Deregulation of the G1 cyclin-dependent kinase (CDK)-retinoblastoma protein (Rb) pathway is well established in virtually all human cancers. Different cancers exhibit selective alterations of the pathway, whether by inactivation of the CDK inhibitor p16Ink4a, by overexpression of the cyclin D1 gene, or by direct mutational inactivation of the Rb gene. The p16Ink-4a-CDK-Rb pathway is often considered to be a linear pathway, with mutation of one component abrogating the need to mutate another. Still, it is unclear why different paths to cancer preferentially select for mutations in one component over another. A paper in this issue of Molecular Cellular Biology by Williams, Classon, and colleagues reveals a surprising requirement for Rb in proliferation and transformation mediated by the Ras oncogene (50). This study provides a rationale for why mutational activation of Ras and genetic disruption of Rb are rarely found together in human cancers. Thus, different cancers appear to select for different disruptions in the CDK-Rb pathway in part based on the spectrum of other mutations, such as in Ras, which also occur during tumorigenesis.
Rb was the first cloned tumor suppressor gene, isolated by virtue of its association with hereditary retinoblastoma (11). The Rb protein is a key component that regulates cell cycle entry and progression in mammalian cells, and Rb is a member of a gene family encoding structurally and functionally similar proteins, which also includes the p107 and p130 proteins (42). Like Rb, p107 and p130 are regulated during the cell cycle by CDK phosphorylation. Rb family members associate with the transcription factor E2F, negatively regulating E2F dependent transcription. E2F activity plays critical roles in cell cycle progression by regulating the transcription of genes with critical roles in cell cycle progression, including genes involved in cell cycle regulation, DNA replication, and mitosis (8, 45).
Rb plays distinct roles in transcriptional regulation relative to p107 and p130, in terms of both controlling E2F-dependent and differentiation-specific transcription (5). While Rb loss in mouse embryo fibroblasts (MEFs) is associated with the increased expression of E2F targets such as cyclin E1 and p107, the combined inactivation of p107 and p130 results in the upregulation of different E2F targets, including B-Myb and Cdc2 (13). Rb in particular has also been shown to promote the transcription of differentiation mediators (44), such as following recruitment by the CBFA1 transcription factor to osteogenic gene promoters (43).
Importantly, Rb mutation uniquely contributes to tumor suppression in mice and humans, despite the facts that tumor suppressor activity for p107 is evident in the context of Rb mutation and that the three Rb family members display functional redundancy in controlling G1- to S-phase progression (5). On the other hand, Rb and p107/p130 have been shown to exert opposing influences on differentiation in cell cultures (6). Finally, while somatic mutations in the RB gene are associated with almost all sporadic retinoblastomas and small cell lung cancers (SCLCs), mutation of RB is much less common in other human cancers.
Ras oncoproteins, encoded by three genes (N-ras, K-ras, and H-Ras), are integral components of signal transduction pathways, linking extracellular signals directed by tyrosine kinase receptors to intracellular signaling cascades, and ultimately to changes in transcription (20). Signal-dependent activation of Ras results in exchange of bound GDP for GTP, and GTP-bound Ras then interacts with effectors such as the Raf serine/threonine kinase, which activates a mitogen-activated protein kinase (MAPK) signaling cascade. Once activated, Ras is quickly down-regulated by the action of a GTPase-activating protein, which promotes GTP hydrolysis. Oncogenic mutations of Ras prevent interactions with GTPase-activating proteins and thus greatly decrease GTP hydrolysis, resulting in constitutively active Ras. Oncogenic Ras mutations (henceforth denoted Ras*) are associated with almost a third of human cancers (20).
The importance of the Ras-MAPK pathway in the growth factor-dependent upregulation of cyclin D-dependent kinase activity is well established (40). In fact, growth factor-dependent Ras activation is necessary for G1 CDK activation, Rb phosphorylation, E2F activation, and S-phase entry. Accordingly, inhibition of Ras prevents entry into S phase in wild-type but not Rb−/− MEFs (18, 22, 25), which seems to place Ras squarely upstream of Rb. The decreased sensitivity of Rb mutant cells to Ras inhibition could result at least in part from the substantially increased Ras activation, at the level of GTP binding, observed in Rb−/− cells (17). Thus, a mutual antagonism between Rb and Ras appears well established. However, as discussed below, Williams et al. now contribute another twist, a functional dependency of Ras* for Rb (50).
In the first demonstrations of oncogene collaboration over two decades ago, Ras* and E1A or Myc were shown to synergize in transforming primary rodent cells (16, 32). The ability of E1A to cooperate with Ras* depends at least in part on its ability to disrupt complexes containing Rb family members, relieving Rb-dependent repression of transcription (23, 27, 49). Similarly, the ability of Myc and Ras* to collaborate in S-phase entry coincides with cyclin E/Cdk2 activation, Rb phosphorylation, and E2F activation (18). Ras* also cooperates with p16Ink4a loss to confer a transformed phenotype (21, 37, 38). In fact, Ras* overexpression results in upregulation of p16Ink4a, which contributes to premature senescence (39). Thus, the inactivation of the CDK-Rb pathway would appear, at face value, to be important for transformation by Ras. However, in these scenarios, genetic deregulation occurs either upstream of G1 CDKs (p16Ink4a loss or Myc overexpression) or via viral oncoproteins, such that inactivation of all three Rb family members occurs. Importantly, while mutations that disrupt the Rb pathway, such as in p16Ink4a, are commonly associated with activated Ras or B-Raf in human tumors (14, 47), direct mutations of the Rb gene and of Ras* are rarely found together in human tumors. Why is this?
Williams and colleagues now shed light on this issue (50). They demonstrate that, quite in contrast to Rb's usual tumor suppressor role and in marked contrast to p107/p130 loss, Rb loss actually impedes the transformation of mouse NIH 3T3 fibroblasts by Ras*. In fact, cotransfection of Rb with Ras* actually increases the number and size of transformed foci. Furthermore, RNA interference-mediated knockdown of Rb inhibits the proliferation of Ras*-transformed NIH 3T3 cells, eliminating concerns that the Rb dependence of Ras* transformation is due to developmental compensation effects in Rb−/− embryos. The authors go on to show that the requirement for Rb in Ras*-mediated transformation extends to human cancer cells, as transfection of Ras* into Rb-deficient Saos-2 osteosarcoma cells inhibits proliferation and soft agar colony formation. In fact, while Rb-deficient cells are resistant to pharmacological inhibition of the Ras-Erk pathway (7), Williams et al. show that pharmacological inhibition of the MAPK pathway restores proliferation in Ras*-transfected Saos-2 cells. Amazingly, reexpression of Rb in these cells partially rescues Saos-2 cells from Ras*-mediated growth inhibition.
Since epithelial lineage cells are the targets for the majority of human cancers, it was important to extend the positive role for Rb in Ras*-dependent transformation to human carcinomas. Human carcinomas such as colorectal cancers with activated Ras or Raf mutations frequently exhibit high-level expression of Rb (10, 12, 51), at least in part due to a deregulated Ras-MAPK pathway. While high Rb levels could simply reflect a futile negative feedback loop resulting from deregulated E2F activation, Williams et al. rather demonstrate that RNA interference-mediated knockdown of Rb in several such Ras*-bearing carcinomas inhibits their proliferation and soft agar colony formation. Analogously, Yamamoto et al. have shown that increased Rb expression in colon cancer cells protects these cells from apoptosis (51). Thus, Rb actually stimulates proliferation in the presence of activated Ras*, and conversely, Ras* inhibits proliferation when Rb is absent.
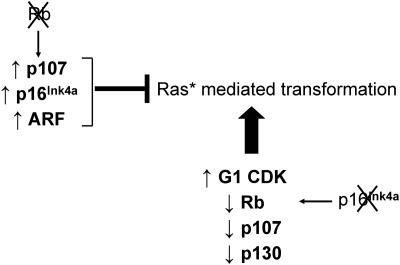
The authors also provide a possible mechanistic explanation for why cells with Ras* need Rb: Rb, presumably in conjunction with E2F, directly limits the transcription of p107. In the absence of Rb, increased levels of p107 may be responsible for inhibiting Ras*-dependent proliferation and transformation (Fig. 1, top). Still, in the context of Ras activation, targeting Rb can be advantageous as long as p107 is also inhibited. In this regard, loss of both Rb and p107, but not either alone, overcomes Ras* inhibition of proliferation (24), and disruption of all three Rb family members in MEFs facilitates partial transformation by Ras (35). Thus, not all paths to Rb inactivation are equivalent, and prior Ras* mutation should select for events that disrupt all Rb family members, such as occurs following inactivation of p16Ink4d or overexpression of cyclin D1 (Fig. 1, bottom).
FIG. 1.
Differential effects of Rb mutation versus G1 CDK deregulation on Ras*-dependent transformation. Mutational inactivation of the Rb gene in a cell leads to increased expression of p107, p16Ink4a, and ARF, which prevents Ras*-dependent transformation. In contrast, inactivation of p16 leads to higher G1 CDK activity, resulting in reduced function (i.e., increased phosphorylation) of all three Rb family members without ARF upregulation, which promotes Ras*-dependent transformation.
Rb loss has been shown to result in increased p107 expression in MEFs (13), and increased p107 is thought to compensate for Rb loss, restoring growth factor dependency (34). In fact, while inheritance of one mutated Rb allele results in high penetrance retinoblastoma in humans, retinoblastoma development in mice requires either mutation of both Rb and p107 or expression of viral oncoproteins that inhibit the activities of all three Rb family members (5).
Importantly, p107 probably does not act alone to inhibit Ras*-mediated transformation. Inactivation of Rb by mutation is also associated with substantial upregulation of p16Ink4a levels and cyclin D-dependent kinase inactivation (14), which should lead to further activation of p107 and p130. Moreover, Rb inactivation and E2F1 overexpression have been shown to promote the transcription of p14ARF (p19ARF in rodents, p14ARF in humans; referred to here simply as ARF), a positive regulator of p53, while normal growth stimulation or deregulated G1 CDK activation fails to upregulate ARF (15). Finally, while perhaps not unique to Rb gene mutation, deregulated E2F activity resulting from Rb inactivation leads to increased expression of proapoptotic genes such as p73 and APAF-1 (8, 42). Therefore, a cancer with direct mutation of the Rb gene must be inherently resistant to or acquire the means to escape these negative feedback loops, which could in part account for cell type dependency for tumors induced by Rb loss. But in cancer cells with Ras*, the feedback loops (p107, p16Ink4a, and/or ARF dependent) following Rb loss appear to more than compensate, impairing proliferation. Thus, p107, p16Ink4a, and ARF upregulation following Rb mutation appear to represent important tumor-suppressive mechanisms that have evolved to disfavor oncogenic mutations.
It is likely that Rb dependency and these negative feedback loops are not limited to tumors with Ras*. For example, cancers with deregulated signaling pathways that lead to increased Ras activation, such as with activation of epidermal growth factor receptor (EGF-R) family members (for example, Her2/Neu in breast cancers) may also exhibit Rb dependency. In fact, Rb gene inactivation is rarely found to occur in breast cancers with Her2 overexpression or amplification (2, 3, 19, 46), but is instead found in breast cancers exhibiting high cyclin E and low cyclin D1 expression without Her2 overexpression. The rarity of Ras* mutations in the context of Rb gene loss has been proposed previously (14) to relate to low cyclin D1 expression in Rb mutant tumors, together with the demonstrated dependency of Ras*- and Neu-dependent tumorigenesis on cyclin D1 in mice (28, 52). The studies by Williams et al. now indicate that Ras*-dependent transformation actually depends on the presence of Rb itself.
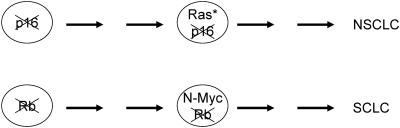
Lung cancers present an interesting case of cell type dependency for specific mutations within the CDK-Rb pathway. The Rb gene is frequently mutated in SCLC, but INK4A inactivation (without Rb gene mutation) is implicated in the genesis of non-small cell lung cancer (NSCLC) (14, 36). Notably, Ras* mutations are not found in SCLC, but K-Ras* mutations are common in NSCLC. Furthermore, overexpression of EGF-R (which should promote Ras activation) is not detected in SCLC, but most NSCLC overexpress EGF-R (4). Inactivation of Rb and INK4A is an early event in these lung cancers, while Ras* mutations occur later (14). Rb inactivation may be selected for in progenitor cells for SCLC, which then alters the clonal evolutionary pathway such that Ras* mutations will not be favored (Fig. 2). In contrast, preferential inactivation of INK4A (frequently by gene methylation) in precursors for NSCLC may provide a positive selective pressure for Ras*. Thus, the selection for late mutations will depend on the platform of genomic alterations already present.
FIG. 2.
Different paths to SCLC and NSCLC cancers. (Top) Should p16 inactivation provide a competitive advantage to the appropriate progenitor cell, then subsequent Ras* mutation may provide a further selective advantage, which together with other genomic alterations leads to NSCLC. (Bottom) In contrast, according to the studies by Williams et al., early mutation of the Rb gene in a progenitor will disfavor subsequent Ras* mutation, but instead may be conducive to other mutations that are found in SCLC, such as those leading to increased N-Myc expression. Similar selections against combined mutations in Rb and Ras, and for combined mutations in Ras with those that deregulate G1 CDK activity, probably function for other tumor types. For both NSCLC and SCLC, tumorigenesis frequently involves inactivation of the p53 pathway.
The order of mutations is certainly also important depending on the target cell. For example, Ras* in a target cell for NSCLC will presumably be disfavored if not preceded by p16Ink4a loss. What is unknown is whether Rb or INK4A inactivation helps determine the type of lung cancer, or whether the target progenitor cell type in the lung determines whether INK4A or Rb inactivation is selected for. Of note, progenitors for tumors resulting from mutation of the Rb gene, in both humans and mice, may share a neuroendocrine origin (14). Regardless, as highlighted in the study by Williams et al., the CDK-Rb pathway is clearly not linear. While mutations at one point in the pathway (such as in Rb) do prevent selection for alterations in other components (such as p16Ink4a inactivation), deregulation at distinct points of the pathways can clearly have different impacts on tumorigenesis depending on cell type and previous mutations.
Beyond an intriguing correlation in human tumors, can the relevance of the Rb dependence for Ras*-mediated transformation be demonstrated in vivo? Fortuitously, a just-published study has shown that epidermis-specific Rb mutation in mice actually results in fewer and smaller papillomas in a two-stage mouse skin carcinogenesis model (30). In this model, tumor initiation occurs via treatment with 7,12-dimethylbenz[a]anthracene (DMBA), which results in a predictable Ras* mutation. Tumor promotion is then provided by repeated phorbol ester treatment. Thus, as predicted by the Williams et al. study, Rb loss actually impairs Ras*-dependent tumorigenesis (30). Papillomas forming in Rb−/− skin exhibit increased E2F expression and activation, increased ARF expression and p53 activation, and decreased expression and activation of NF-κB components. In addition, increased apoptosis is observed in Rb−/− papillomas, which may contribute to suppressed papilloma formation.
As a further parallel to the Williams et al. study, p107 is upregulated in the Rb−/− epidermis (31). Still, Rb−/− papillomas display enhanced conversion to squamous cell carcinomas. Thus, while Rb loss may select against Ras* (or vice versa), rare or forced mutation of both Ras* and Rb could push tumor evolution towards a more malignant state, perhaps by selecting for mutations that disrupt apoptosis (such as in ARF or p53). Of note, there have been reports of combined K-Ras* and Rb gene mutations in human cancers (1, 9), and thus while disfavored under most contexts, activation of Ras and inactivation of Rb may contribute to tumor evolution in some cases.
Demonstrations that Ras*-mediated tumorigenesis is more efficient in the presence of Rb stand in marked contrast to studies using gene-disrupted mice, which have shown that cyclin D1 and Cdk4 are required for efficient skin carcinogenesis (28, 29), and that mice expressing a p16Ink4a-resistant variant Cdk4 gene display enhanced susceptibility to carcinogen-induced melanomas (41). Clearly, the requirement for cyclin D-dependent kinase activation for Ras*-mediated skin tumorigenesis is not mediated solely through Rb, but probably also involves inactivation of p107 and other possible Cdk4 targets (including p130), as well as avoiding p16Ink4a and ARF upregulation.
Interestingly, the dependency of Ras* on Rb may help explain studies showing the ability of transgenic expression of E2F1 to potently suppress papilloma formation in the two-stage skin cancer model or by transgenic Ras*, dependent on ARF/p53 function but apparently independent of promoting apoptosis (26, 33). Thus, in this scenario transgenic E2F1 expression may mimic Rb loss. In contrast, E2F4 expression promotes carcinogenesis in the same two-stage model (48), which could relate to the ability of E2F4 to antagonize p107 and p130 functions and the lack of ARF upregulation by E2F4 overexpression (all in contrast to E2F1).
In all, it appears that for cancers where Ras* mutations are early events, Rb mutations will be rare. Further, where Rb mutations are early events, Ras* mutations will be rare, and this may be due to upregulation of inhibitors of proliferation, including p107, p16Ink4a, and ARF, following Rb loss. In contrast, deregulation at the level of G1 CDK activity, such as by p16Ink4a inactivation or cyclin D1 overexpression, can collaborate with Ras* to promote tumorigenesis by inactivating all Rb family members without ARF activation. The paper by Williams et al., together with the other studies discussed here, provides a rationale for the concordance of distinct disruptions in the CDK-Rb pathway with Ras*.
Acknowledgments
J.D. is supported by a grant from the National Institutes of Health (RO1 CA77314).
Thanks go to Bob Sclafani and David Johnson for critical review of the manuscript.
REFERENCES
- 1.Bautista, D., J. R. Emanuel, C. Granville, R. Howard, and J. Costa. 1996. Identification of mutations in the Ki-ras gene in human retinoblastoma. Invest. Ophthalmol. Vis. Sci. 37:2313-2317. [PubMed] [Google Scholar]
- 2.Berns, E. M., A. de Klein, W. L. van Putten, I. L. van Staveren, A. Bootsma, J. G. Klijn, and J. A. Foekens. 1995. Association between RB-1 gene alterations and factors of favourable prognosis in human breast cancer, without effect on survival. Int. J. Cancer 64:140-145. [DOI] [PubMed] [Google Scholar]
- 3.Borg, A., Q. X. Zhang, P. Alm, H. Olsson, and G. Sellberg. 1992. The retinoblastoma gene in breast cancer: allele loss is not correlated with loss of gene protein expression. Cancer Res. 52:2991-2994. [PubMed] [Google Scholar]
- 4.Bunn, P. A., Jr., and W. Franklin. 2002. Epidermal growth factor receptor expression, signal pathway, and inhibitors in non-small cell lung cancer. Semin. Oncol. 29:38-44. [DOI] [PubMed] [Google Scholar]
- 5.Classon, M., and E. Harlow. 2002. The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer 2:910-917. [DOI] [PubMed] [Google Scholar]
- 6.Classon, M., B. K. Kennedy, R. Mulloy, and E. Harlow. 2000. Opposing roles of pRB and p107 in adipocyte differentiation. Proc. Natl. Acad. Sci. USA 97:10826-10831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.D'Abaco, G. M., S. Hooper, H. Paterson, and C. J. Marshall. 2002. Loss of Rb overrides the requirement for ERK activity for cell proliferation. J. Cell Sci. 115:4607-4616. [DOI] [PubMed] [Google Scholar]
- 8.DeGregori, J. 2002. The genetics of the E2F family of transcription factors: shared functions and unique roles. Biochim. Biophys. Acta 1602:131-150. [DOI] [PubMed] [Google Scholar]
- 9.Enomoto, T., M. Fujita, M. Inoue, A. Nakazawa-Miyamoto, O. Tanizawa, and T. Nomura. 1993. Alterations of the Rb gene and its association with Ki-ras activation and p53 inactivation in endometrial adenocarcinoma. Mol. Carcinog. 8:132-137. [DOI] [PubMed] [Google Scholar]
- 10.Gope, R., and M. L. Gope. 1992. Abundance and state of phosphorylation of the retinoblastoma susceptibility gene product in human colon cancer. Mol. Cell. Biochem. 110:123-133. [DOI] [PubMed] [Google Scholar]
- 11.Hinds, P. W., and R. A. Weinberg. 1994. Tumor suppressor genes. Curr. Opin. Genet. Dev. 4:135-141. [DOI] [PubMed] [Google Scholar]
- 12.Horowitz, J. M., S.-H. Park, E. Bogenmann, J. C. Cheng, D. W. Yandell, F. J. Kaye, J. D. Minna, T. P. Dryja, and R. A. Weinberg. 1990. Frequent inactivation of the retinoblastoma anti-oncogene is restricted to a subset of human tumor cells. Proc. Nat'l. Acad. Sci. USA. 87:2775-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurford, R. K., Jr., D. Cobrinik, M.-H. Lee, and N. Dyson. 1997. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 11:1447-1463. [DOI] [PubMed] [Google Scholar]
- 14.Kaye, F. J. 2002. RB and cyclin dependent kinase pathways: defining a distinction between RB and p16 loss in lung cancer. Oncogene 21:6908-6914. [DOI] [PubMed] [Google Scholar]
- 15.Komori, H., M. Enomoto, M. Nakamura, R. Iwanaga, and K. Ohtani. 2005. Distinct E2F-mediated transcriptional program regulates p14(ARF) gene expression. EMBO J. 24:3724-3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Land, H., L. F. Parada, and R. A. Weinberg. 1983. Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature 304:596-602. [DOI] [PubMed] [Google Scholar]
- 17.Lee, K. Y., M. H. Ladha, C. McMahon, and M. E. Ewen. 1999. The retinoblastoma protein is linked to the activation of Ras. Mol. Cell. Biol. 19:7724-7732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leone, G., J. DeGregori, R. Sears, L. Jakoi, and J. R. Nevins. 1997. Myc and Ras collaborate in inducing accumulation of active cyclin E/Cdk2 and E2F. Nature 387:422. [DOI] [PubMed] [Google Scholar]
- 19.Loden, M., F. Perris, N. H. Nielsen, S. O. Emdin, and G. Landberg. 2003. C-erbB2, p27 and G1/S aberrations in human primary breast cancer. Anticancer Res. 23:2053-2061. [PubMed] [Google Scholar]
- 20.Malumbres, M., and A. Pellicer. 1998. RAS pathways to cell cycle control and cell transformation. Front. Biosci. 3:d887-912. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell, P. J., E. Perez-Nadales, D. S. Malcolm, and A. C. Lloyd. 2003. Dissecting the contribution of p16INK4A and the Rb family to the Ras-transformed phenotype. Mol. Cell. Biol. 23:2530-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mittnacht, S., H. Paterson, M. F. Olson, and C. J. Marshall. 1997. Ras signalling is required for inactivation of the tumour suppressor pRb cell-cycle control protein. Curr. Biol. 7:219-221. [DOI] [PubMed] [Google Scholar]
- 23.Nevins, J. R. 1992. E2F: a link between the Rb tumor suppressor protein and viral oncoproteins. Science 258:424-429. [DOI] [PubMed] [Google Scholar]
- 24.Peeper, D. S., J. H. Dannenberg, S. Douma, H. te Riele, and R. Bernards. 2001. Escape from premature senescence is not sufficient for oncogenic transformation by Ras. Nat. Cell Biol. 3:198-203. [DOI] [PubMed] [Google Scholar]
- 25.Peeper, D. S., T. M. Upton, M. H. Ladha, E. Neuman, J. Zalvide, R. Bernards, J. A. DeCaprio, and M. E. Ewen. 1997. Ras signalling linked to the cell-cycle machinery by the retinoblastoma protein. Nature 386:177-181. [DOI] [PubMed] [Google Scholar]
- 26.Pierce, A. M., R. Schneider-Broussard, I. B. Gimenez-Conti, J. L. Russell, C. J. Conti, and D. G. Johnson. 1999. E2F1 has both oncogenic and tumor-suppressive properties in a transgenic model. Mol. Cell. Biol. 19:6408-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raychaudhuri, P., S. Bagchi, S. H. Devoto, V. B. Kraus, E. Moran, and J. R. Nevins. 1991. Domains of the adenovirus E1A protein required for oncogenic activity are also required for dissociation of E2F transcription factor complexes. Genes Dev. 5:1200-1211. [DOI] [PubMed] [Google Scholar]
- 28.Robles, A. I., M. L. Rodriguez-Puebla, A. B. Glick, C. Trempus, L. Hansen, P. Sicinski, R. W. Tennant, R. A. Weinberg, S. H. Yuspa, and C. J. Conti. 1998. Reduced skin tumor development in cyclin D1-deficient mice highlights the oncogenic ras pathway in vivo. Genes Dev. 12:2469-2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodriguez-Puebla, M. L., P. L. Miliani de Marval, M. LaCava, D. S. Moons, H. Kiyokawa, and C. J. Conti. 2002. Cdk4 deficiency inhibits skin tumor development but does not affect normal keratinocyte proliferation. Am. J. Pathol. 161:405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruiz, S., M. Santos, M. F. Lara, C. Segrelles, C. Ballestin, and J. M. Paramio. 2005. Unexpected roles for pRb in mouse skin carcinogenesis. Cancer Res. 65:9678-9686. [DOI] [PubMed] [Google Scholar]
- 31.Ruiz, S., M. Santos, C. Segrelles, H. Leis, J. L. Jorcano, A. Berns, J. M. Paramio, and M. Vooijs. 2004. Unique and overlapping functions of pRb and p107 in the control of proliferation and differentiation in epidermis. Development 131:2737-2748. [DOI] [PubMed] [Google Scholar]
- 32.Ruley, H. E. 1983. Adenovirus early region 1A enables viral and cellular transforming genes to transform primary cells in culture. Nature 304:602-606. [DOI] [PubMed] [Google Scholar]
- 33.Russell, J. L., R. L. Weaks, T. G. Berton, and D. G. Johnson. 3October2005, posting date. E2F1 suppresses skin carcinogenesis via the ARF-p53 pathway. Oncogene [Online.] doi: 10.1038/sj.onc.1209120. [DOI] [PubMed]
- 34.Sage, J., A. L. Miller, P. A. Perez-Mancera, J. M. Wysocki, and T. Jacks. 2003. Acute mutation of retinoblastoma gene function is sufficient for cell cycle reentry. Nature 424:223-228. [DOI] [PubMed] [Google Scholar]
- 35.Sage, J., G. J. Mulligan, L. D. Attardi, A. Miller, S. Chen, B. Williams, E. Theodorou, and T. Jacks. 2000. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 14:3037-3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schauer, I. E., S. Siriwardana, T. A. Langan, and R. A. Sclafani. 1994. Cyclin D1 overexpression vs. retinoblastoma inactivation: implications for growth control evasion in non-small cell and small cell lung cancer. Proc. Natl. Acad. Sci. USA 91:7827-7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serrano, M., E. Gomez-Lahoz, R. A. DePinho, D. Beach, and D. Bar-Sagi. 1995. Inhibition of Ras-Induced proliferation and cellular transformation by p16Ink4. Science 267:249-252. [DOI] [PubMed] [Google Scholar]
- 38.Serrano, M., H. Lee, L. Chin, C. Cordon-Cardo, D. Beach, and R. A. DePinho. 1996. Role of the INK4a locus in tumor suppression and cell mortality. Cell 85:27-37. [DOI] [PubMed] [Google Scholar]
- 39.Serrano, M., A. W. Lin, M. E. McCurrach, D. Beach, and S. W. Lowe. 1997. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88:593-602. [DOI] [PubMed] [Google Scholar]
- 40.Sherr, C. J. 2002. D1 in G2. Cell Cycle 1:36-38. [PubMed] [Google Scholar]
- 41.Sotillo, R., J. F. Garcia, S. Ortega, J. Martin, P. Dubus, M. Barbacid, and M. Malumbres. 2001. Invasive melanoma in Cdk4-targeted mice. Proc. Natl. Acad. Sci. USA 98:13312-13317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stevaux, O., and N. J. Dyson. 2002. A revised picture of the E2F transcriptional network and RB function. Curr. Opin. Cell Biol. 14:684-691. [DOI] [PubMed] [Google Scholar]
- 43.Thomas, D. M., S. A. Carty, D. M. Piscopo, J. S. Lee, W. F. Wang, W. C. Forrester, and P. W. Hinds. 2001. The retinoblastoma protein acts as a transcriptional coactivator required for osteogenic differentiation. Mol. Cell 8:303-316. [DOI] [PubMed] [Google Scholar]
- 44.Thomas, D. M., H. S. Yang, K. Alexander, and P. W. Hinds. 2003. Role of the retinoblastoma protein in differentiation and senescence. Cancer Biol. Ther. 2:124-130. [PubMed] [Google Scholar]
- 45.Trimarchi, J. M., and J. A. Lees. 2002. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell. Biol. 3:11-20. [DOI] [PubMed] [Google Scholar]
- 46.Varley, J. M., J. Armour, J. E. Swallow, A. J. Jeffreys, B. A. Ponder, A. T'Ang, Y. K. Fung, W. J. Brammar, and R. A. Walker. 1989. The retinoblastoma gene is frequently altered leading to loss of expression in primary breast tumours. Oncogene 4:725-729. [PubMed] [Google Scholar]
- 47.Walker, G. J., and N. K. Hayward. 2002. Pathways to melanoma development: lessons from the mouse. J. Investig. Dermatol. 119:783-792. [DOI] [PubMed] [Google Scholar]
- 48.Wang, D., J. L. Russell, and D. G. Johnson. 2000. E2F4 and E2F1 have similar proliferative properties but different apoptotic and oncogenic properties in vivo. Mol. Cell. Biol. 20:3417-3424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whyte, P., H. E. Ruley, and E. Harlow. 1988. Two regions of the adenovirus early region 1A proteins are required for transformation. J. Virol. 62:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Williams, J. P., T. Stewart, L. Bihua, R. Mulloy, D. Dimova, and M. Classon. 2006. The retinoblastoma protein is required for Ras-induced oncogenic transformation. Mol. Cell. Biol. 26:1169-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yamamoto, H., J. W. Soh, T. Monden, M. G. Klein, L. M. Zhang, H. Shirin, N. Arber, N. Tomita, I. Schieren, C. A. Stein, and I. B. Weinstein. 1999. Paradoxical increase in retinoblastoma protein in colorectal carcinomas may protect cells from apoptosis. Clin. Cancer Res. 5:1805-1815. [PubMed] [Google Scholar]
- 52.Yu, Q., Y. Geng, and P. Sicinski. 2001. Specific protection against breast cancers by cyclin D1 ablation. Nature 411:1017-1021. [DOI] [PubMed] [Google Scholar]