Abstract
We demonstrate that the expression of hem genes in Rhodobacter capsulatus is transcriptionally repressed in response to the exogenous addition of heme. A high-copy suppressor screen for regulators of hem gene expression resulted in the identification of an LysR-type transcriptional regulator, called HbrL, that regulates hem promoters in response to the availability of heme. HbrL is shown to activate the expression of hemA and hemZ in the absence of exogenous hemin and repress hemB expression in the presence of exogenous hemin. Heterologously expressed HbrL apoprotein binds heme b and is purified with bound heme b when expressed in the presence of 5-aminolevulinic acid. Electrophoretic gel shift analysis demonstrated that HbrL binds the promoter region of hemA, hemB, and hemZ as well as its own promoter and that the presence of heme increases the binding affinity of HbrL to hemB.
The cofactor heme is an integral part of the electron transport chain, where it serves as an electron carrier for both membrane-bound and soluble cytochromes. Heme is also commonly employed as a prosthetic group to bind diatomic gases for respiration or signaling, as in the case of FixL (27, 28) and the α subunit of eukaryotic initiation factor 2 (19). However, heme that is not bound to apoproteins is toxic to cells, so its biosynthesis is tightly regulated such that free heme does not significantly accumulate in cells. Despite the importance of heme in biological chemistry, little is known about how cells regulate the synthesis of heme relative to synthesis of apoproteins that bind this cofactor.
The regulation of heme synthesis is additionally complex in photosynthetic organisms that synthesize chlorophyll or bacteriochlorophyll since this utilizes intermediates common to the heme branch, from the first step of the pathway up to protoporphyrin IX. Despite heavy metabolic demands placed on this pathway, particularly during photosynthetic growth, there are no free tetrapyrrole end products (heme or bacteriochlorophyll) or intermediates known to accumulate inside the cell. In this study, we set out to investigate this apparent paradox by identifying and characterizing transcriptional regulators of the heme biosynthetic pathway.
The biosynthetic end product heme has been shown to be an important regulatory molecule in eukaryotes. For example, nuclear localization and activity of the transcriptional activator HAP1 (heme-activated protein) from yeast is controlled by heme (24, 61). Similarly, the mammalian transcription factor Bach1 modulates its DNA-binding activity by specifically binding heme (29). Heme also regulates the transcription of oxygen-induced genes in yeast (22) as well as globin (30) and phosphatase (37) genes in human cell lines. In prokaryotes, the iron response regulator Irr from Bradyrhizobium japonicum has been shown to be conditionally degraded when it binds heme (35). Irr is a member of the GntR family of transcriptional regulators and is thought to repress the transcription of hemB (15). Binding of Irr to the hemB promoter, however, has not been directly demonstrated.
To obtain an understanding of the regulation of heme biosynthesis, it is important that both transcriptional and posttranscriptional regulatory events in this pathway are studied. Towards this goal, we have recently described the expression pattern of heme biosynthesis genes in Rhodobacter capsulatus in response to a reduction in oxygen tension (44), which is a condition that induces high amounts of bacteriochlorophyll biosynthesis. We also demonstrated that the redox-responding transcription factors AerR, FnrL, RegA, and CrtJ control the expression of heme biosynthesis genes in response to changes in oxygen tension (44). These same transcription factors also regulate the transcription of genes that code for apoproteins of cytochromes (46, 47) and apoproteins of the photosystem that bind bacteriochlorophyll (12, 34, 43). In the present study, we report the identification and characterization of an additional transcriptional regulator of heme biosynthesis genes, HbrL, which is a LysR-type transcriptional regulator (LTTR). HbrL (heme-binding regulator, LysR type) exhibits many attributes common to this family of regulators, including binding the end product (heme) of the pathway that it transcriptionally controls. Heme- and DNA-binding characteristics of HbrL are reported.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and culture additions.
Bacterial strains and plasmids used in this study are described in Table 1. Escherichia coli strains were grown aerobically in LB medium (41). R. capsulatus was grown aerobically and semiaerobically, as described previously (8), in PYS medium (60). The antibiotics ampicillin and kanamycin were used in E. coli at 200 μg/ml and 50 μg/ml, respectively. Kanamycin was used in R. capsulatus at 10 μg/ml. Hemin addition to semiaerobic cultures was accomplished by the preparation of a 1 mM hemin stock solution in 50% ethanol-0.05 M sodium bicarbonate that was filter sterilized and added to the culture to a final concentration of 30 μM, as described previously (25). An equal volume of ethanol-bicarbonate containing no hemin was added to control cultures.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | supE44 lacU169(φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 41 |
| BL21(DE3) | C600::RP-4 2-Tc::Mu-Km::Tn7 hsdR hsdM recA thi pro | Novagen |
| R. capsulatus | ||
| SB1003 | rif-10 | 59 |
| Y262 | rif-10 | 58 |
| JS157K | rif-10 gntR::Km | This study |
| JS163K | rif-10 hbrL::Km | This study |
| Plasmids | ||
| pET20b | Expression vector | Novagen |
| pBluescript | Cloning vector | Stratagene |
| pUC4K::KIXX | Kanamycin cassette | 4 |
| pJS147 | pBluescript::gntR-hbrL | This study |
| pJS156 | pBluescript::1.2-kb BamHI-XhoI fragment of pJS147 | This study |
| pJS157K | pJS156::KIXX | This study |
| pJS161 | pBluescript::0.8-kb XhoI fragment of pJS147 | This study |
| pJS163K | pJS161::KIXX | This study |
| pJS174 | pET20b::hbrL::H6 | This study |
| pCAP69 | Translational fusion of hemA to lacZ | 55 |
| pJS123 | Translational fusion of hemZ to lacZ | 44 |
| pJS125 | Translational fusion of hemB to lacZ | 44 |
| pJS145 | Translational fusion of hemH to lacZ | 44 |
| pJS146 | Translational fusion of hemC to lacZ | 44 |
| pJS153 | Translational fusion of hemE to lacZ | 44 |
| pJS152 | pBluescript::hemCE promoter | This study |
Plasmid and strain construction.
Wild-type library elements (43) were conjugated into hemB::lacZ reporter strain SB1003-pJS125 (44), and exconjugates were plated at a nonconfluent density on selective medium. Colonies were transferred onto nitrocellulose filters, which were plated onto PYS medium containing 2% (wt/vol) X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside). Colonies that lacked blue pigmentation were selected as potential high-copy suppressors. Cosmids from these colonies were isolated, and the library element insert was subsequently excised by BamHI digestion. This fragment was subcloned into similarly digested pBluescript (Stratagene) to produce plasmid pJS147. Digestion of pJS147 with XhoI and BamHI and ligation of the resultant 1.2-kb fragment into similarly digested pBluescript produced plasmid pJS156. Digestion of pJS147 with XhoI and ligation of the resultant 0.8-kb fragment into similarly digested pBluescript produced plasmid pJS161.
We created Kmr insertions in the two complete open reading frames that exhibited BLAST matches to putative genetic regulators. The KIXX Kmr fragment (4) was released by EcoRI digestion and ligated into the unique EcoRI site in pJS156 to produce pJS157K. The second insertion strain was constructed similarly; the KIXX fragment was released by EcoRI, blunt ended by treatment with T4 polymerase, and inserted into the unique NruI site in pJS161 to produce pJS163K. Plasmids pJS157K and pJS163K were mobilized to R. capsulatus strain R121, and the inserts were then recombined into the chromosome by gene transfer agent transduction, as described previously (56), to produce two strains, JS157K and JS163K, respectively.
β-Galactosidase reporter assays.
Transcription activities of various heme promoters were assayed using hem reporter constructs pCAP69 (hemA) (55), pJS125 (hemB), pJS146 (hemC), pJS153 (hemE), pJS123 (hemZ), and pJS145 (hemH) (44). These were mated into wild-type (SB1003) and hbrL (JS163K) strains and assayed for β-galactosidase enzyme activity, as described previously (47). β-Galactosidase activity represents averages of triplicate measurements of biological replicates, with error bars representing standard deviations. Units represent micromoles of orthonitrophenyl-β-d-galactoside (ONPG) hydrolyzed per minute per milligram of cellular protein.
Cloning, overexpression, and purification of HbrL.
An HbrL-His6 protein was constructed by PCR amplification of the hbrL coding sequence using pJS147 as a template with primers HbrLSTART (5′-GGAATTCATATGACCCGCGCCGCCG-3′) and HbrLSTOP (5′-CGGGATCCTCGAGGCCCGCCGCTTCGCGGC-3′). This PCR product was digested with EcoRI and BamHI (underlined in primer sequences described above) and ligated into pCRscript (Stratagene). The HbrL coding sequence was verified by dideoxy sequence, excised with NdeI (in boldface type above) and BamHI, and ligated into similarly digested pET20b (Novagen) to produce the HbrL expression plasmid pJS174.
Overexpression of the HbrL-H6 fusion was accomplished as follows. BL21(DE3) cells bearing pJS174 were grown in Terrific broth at 37°C to mid-log phase (A600, 0.6), and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were harvested by centrifugation after 2 h of growth at 37°C and stored at −20°C. Overexpressed cells were resuspended in a minimal volume of buffer H, composed of 50 mM Tris-Cl (pH 8.0) and 500 mM NaCl, and lysed by sonication. Unbroken cells and insoluble material were removed by centrifugation at 5,000 × g for 10 min. Soluble lysate was applied to Ni2+-nitrilotriacetic acid resin (Novagen), washed with 10 column volumes of buffer H, and eluted with 2 column volumes of buffer H containing 100 mM EDTA. This eluant was applied to a Microcon 100kD centrifugal filter (Amicon), and flowthrough was dialyzed overnight into buffer H plus 50% (vol/vol) glycerol, aliquoted, and stored at −20°C.
Heme binding.
Hemin titrations were performed with HbrL-His6 using equivalent concentration of lysozyme (Sigma) as a control. Hemin stock solutions were freshly prepared in dimethyl sulfoxide (DMSO) at 10 mg/ml and then diluted in buffer H to a molar concentration 10 times the protein concentration. This diluted hemin was added in increments of 0.2 mol equivalents (1/50 volume), assuming a 1:1 binding stoichiometry. Visible absorbance spectra of affinity-purified protein samples were collected with a Beckman DU 640 spectrophotometer. Pyridine hemochrome samples were prepared as previously described (6).
DNA binding.
Templates for the generation of hem promoter fragments were generated as follows. A hemA promoter fragment was generated by PCR using primers hemAforward (5′-GCG TCG AGT GTC ATT GAAG-3′) and hemAreverse (5′-AGT CCA TGT GCG TCA CC-3′). A hemB promoter fragment was generated by PCR using primers hemBforward (5′-GCC AGG GCG ATC CAA TAC-3′) and hemBreverse (5′-CAT GGG CGG TCT CCT TC-3′). A 353-bp hemZ promoter fragment was generated by PCR using primers HEMZ (5′-CGG GAT CCA AGT TGC GAT TCA TGT TTC ATG G-3′) and FNRL (5′-GGA ATT CTT CCA ACG AAA TCA GAG GG-3′). A 59-bp hbrL probe was generated by annealing two primers, 5′-CGC ACC TGA CCC CGG CGC AGG TGC GCG AGG CCG ACC GGC TGC GCG AGG CCT GCG ATC AG-3′ and its reverse complement. A 59-bp hemH probe was generated by annealing two primers, 5′-CTG CGC CCG GGC TCG GCC GCC TGT TCC TTG CCG AAA TTC TGG GCA AAA CGC CGC AGG GG-3′ and its reverse complement. A 758-bp DNA fragment encoding the hemC and hemE promoters was generated by PCR from pJS152 using primers specific to the T3 (5′-CAA TTA ACC CTC ACT AAA GGG-3′) and T7 (5′-TAA TAC GAC TCA CTA TAG GGC-3′) promoters.
5′ 32P labeling of one primer was carried out at 37°C for 1 h in a 10-μl reaction mixture volume containing 50 pmol primer, 1× T4 kinase buffer (New England Biolabs), 10 units of T4 polynucleotide kinase (New England Biolabs), and 500 μCi of [γ-32P]ATP (specific activity, 7,000 Ci/mmol; MP Biomedicals). The enzyme was deactivated by heat treatment for 10 min at 80°C. Probes were then generated by PCR, purified by agarose gel electrophoresis, excised from the gel, and purified with the QIAquick PCR purification kit (QIAGEN).
DNA-binding characteristics of HbrL overexpression cell lysates were determined by electrophoretic mobility shift as described previously (33), except that dilutions of cell lysate were prepared in buffer H, and electrophoresis was conducted in 8% Tris-acetate-EDTA. Hemin was added to cell lysate samples from a stock solution (10 mg/ml in DMSO); an equal volume of DMSO was added to cell lysate samples lacking heme.
RESULTS
hem gene expression is regulated by heme.
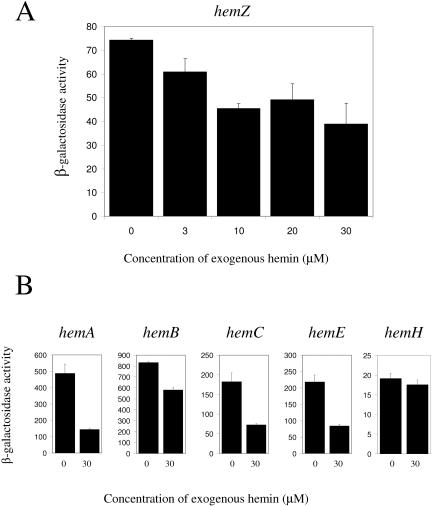
We addressed whether expression of hem genes is sensitive to end product control by assaying expression of the hemZ::lacZ reporter pJS123 in the presence of increasing concentrations of exogenously added hemin. Previous studies have demonstrated that R. capsulatus can effectively transport heme, as evidenced by the suppression of a 5-aminolevulinic acid (ALA) synthase mutation by the exogenous addition of hemin (54). The results presented in Fig. 1A show that the expression (54) of hemZ decreases as hemin addition increases, with maximal repression occurring at approximately 30 μM. Addition of hemin to 100 μM reduces the growth rate of cells, but this effect is not observed at 30 μM (data not shown). Hence, we chose 30 μM as the experimental hemin concentration since this amount of hemin maximally repressed hem reporter activity without noticeably affecting the growth rate. The repressing effect of hemin addition occurs with hemA, hemB, hemC, hemE, and hemZ, ranging from a modest 1.4-fold repression for hemB to a 3.4-fold repression for hemA (Fig. 1B). The only exception was hemH, which had insignificant repression of transcription upon hemin addition. From these results, we conclude that expression of most hem genes is subject to feedback control.
FIG. 1.
Effect of hemin addition on hemZ reporter activity. (A) Wild-type expression of the hemZ reporter under semiaerobic conditions in the presence of various concentrations of exogenous hemin is shown. (B) Wild-type expression of hem reporters under semiaerobic conditions in the presence and absence of 30 μmol hemin is shown. Values presented for each reporter are the means and standard deviations of at least three independent assays and are reported as micromoles of ONPG hydrolyzed per minute per milligram of cellular protein.
Isolation of hem regulators by high-copy-number suppression.
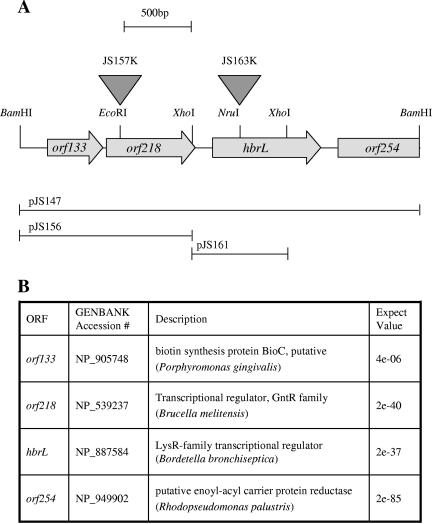
We used the technique of high-copy-number suppression as a screen for transcription factors that control heme gene expression. The previously described hemB::lacZ reporter pJS125 (44) was used as a reporter to visually identify cosmids from a wild-type R. capsulatus library (43) that reduced hemB expression. Colonies that arose from mating with a cosmid library donor strain were lifted from an agar surface with a filter and then placed onto an agar surface that contained 2% X-Gal. The majority of colonies turned blue after 5 min of incubation in the presence of X-Gal; however, a few colonies that did not produce a blue color even after overnight incubation were isolated (data not shown). Several hemB suppressing cosmids that contained a common 2.5-kb BamHI fragment were isolated from this screen (Fig. 2). Inspection of the DNA sequence of this fragment indicated the presence of three open reading frames. The first open reading frame, orf133, coded for an enzyme in the biotin biosynthetic pathway; the second open reading frame, orf218, coded for a transcriptional regulator of the GntR family; and the third open reading frame, designated hbrL, coded for a transcriptional regulator in the LysR (LTTR) family (Fig. 3). The GntR family has about 270 members that regulate a wide variety of metabolic processes such as amino acid biosynthesis, carbon metabolism, etc. (38). The LTTR family is also widely disseminated, with over 1,000 known members that also regulate many different metabolic processes, including amino acid biosynthesis (10); oxidative stress response (49); control of cell growth (23) and virulence (40); carbon (13), nitrogen (57), and sulfur (45) metabolism; and the degradation of aromatic compounds (31, 53).
FIG. 2.
HbrL coding region. (A) Schematic representation of plasmid pJS147, a 2.5-kb genomic BamHI fragment containing HbrL and neighboring genes. Open reading frames are depicted as filled arrows, and a partial restriction map is given. Triangles depict the locations of transposon insertions in HbrL and orf218. Subclones of pJS147 used for the construction of transposon insertions are depicted below. (B) BLAST matches for open reading frames (ORF) in plasmid pJS147.
FIG. 3.
HbrL alignment. (A) Schematic representation of conserved domain features of LTTRs (42). (B) Alignment of HbrL orthologs prepared using hierarchical clustering (11). The consensus sequence is shown using symbols defined previously (16); the regions of primary sequence corresponding to LTTR domain features are shaded as in A. X. axonopodis, Xanthomonas axonopodis; X. campestris, Xanthomonas campestris; P. syringae, Pseudomonas syringae; P. fluorescens, Pseudomonas fluorescens; A. vinelandii, Azotobacter vinelandii; P. aeruginosa, Pseudomonas aeruginosa; R. metallidurans, Ralstonia metallidurans; P. putida, Pseudomonas putida; E. carotovora, Erwinia carotovora; P. profundum, Photobacterium profundum; S. oneidensis, Shewanella oneidensis; V. parahaemolyticus, Vibrio parahaemolyticus.
Identification of the heme-responsive regulator HbrL.
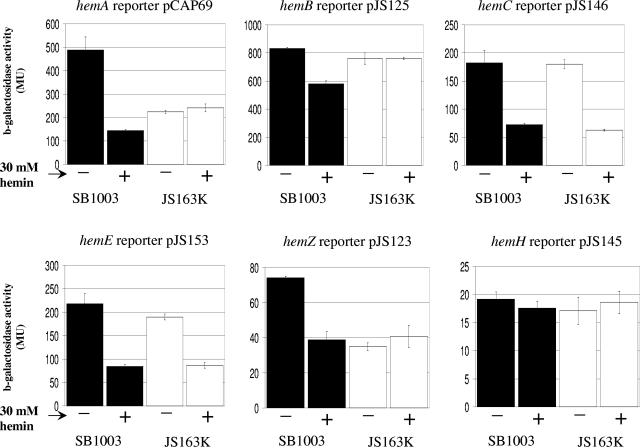
To test for effects on heme gene expression, we constructed chromosomal insertion mutants of the two putative transcriptional regulators, orf218 and hbrL, resulting in strains JS157K and JS163K, respectively. No phenotype was attributed to strain JS157K, so this strain was not further characterized in this study. However, the hbrL-disrupted strain JS163K did exhibit abnormal hem transcription in response to the exogenous addition of hemin. As shown in Fig. 4, the reduction of hemB expression that occurs in wild-type cells upon hemin addition did not occur in the hbrL-disrupted strain JS163K. This indicates that HbrL functions as a heme-dependent repressor of hemB expression.
FIG. 4.
Expression of hem reporters in wild-type and hbrL strains in the presence and absence of exogenous hemin. Expression of hem reporters in the wild type (black bars) and hbrL strain JS163K (white bars) under semiaerobic conditions in the absence (-) and presence (+) of 30 μM hemin is shown. Values presented for each reporter represent triplicate measurements of biological replicates, with error bars representing standard deviations. Units are micromoles of ONPG hydrolyzed per minute per milligram of cellular protein. MU, Miller units.
We also observed that hemA and hemZ expression is abnormal in the hbrL-disrupted strain JS163K. Specifically, expression of hemA and hemZ remains low in the presence and absence of hemin in JS163K (Fig. 4). This result suggests that HbrL functions as an activator of hemA and hemZ in the absence of heme.
Expression of hemC and hemE also remains responsive to exogenous heme as in wild-type cells in JS163K (Fig. 4) as well as in JS157K (data not shown), which indicates that there must be an additional heme response regulator in R. capsulatus for these heme genes.
Overexpression of hbrL.
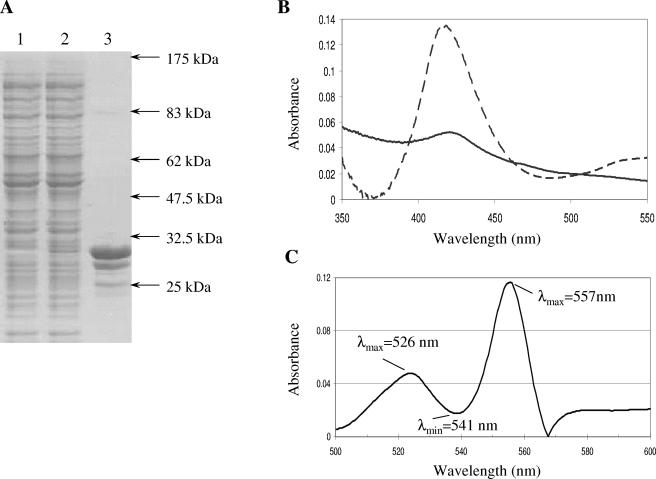
We overexpressed HbrL as a His tag fusion protein in E. coli and affinity purified HbrL to more than 95% purity with Ni2+-nitrilotriacetic acid resin (Fig. 5A, lane 3). When expressed in normal growth medium, the overexpressed protein had a slight absorbance peak at 413 nm (data not shown). When grown in medium that was supplemented with 10 mM 5-aminolevulinic acid, the overexpression lysate had a noticeably brown color. Visible absorption spectroscopy of the overexpression lysate (Fig. 5B, solid line) revealed a clear absorbance peak at 413 nm. Affinity-purified HbrL also exhibited a visible red color with an absorbance peak at 413 nm (Fig. 5B, dashed line). Pyridine hemochrome extraction of tetrapyrroles from affinity-purified HbrL also resulted in a spectrum characteristic of the presence of noncovalently bound b-type heme (Fig. 5C). Collectively, these results indicate that HbrL binds heme noncovalently in vivo, similar to that observed with other heme-binding proteins (7, 20).
FIG. 5.
Overexpression of HbrL protein. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis of uninduced (lane 1) and induced (lane 2) HbrL-H6 whole-cell lysate and affinity-purified HbrL-H6 protein (lane 3). Electrophoretic mobility of protein standards is indicated. (B) Absorbance spectrum of HbrL overexpression lysate (solid line) and affinity-purified HbrL (dashed line) grown in the presence of 5-aminolevulinic acid. (C) Pyridine hemochrome spectrum of affinity-purified HbrL protein. The wavelengths of characteristic heme b maxima and minima are indicated.
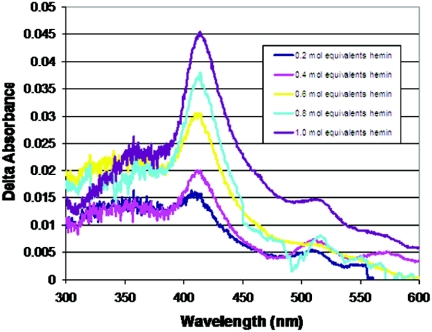
Since HbrL purified from cultures in the absence of 5-aminolevulinic acid has low amounts of heme, it allowed us to undertake heme titration assays to quantitate heme binding (Fig. 6). The addition of hemin to the HbrL apoprotein results in an increase in the Soret band at approximately 413 nm, similar to that observed in overexpression lysates grown in the presence of 5-aminolevulinic acid (Fig. 5B). This demonstrates that HbrL binds heme in vitro. Pyridine hemochrome quantification of bound heme provides an estimation of an E413 of 80,000 M−1 cm−1. From these values, we estimate that HbrL purified from cultures that contain 5-aminolevulinic acid contains approximately 0.4 to 0.5 mol of heme per mole of overexpressed HbrL apoprotein.
FIG. 6.
Titration of HbrL by exogenous addition of hemin. Affinity-purified HbrL-H6 (6.4 μM) was titrated by the addition of hemin (64 μM in DMSO) in 0.2 mol equivalents. Spectra were collected for HbrL-H6 and a control sample of lysozyme (6.4 μM); data presented are the difference spectra (HbrL minus lysozyme).
Gel shift analysis.
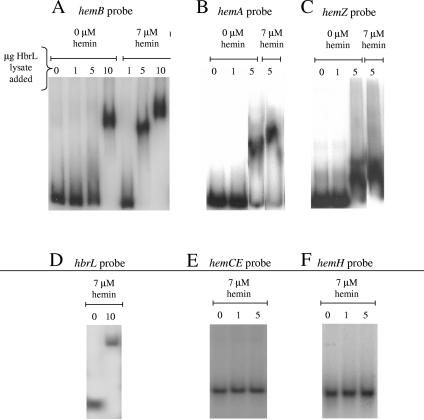
Isolated HbrL frequently aggregated under conditions used for performing DNA-binding assays. Consequently, we assayed for HbrL binding to heme promoters by performing an electrophoretic gel mobility shift assay with HbrL overexpression lysates using probes that span previously defined promoter regions for hemA, hemB, hemZ, and hbrL (44). The addition of HbrL lysate promotes a shift relative to the control cell lysate (Fig. 7). The addition of chymotrypsin or boiling of the lysate sample abolishes the observed shift (data not shown), so we attribute the shift to overexpressed HbrL protein. The addition of hemin to the cell lysate preparation also stimulates binding of HbrL to the hemB probe (Fig. 7A), although a shift is still observed at high concentrations of HbrL lysate in the absence of hemin. This is consistent with genetic evidence that HbrL acts as a repressor of hemB under conditions of heme sufficiency (Fig. 4). In this scenario, hemB expression would be reduced by repressive binding of an HbrL-hemin complex.
FIG. 7.
Electrophoretic mobility shift. Electrophoretic mobility shift assay of HbrL-H6 using hemA, hemB, hemZ, hbrL, hemCE, and hemH probes is shown. (A) Electrophoretic mobility of hemB probe with increasing amounts of HbrL lysate added in the presence and absence of 7 μM hemin is shown. Ten micrograms of Bl21(DE3) lysate was added to lanes marked “0.” (B) Electrophoretic mobility of hemA probe with increasing amounts of HbrL lysate added in the presence and absence of 7 μM hemin is shown. (C) Electrophoretic mobility of hemZ probe with increasing amounts of HbrL lysate added in the presence and absence of 7 μM hemin is shown. (D) Electrophoretic mobility of hbrL probe in the presence of 7 μM hemin with increasing amounts of HbrL lysate added is shown. (E) Electrophoretic mobility of hemCE probe in the presence of 7 μM hemin with increasing amounts of HbrL lysate added is shown. (F) Electrophoretic mobility of hemH probe in the presence of 7 μM hemin with increasing amounts of HbrL lysate added is shown.
The mobility of the hemA and hemZ probes is also decreased by the addition of HbrL lysate; however, this mobility does not appear to be sensitive to hemin addition (Fig. 7B and C). This is also consistent with LTTR family members that function as activators of expression in the presence of coinducers. In this scenario, LTTR members bind DNA in the presence and absence of a coinducer, but the LTTR stimulates transcription only when the coinducer is absent. We conclude that HbrL acts as an activator of hemA and hemZ expression under conditions of heme insufficiency.
As a negative control, we also examined whether HbrL was capable of binding to the hemH and the hemCE promoter regions, since expression of these genes is not affected by the disruption of hbrL in vivo. As shown in Fig. 7E and F, no binding of HbrL to these promoter sequences was observed (Fig. 7).
LTTRs are commonly autoregulatory, so we also evaluated the binding of HbrL protein with a probe for its own promoter region. HbrL lysate decreases the apparent mobility of the hbrL probe, as shown in Fig. 7D. We therefore conclude that HbrL is similarly autoregulatory, as is the case for other LTTRs, reducing its own expression to limit the total number of HbrL proteins in the cell.
DISCUSSION
Regulation of hem gene expression by heme and HbrL.
Heme is known to be an important regulator of heme synthesis, especially at early stages of the heme biosynthetic pathway. For example, heme is an allosteric regulator of ALA synthase activity in many organisms (9). Glutamyl-tRNA reductase is also regulated in response to heme sufficiency in Salmonella enterica serovar Typhimurium (51, 52). The results of our hemin addition study indicate that the regulation of hem gene expression in response to cellular levels of heme is also an important contributor to the regulation of heme biosynthesis. With the exception of hemH (the gene encoding ferrochelatase), the expression of all the tested hem genes is significantly reduced upon the addition of hemin. HbrL is clearly responsible for the regulation of several hem genes in response to heme. We speculate that HbrL acts as a repressor of multiple hem genes, because this provides the cell with a means to rapidly and globally repress tetrapyrrole synthesis when there is a heme surplus. It functions as an activator of hemA and hemZ expression in the absence of heme and as a repressor of hemB expression in the presence of heme (Fig. 4). Other uncharacterized factors are also involved, since hemC and hemE expression is still repressed in response to hemin addition even in an HbrL-disrupted strain.
We previously demonstrated that hem gene expression is regulated in response to changes in cellular redox in R. capsulatus, with many hem genes elevating their expression levels as these cells undergo a growth shift from aerobic to anaerobic environments (44). An anaerobic increase in hem gene expression presumably reflects the need for elevated tetrapyrrole biosynthesis in response to ramping up bacteriochlorophyll biosynthesis. The regulation of hem gene expression appears to be quite complex, involving the redox regulators RegB-RegA, CrtJ, and AerR (44) as well as the heme-specific regulator HbrL. Presumably, the transcriptional control by these transcription factors, coupled with allosteric feedback inhibition of enzyme activity, allows these cells to tightly coordinate the synthesis of heme with that of the other tetrapyrrole end products, cobalamin (B12) and bacteriochlorophyll. Indeed, heme and bacteriochlorophyll that are not complexed to apoproteins are not observed in these cells, suggesting that there is tight control of the synthesis of these end products along with synthesis of tetrapyrrole-binding apoproteins.
The identification of a heme-sensing transcription factor has precedence. In yeast and in mammalian cells, the binding of transcription factors HAP1 and Bach1, respectively, to target DNA is modulated by heme (24, 29, 61). In B. japonicum, hemB transcription is repressed by Irr, which is a conditionally stable protein that is degraded when it binds heme (35). Irr binds heme and is known to be physically associated with ferrochelatase (36). The binding of heme b by HbrL is inferred by the spectral presence of heme when HbrL expressed in the presence of 5-aminolevulinic acid, by a significantly red-shifted heme spectrum when hemin is added to HbrL apoprotein, and by changes in the DNA-binding properties observed upon heme addition.
HbrL is an LTTR regulator.
Inspection of the HbrL sequence indicates similarity to the LTTR family, whose canonical members exhibit several characteristics similar to those observed with HbrL. Most LTTRs regulate the expression of client promoters based on the presence or absence of a coinducer, typically an intermediary metabolite associated with the regulated pathway (42). In the case of HbrL, the end product heme appears to be a cofactor that regulates HbrL activity. Our observation that HbrL activates hemA and hemZ expression in the absence of heme is somewhat unusual, since LTTR proteins typically activate transcription only when a coinducer is bound. There is variability in the mode of regulation by LTTRs, with some LTTRs regulating transcription in the cell without any identified coinducer, as in the case of Nac (5) and SyrM (39). The regulation of hemB is more typical of LTTRs in that HbrL is required to repress hemB in the presence of heme. This type of regulation is common for LTTRs; for example, CysB repression of hslJ expression is enhanced by binding of the coinducer N-acetyl homoserine (21).
LTTRs have been shown to bind client promoters in both the presence and absence of inducers, with the DNase I protection pattern typically exhibiting a slight change, as is observed for OxyR (48), CysB (17, 18), and OccR (1, 2). Indeed, the observed electrophoretic gel mobility shift of probes specific for the hemA and hemZ promoters in the presence of lysate containing overexpressed HbrL is consistent with the binding of HbrL to these promoter regions. The addition of hemin does not affect the binding of HbrL to these promoters as measured with this assay, which is also typical for LTTRs. In contrast, the binding of HbrL to the hemB promoter is stimulated by the addition of heme (Fig. 7B). This is consistent with β-galactosidase reporter activity in which hemB expression is repressed by HbrL in wild-type cells only in the presence of heme (Fig. 4).
A BLAST search (3) for homologs of HbrL in genome databases resulted in the identification of a large number of LTTR family members. A subset of 14 LTTR proteins has a very high degree a similarity throughout the entire length of the protein, with a confidence value of 1e−6 to 1e−40 (Fig. 3). A phylogenetic analysis of the nearest 50 hits indicates that this subset forms a distinct clade with HbrL (data not shown), suggesting that they may be true homologs that have similar regulatory functions in these other species. The highest conserved region is the N terminus, which constitutes a helix-turn-helix domain that is involved in DNA binding (Fig. 3) (26, 32, 42). This is followed by the central domain that is thought to be involved in the binding of a coinducer. The crystal structure for the cofactor-binding fragment of CysB has been solved (50) and shows that the central region folds into two domains that form a cofactor binding pocket. There are numerous conserved aromatic resides in the putative cofactor binding pocket (W85, F93, W178, W181, Y196, H198, F199, Y200, F235, F237, and W260), which could be a potential binding site for a heme coinducer. Of particular note, the highly conserved sequence (Y/F)EHFY that has similarity to known heme b-binding domains utilizes His as a ligand to the centrally coordinated Fe in heme (14). This is clearly a candidate region for heme binding to HbrL.
Additional studies of HbrL, including the definition of the HbrL operator and regulon by DNase I footprint analysis and heme-binding properties of HbrL by site-directed mutagenesis of conserved residues, are under way. Comparative studies of HbrL orthologs from other species of bacteria should also prove useful for elucidating the contribution of HbrL to the regulation of tetrapyrrole synthesis in various prokaryotes.
Acknowledgments
This study was supported by National Institutes of Health grant GM53940.
We thank Aaron Setterdahl for careful reading of the manuscript.
REFERENCES
- 1.Akakura, R., and S. C. Winans. 2002. Constitutive mutations of the OccR regulatory protein affect DNA bending in response to metabolites released from plant tumors. J. Biol. Chem. 277:5866-5874. [DOI] [PubMed] [Google Scholar]
- 2.Akakura, R., and S. C. Winans. 2002. Mutations in the occQ operator that decrease OccR-induced DNA bending do not cause constitutive promoter activity. J. Biol. Chem. 277:15773-15780. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Barany, F. 1985. Single-stranded hexameric linkers: a system for in-phase insertion mutagenesis and protein engineering. Gene 37:111-123. [DOI] [PubMed] [Google Scholar]
- 5.Bender, R. A. 1991. The role of the NAC protein in the nitrogen regulation of Klebsiella aerogenes. Mol. Microbiol. 5:2575-2580. [DOI] [PubMed] [Google Scholar]
- 6.Berry, E. A., and B. L. Trumpower. 1987. Simultaneous determination of hemes a, b, and c from pyridine hemochrome spectra. Anal. Biochem. 161:1-15. [DOI] [PubMed] [Google Scholar]
- 7.Blackmon, B. J., T. A. Dailey, X. Lianchun, and H. A. Dailey. 2002. Characterization of a human and mouse tetrapyrrole-binding protein. Arch. Biochem. Biophys. 407:196-201. [DOI] [PubMed] [Google Scholar]
- 8.Buggy, J., and C. E. Bauer. 1995. Cloning and characterization of senC, a gene involved in both aerobic respiration and photosynthesis gene expression in Rhodobacter capsulatus. J. Bacteriol. 177:6958-6965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burnham, B. F., and J. Lascelles. 1963. Control of porphyrin biosynthesis through a negative-feedback mechanism. Studies with preparations of δ-aminolaevulate synthetase and δ-aminolaevulate dehydratase from Rhodopseudomonas spheroides. Biochem. J. 87:462-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang, M., and I. P. Crawford. 1990. The roles of indoleglycerol phosphate and the TrpI protein in the expression of trpBA from Pseudomonas aeruginosa. Nucleic Acids Res. 18:979-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong, C., S. Elsen, L. R. Swem, and C. E. Bauer. 2002. AerR, a second aerobic repressor of photosynthesis gene expression in Rhodobacter capsulatus. J. Bacteriol. 184:2805-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubbs, P., J. M. Dubbs, and F. R. Tabita. 2004. Effector-mediated interaction of CbbRI and CbbRII regulators with target sequences in Rhodobacter capsulatus. J. Bacteriol. 186:8026-8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esposti, M. D. 1989. Prediction and comparison of the haem-binding sites in membrane haemoproteins. Biochim. Biophys. Acta 977:249-265. [DOI] [PubMed] [Google Scholar]
- 15.Hamza, I., S. Chauhan, R. Hassett, and M. R. O'Brian. 1998. The bacterial irr protein is required for coordination of heme biosynthesis with iron availability. J. Biol. Chem. 273:21669-21674. [DOI] [PubMed] [Google Scholar]
- 16.Henikoff, S., and J. G. Henikoff. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89:10915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hryniewicz, M. M., and N. M. Kredich. 1995. Hydroxyl radical footprints and half-site arrangements of binding sites for the CysB transcriptional activator of Salmonella typhimurium. J. Bacteriol. 177:2343-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hryniewicz, M. M., and N. M. Kredich. 1994. Stoichiometry of binding of CysB to the cysJIH, cysK, and cysP promoter regions of Salmonella typhimurium. J. Bacteriol. 176:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inuzuka, T., B. G. Yun, H. Ishikawa, S. Takahashi, H. Hori, R. L. Matts, K. Ishimori, and I. Morishima. 2004. Identification of crucial histidines for heme binding in the N-terminal domain of the heme-regulated eIF2alpha kinase. J. Biol. Chem. 279:6778-6782. [DOI] [PubMed] [Google Scholar]
- 20.Izadi, N., Y. Henry, J. Haladjian, M. E. Goldberg, C. Wandersman, M. Delepierre, and A. Lecroisey. 1997. Purification and characterization of an extracellular heme-binding protein, HasA, involved in heme iron acquisition. Biochemistry 36:7050-7057. [DOI] [PubMed] [Google Scholar]
- 21.Jovanovic, M., M. Lilic, D. J. Savic, and G. Jovanovic. 2003. The LysR-type transcriptional regulator CysB controls the repression of hslJ transcription in Escherichia coli. Microbiology 149:3449-3459. [DOI] [PubMed] [Google Scholar]
- 22.Kastaniotis, A. J., and R. S. Zitomer. 2000. Rox1 mediated repression. Oxygen dependent repression in yeast. Adv. Exp. Med. Biol. 475:185-195. [PubMed] [Google Scholar]
- 23.Kim, J., J. G. Kim, Y. Kang, J. Y. Jang, G. J. Jog, J. Y. Lim, S. Kim, H. Suga, T. Nagamatsu, and I. Hwang. 2004. Quorum sensing and the LysR-type transcriptional activator ToxR regulate toxoflavin biosynthesis and transport in Burkholderia glumae. Mol. Microbiol. 54:921-934. [DOI] [PubMed] [Google Scholar]
- 24.Lan, C., H. C. Lee, S. Tang, and L. Zhang. 2004. A novel mode of chaperone action: heme activation of Hap1 by enhanced association of Hsp90 with the repressed Hsp70-Hap1 complex. J. Biol. Chem. 279:27607-27612. [DOI] [PubMed] [Google Scholar]
- 25.Lascelles, J. 1960. The synthesis of enzymes concerned in bacteriochlorophyll formation in growing cultures of Rhodopseudomonas spheroides. J. Gen. Microbiol. 23:487-498. [DOI] [PubMed] [Google Scholar]
- 26.Lochowska, A., R. Iwanicka-Nowicka, D. Plochocka, and M. M. Hryniewicz. 2001. Functional dissection of the LysR-type CysB transcriptional regulator. Regions important for DNA binding, inducer response, oligomerization, and positive control. J. Biol. Chem. 276:2098-2107. [DOI] [PubMed] [Google Scholar]
- 27.Monson, E. K., G. S. Ditta, and D. R. Helinski. 1995. The oxygen sensor protein, FixL, of Rhizobium meliloti. Role of histidine residues in heme binding, phosphorylation, and signal transduction. J. Biol. Chem. 270:5243-5250. [DOI] [PubMed] [Google Scholar]
- 28.Monson, E. K., M. Weinstein, G. S. Ditta, and D. R. Helinski. 1992. The FixL protein of Rhizobium meliloti can be separated into a heme-binding oxygen-sensing domain and a functional C-terminal kinase domain. Proc. Natl. Acad. Sci. USA 89:4280-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogawa, K., J. Sun, S. Taketani, O. Nakajima, C. Nishitani, S. Sassa, N. Hayashi, M. Yamamoto, S. Shibahara, H. Fujita, and K. Igarashi. 2001. Heme mediates derepression of Maf recognition element through direct binding to transcription repressor Bach1. EMBO J. 20:2835-2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palma, J. F., X. Gao, C. H. Lin, S. Wu, and W. B. Solomon. 1994. Iron protoporphyrin IX (hemin) but not tin or zinc protoporphyrin IX can stimulate gene expression in K562 cells from enhancer elements containing binding sites for NF-E2. Blood 84:1288-1297. [PubMed] [Google Scholar]
- 31.Park, W., P. Padmanabhan, S. Padmanabhan, G. J. Zylstra, and E. L. Madsen. 2002. nahR, encoding a LysR-type transcriptional regulator, is highly conserved among naphthalene-degrading bacteria isolated from a coal tar waste-contaminated site and in extracted community DNA. Microbiology 148:2319-2329. [DOI] [PubMed] [Google Scholar]
- 32.Parsek, M. R., R. W. Ye, P. Pun, and A. M. Chakrabarty. 1994. Critical nucleotides in the interaction of a LysR-type regulator with its target promoter region. catBC promoter activation by CatR. J. Biol. Chem. 269:11279-11284. [PubMed] [Google Scholar]
- 33.Ponnampalam, S. N., and C. E. Bauer. 1997. DNA binding characteristics of CrtJ. A redox-responding repressor of bacteriochlorophyll, carotenoid, and light harvesting-II gene expression in Rhodobacter capsulatus. J. Biol. Chem. 272:18391-18396. [DOI] [PubMed] [Google Scholar]
- 34.Ponnampalam, S. N., J. J. Buggy, and C. E. Bauer. 1995. Characterization of an aerobic repressor that coordinately regulates bacteriochlorophyll, carotenoid, and light harvesting-II expression in Rhodobacter capsulatus. J. Bacteriol. 177:2990-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi, Z., I. Hamza, and M. R. O'Brian. 1999. Heme is an effector molecule for iron-dependent degradation of the bacterial iron response regulator (Irr) protein. Proc. Natl. Acad. Sci. USA 96:13056-13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi, Z., and M. R. O'Brian. 2002. Interaction between the bacterial iron response regulator and ferrochelatase mediates genetic control of heme biosynthesis. Mol. Cell 9:155-162. [DOI] [PubMed] [Google Scholar]
- 37.Reddy, S. V., O. Alcantara, G. D. Roodman, and D. H. Boldt. 1996. Inhibition of tartrate-resistant acid phosphatase gene expression by hemin and protoporphyrin IX. Identification of a hemin-responsive inhibitor of transcription. Blood 88:2288-2297. [PubMed] [Google Scholar]
- 38.Rigali, S., A. Derouaux, F. Giannotta, and J. Dusart. 2002. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 277:12507-12515. [DOI] [PubMed] [Google Scholar]
- 39.Rushing, B. G., M. M. Yelton, and S. R. Long. 1991. Genetic and physical analysis of the nodD3 region of Rhizobium meliloti. Nucleic Acids Res. 19:921-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Russell, D. A., G. A. Byrne, E. P. O'Connell, C. A. Boland, and W. G. Meijer. 2004. The LysR-type transcriptional regulator VirR is required for expression of the virulence gene vapA of Rhodococcus equi ATCC 33701. J. Bacteriol. 186:5576-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Schell, M. A. 1993. Molecular biology of the LysR family of transcriptional regulators. Annu. Rev. Microbiol. 47:597-626. [DOI] [PubMed] [Google Scholar]
- 43.Sganga, M. W., and C. E. Bauer. 1992. Regulatory factors controlling photosynthetic reaction center and light-harvesting gene expression in Rhodobacter capsulatus. Cell 68:945-954. [DOI] [PubMed] [Google Scholar]
- 44.Smart, J. L., J. W. Willett, and C. E. Bauer. 2004. Regulation of hem gene expression in Rhodobacter capsulatus by redox and photosystem regulators RegA, CrtJ, FnrL, and AerR. J. Mol. Biol. 342:1171-1186. [DOI] [PubMed] [Google Scholar]
- 45.Sperandio, B., P. Polard, D. S. Ehrlich, P. Renault, and E. Guedon. 2005. Sulfur amino acid metabolism and its control in Lactococcus lactis IL1403. J. Bacteriol. 187:3762-3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Swem, D. L., and C. E. Bauer. 2002. Coordination of ubiquinol oxidase and cytochrome cbb3 oxidase expression by multiple regulators in Rhodobacter capsulatus. J. Bacteriol. 184:2815-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swem, L. R., S. Elsen, T. Bird, D. Swem, H. Koch, H. Myllykallio, F. Daldal, and C. Bauer. 2001. The RegB/RegA two-component regulatory system controls synthesis of photosynthesis and respiratory electron transfer components in Rhodobacter capsulatus. J. Mol. Biol. 309:121-138. [DOI] [PubMed] [Google Scholar]
- 48.Toledano, M. B., I. Kullik, F. Trinh, P. T. Baird, T. D. Schneider, and G. Storz. 1994. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell 78:897-909. [DOI] [PubMed] [Google Scholar]
- 49.Turlin, E., O. Sismeiro, J. P. Le Caer, V. Labas, A. Danchin, and F. Biville. 2005. 3-Phenylpropionate catabolism and the Escherichia coli oxidative stress response. Res. Microbiol. 156:312-321. [DOI] [PubMed] [Google Scholar]
- 50.Tyrrell, R., K. H. Verschueren, E. J. Dodson, G. N. Murshudov, C. Addy, and A. J. Wilkinson. 1997. The structure of the cofactor-binding fragment of the LysR family member, CysB: a familiar fold with a surprising subunit arrangement. Structure 5:1017-1032. [DOI] [PubMed] [Google Scholar]
- 51.Wang, L., M. Elliott, and T. Elliott. 1999. Conditional stability of the HemA protein (glutamyl-tRNA reductase) regulates heme biosynthesis in Salmonella typhimurium. J. Bacteriol. 181:1211-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang, L., S. Wilson, and T. Elliott. 1999. A mutant HemA protein with positive charge close to the N terminus is stabilized against heme-regulated proteolysis in Salmonella typhimurium. J. Bacteriol. 181:6033-6041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Watanabe, T., H. Fujihara, and K. Furukawa. 2003. Characterization of the second LysR-type regulator in the biphenyl-catabolic gene cluster of Pseudomonas pseudoalcaligenes KF707. J. Bacteriol. 185:3575-3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wright, M. S., R. D. Cardin, and A. J. Biel. 1987. Isolation and characterization of an aminolevulinate-requiring Rhodobacter capsulatus mutant. J. Bacteriol. 169:961-966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wright, M. S., J. J. Eckert, S. W. Biel, and A. J. Biel. 1991. Use of a lacZ fusion to study transcriptional regulation of the Rhodobacter capsulatus hemA gene. FEMS Microbiol. Lett. 62:339-342. [DOI] [PubMed] [Google Scholar]
- 56.Yang, Z. M., and C. E. Bauer. 1990. Rhodobacter capsulatus genes involved in early steps of the bacteriochlorophyll biosynthetic pathway. J. Bacteriol. 172:5001-5010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yeh, K. C., M. C. Peck, and S. R. Long. 2002. Luteolin and GroESL modulate in vitro activity of NodD. J. Bacteriol. 184:525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yen, H. C., N. T. Hu, and B. L. Marrs. 1979. Characterization of the gene transfer agent made by an overproducer mutant of Rhodopseudomonas capsulata. J. Mol. Biol. 131:157-168. [DOI] [PubMed] [Google Scholar]
- 59.Yen, H. C., and B. Marrs. 1976. Map of genes for carotenoid and bacteriochlorophyll biosynthesis in Rhodopseudomonas capsulata. J. Bacteriol. 126:619-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Young, D. A., C. E. Bauer, J. C. Williams, and B. L. Marrs. 1989. Genetic evidence for superoperonal organization of genes for photosynthetic pigments and pigment-binding proteins in Rhodobacter capsulatus. Mol. Gen. Genet. 218:1-12. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, L., and L. Guarente. 1994. HAP1 is nuclear but is bound to a cellular factor in the absence of heme. J. Biol. Chem. 269:14643-14647. [PubMed] [Google Scholar]