 anti-Myc antibody to immunoprecipitate Spt2-13Myc and the 8WG16 antibody to immunoprecipitate Rpb1. The horizontal bars in the diagram represent the regions assayed by PCR. The fold enrichment is the ratio of percent IP of the indicated region to percent IP of the control region. The values shown represent the averages and standard errors from three independent experiments. UAS, upstream activation sequence. (C) Spt2 is associated with a transcribed region of PDC1. Growth and chromatin immunoprecipitation were performed as described for panel B.
To study further the association of Spt2 with active genes, we used ChIP to analyze the association of Spt2 across two genes constitutively transcribed at a high level, PMA1 and PDC1 (Fig. 3B and C). Again, we observed strong enrichment of Spt2 over the transcribed regions, including the coding regions, 5′UTRs, and 3′UTRs of both genes. In contrast, we did not measure significant enrichment over the untranscribed upstream activation sequence region for either gene. To compare the distributions of Spt2 and RNA polymerase II, we also measured the level of association of RNA polymerase II over PMA1 by ChIP analysis of Rpb1, the largest subunit of RNA polymerase II (Fig. 3B), and found that Rpb1 is distributed similarly to Spt2. Overall, the distribution of Spt2 resembles the pattern previously determined for the elongation factors Spt4, Spt6, and Spt16 (17, 19). It is interesting to note that this pattern is distinct from that of other elongation factors, including the PAF and Tho/Trex complexes, which are not associated with the 3′UTRs of PMA1 and other genes (17, 19).
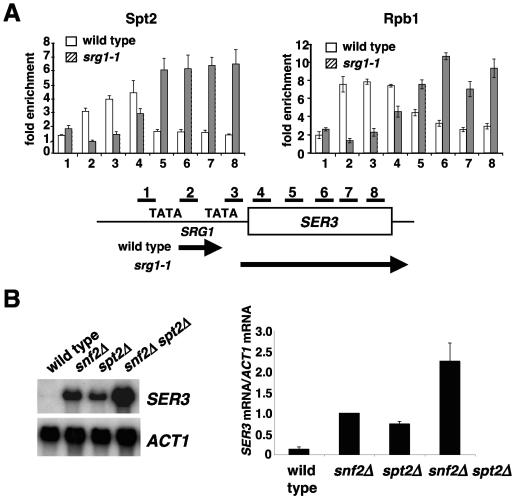
Finally, we examined the association of Spt2 with the SER3 locus. Recent studies have shown that in addition to SER3, this region encodes a second transcript, SRG1, an intergenic RNA that is transcribed across the SER3 promoter, repressing SER3 transcription (32). Using ChIP, we analyzed the association of Spt2 and Rbp1 with eight different regions across the SRG1/SER3 locus (Fig. 4A). This was done with both a wild-type strain and in an srg1-1 mutant (32), in which SRG1 transcription is abolished and SER3 is derepressed. Our results show that the distribution of Spt2 clearly overlaps with that of Rpb1 in both wild-type and srg1-1 strains. That is, in a wild-type strain, Spt2 is physically associated over SRG1, while in the srg1-1 mutant, Spt2 is physically associated over SER3. To test whether Spt2 plays a role in SER3 regulation, we performed Northern analysis of SER3 RNA levels in wild-type and spt2Δ strains. We also included snf2Δ and snf2Δ spt2Δ mutants, as previous studies have shown that Swi/Snf is required for SER3 repression (33, 34). Our results (Fig. 4B) show that spt2Δ causes a large increase in the level of SER3 mRNA. In addition, in the spt2Δ snf2Δ double mutant, the level is further increased, suggesting that in wild-type strains, Spt2 and Swi/Snf repress SER3 by independent mechanisms. While the mechanism by which Spt2 affects SER3 is not yet clear, the association of Spt2 at SRG1 and SER3 and its requirement for SER3 repression support the conclusion that Spt2 is physically associated with actively transcribed regions and plays a role in elongation.
FIG. 4.
Association of Spt2 and Rpb1 with SRG1 and SER3. (A) Cells from a wild-type strain (FY2435) and an srg1-1 mutant (FY2434) with a mutation in the SRG1 TATA element (32) were analyzed by chromatin immunoprecipitation for the association of Spt2-13Myc (using A14 anti-Myc antibody) and Rpb1 (using 8WG16 antibody). The fold enrichment is calculated as described for Fig. 3. The values shown represent the averages and standard errors from three independent experiments. (B) An spt2Δ mutation causes derepression of SER3 transcription. Cells from wild-type (FY2428), snf2Δ (FY2429), spt2Δ (FY2431), and snf2Δ spt2Δ (FY2430) strains were grown in YPD, total RNA was extracted, and Northern hybridization analysis was performed for SER3. ACT1 served as a loading control. The relative expression of the SER3 gene in the different mutants is also shown. The ratio of SER3 mRNA to ACT1 mRNA in the snf2Δ strain was arbitrarily set equal to 1.0. The ratios for the other strains were divided by the snf2Δ ratio. The quantification shown represents the averages and standard errors from three independent experiments.
The transcription elongation factor Spt6 is required for Spt2 recruitment to PMA1.
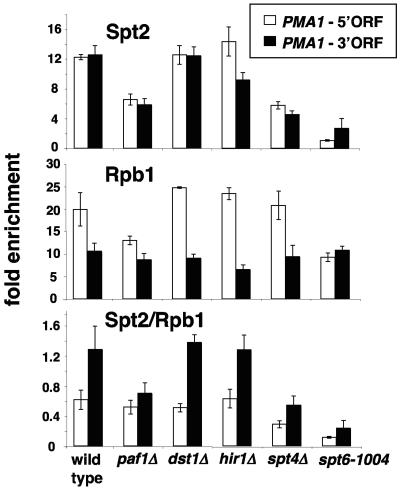
To gain insight into the mechanism of Spt2 recruitment, we analyzed its association with two regions of PMA1 in a set of mutants representing the loss of distinct classes of elongation factors. Our results (Fig. 5) show that the level of Spt2 association was most significantly decreased (5- to 10-fold) in an spt6-1004 mutant. Spt6 plays a role in transcription elongation and chromatin structure and has been proposed to function in the reassembly of nucleosomes in the wake of RNA polymerase II (12, 18). Two other mutants, the paf1Δ and spt4Δ mutants, showed a twofold decrease in Spt2 recruitment, while the other two tested, the hir1Δ and dst1Δ mutants, showed no detectable defect. The loss of Spt2 recruitment in the spt6-1004 mutant is associated with only a twofold decrease in Rpb1 occupancy, indicating that the Spt2 recruitment defect reflects a direct requirement for Spt6 rather than an indirect effect of a change in transcription levels. Importantly, the Spt2-13Myc protein level is not affected in an spt6 mutant (data not shown). These observations suggest that recruitment and activity of Spt2 are components of Spt6-mediated regulation of chromatin structure at PMA1.
FIG. 5.
Recruitment of Spt2 to PMA1 is dependent on Spt6. (A) Spt2 association with PMA1 in different transcription elongation mutants. Chromatin immunoprecipitation of Spt2-13Myc (top panel) and Rpb1 (middle panel) was performed as described for Fig. 3 with wild-type (FY2427), paf1Δ (FY2446), dst1Δ (FY2447), hir1Δ (FY2448), spt4Δ (FY2449), and spt6-1004 (FY2450) strains. Two regions of PMA1 (5′ORF or 3′ORF) were analyzed by PCR. The fold enrichment was calculated as described for Fig. 3. The relative occupancy of Spt2 at PMA1 is shown in the bottom panel. The values represent the averages and standard errors from three independent experiments.
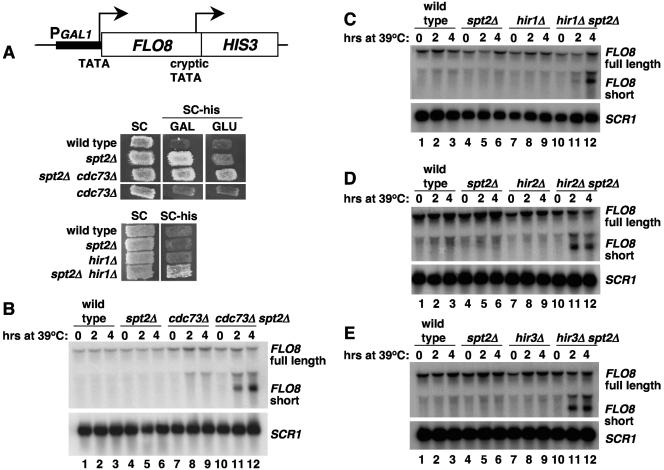
Spt2 cooperates with the PAF and the HIR/HPC complexes to inhibit the initiation of transcription from the FLO8 cryptic promoter.
To test further the role of Spt2 in elongation, we used a reporter system that is sensitive to transcription elongation defects in vivo. This reporter is based on previous studies showing that cryptic promoters exist within the coding regions of certain genes and that in particular transcription elongation mutants, these cryptic promoters can be used (18, 35). This phenotype has been most extensively characterized for the FLO8 gene (18). To test if an spt2Δ mutation allows cryptic initiation, we used a reporter for FLO8 cryptic initiation in which the 3′ coding region of FLO8 has been replaced with the HIS3 coding region such that HIS3 is expressed only when the FLO8 cryptic promoter is active. In addition, the FLO8 promoter was replaced with the GAL1 promoter to allow regulation of initiation from the normal FLO8 initiation site (see Materials and Methods). Using this reporter, we then tested expression of GAL1-FLO8-HIS3 in different mutants by assaying growth on medium lacking histidine, using either glucose or galactose as the carbon source. In these tests, growth on galactose is a more permissive condition to detect cryptic initiation than is growth on glucose. Our results (Fig. 6A) show that spt2Δ allows expression from the FLO8 cryptic promoter when cells are grown on galactose and weak expression when cells are grown on glucose. In both the spt2Δ cdc73Δ and spt2Δ hir1Δ double mutants, this phenotype is considerably stronger, suggesting that the genetic interactions between Spt2, PAF, and HIR/HPC affect transcription elongation.
FIG. 6.
Spt2, the PAF complex, and the HIR/HPC complex collaborate to inhibit transcription from the FLO8 cryptic promoter. (A) Spt2 contributes to the inhibition of transcription initiation from the cryptic promoter of the pGAL1::FLO8-HIS3 reporter gene. The reporter construct is diagrammed in the top part of the figure. Below are shown patches of wild-type (FY2452), spt2Δ (FY2445), spt2Δ cdc73Δ (FY2453), and cdc73Δ (FY2454) strains containing the pGAL1::FLO8::HIS3 reporter construct that were initially grown on a YPD plate and then replica plated to synthetic complete medium (SC) or medium lacking histidine (SC-his) and containing galactose or glucose as the carbon source. The photograph was taken 2 days after incubation at 30°C. In the lower panel, wild-type (FY2452), spt2Δ (FY2445), hir1Δ (FY2503), and spt2Δ hir1Δ (FY2504) strains were replica plated to synthetic complete medium or medium lacking histidine and containing glucose. The photograph was taken after overnight incubation at 30°C. (B) Spt2 and Cdc73 collaborate to inhibit transcription initiation from the FLO8 cryptic promoter. Wild-type (FY1856), spt2Δ (FY2432), cdc73Δ (FY2127), and cdc73Δ spt2Δ (FY2436) strains were grown in YPD at 30°C and then shifted for the indicated times to 39°C. Total RNA was extracted and analyzed by Northern analysis with a probe for FLO8. SCR1 served as a loading control. The FLO8 probe identifies the full-length FLO8 mRNA and the FLO8 short RNA, which has been previously shown to initiate from a cryptic promoter (18). The band between the full-length and short FLO8 RNAs is believed to be an artifact caused by the presence of rRNA. (C) Spt2 and Hir1 collaborate to inhibit initiation of transcription from the FLO8 cryptic promoter. Cells from wild-type (FY2442), hir1Δ (FY1235), spt2Δ (FY2431), and hir1Δ spt2Δ (FY2451) strains were grown and analyzed as described for panel B. (D) Spt2 and Hir2 collaborate to inhibit initiation of transcription from the FLO8 cryptic promoter. Cells from wild-type (FY2442), hir2Δ (FY2443), spt2Δ (FY2431), and hir2Δ spt2Δ (FY2444) strains were treated as described for panel B. (E) Spt2 and Hir3 collaborate to inhibit initiation of transcription from the FLO8 cryptic promoter. Cells from wild-type (FY2442), hir3Δ (FY2441), spt2Δ (FY2431), and hir3Δ spt2Δ (FY2440) strains were treated as described for panel B.
To test cryptic initiation at FLO8 more directly, we also performed Northern hybridization analysis of the wild-type FLO8 gene. These experiments were done both with an spt2Δ single mutant and with double mutants in which spt2Δ was combined with cdc73Δ, hir1Δ, hir2Δ, or hir3Δ. In these experiments, RNA samples were prepared at 0, 2, and 4 h after a shift to the nonpermissive temperature. Our results (Fig. 6B to E) show that in the spt2Δ single mutant, only the full-length FLO8 transcript was observed. Thus, Northern analysis of wild-type FLO8 is a less sensitive assay for cryptic initiation than use of the GAL1-FLO8-HIS3 reporter. Furthermore, in each of the other single mutants tested (cdc73Δ, hir1Δ, hir2Δ, and hir3Δ mutants), the same result was obtained. In contrast, in each double mutant, the FLO8 short transcript was produced at a significant level. These results further suggest that Spt2 and both the PAF and HIR/HPC complexes play roles in regulation of chromatin structure during transcription elongation.
Hir proteins are associated with transcribed regions.
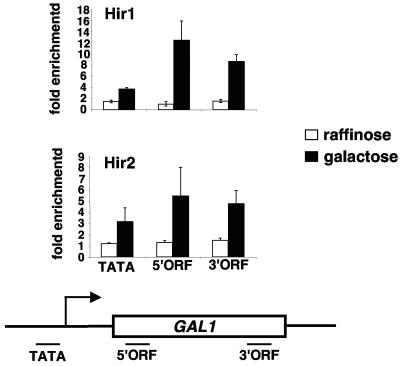
Our results have suggested that Spt2 and the HIR/HPC complex are functionally related in the process of transcription elongation. To test if the Hir proteins are also physically linked to transcription elongation, we analyzed Hir1-13Myc and Hir2-13Myc association with the GAL1 gene under both noninducing and inducing conditions (Fig. 7). Under noninducing conditions (medium containing raffinose as the carbon source), in which GAL1 is not transcribed, we did not observe significant association of Hir1-13Myc and Hir2-13Myc with any of the tested locations at GAL1. However, after induction in galactose, when GAL1 is transcribed at a high level, we found strong association of Hir1-13Myc and Hir2-13Myc over the GAL1 transcribed region. This result shows that the physical association of Hir1-13Myc and Hir2-13Myc to the transcribed regions of GAL1 is dependent upon active transcription. Thus, in addition to its other roles, HIR/HPC likely plays a direct role in transcription elongation.
FIG. 7.
The Hir1 and Hir2 proteins are recruited to the transcribed regions of GAL1 upon galactose induction. Yeast cells expressing Hir1-13Myc (FY2438) or Hir2-13Myc (FY2439) were grown in YPraf medium. Cells were either formaldehyde fixed or shifted to 2% YPgal medium for 2 hours prior to the formaldehyde treatment. Chromatin was extracted and subjected to immunoprecipitation using the anti-Myc antibody. The fold enrichment is calculated as described for Fig. 3. The values shown represent the averages and standard errors from three independent experiments.
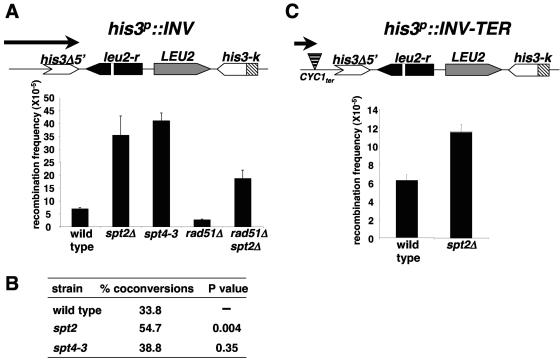
Spt2 inhibits intrachromosomal recombination.
In addition to transcription, chromatin structure can influence other processes related to DNA metabolism. Previously, Malagon and Aguilera (30) showed that spt4 and spt6 mutants exhibit increased intrachromosomal recombination between inverted repeats. In the case of spt6, this phenotype was associated with altered chromatin structure, and under certain conditions it was dependent on transcription (30). To test whether recombination is also altered in spt2Δ mutants, we used the his3p::INV system (30), diagrammed in Fig. 8, in which recombination between inverted repeats results in a His+ phenotype. In these experiments, we constructed an spt2Δ strain bearing the his3p::INV construct and compared the frequency of recombination in this strain to those in wild-type and spt4 mutant strains (Fig. 8). Our results show that the frequency of recombination is increased approximately sevenfold in the spt2Δ mutant, comparable to that in an spt4Δ mutant. In addition, we characterized the recombination events by two other criteria. First, we determined the dependence of the recombination events on Rad51, the RecA-like protein involved in strand exchange during homologous recombination (59). Our results show that recombination in both wild-type and spt2Δ strains is dependent on Rad51 to the same degree (Fig. 8), similar to what was previously observed for an spt6 mutant (30). Second, we determined whether spt2Δ causes an effect on the length of the conversion tract formed during recombination. The his3p::INV system contains the LEU2 gene, which can be used to determine the length of the gene conversion tract in the His+ recombinants (1). If the conversion tract is less than 1.2 kb, the His+ recombinants remain Leu+. If the conversion tract is longer than 1.2 kb, the His+ recombinants are Leu−. It has been previously reported that spt6 mutations increase the long-tract conversion events in the his3p::INV system by approximately 50%. Our results show that an spt2Δ mutation causes a similar increase (by 61%) compared to wild type, indicating that spt2Δ mutants favor long-tract gene conversion events. We did not observe this phenotype in an spt4 mutant. Overall, our results demonstrate that an spt2Δ mutation increases intrachromosomal recombination.
FIG. 8.
An spt2Δ mutation increases recombination between inverted repeats. (A) Measurement of recombination using the hi3p::INV inverted repeat system (1). The structure of the inverted repeats is diagrammed at the top. In each experiment, six separate colonies from wild-type (M137-11B), spt2Δ (L1096), spt4-3 (M236-12D), rad51Δ (L1099), and rad51Δ spt2Δ (L1100) strains were independently grown to saturation in YPD and plated on either synthetic complete medium or medium lacking histidine. The frequency of recombination for each independent culture is equal to the frequency of His+ colonies. The recombination frequencies shown are the averages and standard errors from three experiments. (B) spt2Δ increases coconversion events. The percentage of coconversion was determined by calculating the ratio of His+ Leu− colonies to His+ Leu+ colonies. In each experiment, six separate colonies were tested, and the values shown are the averages and standard errors from three experiments. (C) Reducing transcription through the his3p::INV inverted repeats decreases spt2Δ hyperrecombination. The recombination frequency was calculated for wild-type (ITE-1C) or spt2Δ (L1101) strains containing the his3p::INV TER (diagrammed at the top). This construct has a CYC1 terminator inserted before the his3-5′Δ sequence, reducing transcription through the inverted repeat cassette (30). The recombination frequencies were calculated as described for panel A.
We next asked whether the spt2 hyperrecombination phenotype is dependent on transcription. Previous analysis of the his3p::INV cassette demonstrated that a major 2.2-kb transcript initiates outside of the his3p::INV cassette and terminates inside the cassette around the his3Δ5′ repeat terminator (diagrammed in Fig. 8A) (30). In a derivative of his3p::INV, an insertion of a CYC1 terminator sequence 5′ of his3Δ5′ (diagrammed in Fig. 8C) drastically reduces the level of transcription toward and within the repeats (30). Our results show that the presence of the terminator sequence significantly impairs the hyperrecombination phenotype observed in the spt2Δ mutant (Fig. 8C), similar to previous results for spt6 mutants (30). Therefore, we conclude that the hyperrecombination phenotype of the spt2Δ mutant is dependent on transcription. This suggests that Spt2 plays a role in genome stability, possibly by maintaining proper chromatin structure at transcribed regions.
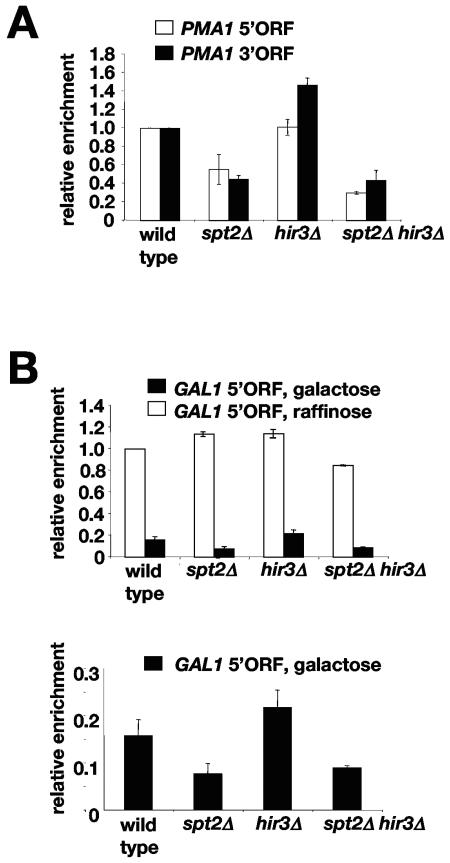
Spt2 is required for normal histone H3 levels across transcribed regions.
Our genetic and molecular data suggest a role for Spt2 in the regulation of chromatin structure during transcription elongation. To gain additional insights into the role of Spt2, we tested whether it affects the levels of histone H3 in the coding region of the PMA1 gene. Our results show that the level of histone H3 is decreased in the spt2Δ mutant at both PMA1 locations tested (Fig. 9A). In contrast, a hir3Δ mutation had no effect on histone H3 levels at PMA1, while an spt2Δ hir3Δ double mutant had a defect similar to that of the spt2Δ single mutant. We also tested by ChIP and Northern analysis the effects of these different mutations on the level of Rpb1 association at PMA1 and on the level of PMA1 transcript, but no significant effects were observed (data not shown). Moreover, the total levels of histone H3 in the different mutants were not different from that in the wild type (data not shown).
FIG. 9.
Spt2 is required for a normal level of histone H3 over the PMA1 and GAL1 transcribed regions. (A) An spt2Δ mutation reduces histone H3 association at PMA1. Cells from wild-type (FY2442), spt2Δ (FY2431), hir3Δ (FY2441), and hir3Δ spt2Δ (FY2440) strains were grown in YPD at 30°C and shifted to 39°C for 4 hours. Chromatin immunoprecipitations were performed using an anti-histone H3 antibody. For each strain, the fold enrichment shown is relative to the fold enrichment calculated for the wild-type strain arbitrarily set at 1.0. The values shown are the averages and standard errors from three independent experiments. (B) An spt2Δ mutation affects histone H3 levels at the GAL1 5′ ORF upon galactose induction. The relative values shown were calculated as described for panel A. The reference value in this case is the fold enrichment for the wild-type strain grown in 2% raffinose. The bottom panel shows only the relative values after a shift to 2% galactose and heat shock treatment for 2 h at 39°C. All values shown are the averages and standard errors from three independent experiments. The P values for these experiments are <0.03.
We next examined the role of Spt2 in maintaining histone H3 levels at the highly regulated GAL1 gene. Interestingly, under noninducing conditions when GAL1 transcription is extremely low, histone H3 occupancy is not significantly affected by spt2Δ (Fig. 9B, top panel). In contrast, under inducing conditions, when GAL1 is transcribed at a high level, the level of histone H3 is significantly reduced in the spt2Δ and spt2Δ hir3Δ mutants compared to the wild type (Fig. 9B, bottom panel). These data support the notion that Spt2 maintains histone H3 levels only at actively transcribed genes. We note that histone H3 levels were dramatically reduced over GAL1 in all strains (Fig. 9B, top panel), indicating that transcription causes a loss of H3 levels, consistent with recently published studies (20, 52, 75). Taken together, these data suggest a role for Spt2 in the maintenance of proper chromatin structure over the transcribed regions of genes.
DISCUSSION
Previous studies have suggested that Spt2 is an HMG-like protein that plays a role in chromatin-mediated transcription initiation (23, 25, 40, 44, 46). In this work we present several new results that strongly suggest that the major role of Spt2 occurs during transcription elongation. First, both genome-wide localization studies and detailed ChIP experiments show that Spt2 is physically associated with the coding regions, but not the regulatory regions, of actively transcribed genes. This association is dependent upon the elongation factor Spt6. Second, double mutant analysis has revealed functional interactions with other elongation factors, including the PAF and HIR/HPC complexes, with respect to both growth and elongation-specific transcriptional effects. Third, Spt2 is required for normal recombination levels in transcriptionally active regions. Finally, Spt2 is important for maintenance of normal histone H3 levels in the coding regions of actively transcribed genes. Taken together, these findings suggest that Spt2 plays roles inmediating chromatin dynamics during transcription elongation.
Spt2 is functionally associated with the PAF and HIR/HPC complexes.
Our double mutant analyses strongly suggest that Spt2 is functionally related to both the PAF and Hir/Hpc complexes. The exact relationship between these factors is unclear, as synthetic phenotypes can be caused by impairment of the same or distinct pathways that affect an important or essential process, in this case transcription elongation. The different interactions observed between spt2Δ and mutations in genes encoding PAF complex members likely reflect the apparent distinct functional roles of different PAF components (5, 37). Two previously studied roles for the Hir/Hpc proteins that are relevant to this process are the regulation of histone gene transcription and nucleosome reassembly (42, 43, 53, 55). While altered histone levels might play a role in transcription elongation, the latter activity appears more directly relevant to a role for Spt2 in elongation. Furthermore, recent genetic evidence has implicated the Hir/Hpc proteins in transcription elongation (12), and we have shown by ChIP analyses that Hir1 and Hir2 are recruited to the GAL1 coding region upon GAL1 activation (Fig. 7). Our observation supports a direct role of the HIR/HPC complex during transcription elongation. These results suggest a possible role for Hir/Hpc in nucleosome reassembly along active templates in the wake of RNA polymerase II transcription. Therefore, it is likely that the synthetic interaction observed between spt2Δ and hir/hpc mutants is related to chromatin dynamics during transcription elongation.
A role for Spt2 in transcription elongation may explain the suppression of transcription initiation defects in spt2 mutants.
One puzzling aspect about Spt2 is that the strongest known spt2 mutant phenotypes affect transcription initiation. Indeed, spt2 mutations were initially isolated as suppressors of the initiation defects caused by insertion mutations (69) and by loss of Swi/Snf function (58). It is important to note that these phenotypes are shared with mutations in SPT4, SPT5, SPT6, and SPT16 (31, 61), genes also believed to encode elongation factors (56). There are several possible reasons for these phenotypes, some of which are described here. First, despite their functional and physical link to elongation, these factors might also directly control initiation. Recent evidence supports such a dual role for Spt16 (6). Second, it is possible that an effect on initiation occurs indirectly via direct effects on chromatin structure in transcribed coding regions. Current evidence suggests that Spt6 and Spt16 both control the chromatin structure of transcribed regions (18, 35), and our results have suggested the same for Spt2. The activation of cryptic promoters in these mutants can explain the suppression of some promoter insertion mutations (18). Furthermore, a defect in chromatin structure that originates in a coding region, such as that caused by reduced nucleosome reassembly, could spread, in a manner analogous to the spreading of transcriptional silencing (51), to nearby promoter regions, thereby suppressing a promoter defect. Finally, in spt2 and other elongation mutants, initiation may be affected due to altered transcription across promoter regions. Recent work has suggested that many intergenic regions are transcribed in S. cerevisiae (15, 32, 72) and other organisms (16). Thus, elongation factors such as Spt2 may play direct roles in this transcription, thereby controlling promoter chromatin structure. We note that early studies showed that spt2 mutations suppress defects in expression of HO-lacZ (46, 58), while a subsequent study showed that such effects are specific to HO-lacZ and do not occur at HO (74). These results may reflect an effect of spt2 mutations on elongation through lacZ, previously noted to pose elongation problems in S. cerevisiae (8, 9).
A functional and physical connection between Spt2 and Spt6.
Our results have shown that the recruitment of Spt2 to transcribed regions is dependent upon Spt6 (Fig. 5). This result suggests that a subset of spt6 mutant phenotypes are caused by a lack of Spt2 association with transcribed regions. Indeed, spt2 and spt6 mutants were initially identified in the same mutant selection and were shown to share many mutant phenotypes (68, 69). In this work, we have added to the common phenotypes by showing that Spt2 plays a role in repression from a cryptic promoter, albeit not as great a role as Spt6, and that Spt2, like Spt6, affects recombination. Furthermore, both play a role in controlling histone levels over transcribed regions. Spt6 clearly has roles beyond those of Spt2, as Spt6 is required for viability, while loss of Spt2 has little effect on growth. Consistent with Spt2 carrying out a subset of Spt6-dependent functions is our finding that an spt2Δ spt6-1004 double mutant does not have any phenotypes beyond those of an spt6-1004 single mutant (data not shown).
Different hypotheses could explain the Spt6 dependence of Spt2 recruitment to transcribed regions. Conceivably, Spt6 might directly recruit Spt2. However, we have been unable to detect coimmunoprecipitation of Spt2 and Spt6, suggesting that there is not a strong physical association between the two factors (data not shown). Alternatively, since Spt6 has nucleosome assembly activity in vitro (7) and controls chromatin structure in vivo (18), it is possible that a defect in Spt6 results in an abnormal chromatin structure that decreases the affinity of Spt2 for transcribed regions. Whatever the nature of the Spt2-Spt6 interaction, a greater understanding of Spt2 will result in a better understanding of Spt6.
Spt2 and repression of recombination.
The enhanced level of recombination in spt2 mutants may be related to the role of Spt2 in maintaining a normal level of histones over transcribed regions. By this model, in the absence of Spt2, a reduced level of histones would result in chromatin that is more permissive for recombination. This model is consistent with the findings that histone H4 depletion in yeast causes an increased level of intrachromosomal recombination (47) and also that an spt6-140 mutation causes a transcription-dependent increase in recombination similar to that caused by spt2Δ (30). Using a different system, earlier studies also showed that there is a transcription-dependent enhancement of recombination at the highly transcribed S. cerevisiae GAL locus (62). Recent work has shown that histone levels over the GAL genes are decreased by transcription, suggesting a change in chromatin structure (20, 52). Our data indicate that Spt2 is important for the protection of transcribed regions from hyperrecombination and genomic instability.
Possible roles for Spt2 in transcription elongation.
Several studies have suggested that elongating RNA polymerase II alters chromatin structure by reducing nucleosome density (12, 13, 18, 20, 52). A number of deleterious consequences might result from such a change, including increased recombination, increased DNA damage, and, as recently shown (18), initiation from cryptic promoters. Based on recent studies, it appears that cells have developed mechanisms to ensure that transcribed regions regain normal chromatin structure in the wake of transcription, at least in part to protect their integrity. These mechanisms likely depend upon the factors FACT, Spt6/Iws1, and Hir/Hpc, all of which may reassemble nucleosomes in the wake of transcription. Our results and those of others strongly support a role for Spt2 in this process. How might Spt2, an HMG-like protein, function in this context? One example of an HMG protein participating in nucleosome reassembly comes from studies of FACT. SSRP1, an HMG protein, is a component of the mammalian FACT complex (41). In S. cerevisiae, the SSRP1 counterpart, Pob3, lacks an HMG motif; however, genetic and biochemical studies show that the HMG factor Nhp6 is required for the binding of S. cerevisiae FACT to nucleosomes (11). By analogy to Nhp6, Spt2 may facilitate the binding of a histone chaperone complex to nucleosomes. Consistent with this idea, Spt2, like other HMG proteins, is capable of binding four-way junction DNA, a structure similar to that found at the entrance-exit point of DNA from a nucleosome (40, 76). Future studies of Spt2 should help to determine its precise role in chromatin-mediated transcription elongation.
Acknowledgments
We thank Mark Hickman for helpful comments on the manuscript. We also thank Lisa Laprade and other members of the Winston lab for helpful suggestions. We are grateful to Louis Lefebvre, Francisco Malagon, and Andres Aguilera for plasmids, strains, and helpful advice and to Elissa Schwartzfarb for construction of the FLO8-HIS3 reporter.
This work was supported by NIH grant GM32967 to F.W. and by a grant from the Canadian Institutes for Health Research (CIHR) to F.R. F.R. holds a new investigator award from the CIHR. A.N. was supported by a Senior Research Fellowship from CIHR.
REFERENCES
- 1.Aguilera, A., and H. L. Klein. 1989. Genetic and molecular analysis of recombination events in Saccharomyces cerevisiae occurring in the presence of the hyper-recombination mutation hpr1. Genetics 122:503-517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1991. Current protocols in molecular biology. Wiley-Interscience, New York, N.Y.
- 3.Baudin, A., O. Ozier-Kalogeropoulos, A. Denouel, F. Lacroute, and C. Cullin. 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21:3329-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baxter, B. K., and E. A. Craig. 1998. Suppression of an Hsp70 mutant phenotype in Saccharomyces cerevisiae through loss of function of the chromatin component Sin1p/Spt2p. J. Bacteriol. 180:6484-6492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Betz, J. L., M. Chang, T. M. Washburn, S. E. Porter, C. L. Mueller, and J. A. Jaehning. 2002. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol. Genet. Genomics 268:272-285. [DOI] [PubMed] [Google Scholar]
- 6.Biswas, D., Y. Yu, M. Prall, T. Formosa, and D. J. Stillman. 2005. The yeast FACT complex has a role in transcriptional initiation. Mol. Cell. Biol. 25:5812-5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bortvin, A., and F. Winston. 1996. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 272:1473-1476. [DOI] [PubMed] [Google Scholar]
- 8.Chavez, S., and A. Aguilera. 1997. The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev. 11:3459-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chavez, S., M. Garcia-Rubio, F. Prado, and A. Aguilera. 2001. Hpr1 is preferentially required for transcription of either long or G+C-rich DNA sequences in Saccharomyces cerevisiae. Mol. Cell. Biol. 21:7054-7064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa, P. J., and K. M. Arndt. 2000. Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics 156:535-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Drovin, S., and F. Roberts. Methods, in press.
- 11.Formosa, T., P. Eriksson, J. Wittmeyer, J. Ginn, Y. Yu, and D. J. Stillman. 2001. Spt16-Pob3 and the HMG protein Nhp6 combine to form the nucleosome-binding factor SPN. EMBO J. 20:3506-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Formosa, T., S. Ruone, M. D. Adams, A. E. Olsen, P. Eriksson, Y. Yu, A. R. Rhoades, P. D. Kaufman, and D. J. Stillman. 2002. Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics 162:1557-1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Green, E. M., A. J. Antczak, A. O. Bailey, A. A. Franco, K. J. Wu, J. R. Yates, and P. D. Kaufman. 2005. Replication-independent histone deposition by the HIR complex and Aaf1. Curr. Biol. 15:2044-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hartzog, G. A., T. Wada, H. Handa, and F. Winston. 1998. Evidence that Spt4, Spt5, and Spt6 control transcription elongation by RNA polymerase II in Saccharomyces cerevisiae. Genes Dev. 12:357-369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95:717-728. [DOI] [PubMed] [Google Scholar]
- 15.Hurowitz, E. H., and P. O. Brown. 2003. Genome-wide analysis of mRNA lengths in Saccharomyces cerevisiae. Genome Biol. 5:R2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson, J. M., S. Edwards, D. Shoemaker, and E. E. Schadt. 2005. Dark matter in the genome: evidence of widespread transcription detected by microarray tiling experiments. Trends Genet. 21:93-102. [DOI] [PubMed] [Google Scholar]
- 17.Kaplan, C. D., M. J. Holland, and F. Winston. 2005. Interaction between transcription elongation factors and mRNA 3′-end formation at the Saccharomyces cerevisiae GAL10-GAL7 locus. J. Biol. Chem. 280:913-922. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan, C. D., L. Laprade, and F. Winston. 2003. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301:1096-1099. [DOI] [PubMed] [Google Scholar]
- 19.Kim, M., S. H. Ahn, N. J. Krogan, J. F. Greenblatt, and S. Buratowski. 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23:354-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kristjuhan, A., and J. Q. Svejstrup. 2004. Evidence for distinct mechanisms facilitating transcript elongation through chromatin in vivo. EMBO J. 23:4243-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krogan, N. J., J. Dover, A. Wood, J. Schneider, J. Heidt, M. A. Boateng, K. Dean, O. W. Ryan, A. Golshani, M. Johnston, J. F. Greenblatt, and A. Shilatifard. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell 11:721-729. [DOI] [PubMed] [Google Scholar]
- 22.Krogan, N. J., M. Kim, A. Tong, A. Golshani, G. Cagney, V. Canadien, D. P. Richards, B. K. Beattie, A. Emili, C. Boone, A. Shilatifard, S. Buratowski, and J. Greenblatt. 2003. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 23:4207-4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kruger, W., and I. Herskowitz. 1991. A negative regulator of HO transcription, SIN1 (SPT2), is a nonspecific DNA-binding protein related to HMG1. Mol. Cell. Biol. 11:4135-4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurdistani, S. K., S. Tavazoie, and M. Grunstein. 2004. Mapping global histone acetylation patterns to gene expression. Cell 117:721-733. [DOI] [PubMed] [Google Scholar]
- 25.Lefebvre, L., and M. Smith. 1993. Mutational and functional analysis of dominant SPT2 (SIN1) suppressor alleles in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:5393-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lieb, J. D., X. Liu, D. Botstein, and P. O. Brown. 2001. Promoter-specific binding of Rap1 revealed by genome-wide maps of protein-DNA association. Nat. Genet. 28:327-334. [DOI] [PubMed] [Google Scholar]
- 27.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 28.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251-260. [DOI] [PubMed] [Google Scholar]
- 29.Malagon, F., and A. Aguilera. 1996. Differential intrachromosomal hyper-recombination phenotype of spt4 and spt6 mutants of S. cerevisiae. Curr. Genet. 30:101-106. [DOI] [PubMed] [Google Scholar]
- 30.Malagon, F., and A. Aguilera. 2001. Yeast spt6-140 mutation, affecting chromatin and transcription, preferentially increases recombination in which Rad51p-mediated strand exchange is dispensable. Genetics 158:597-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malone, E. A., C. D. Clark, A. Chiang, and F. Winston. 1991. Mutations in SPT16/CDC68 suppress cis- and trans-acting mutations that affect promoter function in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:5710-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Martens, J. A., L. Laprade, and F. Winston. 2004. Intergenic transcription is required to repress the Saccharomyces cerevisiae SER3 gene. Nature 429:571-574. [DOI] [PubMed] [Google Scholar]
- 33.Martens, J. A., and F. Winston. 2002. Evidence that Swi/Snf directly represses transcription in S. cerevisiae. Genes Dev. 16:2231-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martens, J. A., P.-Y. J. Wu, and F. Winston. 2005. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae. Genes Dev. 19:2695-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mason, P. B., and K. Struhl. 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 23:8323-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mueller, C. L., and J. A. Jaehning. 2002. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol. 22:1971-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mueller, C. L., S. E. Porter, M. G. Hoffman, and J. A. Jaehning. 2004. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell 14:447-456. [DOI] [PubMed] [Google Scholar]
- 38.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2002. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 16:806-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ng, H. H., F. Robert, R. A. Young, and K. Struhl. 2003. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11:709-719. [DOI] [PubMed] [Google Scholar]
- 40.Novoseler, M., G. Hershkovits, and D. J. Katcoff. 2005. Functional domains of the yeast chromatin protein Sin1p/Spt2p can bind four-way junction and crossing DNA structures. J. Biol. Chem. 280:5169-5177. [DOI] [PubMed] [Google Scholar]
- 41.Orphanides, G., W. H. Wu, W. S. Lane, M. Hampsey, and D. Reinberg. 1999. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400:284-288. [DOI] [PubMed] [Google Scholar]
- 42.Osley, M. A. 1991. The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem. 60:827-861. [DOI] [PubMed] [Google Scholar]
- 43.Osley, M. A., and D. Lycan. 1987. trans-acting regulatory mutations that alter transcription of Saccharomyces cerevisiae histone genes. Mol. Cell. Biol. 7:4204-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perez-Martin, J., and A. D. Johnson. 1998. Mutations in chromatin components suppress a defect of Gcn5 protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 18:1049-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peterson, C. L., W. Kruger, and I. Herskowitz. 1991. A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell 64:1135-1143. [DOI] [PubMed] [Google Scholar]
- 46.Pollard, K. J., and C. L. Peterson. 1997. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol. Cell. Biol. 17:6212-6222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prado, F., and A. Aguilera. 2005. Partial depletion of histone H4 increases homologous recombination-mediated genetic instability. Mol. Cell. Biol. 25:1526-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prochasson, P., L. Florens, S. K. Swanson, M. P. Washburn, and J. L. Workman. 2005. The HIR corepressor complex binds to nucleosomes generating a distinct protein/DNA complex resistant to remodeling by SWI/SNF. Genes Dev. 19:2534-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48a.Ren, B., F. Robert, J. J. Wyrick, O. Aparicio, E. G. Jennings, I. Simon, J. Zeitlinger, J. Schrieber, N. Hannett, E. Kanin, T. L. Volkert, C. J. Wilson, J. P. Bell, and R.A. Young. 2000. Genome-wide location and function of DNA binding proteins. Science 290:2306-2309. [DOI] [PubMed] [Google Scholar]
- 49.Robert, F., D. K. Pokholok, N. M. Hannett, N. J. Rinaldi, M. Chandy, A. Rolfe, J. L. Workman, D. K. Gifford, and R. A. Young. 2004. Global position and recruitment of HATs and HDACs in the yeast genome. Mol. Cell 16:199-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rondon, A. G., M. Gallardo, M. Garcia-Rubio, and A. Aguilera. 2004. Molecular evidence indicating that the yeast PAF complex is required for transcription elongation. EMBO Rep. 5:47-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72:481-516. [DOI] [PubMed] [Google Scholar]
- 52.Schwabish, M. A., and K. Struhl. 2004. Evidence for eviction and rapid deposition of histones upon transcriptional elongation by RNA polymerase II. Mol. Cell. Biol. 24:10111-10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sharp, J. A., E. T. Fouts, D. C. Krawitz, and P. D. Kaufman. 2001. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr. Biol. 11:463-473. [DOI] [PubMed] [Google Scholar]
- 54.Sharp, J. A., A. A. Franco, M. A. Osley, and P. D. Kaufman. 2002. Chromatin assembly factor I and Hir proteins contribute to building functional kinetochores in S. cerevisiae. Genes Dev. 16:85-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sherwood, P. W., S. V. Tsang, and M. A. Osley. 1993. Characterization of HIR1 and HIR2, two genes required for regulation of histone gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 13:28-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sims, R. J., III, R. Belotserkovskaya, and D. Reinberg. 2004. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 18:2437-2468. [DOI] [PubMed] [Google Scholar]
- 57.Squazzo, S. L., P. J. Costa, D. L. Lindstrom, K. E. Kumer, R. Simic, J. L. Jennings, A. J. Link, K. M. Arndt, and G. A. Hartzog. 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21:1764-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sternberg, P. W., M. J. Stern, I. Clark, and I. Herskowitz. 1987. Activation of the yeast HO gene by release from multiple negative controls. Cell 48:567-577. [DOI] [PubMed] [Google Scholar]
- 59.Sung, P., L. Krejci, S. Van Komen, and M. G. Sehorn. 2003. Rad51 recombinase and recombination mediators. J. Biol. Chem. 278:42729-42732. [DOI] [PubMed] [Google Scholar]
- 60.Swanson, M. S., E. A. Malone, and F. Winston. 1991. SPT5, an essential gene important for normal transcription in Saccharomyces cerevisiae, encodes an acidic nuclear protein with a carboxy-terminal repeat. Mol. Cell. Biol. 11:3009-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swanson, M. S., and F. Winston. 1992. SPT4, SPT5 and SPT6 interactions: effects on transcription and viability in Saccharomyces cerevisiae. Genetics 132:325-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas, B. J., and R. Rothstein. 1989. Elevated recombination rates in transcriptionally active DNA. Cell 56:619-630. [DOI] [PubMed] [Google Scholar]
- 63.Thomas, J. O. 2001. HMG1 and 2: architectural DNA-binding proteins. Biochem. Soc. Trans. 29:395-401. [DOI] [PubMed] [Google Scholar]
- 64.Thomas, J. O., and A. A. Travers. 2001. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 26:167-174. [DOI] [PubMed] [Google Scholar]
- 65.Tong, A. H., M. Evangelista, A. B. Parsons, H. Xu, G. D. Bader, N. Page, M. Robinson, S. Raghibizadeh, C. W. Hogue, H. Bussey, B. Andrews, M. Tyers, and C. Boone. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294:2364-2368. [DOI] [PubMed] [Google Scholar]
- 66.Wang, A., S. K. Kurdistani, and M. Grunstein. 2002. Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 298:1412-1414. [DOI] [PubMed] [Google Scholar]
- 67.West, K. L. 2004. HMGN proteins play roles in DNA repair and gene expression in mammalian cells. Biochem. Soc. Trans. 32:918-919. [DOI] [PubMed] [Google Scholar]
- 68.Winston, F., and M. Carlson. 1992. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 8:387-391. [DOI] [PubMed] [Google Scholar]
- 69.Winston, F., D. T. Chaleff, B. Valent, and G. R. Fink. 1984. Mutations affecting Ty-mediated expression of the HIS4 gene of Saccharomyces cerevisiae. Genetics 107:179-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Winston, F., C. Dollard, and S. L. Ricupero-Hovasse. 1995. Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11:53-55. [DOI] [PubMed] [Google Scholar]
- 71.Wood, A., J. Schneider, J. Dover, M. Johnston, and A. Shilatifard. 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278:34739-34742. [DOI] [PubMed] [Google Scholar]
- 72.Wyers, F., M. Rougemaille, G. Badis, J. C. Rousselle, M. E. Dufour, J. Boulay, B. Regnault, F. Devaux, A. Namane, B. Seraphin, D. Libri, and A. Jacquier. 2005. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121:725-737. [DOI] [PubMed] [Google Scholar]
- 73.Xu, H., U. J. Kim, T. Schuster, and M. Grunstein. 1992. Identification of a new set of cell cycle-regulatory genes that regulate S-phase transcription of histone genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:5249-5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu, Y., P. Eriksson, and D. J. Stillman. 2000. Architectural transcription factors and the SAGA complex function in parallel pathways to activate transcription. Mol. Cell. Biol. 20:2350-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang, L., S. Schroeder, N. Fong, and D. L. Bentley. 2005. Altered nucleosome occupancy and histone H3K4 methylation in response to ‘transcriptional stress.’ EMBO J. 24:2379-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zlatanova, J., and K. van Holde. 1998. Binding to four-way junction DNA: a common property of architectural proteins? FASEB J. 12:421-431. [PubMed] [Google Scholar]