Abstract
In this study, we show that exposure of human hepatocellular HepG2 cells to SP600125 rapidly and dramatically reduced global histone H3-Ser10 phosphorylation, without significantly affecting the global acetylation of neighboring lysines. The loss of phosphorylation is not due to changes in cell cycle distribution and/or apoptosis and is mediated independent of either p46/54JNK or MSK-1/2 inhibition. Moreover, SP600125 repressed the basal expression of the endogenous LDL receptor in a gene-specific manner, whereas the expression of squalene synthase, sterol response element-binding protein-1, and β-actin was not altered by SP600125. Finally, chromatin immunoprecipitation and in vivo footprinting assays provided direct evidence that localized histone H3-Ser10 dephosphorylation at the low-density lipoprotein receptor promoter was associated with a significant decrease in the occupancy of the Sp1 binding site, with a slight reduction in the occupancy of RNA polymerase II. Together, our findings show that SP600125 is an efficient inhibitor of histone H3-Ser10 phosphorylation in vivo, and our results led us to hypothesize that this modification plays a novel role in regulating transcriptional control by modulating promoter accessibility to maintain basal expression in a gene-specific manner.
The role played by histone modifications in transcriptional regulation is one recent area of interest in the study of gene expression. There is plenty of evidence suggesting that histone H3 modifications are of fundamental importance in gene regulation (3, 44, 52). Core histones are known to undergo a diverse array of posttranslational modifications, including phosphorylation of serines and acetylation and methylation of lysines. Moreover, these modifications collectively influence a web of regulatory events, and their interconnectedness has led to the hypothesis that there is a “histone code” controlling chromatin dynamics (26, 45). Such a code would allow posttranslational modifications of various amino acids within the core histones to carry informational content and instructions that help specify which genes are to be activated or repressed during development. Many types of cancer are associated with translocations or mutations in chromatin-modifying enzymes and regulatory proteins (20, 33, 38, 49).
In contrast to the many studies on the structural and functional consequences of histone acetylation, the impact of the phosphorylation of core histones is relatively unexplored. The phosphorylation of histone H3 at serine 10 (histone H3-Ser10) is conserved throughout eukaryotes, and an increase in phosphorylation has been shown to correlate with gene activation in mammalian cells (32) and Drosophila (36). Coincident with the induction of early response genes such as c-fos, quiescent fibroblasts treated with epidermal growth factor undergo rapid histone H3-serine 10 phosphorylation (32). The phosphorylation of histone H3-Ser10 is mediated by several specific kinases, which are activated by distinct pathways (5, 11, 47). For example, mammalian mitotic histone H3 phosphorylation is associated with Aurora B kinases (14), histone H3 phosphorylation by IKK-α is important for the activation of NF-κB (53), and the immediate-early gene response is mediated mainly by MSK-1 and MSK-2 (43), although RSK-2 has also been implicated in this process (40a). Mitotic histone H3-Ser10 phosphorylation is observed extensively throughout condensed chromosomes, whereas the inducible phosphorylation affects a minute fraction of chromosomes and is associated with induced genes. However, the role of basal histone H3-Ser10 phosphorylation in transcriptional initiation has never been explored.
Intracellular lipid contents in mammalian cells are regulated by the integration of two pathways that govern the synthesis of endogenous lipids and the uptake of extracellular lipids (9). The expression of genes involved in the production and uptake of cholesterol, fatty acids, triglycerides, and phospholipids is regulated by a group of membrane-bound transcription factors called sterol regulatory element-binding proteins (SREBPs) (10). There are three isoforms of SREBP, designated SREBP-1a, SREBP-1c, and SREBP-2. Although SREBP-1a can stimulate all SREBP-responsive genes, SREBP-1c preferentially activates fatty acid synthetic genes, and SREBP-2 activates mainly cholesterol synthesis genes and the low-density lipoprotein (LDL) receptor in response to the depletion of cholesterol. The LDL receptor is an early gene whose basal expression is regulated by transcription factor Sp1 and requires the cooperation of Sp1 and SREBP-2 for optimal induction in response to the depletion of sterols (16, 40). Signals from a variety of extracellular agents converge on the p42/44MAPK-dependent transactivation of the above-described transcription factors to induce LDL receptor transcription (35). In fact, the degree of p42/44MAPK activation determines the extent of induction (18). In contrast, stress-activated p38MAPK negatively regulates LDL receptor expression (42). Recently, we observed that phorbol ester-dependent LDL receptor induction was associated with an increase in local histone H3-Ser10 phosphorylation independent of p42/44MAPK in HepG2 cells (25). However, information is lacking regarding precisely how this modification integrates with the transcriptional machinery responsible for regulating LDL receptor induction. Furthermore, while the majority of histone H3-Ser10 phosphorylation studies have focused on the mechanisms of gene activation, the results obtained have been limited in their application to the transcriptional initiation process.
During our investigation of the role of c-Jun N-terminal kinase (p46/54JNK) in cytokine-induced LDL receptor transcription, we made an observation that treating human hepatoma HepG2 cells with SP600125 {anthra[1,9-cd]pyrazol-6(2H)-one} (2) alone blocked global histone H3-Ser10 phosphorylation. In the present study, we expanded on this early observation and directly evaluated the influence of global histone H3-Ser10 dephosphorylation on localized histone H3-Ser10 phosphorylation and the basal expression of genes involved in cholesterol homeostasis. We show that SP600125 strongly and specifically inhibited global histone H3-Ser10 phosphorylation, possibly independent of the p46/54JNK pathway. Moreover, SP600125 reduced the basal endogenous levels of LDL receptor expression without affecting the expression of other genes involved in cholesterol homeostasis. Lastly, using the techniques of chromatin immunoprecipitation (ChIP), a direct correlation was observed between the levels of histone H3-Ser10 dephosphorylation, the loss of Sp1 site occupancy, and LDL receptor transcription. These results suggest a model whereby histone H3-Ser10 phosphorylation regulates the accessibility and activity of basal transcriptional machinery for specific genes and enable us to discuss models by which repression may be mediated. Our findings reveal important differences in the precise ways by which the LDL receptor and other genes involved in cholesterol homeostasis are regulated, and they suggest that the existence of subtle differences in the nature of histone H3-Ser10 phosphorylation may be responsible for an efficient and gene-specific transcriptional control. Thus, the LDL receptor represents one of the few known natural model systems with which to study the effects of histone H3-Ser10 phosphorylation in vivo.
MATERIALS AND METHODS
Materials.
SP600125 (2) was purchased from Calbiochem, CA. The polypeptide Jun N-terminal protein kinase (JNK) inhibitor JNK-1 (7) was initially obtained from Tsonwin Hai, The Ohio State University (21), and was later purchased from Calbiochem, CA. TRIzol and tissue culture supplies were purchased from Invitrogen Corp, CA. Phospho-specific antibodies to the activated forms of p42/44MAPK (Thr202/Tyr204), p38MAPK (Thr180/Tyr182), p46/54JNK (Thr183/Tyr185), MSK-1 (Ser376), and pp90RSK (Ser380) and the stress-activated protein kinase/JNK assay kit were purchased from Cell Signaling Technology, Inc., MA. Anti-phospho-Ser10- and anti-acetyl-Lys14-histone H3 were purchased from Upstate Biotechnology, NY. Anti-histone H3, anti-MSK-1/2, anti-Sp1, and anti-RNA polymerase II (RNA Pol II) were purchased from Santa Cruz Technology, CA. [α-32P]dCTP, [γ-32P]ATP, and the enhanced chemiluminescence (ECL) detection kit were purchased from Amersham Pharmacia Biotech, NJ. The Gal4-Sp1/Gal4-5xLuciferase system described previously (37) was used to monitor the effect of SP60012 on the transactivation potential of Sp1.
Cell culture.
Human hepatocellular carcinoma (HepG2) cells were maintained as monolayer cultures in Eagle's minimum essential medium (BioWhittaker, Inc., NY) supplemented with 10% fetal bovine serum (FBS) (Invitrogen Corp., CA) (27, 28). Cells were grown at 37°C in a humidified 5% carbon dioxide-95% air atmosphere. SP600125 treatments of HepG2 cells were done in medium containing 0.5% FBS.
Kinase assays.
Whole-cell extracts used for the evaluation of JNK activity were prepared by suspending cells in 0.5 ml of lysis buffer (25 mM HEPES, pH 7.5, 0.3 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM dithiothreitol, 20 mM β-glycerophosphate, 1 mM Na3VO4, 0.1% Triton X-100, 20 μg/ml aprotinin, 50 μg/ml leupeptin, 10 μM pepstatin, 0.1 μM okadaic acid, and 1 mM phenylmethylsulfonyl fluoride). After 20 min on ice, insoluble material was removed by centrifugation for 20 min at 100,000 × g. p46/54JNK was assayed by a glutathione bead pull-down method using a kit from Cell Signaling Technology (Beverly, MA) according to the manufacturer's directions using 100 μM ATP. Quantitation of the inhibition of JNK by SP600125 was performed by densitometric scanning.
Flow cytometric analysis.
Cells were grown as described earlier (27), and cell cycle/apoptosis analyses were carried out using propidium iodide staining with subsequent fluorescence-activated cell sorting at the Ohio State University College of Medicine Core facility.
Western blot analysis.
Whole-cell extracts were prepared by boiling in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer. Equal amounts of protein were analyzed by 12% acrylamide SDS-PAGE and immunoblotted as described earlier (28). To quantify the signals, membranes were scanned with a Storm 860 phosphorimager (Molecular Dynamics), and image and quantification analyses were carried out with ImageQuant 5.0 software.
Quantitative PCR analysis.
Total RNA was isolated from HepG2 cells by using TRIzol, and cDNA was synthesized from 1 μg of total RNA by using oligo(dT)12-18 primer with the SuperScript first-strand synthesis system (Invitrogen Corp., CA) (27, 28). The sequences of the primers used for quantitating the LDL receptor expression were 5′-AGGCTGTGGGCTCCATCGCCTA-3′ and 5′-AGTCAGTCCAGTACATGAAGCCA-3′, primers for squalene synthase (SS) were 5′-TGGAGTTCGTGAAATGCCTTG-3′ and 5′-ACCGCCAGTCTGGTTGGTAAAG-3′, primers for SREBP-1 were 5′-ATGGACGAGCCACCCTTCAGCGA-3′ and 5′-TGCAGGATGCTCAGTGGCACT-3′, primers for β-actin were 5′-ACTATGACTTAGTTGCGTTA-3′ and 5′-GGGCACGAAGGCTCATCATT-3′, and primers for c-Jun were 5′-GAGCTAGCGCCTGTGGCTCC-3′ and 5′-CTCTGCCACTTGTCTCCGGTC-3′. [32P]dCTP was incorporated into the PCR products for visualization and quantitation. The linear range for each primer set was determined empirically using different amounts of cDNA. Subsequent PCR analyses were carried out using the optimum cycle number determined for each primer set. The LDL receptor mRNA was normalized to β-actin mRNA, and data for each point were plotted as the percentage of LDL receptor mRNA compared with controls.
ChIP assays.
ChIP assays were performed as described earlier (25). For PCR analysis, either 1/100 (input) or 1/10 (immunoprecipitates) of the immunoprecipitated DNA was amplified by using 20 pmol of the indicated primers in 20-μl reaction mixtures containing 200 μM deoxynucleoside triphosphates, 2.7 mM MgCl2, and 0.25 U of AmpliTaq. After 4 min at 94°C, 28 cycles of 1 min at 94°C, 1 min at 60°C, and 1 min at 72°C were performed. PCR products were resolved on 5% polyacrylamide gels, dried, visualized by autoradiography, and quantified. For the results depicted in Fig. 5C, bands were quantified, corrected for the slight variation in input, and presented in arbitrary units. All ChIP assays were performed at least three times. The primers used were as follows: LDL receptor promoter region, 5′-TGTTAACAGTTAAACATCGAGAA-3′ and 5′-CCCGCGATTGCACTCGGGGC-3′ (344-bp-long amplified fragment); LDL receptor exon 10 region, 5′-CTCAGCACCCAGCTTGACA-3′ and 5′-TGAACAGGATCCACCACGAT-3′ (230-bp-long amplified fragment); β-actin promoter region, 5′-AACACCCCAGCCATGTACG-3′ and 5′-ATGTCACGCACGATTTCCC-3′ (420-bp-long amplified fragment); and c-Jun promoter region, 5′-CACCGTCACTAGACAGTCA-3′ and 5′-GAGCTCAACACTTATCTGCTA-3′ (254-bp-long amplified fragment). The linear range of the primers was determined empirically using different amounts of HepG2 genomic DNA. Subsequent PCR analyses were carried out using the optimum cycle number determined for each primer set. Genomic DNA control reactions were always carried out alongside reactions with the immunoprecipitated DNA samples.
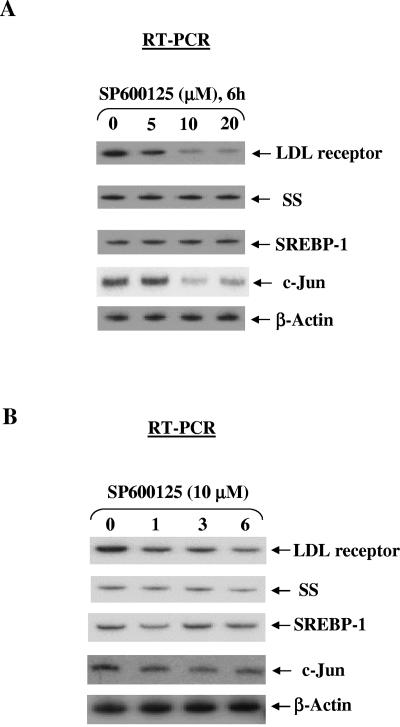
FIG. 5.
SP600125 inhibited basal expression of LDL receptor and c-Jun without significantly affecting the expression of SS, SREBP-1, and β-actin. (A) SP600125 reduced LDL receptor expression in a dose-dependent manner. HepG2 cells were plated on day 0. On day 3, cells were treated with SP600125 for 4 h. Total cellular RNA was extracted and subjected to quantitative reverse transcription-PCR. LDL receptor mRNA levels were normalized to β-actin mRNA levels. A representative autoradiogram is shown. (B) SP600125 reduced LDL receptor expression in a time-dependent manner. HepG2 cells were treated with SP600125, and total RNA obtained at the indicated times was subjected to quantitative reverse transcription-PCR. The experiments were repeated three times with similar results. A representative autoradiogram is shown, and the results of the densitometric analysis of LDL receptor mRNA levels normalized to actin mRNA are shown in Fig. 6C.
Luciferase assays.
The Gal4 luciferase reporter plasmid (0.3 μg) was cotransfected with the control plasmid pRSV β-galactosidase (0.1 μg) and the Gal4-Sp1 expression plasmid (0.6 μg) as indicated (34). Luciferase and β-galactosidase activities were measured 48 h after transfection. Cells were treated with the indicated amounts of SP600125 for 6 h just before lysis. The data are presented as the relative luciferase activity, calculated as the ratio of the luciferase activity to the activity of β-galactosidase (mean ± standard error; n = 4).
In vivo genomic footprinting of the human LDL receptor promoter.
HepG2 cells previously treated for 3 h with 10 μM SP600125 or left untreated were treated with 0.01% dimethyl sulfate (DMS) for 2 min. The methylated genomic DNA was isolated, and the sites of cleavage in the human LDL receptor promoter were mapped using ligation-mediated PCR as described previously (34). Briefly, 5 μg genomic DNA served as the template in a first-strand synthesis reaction using the primer 5′-TGAGGGGGCGTCAGCTCTTCACCGGAG-3′ to examine protein-DNA interactions in the lower strand. Linkers were then ligated onto the blunt-ended products. Next, a linker and a nested gene-specific primer (5′-AAGGACTGGAGTGGG AATCAGAGCTTCA-3′) were used to amplify the cleavage products. The resulting PCR products were end labeled with the 32P-labeled primer 5′-AGTGGGAATCAGAGCTTCACGGGTTA-3′. Reaction products were separated on denaturing 6% polyacrylamide gels and viewed using autoradiography. For examining protein-DNA interactions in the upper strand, a primer for first-strand synthesis (5′-TACCTGCAGTCCCCGCCGCGGCGAGG-3′), a nested gene-specific primer (5′-AATTTCCAGCCCCAGGGCCCCATGCT-3′), and a labeling primer (5′-TGCTGTGTCCTAGCTGGAAACCCT-3′) were used.
RESULTS
Global hypophosphorylation of histone H3-Ser10 by SP600125.
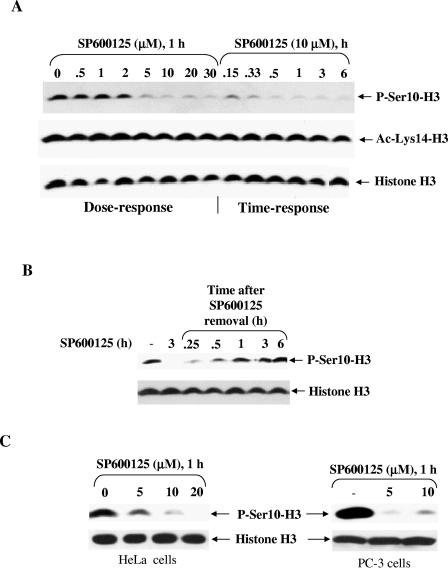
Initial studies were performed to determine the effect of SP600125 on different histone H3 modifications. HepG2 cells were incubated with 10 μM SP600125 for periods of up to 6 h, and cell extracts were probed with a panel of modification-specific antibodies. SP600125 dramatically reduced global histone H3-Ser10 phosphorylation without affecting expression (Fig. 1A). The reduction in phosphorylation started at 10 min, with near-maximal reduction occurring after 20 min of treatment with SP600125, whereas global acetylation of lysine 14 remained largely unaffected. To determine whether SP600125 exhibited dose-dependent effects on histone H3-Ser10 phosphorylation, cells were also treated with increasing doses of SP600125 for 1 h. We observed a gradual decrease in global histone H3-Ser10 phosphorylation, with almost complete dephosphorylation occurring at concentrations of 5 μM and higher (Fig. 1A). We also found that concentrations of SP600125 ranging from 0.1 to 20 μM over a 6-h period did not significantly alter the number of viable cells compared with the control.
FIG. 1.
SP600125 rapidly inhibits histone H3-Ser10 phosphorylation in different mammalian cell types. (A) SP600125 reduced histone H3-Ser10 phosphorylation in a dose-and time-dependent manner in HepG2 cells. Cells (2 × 105) were grown in six-well dishes and treated either with various concentrations of SP600125 for 1 h or with 10 μM SP600125 for different periods prior to harvesting for Western blot analysis. Histone H3-Ser10 phosphorylation, acetylation of histone H3 (lysine 14), and histone H3 expression were detected by Western blot analysis using corresponding antibodies. (B) SP600125 reduced histone H3-Ser10 phosphorylation in a reversible manner. HepG2 cells were incubated for 1 h with 10 μM SP600125 and then switched to a medium lacking SP600125 for the indicated time periods. Phosphorylation of histone H3-Ser10 was detected by Western blot analysis. Probing with an antibody specific for phosphorylation-independent histone H3 revealed equal amounts of histone H3 in all lanes. (C) Effects of SP600125 on histone H3-Ser10 phosphorylation in human cervical carcinoma HeLa cells and human prostate adenocarcinoma PC-3 cells. Cells were treated with SP600125 as described for panel A and analyzed for phospho-histone H3-Ser10 as well as for total histone H3 protein by Western blotting. Equal amounts of histone H3 were observed for both cell lines under all treatment conditions. Results are indicative of those from at least three separate experiments.
To examine whether SP600125 exerted its effect reversibly, cells were maintained for 1 h in a medium containing 10 μM SP600125 and then switched to a medium without inhibitor for various time periods. Total cell extracts were subjected to Western blotting using anti-phospho-Ser10-histone H3. As shown in Fig. 1B, Ser10 phosphorylation increased after SP600125 was removed and by 6 h had reached the level in untreated cells.
We further examined the effect of SP600125 on histone H3-Ser10 phosphorylation in other cell types, i.e., human cervical carcinoma HeLa cells and human prostate carcinoma PC-3 cells (Fig. 1C). In both cell types, SP600125 caused a dramatic reduction in histone H3-Ser10 phosphorylation compared with an untreated control.
SP600125-dependent hypophosphorylation of histone H3-Ser10 is independent of mitogen-activated protein kinases (MAPKs).
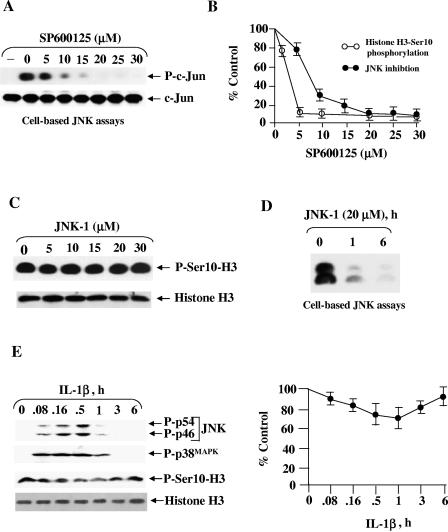
The rapid loss of histone H3-Ser10 phosphorylation by SP600125 possibly indicated that the inhibitor effect on histone kinase was direct, as indirect feedback regulation is expected to take a longer time. We wished to explore the association between histone H3-Ser10 phosphorylation loss and the p46/54JNK inhibition in HepG2 cells. To address this question, we first evaluated the efficacy of p46/54JNK inhibition by SP600125 in a standard immunoprecipitation-based kinase assay. HepG2 cells were pretreated for 1 h with different concentrations of SP600125, and then briefly exposed to interleukin-1β (IL-1β) to stimulate JNK activity. The addition of SP600125 led to a dose-dependent inhibition of IL-1β-stimulated phosphorylation of c-Jun, with a 50% inhibitory concentration of about 8 μM (Fig. 2A), consistent with earlier demonstrations showing 50% inhibitory concentrations in the range of 5 to 15 μM for p46/54JNK inhibition in different mammalian cell lines (15, 19). A comparison of the kinetics of a reduction in histone H3-Ser10 phosphorylation and p46/54JNK inhibition by SP600125 (Fig. 2B) suggests that SP600125-dependent loss of histone H3-Ser10 phosphorylation is possibly independent of the p46/54JNK pathway. A close match would be expected if SP600125 mediated its effect by inhibiting this kinase.
FIG. 2.
SP600125 inhibits histone H3-Ser10 phosphorylation independent of p46/54JNK. (A) Effect of SP600125 on p46/54JNK activity in cultured HepG2 cells. HepG2 cells were pretreated with the indicated concentration of SP600125 for 1 h and then stimulated with IL-1β (5 ng/ml) for 30 min in the presence of inhibitor (28). Cell extracts were prepared and subjected to a p46/54JNK assay with glutathione S-transferase-c-Jun and nonradioactive ATP as substrates. Phosphorylated substrate was detected by immunoblotting with a phospho-specific c-Jun antibody (Ser63) as described in Materials and Methods. To make sure equal amounts of c-Jun were present in all the lanes, samples were also probed using a phosphorylation-independent c-Jun antibody. The results shown are representative of those from more than three independent experiments. (B) Densitometric data expressed as a percentage of the value for the untreated control. Quantitative data are means ± standard errors from three experiments. (C) Inhibition of p46/54JNK by JNK-1 polypeptide inhibitor does not affect histone H3-Ser10 phosphorylation. HepG2 cells were treated with different doses of JNK-1 for 6 h, and total cell extracts were subjected to Western blot analysis using either phosphorylation-independent anti-histone H3 or anti-phospho-Ser10-H3. (D) JNK-1 treatment efficiently blocked p46/54JNK activity of HepG2 cells. Cells treated with 20 μM of JNK-1 for 3 h and 6 h were subjected to p46/54JNK assays as described above. Equal amounts of c-Jun in all the samples were confirmed by probing with nonphosphorylated c-Jun antibody (results not shown). (E) Activation of p46/54JNK does not change histone H3-Ser10 phosphorylation in HepG2 cells. Cells (2 × 105) were grown and treated with 5 ng/ml IL-1β for different periods (28). Total cell extracts were subjected to Western blot analysis to determine the phosphorylation levels of histone H3-Ser10 and different MAPKs. The results are representative of those from three separate experiments. The autoradiograph was quantified, and the results are expressed as fold suppression of histone H3-Ser10 phosphorylation by SP600125 compared with untreated cells (100%).
We further evaluated the consequence of p46/54JNK inhibition on histone H3-Ser10 phosphorylation by inhibiting p46/54JNK with another pharmacological inhibitor. JNK-1 peptide (7), a cell-permeative peptide that inhibits the activation of this pathway, was employed for the experiments shown in Fig. 2C and D. HepG2 cells were treated with different doses of JNK-1, and histone H3-Ser10 phosphorylation and p46/54JNK activity were determined under parallel conditions, as described in Materials and Methods. Treatment with JNK-1 inhibitor had no effect on histone H3-Ser10 phosphorylation even at 30 μM (Fig. 2C), even though SP600125 resulted in the almost complete inhibition of endogenous p46/54JNK activity at 20 μM (Fig. 2D), further supporting the notion that the effect of SP600125 on histone H3-Ser10 phosphorylation is not a direct consequence of p46/54JNK inhibition.
We next performed the converse experiment with the goal of determining whether increased JNK activity could change the level of histone H3-Ser10 phosphorylation. HepG2 cells were treated with IL-1β to activate p46/54JNK in order to examine its effect on histone H3-Ser10 phosphorylation. As expected from our previous study (28), stimulation by IL-1β activated p46/54JNK, and the increase was detected within 5 min, reached a peak after 30 min, and returned to the basal level at 1 h (Fig. 2E). However, p46/54JNK activation was not accompanied by any increase in histone H3-Ser10 phosphorylation (Fig. 2E). In fact, there was a slight and transient decrease in histone H3-Ser10 phosphorylation. This lack of correlation further argues against the involvement of p46/54JNK in SP600125-dependent suppression of histone H3-Ser10 phosphorylation.
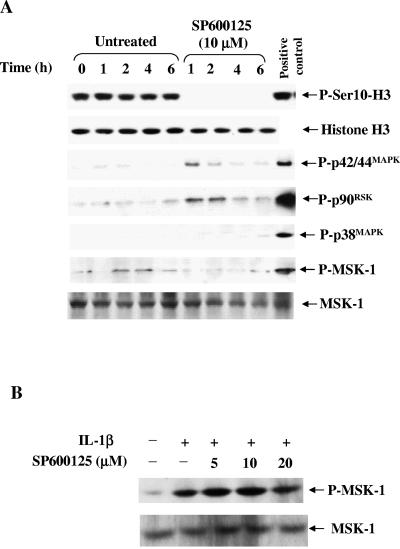
Given the cross talk between MAPKs (41, 42), we were concerned that SP600125 may inhibit the p42/44MAPK cascade and consequently reduce histone H3-Ser10 phosphorylation. To determine the selectivity of SP600125 for p46/54JNK versus other MAPKs, the phosphorylation levels of p42/44MAPK and p38MAPK and downstream kinases pp90RSK and MSK-1 were determined following SP600125 treatment. As shown in Fig. 3A, SP600125 slightly increased the phosphorylation of p42/44MAPK and pp90RSK, without significantly affecting the phosphorylation of p38MAPK or MSK-1. The increase in p42/44MAPK or pp90RSK phosphorylation is in agreement with a recent report suggesting the suppressive effect of JNK on this pathway (41). A slight decrease in basal MSK-1/2 phosphorylation by SP600125 prompted us to investigate the role of this kinase in the dephosphorylation process. We examined the effect of SP600125 on the phosphorylation/activation of MSK-1/2 and found that SP600125 does not affect IL-1β-induced MSK-1/2 phosphorylation (Fig. 3B). This is in line with a recent report showing that SP600125 is a poor inhibitor of MSK-1/2 (1).
FIG. 3.
SP600125-dependent inhibition of histone H3-Ser10 phosphorylation is independent of MAPK pathways, and SP600125 does not inhibit MSK-1 phosphorylation/activation. (A) HepG2 cells were plated on day 0 in medium supplemented with 10% FBS. On day 3, the medium were replaced with 0.5% FBS and the cells were either untreated or treated with 10 μM SP600125 for the indicated times. Total cell extracts were subjected to Western blot analysis for the indicated kinases. (B) Effect of SP600125 pretreatment on IL-1β-induced phosphorylation/activation of MSK-1/2 in HepG2 cells. HepG2 cells were treated with 5 ng/ml IL-1β for 30 min in the absence or presence of 10 μM SP600125. The inhibitor was added 30 min prior to IL-1β addition. Equal amounts of total cell extracts were subjected to SDS-PAGE and immunoblotted with anti-phospho-MSK-1/2 or anti-MSK-1/2. The experiment was repeated two times with similar results.
SP600125-dependent hypophosphorylation of histone H3-Ser10 is also independent of cell cycle changes.
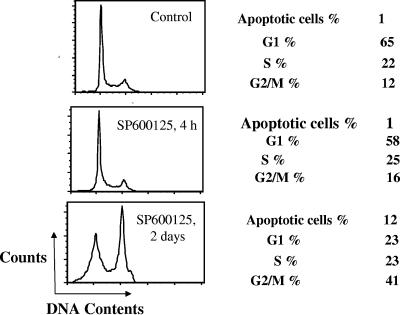
Histone H3-Ser10 phosphorylation has been shown to correlate with cell cycle progression (23). Histone H3-Ser10 phosphorylation initiates during G2, becomes maximal during metaphase, and diminishes during late anaphase and early telophase. Cells arrested in the G1 phase of the cell cycle display predominantly unphosphorylated histone H3-Ser10, whereas cells arrested during mitosis display predominantly phosphorylated histone H3-Ser10. It was thought possible that SP600125 reduced histone H3-Ser10 phosphorylation by arresting entry of cells into mitosis. We therefore determined whether the effect of SP600125 on histone H3-Ser10 phosphorylation was due to an altered cell cycle distribution and/or induced apoptosis. Cells were treated with SP600125 for periods of up to 48 h and subjected to flow cytometry after propidium iodide staining. As shown in Fig. 4, little or no change was noted in the cell cycle distribution after 4 h of treatment. Two days of treatment increased the percentage of apoptotic cells, an observation consistent with previous reports (15, 19). Collectively, these results suggested that SP600125 treatment for a short period did not affect histone H3-Ser10 phosphorylation in HepG2 cells by regulating cell cycle progression.
FIG. 4.
Short treatment of SP600125 does not affect cell cycle distribution. HepG2 cells were untreated or treated with 10 μM SP600125 for 4 h or 2 days, as indicated, and cellular DNA content was determined by propidium iodide staining and flow cytometry. The proportion of nonapoptotic cells in different phases of the cycle is indicated, together with the proportion of sub-G1 apoptotic cells.
Hyphosphorylation of histone H3-Ser10 at the LDL receptor promoter by SP600125 and its correlation with a reduction in LDL receptor expression in a gene-specific manner.
Having demonstrated the ability of SP600125 to block global histone H3-Ser10 phosphorylation, we subsequently focused on the role of this modification in regulating basal gene expression. Histone H3-Ser10 phosphorylation had been shown to influence the transcriptional activation of selected genes; however, the role of this modification in regulating basal gene expression has never been studied due to the lack of a suitable inhibitor for this modification. We recently found high basal levels of histone H3-Ser10 phosphorylation at the LDL receptor promoter chromatin compared to other genes in untreated HepG2 cells (25; W. Huang et al., unpublished results). Since SP600125 significantly reduced global histone H3-Ser10 phosphorylation, we tested whether the phosphorylation is involved in controlling basal LDL receptor expression. Considering these results, we then directly compared expression of LDL receptor and other genes involved in cholesterol homeostasis in SP600125 treated cells. As shown in Fig. 5, SP600125 treatment significantly reduced (three- to fivefold) basal LDL receptor expression in a dose- and time-dependent manner. The suppression of LDL receptor expression was apparent at 1 h, with maximal suppression at 6 h of treatment with SP600125. SP600125-dependent suppression was detected at 5 μM, with maximal suppression occurring at 10 μM. We also measured the basal expression of a number of genes that are either related or completely unrelated to the LDL receptor gene. Interestingly, basal expression of SS, SREBP-1, and β-actin did not change significantly upon treatment with SP600125 (Fig. 5), whereas basal c-Jun expression was repressed by SP600125 (Fig. 5), consistent with an earlier report correlating c-Jun induction with the appearance of phosphorylated histone H3-Ser10 (43).
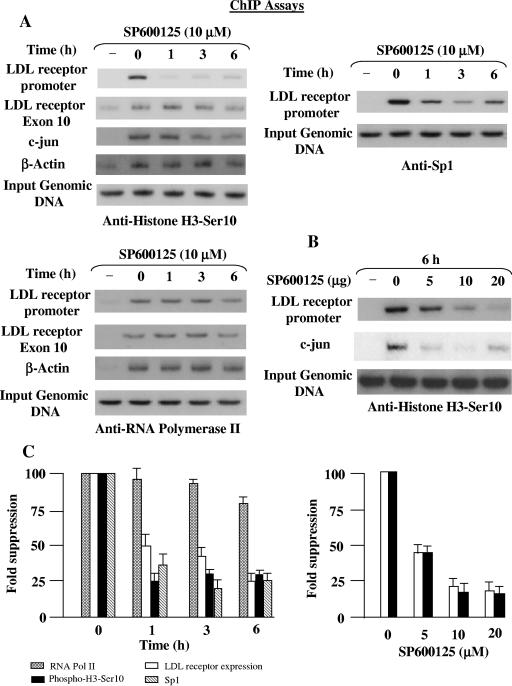
We next sought to determine the correlation between histone H3-Ser10 phosphorylation level and basal gene expression, and we analyzed the promoter of the LDL receptor gene. We performed a time and dose course experiment using ChIP assays to monitor changes in histone H3-Ser10 phosphorylation levels upon SP600125 treatment. The chromatin was immunoprecipitated with antibodies to phosphorylated histone H3-Ser10 from HepG2 cells treated with SP600125 for the indicated time periods. PCR primers specific to the LDL receptor promoter detected a significant reduction (three- to fivefold) in promoter DNA in the phosphorylated chromatin pools from the SP600125-treated cells compared to the untreated cells (Fig. 6A). The significant decrease in histone H3-Ser10 phosphorylation at the LDL receptor promoter also occurred in a dose-dependent manner (Fig. 6B). We found that increasing concentrations of SP600125 caused a reduction in histone H3-Ser10 phosphorylation at the LDL receptor promoter. The concomitant decline in histone H3-Ser10 phosphorylation suggested that this modification might be required for regulating basal LDL receptor expression. In accordance with an expected correlation (43, 47), transcriptional repression of c-Jun was associated with histone H3-Ser10 dephosphorylation (Fig. 6B). In contrast to the LDL receptor, β-actin promoter sequences or the downstream coding region of LDL receptor exon 10 sequences were detected at almost similar levels in the phosphorylated chromatin upon SP600125 treatment (Fig. 6A).
FIG. 6.
Effect of SP600125 on local histone H3-Ser10 phosphorylation and association of Sp1 and RNA Pol II with the LDL receptor gene. (A) HepG2 cells serum starved for 24 h were treated with 10 μM SP600125 for the indicated periods, and ChIP assays were performed as described in Materials and Methods, using anti-phospho-histone H3-Ser10, anti-Sp1, or anti-RNA Pol II antibody. Primers for the LDL receptor promoter, downstream exon 10 region, and β-actin were used to amplify the genomic DNA isolated from the ChIP assays. The status of histone H3-Ser10 phosphorylation at the c-Jun and β-actin promoters was also analyzed for some experiments. Input genomic DNA was used as the loading control, and precipitates from the control immunoglobulin G reaction served as a negative control. A representative autoradiogram is shown for each antibody, and the data shown are representative of those from three separate experiments. (B) SP600125 blocked histone H3-Ser10 phosphorylation at the LDL receptor promoter in a dose-dependent manner. HepG2 cells were treated with different doses of SP600125 and subjected to ChIP assays as described above for anti-phospho-histone H3-Ser10. A representative autoradiogram is shown, and the data shown are representative of those from three separate experiments. (C) Results of the densitometric analysis of the autoradiograms showing changes in the local histone H3-Ser10 phosphorylation, Sp1, and RNA Pol II occupancy at the LDL receptor gene (panels A and 6) and the normalized LDL receptor repression upon SP600125 treatment (from Fig. 5). Results are expressed as means ± standard errors from three different experiments (fold suppression over untreated cells [zero time point]).
Possible requirement of histone H3-Ser10 phosphorylation for the occupancy of the Sp1 site at the LDL receptor promoter.
It is important to know whether basal transcriptional machinery binds to the LDL receptor promoter in vivo in the presence of SP600125. To answer this question, ChIP analysis was performed to compare the association of transcription factor Sp1 and RNA Pol II to the LDL receptor promoter in untreated and SP600125-treated cells. As shown in Fig. 6A, the level of Sp1 decreased at the LDL receptor promoter, and maximal suppression was observed at 3 h of SP600125 treatment, followed by a slight increase at 6 h. A comparison of the kinetics of reductions in histone H3-Ser10 phosphorylation and Sp1 occupancy at the LDL receptor promoter with the decrease in LDL receptor expression indicates that the SP600125-dependent decrease in LDL receptor expression correlates with the status of histone H3-Ser10 phosphorylation and the Sp1 occupancy (Fig. 6C). Importantly, repression of the LDL receptor expression reached significant levels at concentrations that inhibited histone H3-Ser10 phosphorylation and Sp1 recruitment. This suggests involvement of histone H3-Ser10 phosphorylation in the recruitment of Sp1 at the LDL receptor promoter in vivo.
We also considered the possibility that the mechanisms underlying the action of transcription factors and coactivators are their effect on the initiation and/or elongation of transcription by RNA Pol II (4, 8, 54). We extended the ChIP assays to examine the effect of SP600125 on RNA Pol II occupancy at the LDL receptor promoter and the downstream coding region (exon 10). Interestingly, similar efficiencies of association were observed at 1 h and 3 h, whereas a reproducible decrease (∼20%) in the occupancy of RNA Pol II was observed following SP600125 treatment for 6 h at both the promoter and the downstream coding region (Fig. 6A). The occupancy of the β-actin promoter was not significantly different under identical conditions. These results raise an intriguing possibility that loss of histone H3-Ser10 phosphorylation and/or Sp1 occupancy slightly impairs recruitment of RNA Pol II, without affecting transcriptional elongation by Pol II. As a result, the maximal reduction in LDL receptor expression by SP600125 at 6 h (Fig. 5A) may be due to a reduction in the recruitment of both Sp1 and RNA Pol II.
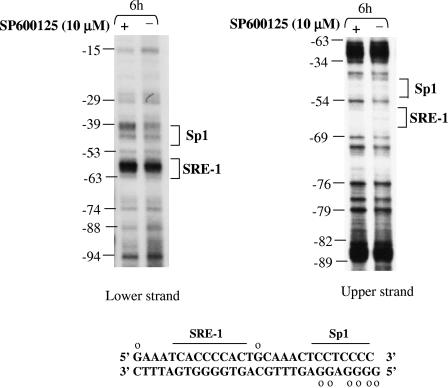
Using in vivo genomic footprinting assays, we also examined the occupancy of the Sp1 and SRE-1 sites of the LDL receptor promoter when cells were treated with SP600125 (Fig. 7). By monitoring the protection from DMS methylation, we observed a significant and reproducible decrease on both strands in the occupancy of the Sp1 site, which has been shown to be involved in controlling basal LDL receptor expression (16). The possibility that reduced occupancy of the Sp1 site by SP600125 is due to reductions in the expression, DNA binding, or transactivation potential of Sp1 was also examined. No significant differences were observed between untreated and SP600125-treated HepG2 cells (results not shown), suggesting that lower accessibility of Sp1, and not changes in expression, DNA binding, or transactivation, is responsible for the compromised interaction of Sp1 at the LDL receptor promoter.
FIG. 7.
In vivo genomic footprinting reveals that SP600125 causes a reduction in Sp1 site occupancy at the LDL receptor promoter. The in vivo DNA methylation patterns between nucleotides −94 and −15 of the human LDL receptor promoter are shown. HepG2 cells were subjected to DMS treatment in the absence or presence of 10 μM SP600125. The modified genomic DNA was chemically cleaved, and the positions of methylated guanines were mapped by ligation-mediated PCR using LDL receptor gene-specific primers as described earlier (34). The positions of binding sites for the transcription factors Sp1 and sterol response element-binding protein are shown. The DNA sequence of this region is also provided, and open circles indicate nucleotides that repeatedly displayed loss of protection upon SP600125 treatment. A similar protection pattern was obtained in two different experiments.
DISCUSSION
The results presented in this paper show that SP600125 is an efficient inhibitor of histone H3-Ser10 phosphorylation in vivo and demonstrate a novel mechanism by which repression of the LDL receptor expression is mediated by this modification.
We report here for the first time, for any cell type, that SP600125 inhibits histone H3-Ser10 phosphorylation and the inhibitory effect appears to be not a direct consequence of JNK inhibition for several reasons. First, SP600125 reduced histone H3-Ser10 phosphorylation with a concentration dependence that does not parallel the inhibition of p46/54JNK, and a close match is expected if SP600125 mediates its effect through this kinase. Second, another selective polypeptide p46/54JNK inhibitor, JNK-1, does not block histone H3-Ser10 phosphorylation at concentrations that completely blocked p46/54JNK. Third, histone H3 is poorly phosphorylated in vitro by the purified p46/54JNK compared to other MAPKs, and a complete deficiency of the p46/54JNK1 or p46/54JNK2 isoform did not affect histone H3-Ser10 phosphorylation (55). Finally, lack of p46/54JNK involvement is consistent with an earlier study showing that SP600125 is capable of inhibiting a variety of kinases (1). The potential therapeutic value, together with the commercial availability of this chemical inhibitor, has made this kinase the subject of intensive studies during the past years (6, 51). Since its first report in 2001 as a selective p46/54JNK inhibitor, SP600125 has become the pharmacological inhibitor of choice for assessing the role of this kinase in a range of biochemical and biological events (6). A perusal of the existing literature on studies using SP600125 for the inhibition of p46/54JNK isoforms in cultured cells indicates that this compound is typically used at concentrations ranging from 1 to 50 μM. Based on the studies reported here, we anticipate that concentrations of SP600125 sufficient to inhibit p46/54JNK activities in cultured cells and tissues would also suppress histone H3-Ser10 phosphorylation. The extent to which histone H3-Ser10 phosphorylation suppression by SP600125 compromises its usefulness as a JNK inhibitor will depend on the nature of the study. In view of our results, data obtained with SP600125 must be interpreted with great caution.
An overwhelming majority of the studies that have led to the present models on the role of histone H3-Ser10 phosphorylation have focused on the mechanisms of gene activation. Therefore, while these studies have been quite valuable for providing the framework within which to study the events that induce gene expression, the results are limited in their application to transcription initiation events. Two of the central questions are how nuclear factors and associated proteins gain access to the chromatin templates and how histone modifications play a role in the initiation process. The LDL receptor promoter is an ideal model system with which to study the role of histone H3-Ser10 phosphorylation in this process. The LDL receptor promoter contains the initiator region and the binding sites for a limited number of transcription factors, including Sp1 and SREBP-2. The Sp1 site adjacent to SRE-1 is crucial for basal LDL receptor transcription, as a mutation of this region in the human gene leads to a dramatic decrease in promoter activity (16). Additionally, maximal LDL receptor induction may require increased histone H3-Ser10 phosphorylation (25). Using ChIPs, we established that even though SP600125 decreased global histone H3-Ser10 phosphorylation, reduced local phospho-histone H3-Ser10 was observed at selective promoters, including LDL receptor and c-Jun promoters, and correlated with their transcriptional repression. Following histone H3-Ser10 dephosphorylation, loss of Sp1 recruitment at the LDL receptor was also observed in a timely manner, and that too correlated with the LDL receptor repression by SP600125, suggesting that histone H3-Ser10 dephosphorylation preceded loss of Sp1 occupancy. This part of the model is also supported by our in vivo footprinting result that revealed reduced occupancy of the Sp1 site upon SP600125 treatment. Notably, though, in contrast to the case for the LDL receptor, histone H3-Ser10 phosphorylation at the β-actin promoter as well as its expression are not affected by SP600125 treatments. These data indicate that inhibition of Sp1 recruitment at the LDL receptor promoter is not a result of the global decrease in the accessibility of Sp1. Since binding of Sp1 and coactivators results in the recruitment of basal transcription machinery and the initiation of transcription, the reduced Sp1 occupancy may also account for the reduction in the occupancy of the RNA Pol II by SP600125. Based on the above results, it is intriguing to speculate about a potential role for this modification in the recruitment of key factors and the optimal transcription initiation at the LDL receptor promoter. Such a model fits well with earlier reports showing that recruitment binding of Sp1 to GC boxes in the cyclin requires histone H3 acetylation (24) and that histone phosphorylation promotes TATA-binding protein recruitment in Saccharomyces cerevisiae (31). Additionally, elevated levels of histone H3-Ser10 phosphorylation have been linked with relaxed chromatin structure (46), and high basal phospho-Ser10-histone H3 levels at the LDL receptor chromatin may contribute to the prompt response of this gene to exogenous stimuli.
The regulation of gene expression is an extremely complex and intricate process involving a multitude of recognized and, most likely, many still-elusive regulatory mechanisms. Perhaps the greatest conundrum lies in the question of specificity; i.e., how is SP600125-mediated repression localized to a particular promoter? At least part of the answer may lie in the striking heterogeneity in the sensitivity of local histone H3-Ser10 phosphorylation to SP600125, akin to that previously described for histone acetylation (50). While numerous studies have correlated gene transactivation with increased acetylation when histone deacetylase activity is inhibited (8), emerging evidence has shown that histone deacetylase inhibitors can also induce local promoter histone deacetylation (48). Most genes are unaffected by histone deacetylase inhibitors, and the number of genes that decrease in expression is approximately equal to the number of genes that are up-regulated (17). For instance, treating a human lymphoid cell line with the histone deacetylase inhibitor trichostatin A revealed a change in the expression of only 8 out of 340 genes examined (50). Consistent with the selective effect of SP600125 on gene expression, global histone H3-Ser10 dephosphorylation certainly does not trigger global repression of gene expression. The expression of other genes involved in cholesterol homeostasis did not show any decrease despite the global reduction in phospho-histone H3-Ser10 following SP600125 treatment. Although a strong correlation between histone H3 dephosphorylation and selective transcriptional repression of the LDL receptor expression suggests that the two processes are mechanistically linked, the exact mechanisms involved for this type of selectivity are still unclear. The mechanism may involve different kinase/lipid and/or protein microenvironments resulting from differences in (i) the transcription factors bound to the promoter; (ii) inherent nucleosome positioning, density, or stability; (iii) the accessibility of the promoter to the transcriptional machinery caused by the participation of chromatin-remodeling machinery; or (iv) the nature of the local histone H3-Ser10 kinase. We favor the last possibility for our results, as several kinases have been implicated as potential histone H3-Ser10 kinase in mammalian cells and are activated by distinct pathways in a cell type-specific manner. These kinases include the MAPKs (p42/44MAPK, p38MAPK, and p46/54JNK) (11, 22, 45), p90RSK2 (40a), MSK-1/2 (43), cyclic AMP-dependent protein kinase A (39), IκB kinase α (53), protein kinase C (25), mixed-lineage triple kinase alpha (12), p21-activated kinase-1 (30), and Fyn (5). The kinase mediating SP600125-dependent histone H3-Ser10 dephosphorylation remains to be characterized. It is interesting to note that there is precedence for the promoter-specific effects of histone H3-Ser10 phosphorylation in both S. cerevisiae (31) and Drosophila (29). Future work will examine whether there are selective histone H3-Ser10 phosphorylation sites that are “marked” for transcriptional regulation and targeted by the SP600125-sensitive kinase in a promoter-specific manner.
In conclusion, this is the first report that demonstrates SP600125 as an effective inhibitor of histone H3-Ser10 phosphorylation. The ability to easily manipulate this modification in vivo by use of SP600125 provides an excellent strategy for elucidating the histone code and to assess the functional role of this modification in various cellular events. Our discovery that histone H3-Ser10 phosphorylation possibly regulates the basal expression of selective genes by modulating their accessibility to the transcription factor(s) is unprecedented and points to this modification as a unique regulatory mechanism for transcription initiation. A better understanding of the underlying mechanisms will help guide the development of more effective agents to treat hypercholesterolemia and cancer.
Acknowledgments
This work was supported by National Institutes of Health research grants HL-742202 and HL-60000355 to K.D.M.
REFERENCES
- 1.Bain, J., H. McLauchlan, M. E. Elliott, and P. Cohen. 2003. The specificities of protein kinase inhibitors: an update. Biochem. J. 371:199-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bennett, B. L., D. Sasaki, B. W. Murray, E. C. O'Leary, S. T. Sakata, W. Xu, J. C. Leisten, A. Motiwala, S. Pierce, Y. Satoh, S. S. Bhagwat, A. M. Manning, and D. W. Anderson. 2003. SP600125, an anthrapyrazolone inhibitor of Jun N-terminal kinase. Proc. Natl. Acad. Sci. USA 98:13681-13686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger, S. L. 2002. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 12:142-148. [DOI] [PubMed] [Google Scholar]
- 4.Blau, J., H. Xiao, S. McCracken, P. O'Hare, J. Greenblatt, and D. Bentley. 1996. Three functional classes of transcriptional activation domain. Mol. Cell. Biol. 16:2044-2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bode, A. M., and Z. Dong. 2005. Inducible covalent posttranslational modification of histone H3. Science's STKE [Online.] www.stke.org/cgi/content/full/sigtrans;2005/281/re4. [DOI] [PubMed]
- 6.Bogoyevitch, M. A., I. Boehm, A. Oakley, A. J. Ketterman, and R. K. Barr. 2004. Targeting the JNK MAPK cascade for inhibition: basic science and therapeutic potential. Biochim. Biophys. Acta 1697:89-101. [DOI] [PubMed] [Google Scholar]
- 7.Bonny, C., A. Oberson, S. Negri, C. Sauser, and D. F. Schorderet. 2001. Cell-permeable peptide inhibitor of JNK. Diabetes 50:77-82. [DOI] [PubMed] [Google Scholar]
- 8.Brown, S. A., Weirich, C. S., Newton, E. M., and R. E. Kingston. 1998. Transcriptional activation domains stimulate initiation and elongation at different times and via different residues. EMBO J. 17:3146-3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, M. S., and J. L. Goldstein. 1986. A receptor mediated pathway for cholesterol homeostasis. Science 232:34-47. [DOI] [PubMed] [Google Scholar]
- 10.Brown, M. S., and J. L. Goldstein. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89:331-340. [DOI] [PubMed] [Google Scholar]
- 11.Cheung, P., D. Allis, and P. Sassone-Corci. 2000. Signaling to chromatin through histone modifications. Cell 103:263-271. [DOI] [PubMed] [Google Scholar]
- 12.Choi, H., Y. Choi, Y. Cho, Z. Feng, A. Bode, and Z. Dong. 2005. Phosphorylation of Ser28 in histone H3 mediated by the mixed-lineage kinase-like mitogen activated protein triple kinase alpha. J. Biol. Chem. 280:13545-13553. [DOI] [PubMed] [Google Scholar]
- 13.Reference deleted.
- 14.Crosio, C., G. M. Fimia, R. Loury, M. Kimura, Y. Okano, H. Zhou, S. Sen, C. D. Allis, and P. Sassone-Corsi. 2002. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian aurora kinases. Mol. Cell. Biol. 22:874-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Curtin, J. F., and T. G. Cotter. 2004. JNK regulates HIPK3 expression and promotes resistance to Fas-mediated apoptosis in DU45 prostate carcinoma cells. J. Biol. Chem. 279:17090-17100. [DOI] [PubMed] [Google Scholar]
- 16.Dawson, P. A., S. L. Hofmann, D. R. van der Westhuyzen, T. C. Sudhof, M. S. Brown, and J. L. Goldstein. 1988. Sterol-dependent repression of low density lipoprotein receptor promoter mediated by 16-base pair sequence adjacent to binding site for transcription factor Sp1. J. Biol. Chem. 263:3372-3379. [PubMed] [Google Scholar]
- 17.Della Ragione, F., V. Criniti, V. Della Pietra, A. Borrielo, A. Oliva, S. Indaco, T. Yamamoto, and V. Zappia. 2001. Genes modulated by histone acetylation as new effectors of butyrate activity. FEBS Lett. 499:199-204. [DOI] [PubMed] [Google Scholar]
- 18.Dhawan, P., A. Bell, A. Kumar, C. Golden, and K. D. Mehta. 1999. Critical role of p42/44MAPK activation in anisomycin and hepatocyte growth factor-induced LDL receptor expression: activation of Raf-1/MEK 1/2/p42/44MAPK cascade alone is sufficient to induce LDL receptor expression. J. Lipid Res. 40:1911-1919. [PubMed] [Google Scholar]
- 19.Du, L., C. S. Lyle, T. B. Obey, W. A. Gaarde, J. A. Muir, B. L. Bennett, and T. C. Chambers. 2004. Inhibition of cell proliferation and cell cycle progression by specific inhibition of basal JNK activity. J. Biol. Chem. 279:11956-11966. [DOI] [PubMed] [Google Scholar]
- 20.Duan, H., C. A. Heckman, and L. M. Boxer. 2005. Histone deacetylase inhibitors down-regulate bcl-2 expression and induce apoptosis in t(14;18) lymphomas. Mol. Cell. Biol. 25:1608-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartman, M. G., D. Lu, M. Kim, G. J. Kociba, T. Shukri, J. Buteau, X. Wang, W. L. Frankel, D. Guttridge, M. Prentki., S. T. Grey, D. Ron, and T. Hai. 2004. Role for activating transcription factor 3 in stress-induced β-cell apoptosis. Mol. Cell. Biol. 24:5721-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He, Z., W. Y. Ma, G. Liu, Y. Zhang, M. Bode, and Z. Dong. 2003. Arsenite-induced phosphorylation of histone H3 at serine 10 is mediated by Akt1, extracellular signal-regulated kinase 2, and p90 ribosomal S6 kinase 2 but not mitogen- and stress-activated protein kinase 1. J. Biol. Chem. 278:10588-10593. [DOI] [PubMed] [Google Scholar]
- 23.Hendzel, M. J., W. Yi, M. A. Mancini, A. Van Hooser, A. Ranalli, B. R. Brinkley, D. P. Bazett-Jones, and C. D. Allis. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106:348-360. [DOI] [PubMed] [Google Scholar]
- 24.Hilton, T. L., Y. Li, E. L. Dunphy, and E. H. Wang. 2005. TAF1 histone acetyltransferase activity in Sp1 activation of the cyclin D1 promoter. Mol. Cell. Biol. 25:4321-4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang, W., V. Mishra, S. Batra, I. Dillon, and K. D. Mehta. 2004. Phorbol ester promotes histone H3-Ser10 phosphorylation at low density lipoprotein receptor promoter in a protein kinase C-dependent manner in human hepatoma HepG2 cells. J. Lipid Res. 45:1519-1527. [DOI] [PubMed] [Google Scholar]
- 26.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 293:1074-1080. [DOI] [PubMed] [Google Scholar]
- 27.Kapoor, G. S., B. A. Atkins, and K. D. Mehta. 2002. Activation of Raf-1/MEK-1/2/p42/44MAPK cascade is sufficient to uncouple LDL receptor expression from cell growth. Mol. Cell. Biochem. 236:13-22. [DOI] [PubMed] [Google Scholar]
- 28.Kumar, A., A. Middleton, T. C. Chambers, and K. D. Mehta. 1998. Differential roles of extracellular signal-regulated kinase-1/2 and p38MAPK in interleukin-1β- and tumor necrosis factor-α-induced low density lipoprotein receptor expression in HepG2 cells. J. Biol. Chem. 273:15742-15748. [DOI] [PubMed] [Google Scholar]
- 29.Labrador, M., and V. G. Corces. 2003. Phosphorylation of histone H3 during transcriptional activation depends on promoter structure. Genes Dev. 17:43-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, F., L. Adam, R. K. Vadlamudi, H. Zhou, S. Sen, J. Chernoff, M. Mandal, and R. Kumar. 2002. p21-activated kinase 1 interacts with and phosphorylates histone H3 in breast cancer cells. EMBO Rep. 3:767-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lo, W., E. R. Gamache, K. W. Henry, D. Yang, L. Pillus, and S. L. Berger. 2005. Histone H3 phosphorylation can promote TBP recruitment through distinct promoter-specific mechanisms. EMBO J. 24:997-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mahadevan, L. C., A. C. Willis, and M. J. Barratt. 1991. Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell 65:775-783. [DOI] [PubMed] [Google Scholar]
- 33.Mariadason, J. M., G. A. Corner, and L. H. Augenlicht. 2000. Genetic reprogramming in pathways of colonic cell maturation induced by short chain fatty acids: comparison with trichostatin A, sulindac, and cucrcumin and implications for chemoprevention of colon cancer. Cancer Res. 60:4561-4572. [PubMed] [Google Scholar]
- 34.Mehta, K. D., R. Chang, J. Underwood, J. Wise, and A. Kumar. 1996. Identification of a novel cis-acting element participating in maximal induction of the human LDL receptor gene transcription in response to low cellular cholesterol levels. J. Biol. Chem. 271:33616-33622. [DOI] [PubMed] [Google Scholar]
- 35.Mehta, K. D., and L. Miller. 1999. Inhibition of stress-activated p38 mitogen-activated protein kinase induces low density lipoprotein receptor expression. Trends Cardiovasc. Dis. 9:201-205. [DOI] [PubMed] [Google Scholar]
- 36.Nowak, S. J., and V. G. Corces. 2000. Phosphorylation of histone H3 correlates with transcriptionally active loci. Genes Dev. 14:3003-3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oliner, J. D., J. M. Andresen, S. K. Hansen, S. Zhou, and R. Tjian. 1996. SREBP transcriptional activity is mediated through an interaction with the CREB-binding protein. Genes Dev. 10:2903-2911. [DOI] [PubMed] [Google Scholar]
- 38.Roth, S. Y., J. M. Denu, and C. D. Allis. 2001. Histone acetyltransferases. Annu. Rev. Biochem. 70:81-120. [DOI] [PubMed] [Google Scholar]
- 39.Salvador, L. M., Y. Park, J. Cottom, E. T. Maizels, J. C. Jones, R. V. Schillace, D. W. Carr, P. Cheung, C. D. Allis, J. L. Jameson, and M. Hunzicker-Dunn. 2001. Follicle-stimulating hormone stimulates protein kinase A-mediated histone H3 phosphorylation and acetylation leading to select gene activation in ovarian granulosa cells. J. Biol. Chem. 276:40146-40155. [DOI] [PubMed] [Google Scholar]
- 40.Sanchez, H. B., L. Yieh, and T. F. Osborne. 1995. Cooperation by SREBP and Sp1 in sterol regulation of low density lipoprotein receptor gene. J. Biol. Chem. 270:1161-1169. [DOI] [PubMed] [Google Scholar]
- 40a.Sassone-Corsi, P., C. A. Mizzen, P. Cheung, C. Crosio, L. Monaco, S. Jacquot, A. Hanauer, and C. D. Allis. 1999. Requirement of Rsk-2 for epidermal growth factor activated phosphorylation of histone H3. Science 285:886-891. [DOI] [PubMed] [Google Scholar]
- 41.Shen, Y. H., J. Godlewski, J. Zhu, P. Sathyanarayana, V. Leaner, M. J. Birrer, A. Rana, and G. Tzivion. 2003. Cross-talk between JNK/SAPK and ERK/MAPK pathways. J. Biol. Chem. 278:26715-26721. [DOI] [PubMed] [Google Scholar]
- 42.Singh, R. P., P. Dhawan, C. Golden, G. S. Kapoor, and K. D. Mehta. 1999. One-way cross-talk between p38MAPK and p42/44MAPK. Inhibition of p38MAPK induces low density lipoprotein receptor expression through activation of p42/44MAPK cascade. J. Biol. Chem. 274:19593-19600. [DOI] [PubMed] [Google Scholar]
- 43.Soloaga, A., S. Thomson, G. R. Wiggin, N. Rampersaud, M. H. Dyson, C. A. Hazzalin, L. C. Mahadevan, and J. S. C. Arthur. 2003. MSK2 and MSK1 mediate the mitogen- and stress-induced phosphorylation of histone H3 and HMG-14. EMBO J. 22:2788-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spencer, V. A., and J. R. Davie. 1999. Role of covalent modifications of histones in regulating gene expression. Gene 240:1-12. [DOI] [PubMed] [Google Scholar]
- 45.Strahl, B. D., and C. D. Allis. 2000. The language of covalent modifications. Nature 403:41-45. [DOI] [PubMed] [Google Scholar]
- 46.Strelkov, I. S., and J. R. Davie. 2002. Ser-10 phosphorylation of histone H3 and immediate early gene expression in oncogene-transformed mouse fibroblasts. Cancer Res. 62:75-78. [PubMed] [Google Scholar]
- 47.Thomson, S., L. C. Mahadevan, and A. L. Clayton. 1999. MAP kinase-mediated signaling to nucleosomes and immediate-early gene induction. Semin. Cell Biol. 10:205-214. [DOI] [PubMed] [Google Scholar]
- 48.Tong, X., L. Yin, S. Joshi, D. W. Rosenberg, and C. Giardina. 2005. Cyclooxygenase-2 regulation in colon cancer cells. J. Biol. Chem. 280:15503-15509. [DOI] [PubMed] [Google Scholar]
- 49.Turner, B. M. 2000. Histone acetylation and an epigenetic code. Bioessays 22:836-845. [DOI] [PubMed] [Google Scholar]
- 50.Van Lint, C., S. Emiliani, and E. Verdin. 1996. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr. 5:245-253. [PMC free article] [PubMed] [Google Scholar]
- 51.Vincenti, M. P., and C. E. Brinckerhoff. 2001. The potential of signal transduction inhibitors for the treatment of arthritis: is it all just JNK? J. Clin. Investig. 108:181-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolffe, A. P. 1998. Chromatin: structure and function, p. 97-108. Academic Press, San Diego, Calif.
- 53.Yamamoto, Y., U. N. Verma, S. Prajapati, Y. T. Kwak, and R. B. Gaynor. 2003. Histone H3 phosphorylation by IKK-alpha is critical for cytokine-induced gene expression. Nature 423:655-659. [DOI] [PubMed] [Google Scholar]
- 54.Yankulov, K., J. Blau, T. Purton, S. Roberts, and D. L. Bentley. 1994. Transcriptional elongation by RNA polymerase II is stimulated by transactivators. Cell 77:749-759. [DOI] [PubMed] [Google Scholar]
- 55.Zhong, S., W. Ma, and Z. Dong. 2000. ERKs and p38 kinases mediate ultraviolet B-induced phosphorylation of histone H3 at serine 10. J. Biol. Chem. 275:20980-20984. [DOI] [PubMed] [Google Scholar]