Abstract
We report the initial characterization of adeno-associated virus type 5 (AAV5) RNAs generated following viral infection and the construction of a replicating infectious clone of AAV5. While the basic transcription profile of AAV5 was similar to that of AAV2, there were also significant differences. Mapping of the AAV5 transcripts demonstrated an efficient transcription initiation site within the AAV5 inverted terminal repeat (ITR), and mapping of the AAV5 intron revealed that it is considerably smaller than that of AAV2. Furthermore, in contrast to the case for AAV2, neither the Rep protein nor additional adenovirus gene products were required to achieve efficient promoter activity and pre-mRNA splicing following transfection of an AAV5 rep/cap plasmid clone lacking the ITRs into 293 cells. Perhaps most surprisingly, RNAs generated from both the AAV5 P7 and P19 promoters were efficiently polyadenylated at a site lying within the intronic region in the center of the genome. Because P7- and P19-generated transcripts are polyadenylated at this site and not spliced, Rep78 and Rep52 were the only Rep proteins detected during AAV5 infection.
The human adeno-associated viruses (AAV) are small, nonenveloped, single-stranded DNA viruses that replicate in mammalian cells best in the presence of larger helper DNA viruses, e.g., adenovirus or herpesvirus (36). Six different serotypes of AAV (AAV type 1 [AAV1] to AAV6) have been characterized. AAV2, the prototypical strain, as well as AAV3 and AAV5 have been isolated directly from human clinical specimens (5, 12, 26). AAV1 and AAV4 have been suggested to be originally of simian origin (5, 26). The more distantly related AAV5 was isolated was from a penile flat condylomatous lesion (2), and epidemiologically, AAV5 transmission appears to follow acquisition of herpesviruses rather than adenovirus (12).
At least portions of the genomes of all six serotypes of AAV have been cloned and sequenced (9, 10, 22, 31, 37, 43). The inverted terminal repeats (ITRs) of AAV1, -2, -3, -4, and -6 are >95% identical, and the rep genes of these different isolates are approximately 85% identical. In contrast, the rep gene and ITR of AAV5 are only 60% similar to those of other serotypes (9).
The Rep proteins of AAV1, -2, -3, -4, and -6 can each support the production of recombinant AAV2 vectors (9, 30), suggesting that they share significant functional homology for replication. In contrast, AAV5 is unable to support replication of AAV2-based vectors (9, 30), most likely because AAV5 Rep processes a novel terminal resolution site (TRS) present only on AAV5 ITR (8).
AAV2 is the best characterized of the AAV serotypes. The genome contains three promoters, P5, P19, and P40. The large Rep proteins (Rep78 and Rep68) and the small Rep proteins (Rep52 and Rep40), encoded from a large open reading frame in the left half of the genome, are generated from RNAs which derive from P5 and P19, respectively, and which polyadenylate near the right-hand ITR. Rep78 and -52 are generated from unspliced RNAs, and Rep68 and -40 are generated from RNAs alternatively spliced at the single AAV2 intron in the center of the genome. The AAV2 capsid proteins are encoded from a large open reading frame in the right half of the genome from spliced RNAs which derive from the P40 promoter and which polyadenylate at the same site near the right-hand ITR. AAV2 Rep78 and Rep68 are site-specific DNA-binding phosphoproteins that play essential roles during viral DNA replication (11, 14, 19, 25, 34, 38), whereas Rep52 and Rep40 are required for packaging of the AAV genome into the capsid (7, 16). AAV2 Rep78 and -68 also play important roles in the splicing of AAV RNAs (29), as well as in regulating transcription initiation of all three viral promoters, following binding to the Rep-binding element (18, 27).
Adenovirus has multiple roles in supporting AAV2 infection. Five adenovirus gene products (E1A, E1B, E2a, E4ORF6, and VA RNA) have been shown to be required for productive AAV2 replication, and the roles of E2a and E4ORF6 in viral DNA replication have been well characterized (3, 4). Adenovirus also plays important roles in the activation of the viral promoters and in splicing of AAV2 pre-mRNAs (21, 35, 39, 40); however, other than the well-defined role of E1A in the regulation of expression of AAV2 P5, the role that adenovirus plays in these processes is as yet only partially understood.
In this study, we report the initial characterization of the AAV5 transcription map following viral infection and the construction of an infectious clone of AAV5. While the basic transcription profile of AAV5 showed similarity to that of AAV2, there were also significant differences. First, following transfection of either the AAV5 infectious clone or a rep/cap AAV5 clone lacking the ITRs into 293 cells, neither the Rep protein nor additional adenovirus gene products were required to achieve efficient promoter activity and pre-mRNA splicing. Second, mapping of the AAV5 transcripts revealed an efficient transcription initiation site within the AAV5 ITR. Third, the AAV5 intron was shown to be considerably smaller than that of AAV2. Fourth, and perhaps most surprisingly, RNAs generated from the P7 and P19 promoters were efficiently polyadenylated at a site lying within the intronic region in the center of the genome. These results suggest that there may be considerable variability in expression profiles among the various AAV serotypes.
MATERIALS AND METHODS
Cell and virus.
AAV5 virus was a gift from Ursula Bantel-Schaal (Deutsches Krebsforschungzentrum, Heidelberg, Germany) through Jay Chiorini (National Institutes of Health). For viral RNA isolation, 293 or HeLa cells were coinfected with AAV5 (approximately 200 genomic copies equivalent to approximately 15 infectious center-forming units [IU]/cell) and adenovirus type 5 dl309 (Ad5) (32) at 5 PFU/cell.
Plasmid constructs. (i) AAV5 ITR-containing and full-length AAV5 constructs.
pAV5leftITR was constructed by first cloning the EcoRI-SmlI fragment from an AAV5 clone containing the right-hand ITR (pAV5rightITR [9], a gift from Jay Chiorini), which contains the corresponding AAV5 right ITR sequence from nucleotide (nt) 4492 to 4642, together with a SmlI-HindIII linker with identical AAV5 sequences from nt 148 to 183 of AAV5, into EcoRI-HindIII-digested pSK+ (Stratagene). The final pAV5leftITR plasmid contains the authentic AAV5 sequence from nt 1 to 183 in pSK+ and was used for generation of the following ITR-containing constructs. All the nucleotide numbers for AAV5 sequences follow the AAV5 sequence deposited in GenBank (accession no. AF085716). The pAV5RepCap plasmid, which contains the AAV5 rep gene and cap gene from nt 185 to 4448, was made from pSKAV5CapRep (44) (a gift from Ziying Yan and John Engelhardt, University of Iowa) by replacing the XbaI site in front of the P7 promoter with a unique PstI site to facilitate future constructions. To generate pAV5RepCapP7mATG, the initiating ATG for the large Rep proteins at nt 360 was mutated to CGG. The pAV5RepCaprightITR plasmid was constructed by inserting the PstI-BstEII Rep-Cap fragment from pAV5RepCap (nt 185 to 4205) into PstI-BstEII-digested pAV5rightITR (9). pAV5, a full-length AAV genome construct, was constructed by ligating the PstI-SmlI fragment (nt 1 to 147) from pAV5leftITR (which contains a PstI site prior to the beginning of the AAV5 left-hand ITR sequence) and a SmlI-SwaI fragment spanning AAV5 nt 148 to 486 into PstI-SwaI-digested pAV5RepCaprightITR. To generate pAV5Δ752C, the C at nt 752 in the pAV5 infectious clone was deleted, causing a frameshift mutation such that only the first 129 amino acids of Rep78 were expressed.
(ii) Minimal AAV5 constructs.
pAV5P7P41Cap was constructed by fusing the AAV5 P7 promoter (nt 185 to 358) to AAV5 nt 1637 to 4448, which contain the AAV5 P41 promoter, AAV5 intron, and capsid gene. pAV5ITRP41Cap contains the AAV5 left ITR (nt 1 to 183) fused to AAV5 nt 1637 to 4448.
(iii) AAV5 Rep-supplementing plasmid.
pHIVAV5RepSM was constructed by cloning the AAV5 rep gene (nt 305 to 4448, but containing a capsid gene deletion from nt 2289 to 3610) under control of the human immunodeficiency virus long terminal repeat promoter and inserting silent mutations within the region protected by the AV5RP probe (nt 1843 to 2034) so that its expression would not interfere with RNase protections of the reporter constructs in question.
(iv) Constructs for RNA probe generation.
Probe clones for mapping transcripts initiating in the left-hand ITR, InrP1, InrP2, and InrP3, were constructed by cloning AAV5 nt 101 to 313, 101 to 341, and 101 to 480, respectively, into EcoRI-BamHI-digested vector pGEM3Z (Promega, Madison, Wis.). For the P41 transcript initiation site and splice donor identification, pAV5RP was created by cloning AAV5 nt 1843 to 2034 into the BamHI-HindIII sites of pGEM3Z. For acceptor definition, pAV5DH was created by cloning AAV5 nt 1994 to 2341 between the EcoRI and BamHI sites of pGEM3Z. The P19 transcript initiation site was mapped by using the AV5SB probe. pAV5SB was created by cloning AAV5 nt 801 to 1023 into the EcoRI-BamHI sites of pGEM3Z. The P7 transcript initiation site was mapped by using the AV5IP probe. pAV5IP was creating by ligating nt 269 to 480 into the EcoRI-HindIII sites of pGEM3Z. The distal polyadenylation cleavage site was mapped with probe pAV5PA, which was constructed by cloning AAV5 nt 4294 to 4498 into the EcoRI-BamHI sites of pGEM3Z.
(v) RNase protection assay (RPA) size markers for protected bands.
Templates for in vitro transcription by T7 polymerase were generated by PCR from pGEM3Z; these were transcribed and pooled to yield a radioactive ladder spanning nt 50 to 350.
RPA and Northern assay.
Plasmid DNA (2 μg/60-mm-diameter dish) or linear PCR-generated DNA (5 μg/60-mm-diameter dish) was transfected into 60 to 80% confluent 293 cells by using Lipofectamine and Plus reagent (Gibco BRL, Gaithersburg, Md.) as previously described (29). In some samples the Rep-supplementing plasmid pHIVAV5RepSM (0.5 μg/60-mm-diameter dish) was included. When indicated, Ad5 was added at 5 PFU/cell immediately following transfection. Total RNA was isolated 36 to 40 h later by using guanidine isothiocyanate as previously described (33).
RPAs were performed as previously described (21, 24, 33). Probes were generated from linearized templates by in vitro transcription with T7 or SP6 polymerase, depending on the respective probe clones, as previously described (33). RNA hybridizations for RPAs were done with a substantial probe excess, and RPA signals were quantified with the Molecular Imager FX and Quantity One (version 4.2.2 image software; Bio-Rad, Hercules, Calif.). Relative molar ratios of individual species of RNAs were determined after adjusting for the number of 32P-labeled uridines in each protected fragment as previously described (33).
Northern analyses were done exactly as previously described (28), using total RNA samples and 32P-labeled DNA probes as indicated.
Immunoblot and immunofluorescence techniques.
AAV5-infected (15 IU/cell) or pAV5- or PCR-generated linear AAV5 DNA-transfected 293 cells were coinfected with Ad5 at 5 PFU/cell, and lysates were subjected to immunoblot analysis as previously described (20), using monoclonal antibody 303.9 raised to an amino-terminally truncated AAV2 Rep78 (42) (American Research Products, Inc., Belmont, Mass.). For immunofluorescence analysis, cells and media taken 40 h after transfection of 293 cells with pAV5 (2 μg/60-mm-diameter dish) cells were combined and frozen and thawed three times, and 2 μl was used to infect HeLa cells on chamber slides with Ad5 coinfection (5 PFU/cell). Infection with AAV5 plus Ad5 was done in parallel as a positive control. Immunofluorescence analyses were performed as previously described (23) with the same primary anti-Rep antibody as described above and a fluorescein isothiocyanate-conjugated anti-mouse secondary antibody (ICN Biochemicals, Irvine, Calif.). Images were generated with a SPOT digital camera; all images were obtained at the same exposure time.
RT-PCR.
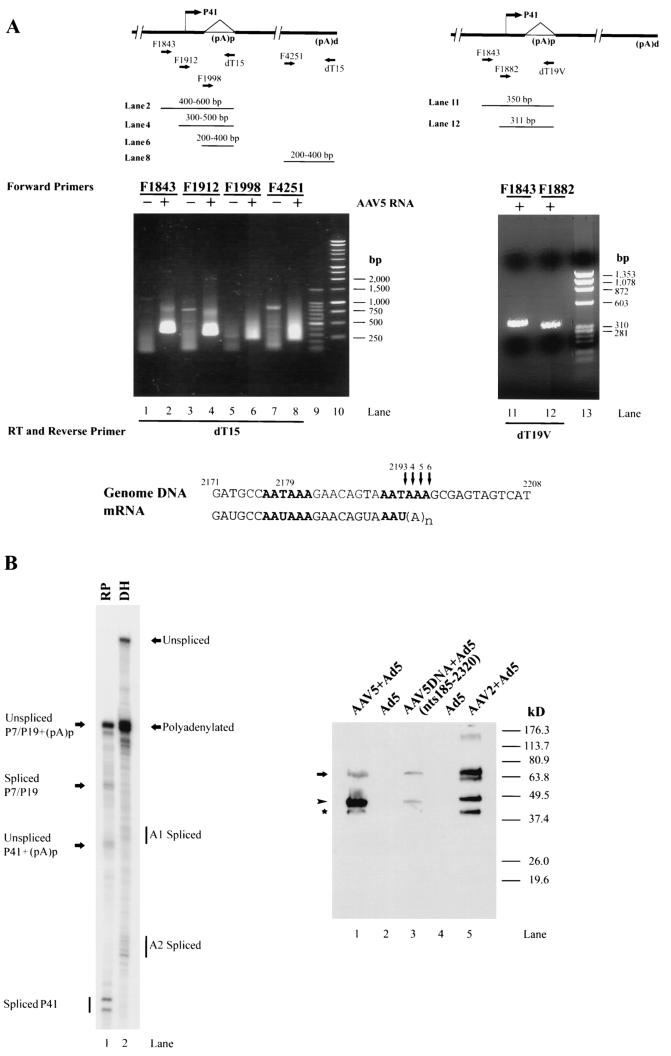
For oligo(dT)-primed reverse transcription-PCR (RT-PCR), single-tube RT-PCR (Titan-One; Roche, Nutley, N.J.) was performed with polyadenylated RNA (50 ng), which was isolated from either uninfected 293 cells or 293 cells coinfected with both AAV5 (15 IU/cell) and Ad5 (5 PFU/cell), using Dynabeads oligo(dT25) (Dynal) and a combination of primer SbfNotT15 [GTAATCCTGCAGGCGGCCGC(T)15] and one of the following four forward primers: F1843 (5′-CGCGGATCCTCTAAAACGCCCACTGGGTGA-3′, containing AAV5 nt 1843 to 1863), F1912 (5′-GTAATGGATCCGATTTGTTCCCGAGACGCCTCGC-3′, containing AAV5 nt 1912 to 1933), F1998 (5′-GCAGAATTCGCAAATGTGACTATCATG-3′, containing AAV5 nt 1998 to 2015), and F4251 (5′-CCGGAATTCGGTGGAAACCAGAGATCCAG-3′, containing AAV5 nt 4251 to 4270). The RT reaction was carried out at 50°C for 30 min, and then the samples were denatured at 94°C for 5 min before beginning the PCR (35 cycles of 94°C for 2 min, 55°C for 2 min, and 68°C for 2 min.) in a GeneAmp thermal cycler (Perkin-Elmer).
For some experiments a pooled oligo(dT)19V (where V is A, G, or C) was used both for first-strand cDNA synthesis and with a reverse primer in subsequent single-tube RT-PCR. Two forward primers, F1843 and F1882 (5′-AAAAGTCTGGAGAAGCGGGCCAG-3′, containing AAV5 nt 1882 to 1904) were used to specifically amplify cDNAs from AAV5 total RNA. PCR was performed with 35 cycles of 94°C for 30 s, 55°C for 30 s, and 68°C for 30 s. Direct sequencing of the PCR fragments was performed at the University of Missouri DNA core facility.
Analysis of intracellular AAV5 DNA.
293 cells were propagated in Dulbecco's modified Eagle's medium containing 10% bovine serum. 293 cells were seeded at 4 × 105 cells/well in six-well dishes (35-mm diameter) 48 h prior to the experiment. Cells were transfected with either pAV5Δ752C (0.2 pmol/well) or pAV5 (0.2 pmol/well) by the calcium phosphate method as previously described (41) or were infected with AAV5 (5 IU/cell) for 1 h at 37°C. Some wells were infected with Ad5 (3 PFU/cell) for 1 h before transfection or infection with AAV5. At 48 h postinfection, the cells were suspended into the spent tissue culture medium and transferred to centrifuge tubes. Cells were separated from the medium by centrifugation (11,000 × g for 3 min), and cell pellets were resuspended in 2% sodium dodecyl sulfate-150 mM NaCl-10 mM Tris-HCl (pH 8)-1 mM EDTA and incubated at 55°C for 2 h to lyse the cells. Cellular DNA was sheared by passing the samples through a syringe fitted with a 25-gauge needle. The tissue culture medium was adjusted to 2% sodium dodecyl sulfate and 0.15 M NaCl. Both cell lysates and spent medium were digested with proteinase K (0.5 mg/ml) at 37°C for 12 h. Approximately equal amounts of cell lysates and spent medium (104 cell equivalents/lane) were subjected to electrophoresis in an agarose gel (1%) in 40 mM Tris-HCl (pH 8.5)-50 mM sodium acetate-10 mM EDTA. DNA was transferred to a nitrocellulose membrane and hybridized to a random-primed probe as described previously (41). The probe was generated from a 2.3-kb Aat II-BstEII fragment of AAV5 (nt 1867 to 4205).
RESULTS
Mapping of AAV5 transcription units.
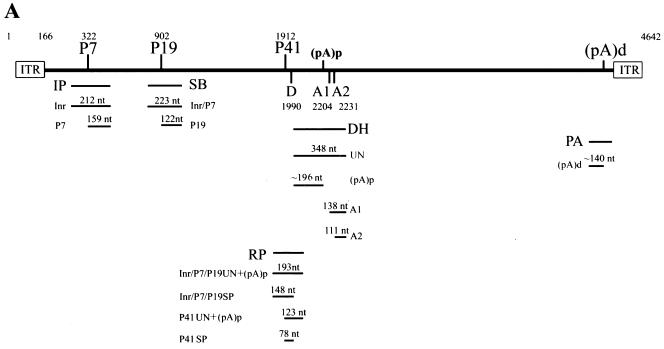
We initially sought to develop a map of AAV5 transcripts following AAV5 coinfection of 293 cells with adenovirus, using the transcriptional landmarks of AAV5 inferred from the original sequence determination (1, 9). A schematic diagram of the AAV5 genome and five antisense RPA probes (IP, SB, RP, DH, and PA) is shown in Fig. 1A. Probe IP, which spans the putative P7 promoter region, protected a band of approximately 159 nt (Fig. 1B, lane 3). This mapped the start site of the P7-generated RNA to approximately nt 322. Surprisingly, another abundant protected band of approximately 212 nt which spanned the entire AAV-specific region of the probe was detected (Fig. 1B, lane 3). This suggested that during infection viral RNA was also generated from a region upstream of the P7 promoter. The derivation of this RNA, which initiates within the ITR, is addressed below. The SB probe, which spans the putative P19 promoter, protected two bands (Fig. 1B, lane 4). The band at nt 223 was generated from read-through transcripts originating upstream from the P7 promoter and from the promoter in the ITR; the band at nt 122 mapped the P19 start site to approximately nt 902.
FIG. 1.
(A) Schematic diagram of the AAV genome and the probes used in this study. The ITRs, the promoters (P7, P19, and P41), the intron donor (D) and acceptors (A1 and A2), and the proximal [(pA)p] and distal [(pA)d] polyadenylation sites are shown. The locations of the IP (nt 269 to 480), SB (nt 801 to 1023), RP (nt 1843 to 2034), DH (nt 1994 to 2341), and PA (nt 4294 to 4498) probes used in this study are shown. Below each probe is a depiction of the bands expected following protection by each of the designated RNA species. (B) Mapping of the AAV5 transcription units by RPA. 293 cells were infected with AAV5 (15 IU/cell) and Ad5 (5 PFU/cell). Ten micrograms of total RNA was isolated 36 to 40 h after coinfection and protected by the IP, SB, RP, DH, and PA probes as indicated. Lane 1, a 32P-labeled RNA ladder with the respective sizes indicated to the left. Lane 2, protection of total RNA generated following AAV2 infection of 293 cells by a homologous RP probe which serves as an additional marker. The sizes of these bands are 192, 139, 105, and 53 nt, respectively. The origins of the protected bands in lanes 3, 4, 6, and 7 are indicated. For lane 5, the top arrow designates unspliced and proximally polyadenylated [(pA)p] RNAs from the upstream promoter within the ITR (Inr), P7, and P19 (top), and the lower arrow indicates unspliced RNA and RNAs utilizing (pA)p generated from the P41 promoter. The top arrowhead shows spliced RNA from P7 plus P19, and perhaps from the ITR Inr, and the lower arrowhead identifies bands protected by spliced P41 RNA. The band designated by a star in lane 3 is undigested probe. The band designated by a diamond in lane 7 is likely transcription through the distal polyadenylation site (pA)d.
The RP probe, which spans the putative P41 promoter and intron donor site, was protected to generate bands of approximately 193, 148, 123, and 78 nt (Fig. 1B, lane 5). The 193- and 148-nt bands represent unspliced and spliced versions, respectively, of transcripts generated upstream (from P19, P7, and the ITR) and mapped the intron donor to nt 1990, which is slightly different than sequence inspection had previously suggested (1, 9). The 112- and 78-nt bands represent unspliced and spliced versions, respectively, of RNAs initiated at the P41 promoter; they map the P41 initiation site to approximately nt 1912. The relative amount of spliced RNAs generated from the upstream P7-, P19-, and ITR-generated promoters was significantly less, and the ratio of spliced to unspliced P41-generated RNA was somewhat less, than observed for RP probe protection of AAV2 RNA (Fig. 1B, lane 2) (29). However, as discussed further below, for AAV5, the unspliced RNAs detected by the RP probe include a combination of RNAs reading through to the distal polyadenylation site (pA)d at the right-hand end of the genome and RNAs polyadenylated at a proximal site within the intron (pA)p.
The PA probe yielded a major band of approximately 140 nt, confirming usage of the predicted right-hand end polyadenylation site at approximately nt 4434 (Fig. 1B, lane 7). The other protected band of approximately 212 nt, which spans the entire AAV-specific region of the probe, likely derived from transcripts that had not yet been cleaved and polyadenylated.
Protection by the DH probe gave an unexpected result (Fig. 1B, lane 6). In addition to the bands expected from protection of unspliced RNAs that presumably read through to the terminal polyadenylation site (348 nt) and RNAs using the predicted acceptors A1 at nt 2204 (138 nt) and A2 at nt 2231 (111 nt), an additional abundant band of approximately 196 nt in length was detected. The size of this band is consistent with a probe fragment protected by an RNA that was polyadenylated within the intron region. There is a highly consensus polyadenylation signal at this position in the genome, and further characterization of this RNA is addressed below. The ratio of usage of the A1 and A2 acceptors for AAV5 RNA in these assays with total RNA preparations was approximately 1:1, in contrast to the 1:10 ratio seen for AAV2 RNA; however, as explained below, this ratio varied under different experimental circumstances. Also, these assays revealed that the AAV5 intron is 81 nt smaller than the intron of AAV2.
Assays using total RNA isolated from AAV5-plus-Ad5 coinfection of HeLa cells gave results similar to those presented above (data not shown). Assays using total RNA isolated from an AAV5-plus-herpes simplex virus coinfection of HeLa cells or poly(A)-selected RNA isolated from AAV5-plus-Ad5 coinfection of 293 cells also showed similar levels of all AAV5 RNAs (including the internally polyadenylated species); however, for reasons not yet clear, the ratio of A1 to A2 in these coinfections was closer to the 1:10 ratio that has been reported for AAV2 spliced products (data not shown).
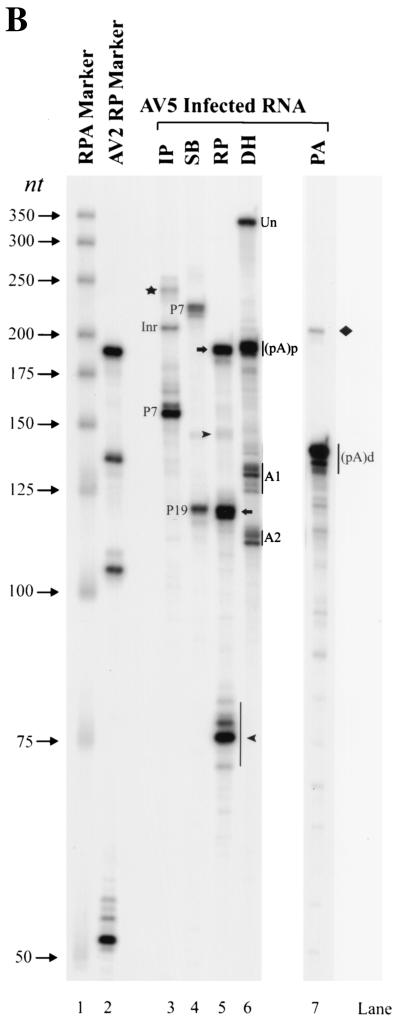
Northern analysis of AAV5 RNA confirmed that the transcription profile of AAV5 was significantly different from that of AAV2. Hybridization of total AAV5 RNA with a whole AAV5 genomic probe detected a major band of approximately 2.3 kb and a minor band of approximately 4.3 kb (Fig. 2A, lanes 1; compare with Fig. 2A, lanes 3, which show AAV2 RNA detected with an AAV2 genomic probe). As expected, when visualized using a probe comprising the rep region of AAV2, the AAV2 P5 spliced and unspliced RNAs (which do not separate under these conditions) were detected at approximately 4.2 kb, and the P19-generated RNAs were detected at 3.6 (unspliced) and 3.3 (spliced) kb (Fig. 2A, lanes 5). In contrast, an AAV5 probe comprising the rep region hybridized to RNAs of 1.9 and 1.3 kb (Fig. 2B, lane 2). The 2.3-kb RNA hybridized to a probe from the cap region and not to a probe from the rep region (Fig. 2B, lane 3). The more scarce 4.3-kb RNA hybridized to both the rep and cap probes (Fig. 2B, lanes 2 and 3) and, as shown below (see Fig. 4C), hybridized with a probe upstream of P7, indicating that it corresponds to RNA that initiates within the ITR. These results are consistent with a model in which RNAs derived from the AAV5 P7 and P19 promoters are predominately polyadenylated at a site within the central intron [the proximal polyadenylation site (pA)p] and RNAs derived from P41 and the ITR promoter predominately extend through to the polyadenylation signal at the right-hand end of the molecule [the distal polyadenylation site (pA)d]. Since the RP probe used above terminates upstream of the polyadenylation signal in the intron, bands therein designated unspliced contain a significant, although as-yet unquantified, amount of RNA polyadenylated at the internal site.
FIG. 2.
Northern blot analysis of AAV5 RNA. (A) RNA was isolated at 36 to 40 h postinfection from 293 cells infected (I) with AAV5 (15 IU/cell) and Ad5 (5 PFU/cell) (lane 1) or with AAV2 (10 IU/cell) plus Ad5 (5 PFU/cell) (lanes 3 and 5) or from uninfected (U) 293 cells (lanes 2, 4, and 6). Northern blots were hybridized to either an AAV5 whole-genome probe (lanes 1 and 2), an AAV2 whole-genome probe (lanes 3 and 4), or an AAV2 rep gene probe spanning AAV2 nt 810 to 965, which overlap the P19 promoter (lanes 5 and 6), as indicated. The sizes of the RNA species are shown to the left of each panel. The identities of the AAV2 transcripts are as follows: unspliced P5-generated RNA (4.3 kb), unspliced P19-generated RNA (3.6 kb), spliced P19-generated RNA (3.3 kb), unspliced P40-generated RNA (2.6 kb), and spliced P40-generated RNA (2.3 kb). (B) RNA was isolated at 36 to 40 h postinfection from 293 cells infected (I) with AAV5 (15 IU/cell) plus Ad5 (5 PFU/cell) (lane 1). Northern blots were hybridized to either a whole AAV5 genomic probe (lane 1), a subfragment of the AAV5 rep gene spanning AAV5 nt 1434 to 1654 (lane 2), or a subfragment of the AAV5 cap gene spanning AAV5 nt 3359 to 3599 (lane 3), as indicated. The sizes of the bands are shown on the left. AAV5 rep and cap gene probes contain similar number of labeled adenosines.
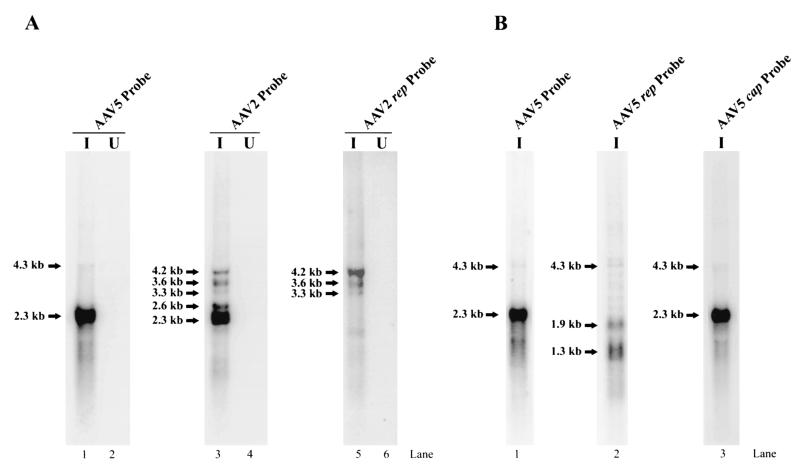
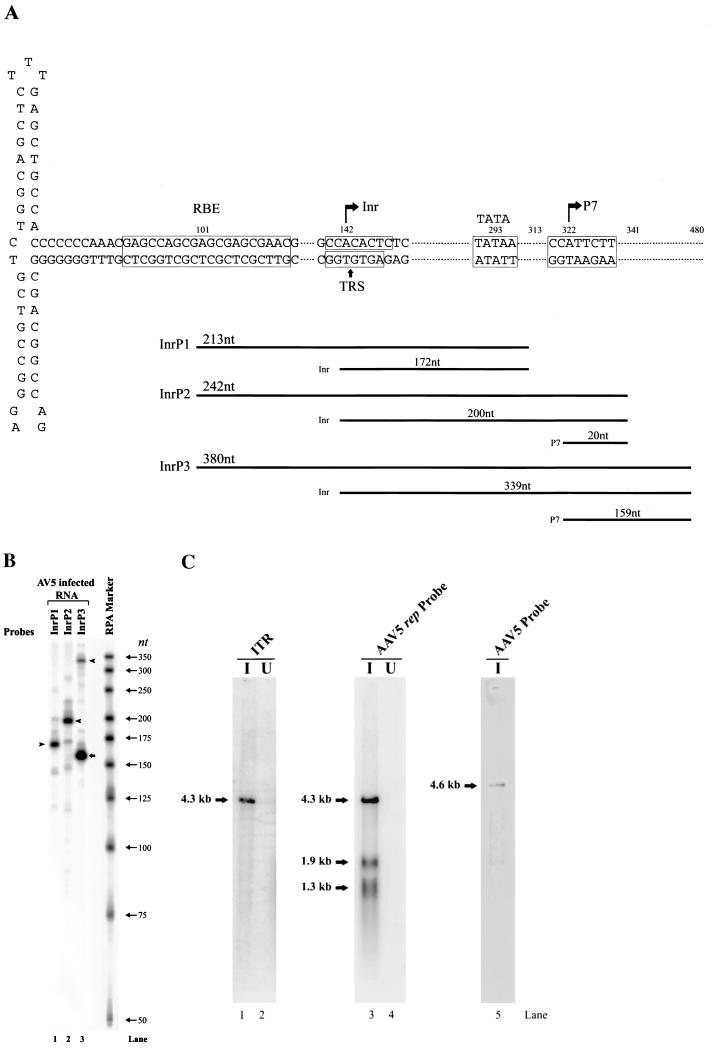
FIG. 4.
(A) Schematic diagram of AAV5 ITR structure and probes used for determination of the origin of the ITR-generated transcript. The locations of the Rep-binding element (RBE), TRS, TATA box, and P7-generated initiation site are shown. The three probes (InrP1, InrP2, andInrP3) used in this analysis and the bands that they are predicted to protect are diagramed under each probe. (B) Total RNA (10 μg) from AAV5-infected 293 cells was protected by the InrP1, InrP2, and InrP3 probes, as indicated. Arrowheads show the specific band protected by the ITR-generated RNA, which became longer as the 5′ end of the probe used extended toward the P7 promoter (see diagram in panel A). The arrow shows the band protected by the transcript generated from the P7 promoter. A 32P-labeled RNA ladder was used as a maker; sizes are shown to the right. (C) Northern blot analysis of RNA isolated at 36 to 40 h postinfection from 293 cells infected with AAV5 (15 IU/cell) plus Ad5 (5 PFU/cell) (I) (lanes 1 and 3) or RNA isolated from 293 cells infected only with Ad5 (5 PFU/cell) (U) (lanes 2 and 4). As a marker, AAV5 single-stranded DNA was also loaded on the gel (lane 5). Northern blots were hybridized either to a subfragment of AAV5 upstream of P7 (nt 168 to 321) (lanes 1 and 2), to the AAV5 rep probe downstream of P7 (spanning AAV5 nt 632 to 1376) (lanes 3 and 4), or to the whole AAV5 genomic probe (lane 5), as indicated.
Characterization of transcripts that polyadenylate at a site within the AAV5 intron.
Polyadenylated RNA utilizing the site within the intron (pA)p was also detected by anchored RT-PCR (Fig. 3A). RT-PCR with poly(A)-selected RNA as a template, an oligo(dT15) primer, and three AAV5-specific primers (F1843, F1912, and F1998) generated PCR products whose sizes were as expected for RNAs using the internal polyadenylation site (pA)p (Fig. 3A, lanes 1 to 6). A fourth primer (F4251) upstream of the 3′ terminal polyadenylation site (pA)d served as a positive control (Fig. 3A, lanes 7 and 8). The variation in the sizes of the RT-PCR products is consistent with the oligo(dT15) primer hybridizing to various locations within the poly(A) tail. When an oligo(dT) primer was anchored with a random 3′ nucleotide [G, A, C, or d(T)19V], the RT-PCR product was resolved into a single species (Fig. 3B, lanes 11 and 12). Sequencing of those products identified the primary (pA)p cleavage and polyadenylation addition site to be at or near the U at nt 2193, approximately 11 nt downstream of the first AAUAAA signal (and within the second AAUAAA signal). Because three A residues follow the U at nt 2193, the actual cleavage site could be anywhere between nt 2193 and 2196 (as diagrammed in Fig. 3A, bottom). Cleavage of approximately 70% of vertebrate mRNAs follows an adenosine (45).
FIG. 3.
(A) AAV5 utilizes a polyadenylation site within the intron. Left panel, RT-PCR of polyadenylated RNA isolated from either uninfected 293 cells (lanes 1, 3, 5, and 7) or 293 cells infected with both AAV5 (15 IU/cell) and Ad5 (5 PFU/cell) (lanes 2, 4, 6, and 8), using an oligo(dT)15primer (dT15) and one of four forward primers: F1843 (lanes 1 and 2), F1912 (lanes 3 and 4), F1998 (lanes 5 and 6), and F4250 (lanes 7 and 8). The positions of the primers are diagrammed above the gel. The sizes of DNA fragments in the 100-bp ladder (Promega) (lane 9) are 1,500, 1,000, 900, 800, 700, 600, 500, 400, 300, 200, and 100 bp. The sizes of the DNA fragments in the 1-kb ladder (Promega) (lane 10) are 10, 8, 6, 5, 4, 3, 2.5, 2, 1.5, 1, 0.75, 0.5, and 0.25 kb. Expected sizes of the PCR products for transcripts using the proximal poly(A) site (pA)p, including the poly(A) tail, are shown above the gel. The PCR conditions used did not efficiently amplify the longer RNA products using the distal polyadenylation site. Right panel, Total RNA (1 μg) from AAV5-infected 293 cells was subjected to anchored RT-PCR using oligo(dT)19V (V = A, G, or C) (dT19V), both for the first-strand cDNA synthesis and with a forward primer for subsequent PCR. Two different forward primers, F1843, which starts at nt 1843, and F1882, which starts at nt 1882 (diagrammed above the gel), were used and resulted in bands of different mobilities (compare lane 11 with lane 12). Sequencing of the PCR products from lanes 11 and 12 yielded an identical sequence prior to the U at nt 2293. However, because there are three A residues after the U at nt 2193, the cleavage site may range from nt 2193 to 2196 (diagramed at the bottom). (B) Left panel, a PCR product containing only AAV5 DNA nt 185 to 2320 was transfected into 293 cells in the presence of Ad5 (5 PFU/cell), and total RNA (10 μg) from transfected cells was protected by the RP (lane 1) and DH (lane 2) probes. The designations of the protected bands are shown. Right panel, immunoblot analysis of AAV5 rep gene products in cell lysates of 293 cells infected with AAV5 (15 IU/cell) plus Ad5 (5 PFU/cell) (lane 1) or transfected with an AAV5 PCR product comprising nt 185 to 2320 (5 μg/60-mm-diameter dish) with Ad5 coinfection (lane 3), infected with AAV2 (10 IU/cell) plus Ad5 (lane 5), or infected with Ad5 alone as a negative control (lanes 2 and 4). A monoclonal antibody raised to NH2-terminally truncated AAV2 Rep proteins (monoclonal antibody 303.9; American Research Products) was used to identify AAV2 and AAV5 Rep proteins. Binding of 303.9 to the blot was detected by using a horseradish peroxidase-conjugated goat anti-mouse antibody. The migration of the AAV5 Rep protein products was compared to that of a series of prestained proteins (Invitrogen) The positions and apparent molecular masses of the markers are shown on the right of the immunoblot. Lane 5 shows the AAV2 Rep proteins, Rep78, -68, -52, and -40. The arrow and arrowhead identify AAV5 Rep78 and -52, respectively. The band designated by the star is most likely a degradation product, as its abundance increases upon sample storage; its derivation is not known.
As would be expected if the P7 and P19 RNAs were predominately polyadenylated at the proximal site (and not spliced), only Rep78 and Rep52 were detected in AAV5 coinfection (Fig. 3B, right panel, lane 1). The relative ratio of Rep52 to Rep78 is higher during AAV5 infection than seen during AAV2 infection (Fig. 3B, right panel, lane 5). The reason for this difference is not yet clear. There is more P19-generated RNA relative to P7-generated RNA during AAV5 infection (Fig. 1B, lane 4, and 2B, lane 2); however, whether this difference is enough to account for the higher relative levels of Rep52 has not yet been determined. In addition, following transfection into 293 cells of an isolated portion of AAV5 (nt 185 to 2320) that contains only the rep gene and the (pA)p polyadenylation signals within the AAV5 intron, a stable RNA product that was polyadenylated at that site (Fig. 3B, left panel, lane 2) and was capable of programming the production of Rep78 and Rep52 (Fig. 3B, right panel, lane 3) was generated. Interestingly, an increased ratio of Rep52 to Rep78 is not seen following transfection of this PCR product.
Mapping of the transcription initiation site within the AAV5 ITR.
A number of studies have detected promoter activity from the ITR of AAV2, and one of those studies mapped this activity to a 37-nt stretch (AAV2 nt 109 to 145) in the A/D elements of the ITR (13). At least under some circumstances, transcripts arising from the AAV5 ITR were seen to be more abundant than those previously detected from the AAV2 ITR, which allowed a more precise identification of its initiation site. Figure 4A shows a map of the AAV5 ITR and the RPA probes used to map the ITR-derived transcripts. In contrast to the case for AAV2, the sequence of the AAV5 putative TRS contains a pyrimidine-rich putative transcription initiator element (Inr), 5′Y-Y-A-N-T/A-Y-Y-Y3′ (where Y is C/T and N is any nucleotide) (15). Three probes were designed to overlap the potential initiator site, and each had different right-hand (5′) ends. Therefore, when protected by the ITR-generated transcripts, they should each yield bands of different sizes and accurately identify the initiation site. RPA with probe InrP1 yielded a band of approximately 172 nt (Fig. 4B, lane 1), protection of probe InrP2 yielded a band of approximately 200 nt (Fig. 4B, lane 2 [this gel would not resolve a protected band from P7 of 20 nt]), and protection of probe InrP3 yielded bands of approximately 339 and 159 nt (the band of 159 nt is protected by P7-generated RNA) (Fig. 4B, lane 3). These results localize the 5′ end of the ITR-generated transcripts to the consensus initiator sequence (which overlaps the putative TRS) at approximately nt 142.
Northern analysis of the ITR-generated transcript, using a DNA probe upstream of P7, indicated that this transcript is approximately 4.3 kb, indicating that it extends to near the right-hand end of the genome (Fig. 4C, lane 1). This band was also detected in poly(A)-selected RNA by RPA (data not shown), suggesting that it may be polyadenylated at the right-hand site at nt 4434. It is not known yet whether this transcript is spliced or not.
Construction of an infectious plasmid clone of AAV5.
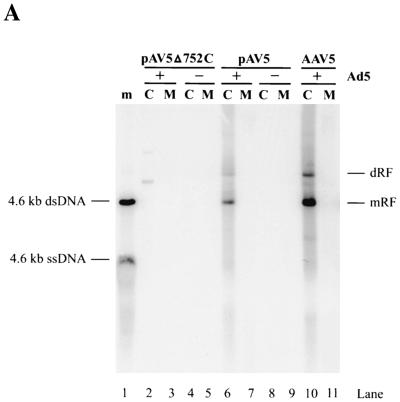
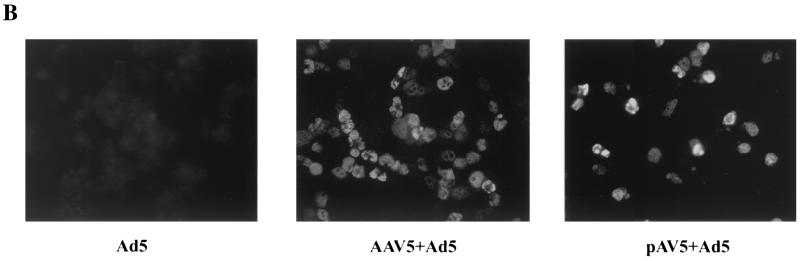
A full-length plasmid clone of AAV5 was constructed (see Materials and Methods for details). Following transfection of pAV5 into Ad5-infected 293 cells, AAV5 excised from the plasmid and replicated (Fig. 5A, lane 6). In the presence of Ad5, AAV5 accumulated double-stranded replicative DNA, similar to the case for other parvoviruses. The difference in the amount of replicative DNA that accumulated following pAV5 transfection and AAV5 infection correlates with the difference in transfection efficiency (∼20%) and the infection efficiency (∼95%) in this experiment. Therefore, on a per-cell basis, pAV5 produces amounts of AAV5 replicative DNA forms similar to those produced in a viral infection. Although progeny single-strand DNA was not detected by Southern blotting, infectious AAV5 virions were detected in pAV5-transfected cell lysates by immunofluorescence with an anti-Rep monoclonal antibody following infection of HeLa cells with this material (Fig. 5B, pAV5+Ad5).
FIG. 5.
(A) Southern blot of AAV5 DNA replicative forms that derive from either pAV5 or AAV5. 293 cells were transfected with either pAV5Δ572C (to display background from a nonreplicating plasmid) (lanes 2 to 5) or pAV5 (lanes 6 to 9) or infected with AAV5 (5 IU/cell) (lanes 10, 11). Cells were either infected with Ad5 at a 3 PFU/cell (+) (lanes 2, 3, 6, 7, 10, and 11) or not (−) (lanes 4, 5, 8, and 9). Cells were harvested at 48 h postinfection by being resuspended in spent tissue culture medium (lanes M) and pelleted by microcentrifugation. Cell pellets (lanes C) were lysed as described previously (41), and all samples were digested with proteinase K (0.5 mg/ml) prior to electrophoresis. The Southern blot was hybridized to a 2.3-kb AatII-BstEII fragment of AAV5 (nt 1867 to 4205) that had been labeled by using random nonamers. Markers (m; lane 1) are a mixture of untreated and denatured (100°C, 3 min) 4.6-kb Acc65I fragments from pAV5, which corresponds to the full-length AAV5 genome. Monomer-length and dimer-length replicative forms (mRF and dRF, respectively) are indicated on the right. dsDNA, double-stranded DNA; ssDNA, single-stranded DNA. (B) HeLa cells in chamber slides were infected with Ad5 (5 PFU/cell), AAV5 (15 IU/cell) plus Ad5 (5 PFU/cell), or 2 μl of cell lysates from pAV5-transfected 293 cells plus Ad5 (5 PFU/cell), as indicated. Immunofluorescence assays were performed at 40 h postinfection, using an AAV Rep monoclonal antibody that recognizes both AAV2 and AAV5 Rep proteins.
In 293 cells, efficient transcription and splicing of AAV RNA requires neither additional Ad5 gene products nor Rep.
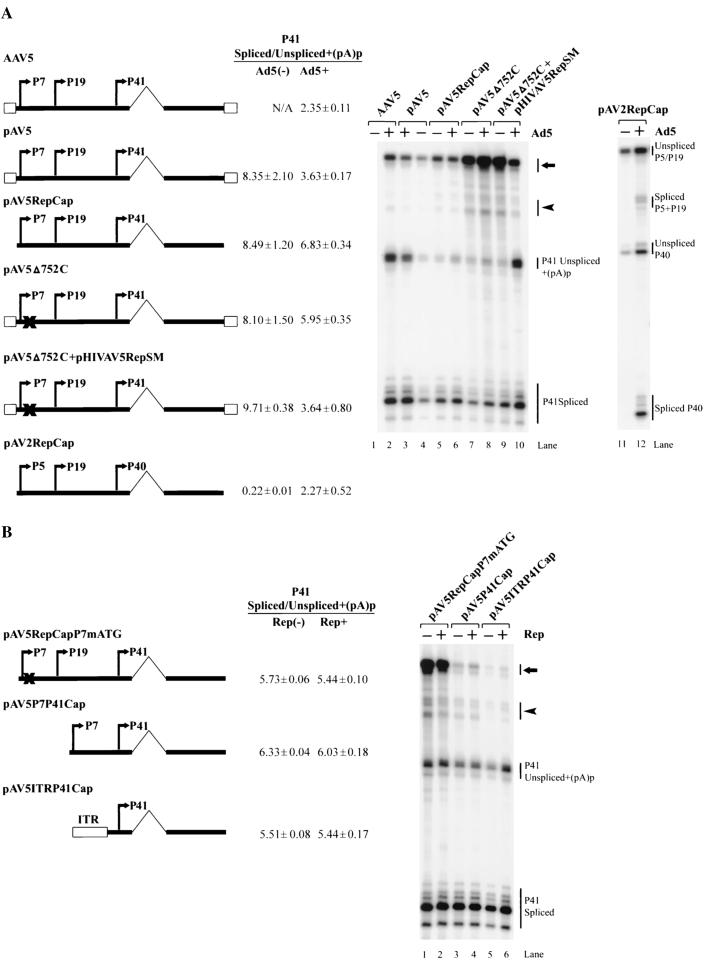
In dramatic contrast to those of AAV2 (29), the basal levels of expression and splicing of AAV5 RNA generated from pAV5RepCap transfected into 293 cells in the absence of Ad5 were high and were not further stimulated significantly by Ad5 coinfection (Fig. 6A, compare lanes 5 and 6 to lanes 11 and 12). The relative levels of spliced products obtained following the transfection of pAV5RepCap into HeLa cells in the absence of Ad5 were similar to those seen in 293 cells, although the overall levels of expression were much lower (data not shown).
FIG. 6.
(A) Splicing of AAV5 RNA is independent of Ad5 and Rep. 293 cells were either infected with AAV5 virus (15 IU/cell) (lanes 1 and 2) or transfected, in the presence (+) or absence (−) of Ad5 (5 PFU/cell), with the following plasmids, which are diagramed on the left: a replicating full-length AAV5 genomic plasmid (pAV5) (lanes 3 and 4); an AAV5 plasmid which contains the AAV5 rep and cap genes and lacks the ITRs (pAV5RepCap) (lanes 5 and 6); a full-length plasmid, pAV5Δ752C, that produces a mutant Rep protein (lanes 7 and 8); pAV5Δ752C cotransfected with a Rep-supplementing plasmid pHIVAV5RepSM (lanes 9 and 10); or an AAV2 rep and cap gene-containing plasmid (pAV2RepCap) (lanes 11 and 12). Total RNA (10 μg), taken 36 to 40 h after transfection or infection, was protected by either the AAV5 RP or AAV2 RP probe, and the gel from a representative experiment is shown. The identities of the bands are shown on the right. The ratio of spliced P41 (or P40) transcripts to transcripts that are either unspliced or polyadenylated at the proximal site [(pA)p] are shown and are averages of theresults of at least three separate experiments with standard deviations. The arrow and arrowhead show unspliced RNA and RNA utilizing (pA)d, or spliced RNA, respectively, generated from promoters upstream of P41. (B) 293 cells were transfected with the AAV5 plasmids diagramed on the left, either with (Rep+) or without (Rep−) cotransfection of the AAV5 Rep-supplementing plasmid (pHIVAV5RepSM) in the presence of Ad5 coinfection (5 PFU/cell). Total RNA (10 μg), taken 36 to 40 h posttransfection, was protected by the RP probe, and the image shown is from a representative experiment. The ratio of spliced P41 transcripts to transcripts that are either unspliced or polyadenylated at the proximal site [(pA)p] are shown and are averages of the results of at least three separate experiments with standard deviations. The arrow and arrowhead show unspliced RNA and RNA utilizing the proximal polyadenylation site [(pA)p], or spliced RNA, respectively, generated from promoters upstream of P41.
The ITR-containing replicating clone of AAV5 (pAV5), however, showed an increase in relative levels of unspliced plus internally polyadenylated P41 RNA in the presence of Ad5 (Fig. 6A, lanes 3 and 4), similar to what was seen during viral coinfection (Fig. 6A, lane 2). This increase was likely due to viral replication rather than merely the presence in cis of the ITR. The relative levels of spliced P41-generated RNA following transfection of pAV5Δ752C (Fig. 6A, lanes 7 and 8), which has a single-nucleotide frameshift deletion at nt 752 of the AAV5 genome, were similar to those seen following transfection of pAV5RepCap; however, when Rep was added in trans, Ad5 coinfection resulted in an increase in unspliced plus internally polyadenylated P41 products (Fig. 6A, lanes 9 and 10), similar to the amount observed for the replicating clone in the presence of Ad5 and for AAV5-plus-Ad5 coinfection (Fig. 6A, lanes 2 and 3). As mentioned before, the RP probe used in this experiment terminates prior to the internal polyadenylation site, and so it did not distinguish between unspliced P41-generated RNA that extended to the right-hand end polyadenylation site (pA)d and P41-generated RNA that may have used the polyadenylation site in the intron, (pA)p. Therefore, the relative extent to which these two species accounted for the increase in this band following replication is unknown. A role for P41-generated RNA that utilizes (pA)p is not readily apparent.
Although the large Rep proteins of AAV2 are necessary for both efficient expression and splicing of AAV2 P40-generated RNA (18, 27, 29), this was not the case for AAV5. In the presence of Ad5, added AAV5 Rep has no effect upon the expression or splicing of P41-generated RNA produced by pAV5RepCapP7mATG, which expresses no large Rep proteins itself (Fig. 6B, lanes 1 and 2), or upon P41-generated RNA produced from minimal constructs in which the AAV5 P7 promoter (Fig. 6B, lanes 3 and 4) or ITR (Fig. 6B, lanes 5 and 6) was linked just upstream of the AAV5 P41 transcription unit. Similar minimal constructs composed of analogous AAV2 elements show dramatic increases in both expression and levels of splicing of AAV2 RNA following addition of AAV2 Rep78 and -68 (29).
DISCUSSION
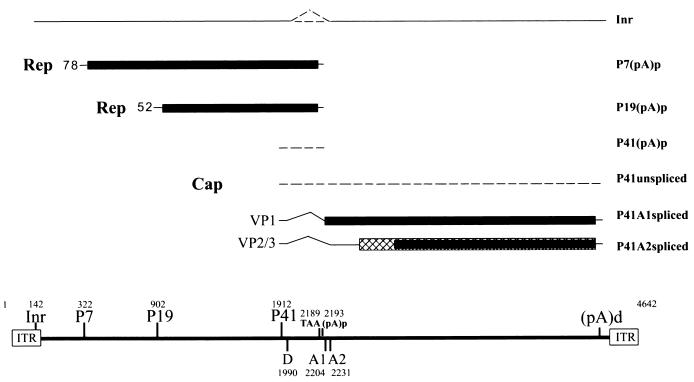
In this paper we report the initial characterization of the transcription profile of AAV5 (Fig. 7) and the construction of an infectious AAV5 plasmid clone.
FIG. 7.
Transcription map of the AAV5. The 4,642-nt AAV5 genome is shown to scale with the major transcription landmarks, including the ITRs, promoters, initiator (Inr) site within the ITR, the initiation sites for the various RNAs, splice donor (D) and acceptors (A1 and A2), and the proximal [(pA)p] and distal [(pA)d] polyadenylation sites. The major transcripts observed in this study are listed on the right; however, only two Rep proteins were detected. The extent to which P41-generated transcripts use the proximal polyadenylation sites and whether the ITR-initiated product is spliced have not yet been determined, and so these RNA species are indicated as dashed lines.
The transcription profile of AAV5 shows some similarities to that of AAV2, but there are also significant differences. First, AAV5 generates an abundant transcript from its ITR. This RNA uses an initiation site that maps to the AAV5 ITR TRS, which in AAV5 is similar to a consensus transcription initiator sequence, and extends to the right-hand end of the genome. Although this 4.3-kb RNA is polyadenylated, it is not yet clear whether it is spliced. We have seen different accumulated levels of this transcript under different experimental conditions, but at its most abundant it is nearly as prevalent as the P7-generated transcript, making it significantly more abundant that the ITR-generated product previously identified for AAV2. Whether this RNA encodes a protein product and what role it may play during viral infection are currently being investigated.
Another striking difference between the AAV5 and AAV2 transcription maps is that RNAs generated by the AAV5 P7 and P19 promoters predominately polyadenylate at a site within the AAV5 intron. Inspection of the intron region of AAV5 shows the presence of two relatively consensus AAUAAA signals at nt 2177 and 2191, which are immediately upstream of the first intron acceptor A1 at nt 2205. RNA cleavage and polyadenylation occur 11 to 14 nt downstream of the first AAUAAA motif within the second AAUAAA. Although AAV2 contains a single AAUAAA polyadenylation signal in this region, its use has not been previously detected (37). P41 RNAs seem to utilize this site with significantly reduced efficiency. However, accurate quantification of the relative usage of this site by P41-generated RNA is difficult; it requires the use of a long RPA probe which spans from upstream of P41 to beyond the intron acceptors. The use of such a probe is currently being developed. Why the P7- and P19-generated RNAs are predominately polyadenylated at the internal polyadenylation site rather than spliced and polyadenylated downstream, while P41-generated RNAs use the internal site less frequently and the ITR-generated products almost exclusively read through, is currently under investigation. A consequence of the internal polyadenylation of the P7- and P19-generated RNAs is that only two Rep proteins, Rep78 and Rep52, are generated during infection. Human B19-generated RNA also utilizes an internal polyadenylation site, and differential 3′-end processing has been suggested to be a focus of regulation that influences viral tropism (6, 17).
AAV2 is highly dependent upon helper virus coinfection for numerous steps in its life cycle. In addition to genome replication, expression from the AAV2 promoters and the splicing of AAV2 RNA are dramatically stimulated by adenovirus, even in 293 cells that express adenovirus E1A and E1B (21, 35, 39, 40). Our studies show that, in contrast to the case for AAV2, following transfection of an AAV5 rep/cap plasmid into 293 cells, the basal levels of expression and splicing of AAV5 P41-generated RNA were high and were not further stimulated by Ad5 infection. The splicing of AAV5 P41-generated RNA was also efficient and was not further stimulated by Ad5 following transfection of pAV5RepCap into HeLa cells, although the overall levels of expression were considerably lower. These results suggest that the expression of AAV5 is considerably less dependent on helper virus than is that of AAV2. Although the requirement that the AAV5 life cycle has for Ad5 may be more limited, helper virus is still required for AAV5 replication following either infection of AAV5 or transfection of the AAV5 infectious clone (Fig. 5A).
Also in contrast to that of AAV2 (29), splicing of AAV5 RNA is not stimulated by the AAV5 Rep protein, in either 293 or HeLa cells. The AAV5 intron is 81 nt smaller than the AAV2 intron, and it will be interesting to determine whether the independence of AAV5 splicing is related to its smaller size and whether its constitutive activity is governed merely by the strength of its 3′ and 5′ splice sites (often not easily determined merely by inspection) or by other cis-acting elements within the AAV5 intron.
Our characterization of AAV5 suggests that there is significant variability in the expression profiles of the various serotypes of AAV. In addition, the relatively independent expression of AAV5 and the presence of a strong promoter within the AAV5 ITR may be features that are useful for the development of AAV5-based vectors for gene therapy.
Acknowledgments
We thank Ursula Bantel-Schaal (DKFZ, Heidelberg, Germany) for AAV5 virus and Jay Chiorini (NIH) and Ziying Yan and John Englehardt (University of Iowa) for AAV5 plasmid constructs and for sharing information prior to publication. We thank Lisa Burger for excellent technical assistance and Dave Farris for his participation in the initial phases of this work.
R.N. and J.Q. were supported by the University of Missouri Life Sciences Mission Enhancement Program and Molecular Biology Program, respectively, during a portion of this work. The work was supported by Public Health Service grants RO1 AI46458 and RO1 AI21302 from the National Institute of Allergy and Infectious Diseases to D.J.P.
REFERENCES
- 1.Bantel-Schaal, U., H. Delius, R. Schmidt, and H. zur Hausen. 1999. Human adeno-associated virus type 5 is only distantly related to other known primate helper-dependent parvoviruses. J. Virol. 73:939-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bantel-Schaal, U., and H. zur Hausen. 1984. Characterization of the DNA of a defective human parvovirus isolated from a genital site. Virology 134:52-63. [DOI] [PubMed] [Google Scholar]
- 3.Berns, K. 1996. Parvoviridae: the viruses and their replication, p. 1017-1041. In B. N. Fields, D. M. Knipe, and P. M. Howley (ed.), Fundamental virology. Lippincott-Raven, Philadelphia, Pa.
- 4.Berns, K. I., and C. Giraud. 1996. Biology of adeno-associated virus. Curr. Top. Microbiol. Immunol. 218:1-23. [DOI] [PubMed] [Google Scholar]
- 5.Blacklow, N. R., M. D. Hoggan, A. Z. Kapikian, J. B. Austin, and W. P. Rowe. 1968. Epidemiology of adenovirus-associated virus infection in a nursery population. Am. J. Epidemiol. 88:368-378. [DOI] [PubMed] [Google Scholar]
- 6.Brunstein, J., M. Söderlund-Venermo, and K. Hedman. 2000. Identification of a novel RNA splicing pattern as a basis of restricted cell tropism of erythrovirus B19. Virology 274:284-291. [DOI] [PubMed] [Google Scholar]
- 7.Chejanovsky, N., and B. J. Carter. 1989. Mutagenesis of an AUG codon in the adeno-associated virus rep gene: effects on viral DNA replication. Virology 173:120-128. [DOI] [PubMed] [Google Scholar]
- 8.Chiorini, J. A., S. Afione, and R. M. Kotin. 1999. Adeno-associated virus (AAV) type 5 Rep protein cleaves a unique terminal resolution site compared with other AAV serotypes. J. Virol. 73:4293-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiorini, J. A., F. Kim, L. Yang, and R. M. Kotin. 1999. Cloning and characterization of adeno-associated virus type 5. J. Virol. 73:1309-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chiorini, J. A., L. Yang, Y. Liu, B. Safer, and R. M. Kotin. 1997. Cloning of adeno-associated virus type 4 (AAV4) and generation of recombinant AAV4 particles. J. Virol. 71:6823-6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiorini, J. A., L. Yang, B. Safer, and R. M. Kotin. 1995. Determination of adeno-associated virus Rep68 and Rep78 binding sites by random sequence oligonucleotide selection. J. Virol. 69:7334-7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georg-Fries, B., S. Biederlack, J. Wolf, and H. zur Hausen. 1984. Analysis of proteins, helper dependence, and seroepidemiology of a new human parvovirus. Virology 134:64-71. [DOI] [PubMed] [Google Scholar]
- 13.Haberman, R. P., T. J. McCown, and R. J. Samulski. 2000. Novel transcriptional regulatory signals in the adeno-associated virus terminal repeat A/D junction element. J. Virol. 74:8732-8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hermonat, P. L., M. A. Labow, R. Wright, K. I. Berns, and N. Muzyczka. 1984. Genetics of adeno-associated virus: isolation and preliminary characterization of adeno-associated virus type 2 mutants. J. Virol. 51:329-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Javahery, R. A., K. K. Lo, B. Zenzie-Gregory, and S. Smale. 1994. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol. Cell. Biol. 14:116-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.King, J. A., R. Dubielzig, D. Grimm, and J. A. Kleinschmidt. 2001. DNA helicase-mediated packaging of adeno-associated virus type 2 genomes into preformed capsids. EMBO J. 20:3282-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu, J. M., S. W. Green, T. Shimada, and N. S. Young. 1992. A block in full-length transcript maturation in cells nonpermissive for B19 parvovirus. J. Virol. 66:4686-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarty, D. M., M. Christensen, and N. Muzyczka. 1991. Sequences required for coordinate induction of adeno-associated virus p19 and p40 promoters by Rep protein. J. Virol. 65:2936-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCarty, D. M., D. J. Pereira, I. Zolotukhin, X. Zhou, J. H. Ryan, and N. Muzyczka. 1994. Identification of linear DNA sequences that specifically bind the adeno-associated virus Rep protein. J. Virol. 68:4988-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller, C. L., and D. J. Pintel. 2001. The NS2 protein generated by the parvovirus minute virus of mice is degraded by the proteasome in a manner independent of ubiquitin chain elongation or activation. Virology 285:346-355. [DOI] [PubMed] [Google Scholar]
- 21.Mouw, M., and D. J. Pintel. 2000. Adeno-associated virus RNAs appear in a temporal order and their splicing is stimulated during coinfection with adenovirus. J. Virol. 74:9878-9888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muramatsu, S., H. Y. N. S. Mizukami, and K. E. Brown. 1996. Nucleotide sequencing and generation of an infectious clone of adeno-associated virus 3. Virology 221:208-217. [DOI] [PubMed] [Google Scholar]
- 23.Naeger, L. K., J. Cater, and D. J. Pintel. 1990. The small nonstructural protein (NS2) of minute virus of mice is required for efficient DNA replication and infectious virus production in a cell-type-specific manner. J. Virol. 64:6166-6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Naeger, L. K., R. V. Schoborg, Q. Zhao, G. E. Tullis, and D. J. Pintel. 1992. Nonsense mutations inhibit splicing of MVM RNA in cis when they interrupt the reading frame of either exon of the final spliced product. Genes Dev. 6:1107-1111. [DOI] [PubMed] [Google Scholar]
- 25.Narasimhan, D., R. Collaco, V. Kalman-Maltese, and J. P. Trempe. 2002. Hyper-phosphorylation of the adeno-associated virus Rep78 protein inhibits terminal repeat binding and helicase activity. Biochim. Biophys. Acta 1576:298-305. [DOI] [PubMed] [Google Scholar]
- 26.Parks, W. P., A. M. Casazza, J. Alcott, and J. L. Melnick. 1968. Adeno-associated satellite virus interference with the replication of its helper adenovirus. J. Exp. Med. 127:91-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pereira, D. J., D. M. McCarty, and N. Muzyczka. 1997. The adeno-associated virus (AAV) Rep protein acts as both a repressor and an activator to regulate AAV transcription during a productive infection. J. Virol. 71:1079-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pintel, D. J., D. Dadachanji, C. R. Astell, and D. C. Ward. 1983. The genome of minute virus of mice, an autonomous parvovirus, encodes two overlapping transcription units. Nucleic Acids Res. 11:1019-1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiu, J., and D. J. Pintel. 2002. The adeno-associated virus type 2 Rep protein regulates RNA processing via interaction with the transcription template. Mol. Cell. Biol. 22:3639-3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rabinowitz, J. E., F. Rossing, C. Li, H. Conrath, W. Xiao, X. Xiao, and R. J. Samulski. 2002. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 76:791-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rutledge, E. A., C. L. Halbert, and D. W. Russell. 1998. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J. Virol. 72:309-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samulski, R. J., L. S. Chang, and T. Shenk. 1989. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J. Virol. 63:3822-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schoborg, R. V., and D. J. Pintel. 1991. Accumulation of MVM gene products is differentially regulated by transcription initiation, RNA processing and protein stability. Virology 181:22-34. [DOI] [PubMed] [Google Scholar]
- 34.Senapathy, P., J. D. Tratschin, and B. J. Carter. 1984. Replication of adeno-associated virus DNA, complementation of naturally occurring rep− mutants by a wild-type genome or an ori− mutant and correction of terminal palindrome deletions. J. Mol. Biol. 179:1-20. [DOI] [PubMed] [Google Scholar]
- 35.Seto, E., Y. Shi, and T. Shenk. 1991. YY1 is an initiator sequence-binding protein that directs and activates transcription in vitro. Nature 354:241-245. [DOI] [PubMed] [Google Scholar]
- 36.Siegl, G., R. C. Bates, K. I. Berns, B. J. Carter, D. C. Kelly, E. Kurstak, and P. Tattersall. 1985. Characteristics and taxonomy of Parvoviridae. Intervirology 23:61-73. [DOI] [PubMed] [Google Scholar]
- 37.Srivastava, A., E. W. Lusby, and K. I. Berns. 1983. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 45:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tratschin, J. D., I. L. Miller, and B. J. Carter. 1984. Genetic analysis of adeno-associated virus: properties of deletion mutants constructed in vitro and evidence for an adeno-associated virus replication function. J. Virol. 51:611-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trempe, J. P., and B. J. Carter. 1988. Alternate mRNA splicing is required for synthesis of adeno-associated virus VP1 capsid protein. J. Virol. 62:3356-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trempe, J. P., and B. J. Carter. 1988. Regulation of adeno-associated virus gene expression in 293 cells: control of mRNA abundance and translation. J. Virol. 62:68-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tullis, G. E., and T. Shenk. 2000. Efficient replication of adeno-associated virus type 2 vectors: a cis-acting element outside of the terminal repeats and a minimal size. J. Virol. 74:11511-11521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wistuba, A., S. Weger, A. Kern, and J. A. Kleinschmidt. 1995. Intermediates of adeno-associated virus type 2 assembly: identification of soluble complexes containing Rep and Cap proteins. J. Virol. 69:5311-5319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiao, W., N. Chirmule, S. C. Berta, B. McCullough, G. Gao, and J. M. Wilson. 1999. Gene therapy vectors based on adeno-associated virus type 1. J. Virol. 73:3994-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan, Z., R. Zak, G. W. Luxton, T. C. Ritchie, U. Bantel-Schaal, and J. F. Engelhardt. 2002. Ubiquitination of both adeno-associated virus type 2 and 5 capsid proteins affects the transduction efficiency of recombinant vectors. J. Virol. 76:2043-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63:405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]