Abstract
Glutathione is a tripeptide and antioxidant, synthesized at high levels by cells during the production of reactive oxygen and nitrogen intermediates. Glutathione also serves as a carrier molecule for nitric oxide in the form of S-nitrosoglutathione. Previous studies from this laboratory have shown that glutathione and S-nitrosoglutathione are directly toxic to mycobacteria. Glutathione is not transported into the cells as a tripeptide. Extracellular glutathione is converted to a dipeptide due to the action of transpeptidase, and the dipeptide is then transported into the bacterial cells. The processing of glutathione and S-nitrosoglutathione is brought about by the action of the enzyme γ-glutamyl transpeptidase. The function of γ-glutamyl transpeptidase is to cleave glutathione and S-nitrosoglutathione to the dipeptide (Cys-Gly), which is then transported into the bacterium by the multicomponent ABC transporter dipeptide permease. We have created a mutant strain of Mycobacterium tuberculosis lacking this metabolic enzyme. We investigated the sensitivity of this strain to glutathione and S-nitrosoglutathione compared to that of the wild-type bacteria. In addition, we examined the role of glutathione and/or S-nitrosoglutathione in controlling the growth of intracellular M. tuberculosis inside mouse macrophages.
Mycobacterium tuberculosis accounts for the largest number of deaths caused by a single human pathogen (5). It is estimated that there will be 80 million new cases and 20 million deaths from tuberculosis in the coming decade (5). The appearance of drug-resistant strains of M. tuberculosis and the human immunodeficiency virus pandemic has exacerbated this situation (5).
Glutathione (GSH) is a tripeptide comprised of γ-glutamate, cysteine, and glycine. It is an antioxidant found in all cells of the body, including macrophages, which are considered major phagocytic cells involved in protection against M. tuberculosis (28, 29). However, mycobacteria do not produce GSH. Instead, they have mycothiols for regulating their reduction or oxidation activities (1, 23).
Reactive oxygen intermediates (ROI) and reactive nitrogen intermediates (RNI), generated by phagocytic cells, are thought to play an important role in inhibiting growth of intracellular pathogens (9, 13, 22). When host cells, such as macrophages, generate ROI and RNI, there will be simultaneous synthesis of GSH for protection against the toxic effects ofROI and RNI. Nitric oxide (NO) has been shown to inhibit the growth of M. tuberculosis in the murine system (7, 8, 11, 19). NO also reacts with GSH to form S-nitrosoglutathione (GSNO). GSNO, an NO donor, can then release NO, leading to the death of the pathogen (12, 24). The stability of NO is increased when it is in complex with GSH. We have previously shown that M. tuberculosis is sensitive to 5 mM GSH and 5 mM GSNO (14, 30). The sensitivity of M. tuberculosis to GSNO is due to the bactericidal effects of NO released from the GSNO complex. The mechanism of action of antimycobacterial activity of GSH is uncertain. Since mycobacteria do not synthesize GSH, we predict that the exposure of mycobacteria to high concentrations of GSH may create a redox imbalance leading to growth inhibition.
The mechanisms of processing and transport of GSH/GSNO have been successfully worked out in Salmonella enterica serovar Typhimurium, where GSNO is first processed by the action of the γ-glutamyl transpeptidase (GgtA) enzyme into a dipeptide [Cys(NO)-Gly] and glutamate, after which Cys(NO)-Gly enters the bacterium via dipeptide permease (12, 18). In this study, we have demonstrated that the γ-glutamyl transpeptidase mutant of M. tuberculosis (the GGTA mutant) is resistant to the toxic effects of GSH/GSNO and shows increased survival inside macrophages compared to the wild-type strain.
MATERIALS AND METHODS
Sensitivity of M. tuberculosis H37Rv to GSH and GSNO.
M. tuberculosis H37Rv was grown to mid-log phase in 7H9 medium containing albumin dextrose complex (ADC) and Tween and then diluted to an optical density (OD) of approximately 0.1. The bacterial suspension was then treated with 5 mM GSH (Sigma) or 5 mM GSNO (Sigma) or left untreated. The optical density was measured everyday for 5 days. Each day, an aliquot was taken from every sample, diluted to 100-fold, and plated on 7H11 plates containing ADC to determine the number of CFU. All experiments were performed three times in triplicate.
Maintenance of J774.1 cell line.
The J774.1 cell line was maintained in Dulbecco modified Eagle medium (DMEM; Sigma) containing 10% fetal bovine serum (FBS; Sigma), 2 mM glutamine, and essential amino acids (Gibco BRL).
Construction of mutants.
The ggtA mutant was constructed by means of an allelic exchange via mycobacteriophage delivery (2, 3, 6). Briefly, the N and C termini of the ggtA gene were amplified using the following primers: N-forward, 5′-AGCTATGGTACCGGACGGAGGCAGCAGCTA-3′; N-reverse, 5′-AGCTATTCTAGAACCGTCACGGAGTTCCAG-3′; C-forward, 5′-AGCTATCTCGAGACACCATCCACCAGATACCG-3′; and C-reverse, 5′-AGCTATACTAGTCTCCCGAACTGGCTGTAGTC-3′.
The N and C fragments were cloned into pYUB854 (kindly provided by W. Jacobs) via two multiple cloning sites separated by a hygromycin resistance cassette. The cosmid and self-ligated mycobacteriophage DNA were then linearized with PacI. The cosmid was subsequently ligated into the phage DNA, and the ligation mix was packaged into λ-phage heads using λ-packaging extracts from Stratagene, followed by transduction using Escherichia coli HB101. Phasmid DNA was isolated from hygromycin-resistant clones and electroporated into Mycobacterium smegmatis mc2155. Electroporated cells were mixed with top agar and poured onto 7H11 plates and incubated at 30°C. A single plaque was chosen to confirm the mycobacteriophage temperature-sensitive phenotype and to produce a high-titer lysate (1010 to 1011 PFU/ml). H37Rv was then infected with the mycobacteriophage carrying the allelic exchange construct at a multiplicity of infection (MOI) of 10 at 37°C. The phage cell mix was incubated in a heating block for 4 to 6 h at 39°C. The mix was then centrifuged for 10 min at 10,000 rpm and resuspended in 2 ml 7H9. Cells (0.2 ml) were plated on 7H11 plates supplemented with hygromycin (75 μg/ml). After 3 weeks, colonies were picked and inoculated into 5 ml 7H9 and allowed to grow at 37°C. Genomic DNA was isolated, and confirmation of the mutants was done via PCR. Primers were designed such that the oligonucleotides were outside of the region cloned in pYUB854 for allelic exchange: F1, 5′-CCGGGAAGATGACCAACTG-3′; R1, 5′-TCAGGAACACCTTCTTCACG-3′; F2, 5′-CATGGGAAGCTTCGTGGAT-3′; and R2, 5′-CCTCAGTTCAATTCGGTTGC-3′.
Of the 12 colonies screened, one colony showed amplification products consistent with allelic replacement. The complemented strain (the cGGTA mutant) was constructed by inserting the ggtA gene, with its own transcriptional and translational signals, into plasmid pMV305p, an integrating vector that exploits the L5 mycobacteriophage integration system containing a kanamycin resistance marker (17). The construct was then electroporated into the ggtA-deleted strain, and colonies were selected on 7H11 plates supplemented with both hygromycin (75 μg/ml) and kanamycin (50 μg/ml).
Southern blot analysis.
Genomic DNA was prepared from wild-type H37Rv, the GGTA mutant strain, and the cosmid containing the pYUB854 vector and N- and C-terminal regions of the ggtA gene. One microgram of DNA was cut with AgeI, and the DNA fragments were separated on a 1% agarose gel and transferred onto a nylon membrane. The probe used was a 500-base-pair fragment of the hygromycin resistance cassette or a 200-base-pair region representing the cloned region of the ggtA gene. The primers used to make these probes are HygF (5′-CATAGACGTCGGTGAAGTCG-3′) and HygR (5′-GGCCCTACCTGGTGATGAG-3′) for the 500-bp hygromycin probe and DggtL (5′-CAACGGCTTTCTGGTCTCAC-3′) and DggtR (5′-CTCTCCCCGGTAGAACTCCT-3′) for the 200-bp internal ggtA probe. The probe (100 ng) was radiolabeled with 32P using the HexaLabel DNA labeling kit (Fermentas Life Sciences) and was then used to probe the nylon membrane overnight. The blot was exposed to an autoradiography film and developed.
γ-Glutamyl transpeptidase enzyme assay.
M. tuberculosis (H37Rv and the GGTA and cGGTA mutant strains) was grown to an OD of 1.0, and 1/10 of the volume of cells was added to the reaction buffer for a final A600 of 0.1. The reaction buffer consisted of 20 mM glycine-glycine, 300 μM l-γ-glutamyl-p-nitroanilide, and 60 mM Tris (pH 8.0). Reactions proceeded at 37°C for 60 min, and the release of p-nitroanilide was monitored at A405.
Disk diffusion susceptibility assay.
The Bauer-Kirby disk diffusion method was used to determine the susceptibility of H37Rv and the GGTA and cGGTA mutants to GSH and GSNO (4). Thirty microliters of either 5 mM GSH or 5 mM GSNO was added daily to a 1/4" paper disk placed over a lawn of 106 bacteria spread over a 7H11 agar plate until a zone of inhibition was observed and measured.
Sensitivity assays.
Mycobacterial strains (H37Rv and the GGTA and cGGTA mutant strains) were grown to mid-log phase in 7H9 medium supplemented with ADC and Tween 80 and then diluted to an OD of approximately 0.1. The bacterial suspensions were treated with GSH or GSNO or left untreated. The OD was measured each day for 5 days. An aliquot was taken from every sample, diluted 100-fold, and plated on 7H11 agar for determination of the number of CFU. All experiments were performed three times in triplicate.
Processing of M. tuberculosis for infection.
All three strains of M. tuberculosis (H37Rv and the GGTA and cGGTA mutant strains) were grown in 7H9 medium containing ADC and Tween 80. Static cultures of mycobacteria at peak logarithmic phase of growth (between 0.5 and 0.8 at A600) were used for infection of macrophages. The bacterial suspension was diluted to a 50-ml volume in phosphate-buffered saline (PBS; Sigma) and centrifuged at 3,000 rpm for 15 min at room temperature. The bacterial pellet was resuspended in RPMI medium containing 10% FBS. Bacterial clumps were disaggregated by vortexing five times with 3-mm sterile glass beads (Fisher Scientific). The duration of each cycle was approximately 2 min. The bacterial suspension was passed through a 5-μm filter (Micron Separations, Inc.) to remove any further clumps. Processed mycobacteria were frozen in −80°C as aliquots and plated in parallel on 7H11 medium to determine the number of organisms in the stock. At the time of macrophage infection, frozen stocks of mycobacteria were thawed and used for infection.
Isolation of peritoneal macrophages from wild-type mice.
Peritoneal macrophages were isolated from 8-week-old C57BL/6 mice (Taconic Laboratory). Five milliliters of sterile 3% thioglycolate (Sigma) solution was injected into the peritoneal cavities of the animals. Four days after injection, animals were sacrificed via cervical dislocation and ice-cold PBS was injected into the peritoneal cavity. The abdomens of the animals were gently massaged several times, and peritoneal lavage fluid was recovered.
Macrophage infection.
J774.1 and mouse peritoneal macrophages were maintained in vitro, as described above. Macrophages (5 × 105/well) were infected separately with H37Rv and the GGTA and cGGTA mutants at an MOI of 10:1. Macrophages were incubated with mycobacteria for 2 h (to allow for phagocytosis), after which extracellular organisms were removed by washing with serum-free medium. Macrophages were stimulated for GSNO synthesis as follows. Infected macrophages were maintained in tissue culture medium with and without gamma interferon (IFN-γ) (100 U/ml) and lipopolysaccharide (LPS) (1 μg/ml). IFN-γ and LPS stimulation will result in NO production by macrophages and subsequent formation of GSNO. Infected J774.1 cells were terminated at 1 h and 72 h after infection and infected peritoneal macrophage cultures were terminated at 24 h and 5 days after infection to study the intracellular viabilities of H37Rv and the GGTA and cGGTA mutants. Intracellular viability of mycobacteria inside macrophages was determined by lysing the infected macrophages with sterile distilled water and plating the lysates on 7H11 medium enriched with ADC for mycobacterial colony numeration.
Statistical analysis.
Data in the table are represented as means and standard errors. Statistical analysis of the data was carried out using the StatView program, and the statistical significance was determined using the unpaired t test. Differences were considered significant at a P value of <0.05 and were indicated by asterisks.
RESULTS
Construction and characterization of GGTA and cGGTA mutant strains.
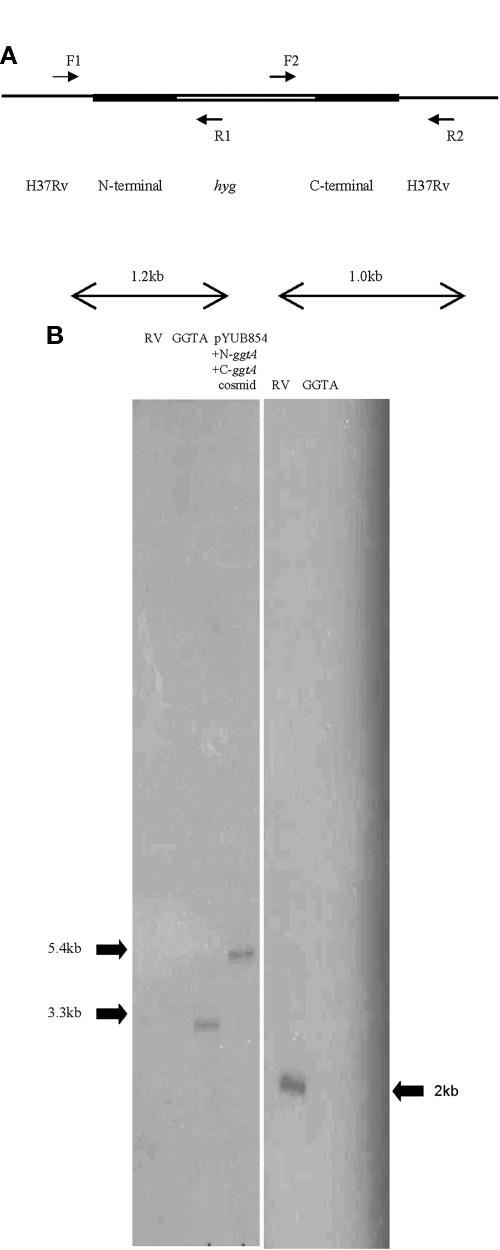
There are two open reading frames (ORF) in the M. tuberculosis genome that encode γ-glutamyl transpeptidase. The ggtB ORF is 2 kb and is designated Rv2394. The ggtA ORF (900 bp; Rv0773c) has 40% identity to the ggt gene of Bacillus subtilis in a 516-amino-acid overlap. We have successfully interrupted the ggtA gene with a hygromycin resistance cassette via a mycobacteriophage delivery system (6). Figure 1A depicts two sets of primers, F1 and R1 and F2 and R2 (Materials and Methods), used to amplify the N and C termini, respectively, of the ggtA gene, including the flanking regions of the crossover event. These primers were designed to include, for either the N or C terminus, wild-type sequences outside of the cloned region and also sequences from the allelic exchange cosmid pYUB854, including a portion of the hygromycin resistance cassette. PCRs resulted in single distinct bands representing the 1.2-kb and 1.0-kb amplified regions as seen on a 1% agarose gel (data not shown). To confirm successful crossover events, Southern analysis was done (Fig. 1B). Genomic DNA from wild-type H37Rv, the GGTA mutant, and the cosmid was digested with AgeI and probed with a 500-bp DNA fragment representing a fragment of the hygromycin cassette (Fig. 1B, left panel). The probe hybridized once to both the fragment from the GGTA mutant DNA and the cosmid DNA but not to the wild-type H37Rv DNA. In addition, AgeI-digested DNA from wild-type H37Rv and the GGTA mutant was probed with a 200-base-pair fragment representing the internal cloned region of the ggtA gene (Fig. 1B, right panel); this is the region deleted by the crossover event. The probe hybridized to the DNA from wild-type H37Rv but not to the DNA from the mutant lacking the ggtA gene. These results indicate that the hygromycin resistance marker recombined successfully onto the chromosome to create the GGTA mutant strain.
FIG.1.
(A) Schematic diagram showing the region of homologous recombination in H37Rv to create a mutant lacking the ggtA gene and the regions amplified using the primers F1 and R1 (N terminus) and F2 and R2 (C terminus). The regions amplified include sequences from the wild-type host DNA and vector DNA. (B) Southern analysis of genomic DNA from H37Rv, the GGTA mutant, or cosmid digested with AgeI and probed with a 500-bp fragment from the hygromycin resistance cassette (left panel) or a 200-bp fragment representing the internal cloned region of ggtA (right panel).
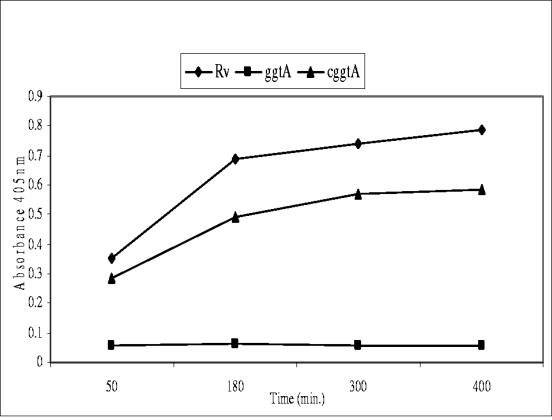
To assess whether the interruption of the ggtA gene alone results in loss of enzyme activity, we employed a γ-glutamyl transpeptidase enzyme assay (Fig. 2) (21). Interruption of the ggtA gene resulted in almost complete loss of enzyme activity compared to that of wild-type H37Rv, while introduction of a wild-type copy of the ggtA gene into the mutant strain restored most of its activity. Since most of the enzyme activity was lost in the GGTA mutant, we did not mutate the second ORF, ggtB.
FIG. 2.
γ-Glutamyl transpeptidase assay measuring p-nitroanilide released when l-γ-glutamyl-p-nitroanilide and glycine-glycine were added to cultures of H37Rv (diamonds) and the GGTA (squares) and cGGTA (triangles) mutants at 37°C. Error bars are too small to be detectable.
Sensitivity of M. tuberculosis H37Rv and the GGTA and cGGTA mutants to GSH and GSNO.
We have reported that 5 mM GSH is bacteriostatic to Mycobacterium bovis BCG and to H37Rv but that 5 mM GSNO is bactericidal (14, 30). In this study, we treated the GGTA and cGGTA mutant strains with 5 mM GSH and 5 mM GSNO and measured the strains' sensitivities to these compounds via OD during growth in liquid medium (Fig. 3A and B), number of CFU (Fig. 3C and D), and a disk diffusion assay (Fig. 4). Figure 3A shows that the GGTA mutant of H37Rv is resistant to 5 mM GSH while the wild-type strain and the complemented mutant are sensitive after 5 days in liquid growth. Similarly, the GGTA mutant is resistant to 5 mM GSNO (Fig. 3B). However, H37Rv and the cGGTA mutant not only exhibited sensitivity to GSNO but also were killed after 5 days. This is not surprising since NO released from GSNO leads to killing of the bacteria (14, 28). Figures 3C and D show a similar trend where the number of CFU increased over 5 days when the GGTA mutant was exposed to 5 mM GSH and 5 mM GSNO, respectively, while there was no growth of H37Rv or the cGGTA mutant under the same treatment. Our third method of measuring sensitivity was by disk diffusion. Figure 4 shows that the GGTA mutant was resistant to both GSH and GSNO compared to the wild-type and complemented mutant strains.
FIG.3.
Growth of M. tuberculosis H37Rv and the GGTA and cGGTA mutants in the presence and absence of GSH (A) and GSNO (B). Each strain was grown to mid-log phase of growth in Middlebrook 7H9 medium with ADC and Tween 80 and then diluted to an OD of approximately 0.1. The bacteria cell suspension was treated with 5 mM GSH or 5 mM GSNO or left untreated. The OD was measured every day for 5 days. Each day, an aliquot was taken from every sample, diluted to 100-fold, and plated on 7H11 plates containing ADC to determine the numbers of CFU for GSH (C) and GSNO (D). All experiments were repeated three times in triplicate.
FIG. 4.
Bauer-Kirby disk diffusion assay measuring the zone of inhibition of H37Rv and the GGTA and cGGTA mutants around a disk inoculated everyday with 30 μl of GSH or GSNO. *, significant at P < 0.05.
Intracellular survival of the GGTA mutant in J774.1 murine macrophages.
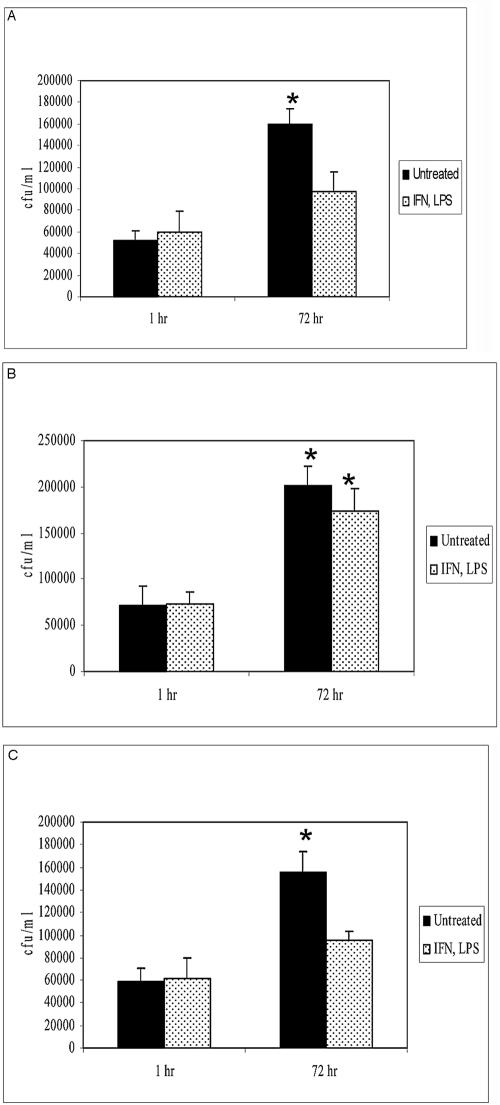
Mammalian cells contain levels of GSH in the range of 0.1 to 10 mM (15). Having demonstrated the GSH/GSNO-resistant phenotype of the GGTA mutant in vitro, we sought to examine the sensitivity to GSH/GSNO within macrophages. We looked at the survival of H37Rv and the GGTA and cGGTA mutants in J774.1 cells under conditions known to induce NO production. J774.1 macrophages were infected with M. tuberculosis strains and treated with IFN-γ (100 U/ml) and LPS (1 μg/ml). Treatment of murine macrophages with IFN-γ alone does not bring about enhanced synthesis of inducible nitric oxide synthase and generation of NO. J774.1 cells require tumor necrosis factor as a second signal; the addition of LPS to macrophage cultures results in tumor necrosis factor production (9). GSH within macrophages complexes with NO to form GSNO. Macrophage cultures were terminated at 1 h and 72 h after infection and treatment to analyze the growth of mycobacteria inside activated and nonactivated macrophages. At each time point, supernatants were collected, and macrophages were lysed with sterile distilled water. Both supernatants and macrophage lysates were plated on 7H11 medium with ADC to recover extracellular and intracellular bacteria, respectively. No significant levels of extracellular bacteria were detected in the macrophage supernatants. Figure 5 shows the results of these experiments. We observed a >3-fold increase in the number of CFU of wild-type H37Rv in unstimulated J774.1 macrophages between 1 h and 72 h, while stimulation with IFN-γ and LPS resulted in inhibition of growth of the intracellular H37Rv (Fig. 5A). In contrast, we observed significant growth of the GGTA mutant inside J774.1 macrophages in both stimulated and unstimulated macrophages (Fig. 5B). Since the GGTA mutant cannot process GSNO, there is no inhibition of bacterial growth in stimulated macrophages. The complemented strain behaved like the wild-type strain and demonstrated significant growth in unstimulated J774.1 macrophages and inhibited growth inside macrophages stimulated with IFN-γ and LPS (Fig. 5C).
FIG. 5.
Growth of H37Rv (A), the GGTA mutant (B), and the cGGTA mutant (C) in untreated and IFN-γ- and LPS-treated J774.1 cells. J774.1 cells were maintained in DMEM containing 10% FBS and then infected with bacteria at an MOI of 10:1. Two hours after infection, unphagocytosed organisms were removed. Mycobacterium-infected macrophages were maintained in DMEM containing 10% FBS in the absence and presence of IFN-γ (100 U/ml) plus LPS (1 μg/ml). Infected macrophages were terminated at 1 h and 72 h to determine intracellular growth of bacteria inside J774.1 cells. Intracellular colony counts were determined by plating lysed cultures on Middlebrook 7H11 medium. *, significant at P < 0.05.
Intracellular survival of the GGTA mutant in primary mouse peritoneal macrophages.
We further investigated the effects of GSH and GSNO in the macrophage killing of intracellular mycobacteria in the murine system by comparing the extents of survival of the GGTA mutant in primary macrophages derived from wild-type C57BL/6 mice. If GSH/GSNO is a primary species responsible for controlling mycobacterial growth in murine macrophages, we predict that the GGTA mutant should demonstrate a hypersurvival phenotype compared to wild-type H37Rv as seen in the J774.1 macrophages. It is evident from Table 1 that H37Rv and the cGGTA mutant show significant growth in unstimulated mouse peritoneal macrophages. However, when the macrophages were stimulated with IFN-γ and LPS, we observed growth inhibition of H37Rv and the cGGTA mutant. As with the results seen in J774.1 macrophages, there is a statistically significant increase in colony counts of the GGTA mutant in both unstimulated and stimulated mouse peritoneal macrophages (Table 1). These results indicate that the GGTA mutant is indeed resistant to the toxic effects of GSH and GSNO generated by the macrophages due to IFN-γ and LPS stimulation.
TABLE 1.
Survival of H37Rv and the GGTA and cGGTA mutants in primary murine macrophagesa
| Strain | Treatment | Colony count
|
|||
|---|---|---|---|---|---|
| Mean at:
|
SE at:
|
||||
| 24 h | 5 days | 24 h | 5 days | ||
| H37Rv | None | 2,541 | 11,533* | 197 | 261 |
| IFN-γ and LPS | 3,127 | 5,185 | 212 | 482 | |
| GGTA mutant | None | 1,982 | 10,320* | 167 | 248 |
| IFN-γ and LPS | 2,650 | 9,892* | 210 | 318 | |
| cGGTA mutant | None | 2,148 | 10,572* | 321 | 497 |
| IFN-γ and LPS | 1,872 | 3,182 | 156 | 219 | |
Thioglycolate (3%) was injected into the peritoneal cavities of wild-type C57BL/6 mice. Four days after injection, animals were sacrificed via cervical dislocation. Peritoneal cavities were washed with ice-cold PBS to recover peritoneal macrophages. Peritoneal macrophages were infected with H37Rv or the GGTA or cGGTA mutant at an MOI of 10:1. Two hours after infection, unphagocytosed organisms were removed. Infected macrophages were maintained in RPMI medium containing 10% fetal bovine serum or in RPMI medium containing 10% fetal bovine serum, IFN-γ (100 U/ml), and LPS (1 μg/ml). Intracellular colony counts were determined by plating macrophage lysates on Middlebrook 7H11 medium. Data are averages from three experiments. *, P < 0.05 by unpaired t test.
DISCUSSION
Most of the glutathione present in cells is present in the cytosol in its reduced form, GSH. It has been shown in S. enterica serovar Typhimurium that GSNO is converted to a dipeptide by the products of the gene ggt (12). Presumably, GSH is also broken down by the enzyme γ-glutamyl transpeptidase. Among a collection of mutants of S. enterica serovar Typhimurium exhibiting resistance to GSNO were strains with mutations mapping to the ggt gene (12). The ggt gene has been cloned and sequenced from Escherichia coli (27), Pseudomonas aeruginosa (16), Bacillus subtilis (31), and Helicobacter pylori (10). Interestingly, for H. pylori, the ggt gene was identified as a virulence factor essential for colonization in the mouse model (10) but only a contributing factor in colonization in the gnotobiotic piglet model (20). Also, there have been no published reports of spontaneous ggt mutants in clinical isolates.
Our laboratory has constructed and characterized a dipeptide permease mutant of BCG resistant to GSH and GSNO (14). The M. tuberculosis dipeptide permease is an ABC transporter encoded by the dpp operon containing Rv1280c to Rv1283c. Interruption of this operon in S. enterica serovar Typhimurium led to GSNO resistance (12). In our present study, we compared the growths of the GGTA mutant and wild-type H37Rv in vitro and in isolated macrophages. As seen in Fig. 3A and B, H37Rv and the GGTA mutant grew similarly inbroth cultures in the absence of GSH or GSNO. However, in the presence of GSH or GSNO, there was inhibition of growth of H37Rv (Fig. 3A to D). In contrast to these results, the GGTA mutant showed resistance to both GSH and GSNO in broth culture over 5 days (Fig. 3A to D). The mutant lacking the Ggt enzyme grew similarly in the absence and presence of these compounds. We propose that the resistance of the GGTA mutant to GSH and GSNO is due to its inability to metabolize the tripeptide into a dipeptide. Previous work in our laboratory has shown that the dipeptide permease mutant of BCG displayed the same degree of resistance to the tripeptide as that to the dipeptide; the dipeptide is toxic to the same extent as the tripeptide (14). Lack of transpeptidase and dipeptide permease enzyme will result in resistance to GSH and GSNO (14). In contrast to GSH, GSNO exhibited bactericidal effects. NO is stabilized when it is complexed with GSH. The subsequent transport of the dipeptide NO complex and release of NO lead to bacterial cell death. The phenotype of the GGTA mutant was further confirmed in the disk diffusion assay in which no zone of inhibition was present when a disk containing GSH or GSNO was placed on a lawn of GGTA mutant cells (Fig. 4). A prominent zone of inhibition was seen when the disk containing GSH or GSNO was placed over a lawn of H37Rv or cGGTA mutant cells (Fig. 4). Since the GGTA mutant lacks the GSH processing enzyme GgtA, the tripeptides GSH and GSNO are unable to enter the cytoplasmic space of the cell, and hence this strain is resistant to GSH and GSNO. The sensitivity of H37Rv and the cGGTA mutant is due to the presence of the γ-glutamyl transpeptidase enzyme.
To examine the effect of diminished GgtA enzyme activity on the survival of M. tuberculosis in the macrophage, we compared the growths of wild-type H37Rv and the GGTA mutant in J774.1 macrophages and in peritoneal macrophages from wild-type C57BL/6 mice. As shown in Fig. 5A, H37Rv replicated efficiently in unstimulated J774.1 macrophages between 1 h and 72 h, and this increase was statistically significant. In contrast, when these macrophages were stimulated with IFN-γ and LPS, there was no change in the number of CFU between 1 h and 72 h (Fig. 5A). When unstimulated J774.1 macrophages were infected with the GGTA mutant, we observed a significant increase in the number of CFU over 72 h (Fig. 5B). In contrast to H37Rv, the GGTA mutant replicated significantly in IFN-γ- and LPS-stimulated J774.1 macrophages (Fig. 5B). When J774.1 macrophages are stimulated with IFN-γ and LPS, there is production of NO and consequently GSNO. Although the differences in the numbers of CFU in this experiment may be statistically significant but with no compelling large effects, we performed further studies with primary mouse peritoneal macrophages.
More convincing results were seen when these strains of bacteria were used to infect unstimulated and IFN-γ- and LPS-stimulated mouse peritoneal macrophages. As seen in Table 1, the number of H37Rv CFU increased significantly between 24 h and 5 days in unstimulated mouse primary macrophages. When these macrophages were treated with IFN-γ and LPS, no significant growth of H37Rv was observed over 5 days (Table 1). The GGTA mutant replicated significantly inside both unstimulated and stimulated primary mouse macrophages, showing a trend similar to that in J774.1 cells (Table 1). However, the GGTA mutant showed similar increases (n-fold) in growth in both unstimulated and stimulated primary mouse macrophages. In all cases, studies with the complemented mutant, cGGTA, gave results similar to those for the wild-type H37Rv, confirming that the GSH/GSNO-resistant phenotype of the GGTA mutant is a result solely from loss of the ggtA gene.
We have previously shown that IFN-γ and LPS stimulation of macrophages (J774.1 and primary murine macrophages) results in increased NO production and GSNO generation (30). IFN-γ and LPS also stimulate macrophages to induce a respiratory burst and synthesis of ROI, leading to an increase in intracellular levels of GSH to protect the macrophages from the toxic effects of ROI (29, 30). We observed that there was significant growth of the GGTA mutant strain in stimulated macrophages despite the production of NO and ROI. This phenotype is due to the loss of the GSH/GSNO processing enzyme in this strain.
The mechanism of GSH toxicity in mycobacteria is not yet known, although several hypotheses have been put forward (25). One theory is that the presence of high concentrations of GSH may result in an imbalance in this bacterium containing an alternative thiol for regulating reduction/oxidation activity (i.e., mycothiol). Secondly, Spallholz has hypothesized that GSH is an evolutionary precursor of antibiotics produced by higher eukaryotes before the emergence of cellular immunity (26): GSH is similar in structure to penicillin precursors produced by Penicillium and Cephalosporium. The mycobacterial cell wall may possess some intrinsic sensitivity to this structure.
In conclusion, we have begun to delineate the metabolic pathway of GSH and its derivative, GSNO, in M. tuberculosis in the context of mycobacterial infection and pathogenesis. The tripeptide is processed to a dipeptide by the action of GgtA, and the dipeptide is then transported into the mycobacterial cell by the dipeptide permease. We continue to study the interaction of M. tuberculosis with the macrophage to determine the role of GSH and GSNO in innate immune regulation.
Acknowledgments
This work is supported by the American Heart Association Scientist Development Grant 0335370T (Vishwanath Venketaraman) and NIH AI34436 (Nancy D. Connell).
We thank Ariel Millman for helpful discussions.
REFERENCES
- 1.Anderberg, S. J., G. L. Newton, and R. C. Fahey. 1998. Mycothiol biosynthesis and metabolism. Cellular levels of potential intermediates in the biosynthesis and degradation of mycothiol in Mycobacterium smegmatis. J. Biol. Chem. 273:30391-30397. [DOI] [PubMed] [Google Scholar]
- 2.Bardarov, S., S. Bardarov, Jr., M. S. Pavelka, Jr., V. Sambandamurthy, M. Larsen, J. Tufariello, J. Chan, G. Hatfull, and W. R. Jacobs, Jr. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007-3017. [DOI] [PubMed] [Google Scholar]
- 3.Bardarov, S., J. Kriakov, C. Carriere, S. Yu, C. Vaamonde, R. A. McAdam, B. R. Bloom, G. F. Hatfull, and W. R. Jacobs, Jr. 1997. Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10961-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bauer, A. W., W. M. Kirby, J. C. Sherris, and M. Turck. 1966. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 45:493-496. [PubMed] [Google Scholar]
- 5.Bloom, B. R., and C. J. Murray. 1992. Tuberculosis: commentary on a reemergent killer. Science 257:1055-1064. [DOI] [PubMed] [Google Scholar]
- 6.Braunstein, M., S. S. Bardarov, and W. R. Jacobs, Jr. 2002. Genetic methods for deciphering virulence determinants of Mycobacterium tuberculosis. Methods Enzymol. 358:67-99. [DOI] [PubMed] [Google Scholar]
- 7.Chan, E. D., J. Chan, and N. W. Schluger. 2001. What is the role of nitric oxide in murine and human host defense against tuberculosis? Current knowledge. Am. J. Respir. Cell Mol. Biol. 25:606-612. [DOI] [PubMed] [Google Scholar]
- 8.Chan, J., and J. L. Flynn. 1999. Nitric oxide in Mycobacterium tuberculosis infection, p. 281-310. In F. C. Fang (ed.), Nitric oxide and infection. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 9.Chan, J., Y. Xing, R. S. Magliozzo, and B. R. Bloom. 1992. Killing of virulent Mycobacterium tuberculosis by reactive nitrogen intermediates produced by activated murine macrophages. J. Exp. Med. 175:1111-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chevalier, C., J. M. Thiberge, R. L. Ferrero, and A. Labigne. 1999. Essential role of Helicobacter pylori gamma-glutamyltranspeptidase for the colonization of the gastric mucosa of mice. Mol. Microbiol. 31:1359-1372. [DOI] [PubMed] [Google Scholar]
- 11.Cooper, A. M., J. E. Pearl, J. V. Brooks, S. Ehlers, and I. M. Orme. 2000. Expression of the nitric oxide synthase 2 gene is not essential for early control of Mycobacterium tuberculosis in the murine lung. Infect. Immun. 68:6879-6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Groote, M. A., D. Granger, Y. Xu, G. Campbell, R. Prince, and F. C. Fang. 1995. Genetic and redox determinants of nitric oxide cytotoxicity in a Salmonella typhimurium model. Proc. Natl. Acad. Sci. USA 92:6399-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding, A. H., C. F. Nathan, and D. J. Stuehr. 1988. Release of reactive nitrogen intermediates and reactive oxygen intermediates from mouse peritoneal macrophages. Comparison of activating cytokines and evidence for independent production. J. Immunol. 141:2407-2412. [PubMed] [Google Scholar]
- 14.Green, R. M., A. Seth, and N. D. Connell. 2000. A peptide permease mutant of Mycobacterium bovis BCG resistant to the toxic peptides glutathione and S-nitrosoglutathione. Infect. Immun. 68:429-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffith, O. W. 1999. Biologic and pharmacologic regulation of mammalian glutathione synthesis. Free Radic. Biol. Med. 27:922-935. [DOI] [PubMed] [Google Scholar]
- 16.Ishiye, M., M. Yamashita, and M. Niwa. 1993. Molecular cloning of the gamma-glutamyltranspeptidase gene from a Pseudomonas strain. Biotechnol. Prog. 9:323-331. [DOI] [PubMed] [Google Scholar]
- 17.Lee, M. H., L. Pascopella, W. R. Jacobs, Jr., and G. F. Hatfull. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis, and bacille Calmette-Guérin. Proc. Natl. Acad. Sci. USA 88:3111-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu, S., P. B. Killoran, F. C. Fang, and L. W. Riley. 2002. The global regulator ArcA controls resistance to reactive nitrogen and oxygen intermediates in Salmonella enterica serovar Enteritidis. Infect. Immun. 70:451-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacMicking, J., Q.-W. Xie, and C. Nathan. 1997. Nitric oxide and macrophage function. Annu. Rev. Immunol. 15:323-350. [DOI] [PubMed] [Google Scholar]
- 20.McGovern, K. J., T. G. Blanchard, J. A. Gutierrez, S. J. Czinn, S. Krakowka, and P. Youngman. 2001. γ-Glutamyltransferase is a Helicobacter pylori virulence factor but is not essential for colonization. Infect. Immun. 69:4168-4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meister, A., S. S. Tate, and O. W. Griffith. 1981. Gamma-glutamyl transpeptidase. Methods Enzymol. 77:237-253. [DOI] [PubMed] [Google Scholar]
- 22.Nathan, C., and M. U. Shiloh. 2000. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc. Natl. Acad. Sci. USA 97:8841-8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Newton, G. L., and R. C. Fahey. 2002. Mycothiol biochemistry. Arch. Microbiol. 178:388-394. [DOI] [PubMed] [Google Scholar]
- 24.Nikitovic, D., and A. Holmgren. 1996. S-nitrosoglutathione is cleaved by the thioredoxin system with liberation of glutathione and redox regulating nitric oxide. J. Biol. Chem. 271:19180-19185. [DOI] [PubMed] [Google Scholar]
- 25.Penninckx, M. J., and M. T. Elskens. 1993. Metabolism and functions of glutathione in micro-organisms. Adv. Microb. Physiol. 34:239-301. [DOI] [PubMed] [Google Scholar]
- 26.Spallholz, J. 1987. Glutathione: is it an evolutionary vestige of the penicillins? Med. Hypotheses 23:253-257. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki, H., H. Kumagai, T. Echigo, and T. Tochikura. 1989. DNA sequence of the Escherichia coli K-12 γ-glutamyltranspeptidase gene, ggt. J. Bacteriol. 171:5169-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Venketaraman, V., and N. D. Connell. 2003. Glutathione and its role in human and bacterial cells, with special reference to tuberculosis infection. Recent Res. Dev. Bacteriol. 1:1-22. [Google Scholar]
- 29.Venketaraman, V., Y. K. Dayaram, A. G. Amin, R. Ngo, R. M. Green, M. T. Talaue, J. Mann, and N. D. Connell. 2003. Role of glutathione in macrophage control of mycobacteria. Infect. Immun. 71:1864-1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Venketaraman, V., Y. K. Dayaram, M. T. Talaue, and N. D. Connell. 2005. Glutathione and nitrosoglutathione in macrophage defense against Mycobacterium tuberculosis. Infect. Immun. 73:1886-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu, K., and M. A. Strauch. 1996. Identification, sequence, and expression of the gene encoding γ-glutamyltranspeptidase in Bacillus subtilis. J. Bacteriol. 178:4319-4322. [DOI] [PMC free article] [PubMed] [Google Scholar]