Among the reasons for the growing interest in studying biofilm formation is the role of these microbial communities in chronic infections (24, 38). Such biofilm-like chronic infections include the respiratory infections caused by Pseudomonas aeruginosa in the cystic fibrosis (CF) lung (7), relapsing otitis media primarily caused by Haemophilus influenzae (41), and staphylococcal lesions in endocarditis (17). It is also important to note, however, that all of these microbes can also contribute to acute infections in human patients and, in fact, are among the most feared nosocomial pathogens (48). So, how do these organisms cause acute infections in some settings and chronic infections in others? A series of recent papers suggests that bacteria can choose which strategy they employ, either causing an acute infection, growing and spreading rapidly in the host, or, alternatively, adopting a chronic, biofilm infection strategy. While replication in the context of the chronic infection is likely less rapid, bacteria involved in these long-term infections can persist for extended periods of time and continue to shed planktonic (e.g., free-swimming) bacteria, as well as biologically active molecules, into the host during the course of the persistent infection (69).
Establishing that acute and chronic infections are distinct processes requires we demonstrate that these are really two different strategies employed by microbes when interacting with a host. That is, do organisms differ in the molecular mechanisms utilized to cause acute versus chronic infections? Furthermore, a single microbe must presumably have the capability to cause both acute and persistent infections.
Take P. aeruginosa as an example. In some cases, P. aeruginosa is capable of causing pneumonia, breaking down lung defenses and disseminating in the bloodstream, leading to death of a patient within hours or days. This organism has a type III secretion system (TTSS) and produces a variety of extracellular toxins that are thought to play a role in acute infections such as pneumonia (4, 22, 35). For example, the TTSS of P. aeruginosa has been shown recently to play a role in survival of P. aeruginosa in the blood and in systemic dissemination (57). Similarly, an exsA mutant of P. aeruginosa, which is defective for expression of the TTSS, and a strain lacking the TTSS effectors ExoT and ExoU were defective in causing acute corneal disease (32). The TTSS has also been characterized by its role in rapidly killing cultured cells in vitro (4). In addition to its TTSS, P. aeruginosa produces a variety of other virulence factors required for pathogenesis, as shown in acute models of burn and corneal infections in mice, in the invertebrate Caenorhabditis elegans, and in the plant model Arabidopsis thaliana, including quorum-sensing molecules (52, 53, 70), elastase (21, 35), hydrogen cyanide (20, 43, 44), type IV pili (TFP) (71), and lipopolysaccharide (42, 67). Historically, studies of pathogenesis have focused on virulence factors required for acute-infection pathways.
Again using P. aeruginosa as an example, this same organism capable of causing the acute infections described above also participates in a chronic infection of the lungs of CF patients. This chronic infection can last for decades, but P. aeruginosa rarely if ever reaches the bloodstream, indicating that the acute versus chronic infections caused by this microbe may be quite distinct. In contrast to acute infections, chronic infections have received less attention, likely due to more-complex animal models and difficulty in modeling these infections in vitro.
The concept that bacterial biofilm growth may be responsible for some chronic in vivo infections has gained recent support (13, 15, 19). What evidence supports the idea that biofilms are analogous to chronic infections? At this stage, the data are intriguing but in most cases not definitive. For example, there are few well-defined markers of in vitro biofilm growth that can be correlated with putative in vivo biofilm-like infections. Chronic infection of the CF lung by P. aeruginosa has been postulated to be a biofilm-type infection. Singh and colleagues (51) reported synthesis profiles of quorum-sensing signals in CF sputum samples that were consistent with in vitro profiles of these molecules observed for biofilm rather than planktonic bacteria. Furthermore, images of polysaccharide-encased bacterial clusters in CF sputum are also consistent with the idea that P. aeruginosa exists in a biofilm or biofilm-like state in the CF lung (51). Another key trait associated with biofilms is their resistance to antibiotics (34). This resistance phenotype is shared by P. aeruginosa in the CF lung; this microbe is notorious for the fact that it cannot be eliminated from the CF lung by use of current antibiotic therapies (10).
Studies of H. influenzae with otitis media model systems also support a role for a biofilm existence for these microbes, including direct microscopic visualization of bacterial communities in the middle ear and recalcitrance to antibiotic treatment (40, 41). Similarly, staphylococcal endocarditis is associated with compact bacterial cell masses surrounded by a matrix and the ability to withstand extended antibiotic treatment (38).
Biofilms have been conclusively demonstrated to form on a variety of medical and surgical implants; like other biofilm infections, these implant-associated microbes are resistant to treatment by standard antibiotic therapy (18). While additional studies will be needed to further build the case for biofilm-based chronic infections, a picture is emerging where, for at least a subset of persistent infectious diseases, bacteria display phenotypes typically associated with biofilm communities.
MAKING A CHOICE
How does a microbe like P. aeruginosa make a decision regarding causing acute versus chronic disease? Several recent studies provide some clues as to how P. aeruginosa makes this complex choice (Fig. 1).
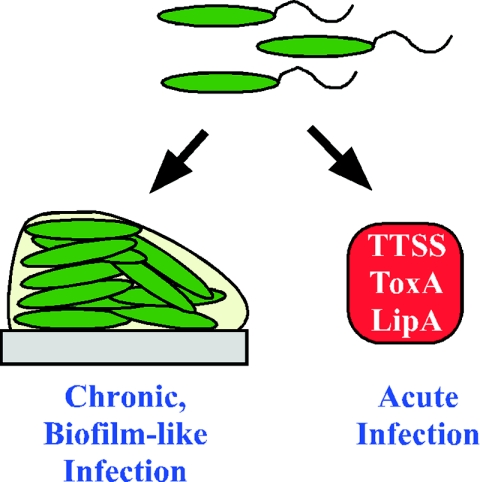
FIG. 1.
Acute versus persistent infections. In response to uncharacterized signals, bacteria either initiate an acute infection utilizing factors like TTSS and various toxins or establish a chronic, biofilm-like infection, wherein bacterial cells (green) are surrounded by a matrix (gray), displaying properties, like antibiotic resistance, typically associated with biofilms.
The model outlined in Fig. 1 is quite simplistic, which is due in part to the fact that we understand very little about the processes that may allow an organism to express factors required for acute versus chronic infections. In the following sections, our discussion largely uses P. aeruginosa as a paradigm of an organism capable of causing both acute and chronic infections.
What might influence expression of factors required for an acute- versus a chronic-infection pathway? For example, the route of entry for the infection may impact whether an acute or chronic infection might be initiated. It is possible that the initiation of an implant-based infection (versus an acute infection) could be the consequence of the response of a microbe to particular signals upon gaining entry to the patient via the catheter insertion site. Similarly, as mentioned above, P. aeruginosa appears to be in a biofilm-like mode of growth when growing in the CF lung. This growth strategy could be a result of the particular environmental signals and/or host factors encountered in the lungs of patients afflicted with this disease. Other factors, such as immune status of the host, tissue integrity, or patient nutrition, might also impact whether microbes initiate an acute versus a chronic infection.
EVIDENCE FOR REGULATING THE CHOICE
Several recent papers report the identification of regulators in P. aeruginosa that may impact whether an organism engages in the acute- versus chronic-disease process. We discuss aspects of these reports below in the context of how they might contribute to the modulation of disease by P. aeruginosa.
Goodman and colleagues identified RetS (PA4856) based on the observation that strains lacking the retS gene made enhanced biofilms compared to the wild-type parent (23). RetS has an unusual structure in that it is predicted to be a protein with seven transmembrane domains at its amino terminus, followed by a sensor kinase-like histidine kinase and ATPase domains in its C terminus, as well as two predicted response regulator (RR)-like receiver domains. Goodman et al. showed that inactivation of retS resulted in the increased ability to form biofilms on both an abiotic surface (glass) and a biotic surface (cultured hamster cells). Interestingly, despite the ability of the retS mutant to adhere to this biotic surface, the mutant P. aeruginosa was less able to damage the hamster cells to which it adhered. Furthermore, the retS mutant strain was also defective for virulence in a mouse model of acute pneumonia (23).
Goodman et al. investigated the genes in P. aeruginosa that are regulated by RetS by using DNA microarrays (23). They found that, in the RetS-deficient strain, the expression of genes required to produce the TTSS (necessary for acute infection) was reduced by as much as 25-fold. The expression levels of other genes associated with acute infections were also reduced, including genes required to produce type IV pili and those that encode toxins such as LipA and ToxA. In contrast, P. aeruginosa genes involved in the formation of the sugar-rich matrix of a biofilm, psl and pel genes (19a, 19b, 28a, 34a), were markedly up-regulated in the retS mutant bacteria. These data suggest that RetS usually represses the genes needed to make a biofilm but is required for inducing the expression of the factors necessary for an acute infection.
Finally, Goodman et al. used transposon mutagenesis in a suppressor screen and identified GacS, its cognate response regulator GacA, and the GacA-dependent small, untranslated RNA rsmZ as additional components of the RetS signaling network. GacA has been shown to play a role in acute infection with several model systems (11, 43-45). The small RNA rsmZ modulates RsmA levels by posttranscriptional regulation, which in turn regulates a variety of virulence factors (9, 25, 39). Phenotypes of a retS mutant were suppressed by second site mutations that increase the supply of free RsmA (23). Based on their findings, the authors proposed the model wherein during acute infection RetS represses the GacA pathway and thus down-regulates biofilm formation and up-regulates factors, such as TTSS, etc., required for acute infections. During a persistent infection, the GacAS pathway is induced, leading to increased biofilm formation and lower levels of expression of acute-infection-related functions. Consistent with their hypothesis, a mutation in gacA is defective for biofilm formation (37).
Concurrently with the report of Goodman et al., Laskowski et al. also described the effects of PA4856 on the regulation of the TTSS and virulence of P. aeruginosa (31). Working with strain PA103, these investigators showed that deletion of the gene they called rtsM results in loss of expression of the TTSS effectors exoU and exoT. Furthermore, loss of RtsM/RetS (PA4856) function in strain PA103 resulted in an ∼50 to 75% decrease in cytotoxicity in vitro when tested against epithelial cells and decreased virulence in an acute-infection model. This virulence phenotype was also observed by Goodman et al. (23) for strain PAK, lacking PA4856 function, thereby showing independently the role of this sensor kinase response regulator in the control of virulence gene expression. Interestingly, overexpression of the regulators ExsA and Vfr rescued the TTSS defect of rtsM in regards to effector production, suggesting that RtsM works upstream of these known regulators of TTSS gene expression.
The biofilm formation phenotype of the PA103 rtsM mutant was not assessed on an abiotic surface; however, Laskowski et al. observed no difference between the wild type and the rtsM mutant in regards to colonizing epithelial cells. This finding is in contrast to the observation of Goodman et al. that loss of rtsM or retS in the PAK strain resulted in an ∼3-fold increase in colonization of epithelial cells in vitro. Perhaps this discrepancy is due to differences in the strains or the epithelial cells used in their respective assays.
As described above, a mutation in rtsM or retS results in a hyper-biofilm formation phenotype (at least in the PAK strain) and decreased expression of genes of the TTSS. Given the observation that expression of the global regulator Vfr can bypass the expression defect of the TTSS genes in an rtsM or a retS mutant background, we would predict that overexpression of Vfr in an rtsM or a retS mutant might also lead to decreased biofilm formation, thereby further confirming the reciprocal regulation of TTSS gene expression and biofilm formation.
Further building the case for bacteria choosing between acute and chronic infection, Kuchma and colleagues reported that the SadARS three-component system of P. aeruginosa participates in both the maturation of a biofilm and controlling expression of TTSS genes (29). The organization of the sadARS locus is reminiscent of the bvg locus of Bordetella pertussis (2, 3), and the respective genes in these loci share 25 to 57% amino acid sequence identity. These investigators found that nonpolar mutations in any of the sadARS genes resulted in a mature biofilm with an altered mature structure but that these mutations did not confer defects in early biofilm formation, growth, swimming, or twitching motility. The sadARS locus is comprised of genes coding for a putative sensor histidine kinase (SadS, PA3946) and two RRs (SadA and SadR). The RR protein SadR (PA3947) has a predicted CheY-like phospho-receiver domain as well as an EAL domain, which is associated with a cyclic diguanylate phosphodiesterase activity (55). The second RR protein, SadA (PA3948), is predicted to have a CheY-like phospho-receiver domain and helix-turn-helix DNA-binding domain and therefore appears to have properties more like those of classical RR proteins.
Kuchma et al. used a DNA microarray approach to identify downstream targets of the SadARS system, and they discovered that expression of the TTSS is negatively regulated by SadARS and that this negative regulation is observed only with biofilm-grown bacteria. Furthermore, mutations in TTSS regulatory proteins, effectors, or structural components of the secretion system all enhanced biofilm formation (29). These data strongly suggest that repression of the TTSS genes by SadARS under biofilm-forming conditions helps facilitate formation of these surface-attached communities and that, moreover, expression of the TTSS may actually be detrimental to biofilm formation. Finally, recall that any mutation in the sadARS system alters formation of the mature biofilm but that early biofilm development, including formation of microcolonies, is unaffected in these mutants (29). Taken together, these data suggest that any commitment to an acute versus persistence pathway may occur downstream of the formation of microcolonies.
Concurrent with the study of Kuchma et al., Kulasekara and colleagues identified a signal transduction system that regulates expression of the cupB and cupC loci in P. aeruginosa PAK (30). This signal transduction system is comprised of proteins designated RocS1, RocR, and RocA1. Genes coding for the Roc system and the SadARS system are at the same genetic loci (PA3946 to PA3948). The rocS1 (sadS, PA3946) gene codes for a sensor histidine kinase, and the rocA1 (sadA, PA3948) and rocR (sadR, PA3947) genes code for RRs. The rocARS genes were identified because some mutations in this locus resulted in up-regulation of cupB-lacZ and cupC-lacZ transcriptional fusions (30). Further studies confirmed that the RocARS (SadARS) system regulates the expression of the cup loci: RocA1 and RocS1 appear to activate cup gene expression and RocR1 to repress expression of the cup loci. Genetic studies indicated that cupA, cupB, and cupC all contribute to pellicle formation at an air-liquid interface (30), and the cupA locus has been shown to play a role in biofilm formation on abiotic surfaces (56). Taken together with the work of Kuchma and colleagues (29), these data suggest that the RocARS (SadARS) system controls the expression of several factors that positively and negatively contribute to biofilm formation and to acute infection. The complex nature of this unusual three-component system may allow the cells to modulate the choice between an acute- and a persistent-infection pathway by integrating a variety of environmental inputs.
Additional evidence for differential expression of factors required for acute versus persistent infection comes from Bleves and colleagues (6), who reported that a quorum-sensing system of P. aeruginosa negatively regulates the TTSS. In particular, a mutation in the rhlI gene results in early expression of the TTSS and, furthermore, RhlR plus C4-homoserine lactone (C4HSL) is required to repress TTSS gene expression. Interestingly, Singh and colleagues showed that for in vitro-grown biofilms and CF sputum samples, the ratio of the synthesis of C4HSL to C12HSL increases relative to the ratio of these molecules in planktonic cultures (51). Therefore, growth in a persistent biofilm would presumably lead to down-regulation of TTSS genes. This phenomenon has been observed for some P. aeruginosa strains isolated from chronically infected CF patients (49). Roy-Burman and colleagues also showed that expression of the TTSS increased the severity of acute disease and that, furthermore, strains of P. aeruginosa from chronic infections were less likely to express TTSS genes (47).
Another recent study also links TTSS and multicellular behavior but in a very different fashion. Yap and colleagues reported that the TTSS of the plant pathogen Erwinia chrysanthemi is required for the formation of pellicles (66). They postulate that the needle-like structure of the TTSS could act as a structural component of the matrix, stabilizing these multicellular aggregates (66). In contrast to the role of this secretion system on pellicle formation, mutations in the TTSS had no apparent effect on the formation of biofilms on an abiotic surface. While the mechanism by which E. chrysanthemi utilizes its TTSS to form a pellicle must still be resolved, another example linking TTSS to the multicellular behavior of bacteria is intriguing and further emphasizes the need for bacterial cells to coordinately regulate their myriad surface factors.
WHAT MOLECULAR MECHANISMS INTEGRATE EXPRESSION OF GENES REQUIRED FOR ACUTE VERSUS PERSISTENT INFECTIONS?
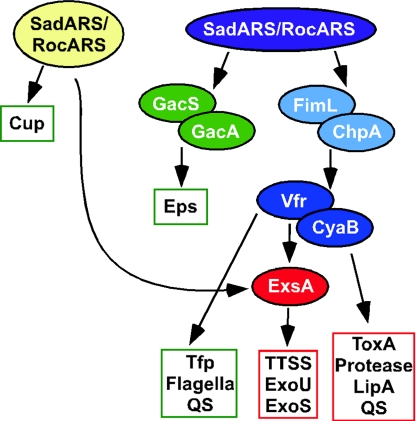
A number of regulatory genes have been implicated in controlling expression of TTSS and biofilm formation. How might these regulatory pathways intersect in P. aeruginosa? One potential model for the coordinate regulation of virulence gene expression and biofilm formation is proposed in Fig. 2.
FIG. 2.
A model for the coordinate regulation of factors required for acute infection and chronic persistence. The model shown here is based largely on genetic and in vivo regulatory studies performed with P. aeruginosa strains PA14, PAO1, PAK, and PA103. The functions boxed in green are involved primarily in biofilm formation and/or chronic persistence. The factors in the red boxes play a role in acute infection. Arrows indicate experimentally demonstrated genetic interactions. Abbreviations: EPS, exopolysaccharide; Cup, cupB and cupC genes; ToxA, endotoxin A; QS, quorum-sensing systems.
Laskowski et al. showed that the TTSS-mediated virulence defects of the rtsM or retS mutant could be bypassed by expressing ExsA and Vfr (31), and it had been shown previously that ExsA could bypass the expression defect observed for the TTSS genes in a vfr mutant (64). Taken together, these genetic data suggest that ExsA functions downstream of RtsM/RetS and Vfr. Vfr has been shown to regulate exotoxin A and protease production (61), up-regulation of lasR expression (1), repression of the flagellar regulator fleQ (16), TFP biogenesis (5), and the expression of genes required for the TTSS (64). Expression of the TTSS genes also requires the adenylate cyclase CyaB (54), which is likely involved in production of the cyclic AMP cofactor of Vfr. These data suggest that Vfr may help coordinate the expression of genes required for biofilm formation (TFP, flagellar motility) and virulence (exotoxin A, protease, TTSS). Given that Vfr appears to be upstream of ExsA in terms of a regulatory cascade and that ExsA has not been shown to regulate any genes required for biofilm formation, it is possible that Vfr is one point wherein the acute-infection versus chronic-persistence pathways diverge.
Recently, Whitchurch and colleagues (62) reported the identification of FimL (PA1822), a protein required for TFP production and TTSS gene expression. The decrease in expression of TFP and TTSS genes in strains lacking a functional FimL is likely indirect and due to the decreased expression of vfr in a fimL mutant (62). FimL has sequence similarity to ChpA, a protein in P. aeruginosa identified as important for twitching motility (63). The N terminus of ChpA contains putative histidine phosphotransfer (Hpt) and threonine phosphotransfer (Tpt) domains (63). Interestingly, the key residues of these domains are replaced with glutamine residues in FimL; Whitchurch et al. proposed that FimL acts as a competitive inhibitor of ChpA. Therefore, ChpA and FimL may act at the same step in this pathway. Given that FimL can regulate Vfr, we placed FimL (and ChpA) downstream of the RetS/RtsM system but upstream of Vfr.
Suppressor analysis studies by Goodman et al. strongly suggest that the GacA/GacS-RsmZ/RsmA system works downstream of RetS/RtsM in reciprocally regulating exopolysaccharide production and expression of virulence factors, including TTSS and exotoxin A (23). Their data also indicate that the positive effects of RetS/RtsM are mediated through GacAS via Vfr; however, repression of exopolysaccharide production is likely Vfr independent (23). The model in Fig. 2 shows GacAS and FimL/ChpA as part of parallel pathways. It is possible that GacAS and FimL/ChpA are in the same pathway, but there are currently no data to differentiate between these two possibilities.
How the SadARS/RocARS systems fit into the overall regulatory cascade shown in Fig. 2 is unclear. Kulasekara et al. demonstrated that this regulatory system controls the expression of the cupB and cupC loci (30); however, it is not clear whether this regulation is via GacAS, FimL/ChpA, or Vfr. In P. aeruginosa strain PA14, mutations in the sadARS loci lead to altered expression of the TTSS genes. Array studies using sadARS mutants indicate that SadARS contributes to the transcriptional regulation of ExsA but has no apparent role in the modulation of GacAS, Vfr, or the other regulators shown in Fig. 2 (29).
HOW DO BACTERIA CHOOSE?
In deciding between acute and persistent infections, what environmental cues do bacteria evaluate? It is clear that this is the next key question in understanding this aspect of bacterial pathogenesis. It is likely that multiple signals are involved in this decision-making process and that they are related to the metabolism of the organism, for example, the availability of iron and other nutrients or aerobic versus anaerobic conditions (50, 65). The nature and identity of such signals are still a wide-open question.
One must also consider the possibility that cells may be “primed” for a certain route of infection. For example, if planktonically derived P. aeruginosa initiates an infection, is it more likely to cause an acute infection? Conversely, is biofilm-grown P. aeruginosa more likely to initiate chronic infections? The physiological state of the infecting organisms may contribute to their infectious fate in vivo, as has been argued for Vibrio cholerae. The fact that this organism can form biofilms on plankton is thought to contribute to disease, based on the increased population of microbes on these small animals and the biofilm-associated protection of these microbes from a variety of environmental stresses, including low pH (12, 26, 27). However, it is also important to consider that the infecting organisms will rapidly adapt their physiology to the environment that they encounter in the host, independent of their environmental reservoir.
ACUTE VERSUS PERSISTENT INFECTIONS
These recent studies suggest that P. aeruginosa has regulatory systems for controlling the choice between an acute infection and persistence (6, 23, 29, 30, 31) and that, furthermore, expression of functions required for acute infection, such as the TTSS or toxins, may be incompatible with efficient surface attachment (28, 29). This concept was first elucidated in studies by Irie and colleagues (28), who investigated the relationship between virulence and biofilm formation in the bacterium Bordetella bronchiseptica, a microbe that is related to the organism which causes whooping cough. The BvgAS (Bordetella virulence gene) two-component signal transduction system regulates gene expression among at least three phases of B. bronchiseptica growth: a virulent (Bvg+) phase, a nonvirulent (Bvg−) phase, and an intermediate (Bvgi) phase (33, 36). In the Bvg+ phase, the virulence factors filamentous hemagglutinin (FHA), fimbriae, and the bifunctional adenylate cyclase/hemolysin (ACY) genes are expressed. In the Bvgi phase, FHA and fimbriae are expressed, but ACY is not significantly expressed under these conditions (14). Irie et al. found that B. bronchiseptica formed biofilms maximally in the Bvgi phase (28). They further showed that FHA is required for maximal biofilm formation and ACY inhibits biofilm formation, presumably via interactions with FHA (68). These data strongly suggest that not only are functions required for acute infection and persistence expressed differentially but, furthermore, the expression of a toxin required for acute infection is actually incompatible with biofilm formation. As mentioned above, a similar phenomenon was observed with P. aeruginosa, wherein mutation of the TTSS, which is required for acute infection, resulted in increased biofilm formation (29).
A GENERAL PHENOMENON?
Do only gram-negative organisms employ this acute versus persistence strategy? A recent microarray study by Resch and colleagues (46) indicates that Staphylococcus aureus may regulate its acute versus persistent virulence functions in a manner analogous to P. aeruginosa and B. bronchiseptica. These investigators noticed that genes coding for toxins, proteases, and other virulence factors were much more highly expressed in planktonic cells than in biofilm cells and that no toxins were expressed at higher levels in biofilm cells than in their planktonic counterparts. These same investigators went on to suggest that nonbiofilm cells (e.g., planktonic cells) would be much more likely to participate in acute infections (46). While functional studies are needed to determine if planktonically expressed virulence factors interfere with biofilm formation by S. aureus, as is the case for B. bronchiseptica (28) and P. aeruginosa (29), these findings are very suggestive and warrant further study. This array study supported an observation made several years ago that a mutation in agr, a global regulator of virulence gene expression required for pathogenesis of S. aureus (8), results in a strain that displays increased biofilm formation (60). A similar increase in biofilm formation was observed for an agr mutant of Staphylococcus epidermidis both in vitro and in vivo (58, 59). These data suggest that Agr may participate in the reciprocal regulation of virulence and biofilm formation in this important group of gram-positive pathogens.
CONCLUSIONS AND FUTURE STUDIES
As is often the case, one motivation for writing this minireview is to stimulate some thought and discussion on a topic we find intriguing and exciting. A key take-home message we hope to convey in this review is the hypothesis that chronic infections more closely resemble a biofilm growth mode, while acute infections may more closely resemble planktonic growth. The studies published to date supporting this hypothesis also allow us to formulate some testable hypotheses for future work. (i) Can we identify mutants that are defective for only acute or for only persistent infections? That is, are these infection pathways genetically distinct? (ii) Is there incompatibility between factors required for acute and persistent infections; for example, does production of the TTSS or certain toxins somehow physically preclude the cell from efficient biofilm formation, or is interference at the level of regulation? (iii) Can in vitro biofilm models serve as surrogates for in vivo chronic infections to help us better understand mechanisms regulating this potential acute versus persistent choice? (iv) Can we identify markers to distinguish between acute and chronic infections in vivo?
At the very least, considering the possibility of distinct pathways for acute versus chronic infections may serve as an experimental framework for designing future studies. In addition, of course, considering the concept of distinct acute- versus persistent-infection pathways has obvious clinical implications. Are different treatment strategies required for these two infections, or should different antibiotics be used? To approach these questions and address the possibility of a biofilm-like persistent state in chronic infections, it is likely that new models of persistent infections will need to be developed.
Acknowledgments
We thank both reviewers for excellent comments and suggestions. Our thanks also go to P. Singh for reading the manuscript and for his helpful comments.
This work was supported by an Overseas Researcher Grant from Nihon University to S.F. and by grants AI555774-2, AI51360-01, and 1 P20 RR018787 from the Institutional Development Award (IDeA) Program of the National Center for Research Resources to G.A.O.
REFERENCES
- 1.Albus, A. M., E. C. Pesci, L. J. Runyen-Janecky, S. E. West, and B. H. Iglewski. 1997. Vfr controls quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3928-3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arico, B., J. F. Miller, C. Roy, S. Stibitz, D. Monack, S. Falkow, R. Gross, and R. Rappuoli. 1989. Sequences required for expression of Bordetella pertussis virulence factors share homology with prokaryotic signal transduction proteins. Proc. Natl. Acad. Sci. USA 86:6671-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arico, B., V. Scarlato, D. M. Monack, S. Falkow, and R. Rappuoli. 1991. Structural and genetic analysis of the bvg locus in Bordetella species. Mol. Microbiol. 5:2481-2491. [DOI] [PubMed] [Google Scholar]
- 4.Barbieri, J. T., and J. Sun. 2004. Pseudomonas aeruginosa ExoS and ExoT. Rev. Physiol. Biochem. Pharmacol. 152:79-92. [DOI] [PubMed] [Google Scholar]
- 5.Beatson, S. A., C. B. Whitchurch, J. L. Sargent, R. C. Levesque, and J. S. Mattick. 2002. Differential regulation of twitching motility and elastase production by Vfr in Pseudomonas aeruginosa. J. Bacteriol. 184:3605-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bleves, S., C. Soscia, P. Nogueira-Orlandi, A. Lazdunski, and A. Filloux. 2005. Quorum sensing negatively controls type III secretion regulon expression in Pseudomonas aeruginosa PAO1. J. Bacteriol. 187:3898-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boucher, R. C. 2004. New concepts of the pathogenesis of cystic fibrosis lung disease. Eur. Respir. J. 23:146-158. [DOI] [PubMed] [Google Scholar]
- 8.Bronner, S., H. Monteil, and G. Prevost. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28:183-200. [DOI] [PubMed] [Google Scholar]
- 9.Burrowes, E., A. Abbas, A. O'Neill, C. Adams, and F. O'Gara. 2005. Characterization of the regulatory RNA RsmB from Pseudomonas aeruginosa PAO1. Res. Microbiol. 156:7-16. [DOI] [PubMed] [Google Scholar]
- 10.Chernish, R. N., and S. D. Aaron. 2003. Approach to resistant gram-negative bacterial pulmonary infections in patients with cystic fibrosis. Curr. Opin. Pulm. Med. 9:509-515. [DOI] [PubMed] [Google Scholar]
- 11.Chieda, Y., K. Iiyama, C. Yasunaga-Aoki, J. M. Lee, T. Kusakabe, and S. Shimizu. 2005. Pathogenicity of gacA mutant of Pseudomonas aeruginosa PA01 in the silkworm, Bombyx mori. FEMS Microbiol. Lett. 244:181-186. [DOI] [PubMed] [Google Scholar]
- 12.Colwell, R. R. 1996. Global climate and infectious disease: the cholera paradigm. Science 274:2025-2031. [DOI] [PubMed] [Google Scholar]
- 13.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 14.Cotter, P. A., M. H. Yuk, S. Mattoo, B. J. Akerley, J. Boschwitz, D. A. Relman, and J. F. Miller. 1998. Filamentous hemagglutinin of Bordetella bronchiseptica is required for efficient establishment of tracheal colonization. Infect. Immun. 66:5921-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Darouiche, R. O. 2004. Treatment of infections associated with surgical implants. N. Engl. J. Med. 350:1422-1429. [DOI] [PubMed] [Google Scholar]
- 16.Dasgupta, N., E. P. Ferrell, K. J. Kanack, S. E. H. West, and R. Ramphal. 2002. fleQ, the gene encoding the major flagellar regulator of Pseudomonas aeruginosa, is σ70 dependent and is downregulated by Vfr, a homolog of Escherichia coli cyclic AMP receptor protein. J. Bacteriol. 184:5240-5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunne, W. M., Jr. 2002. Bacterial adhesion: seen any good biofilms lately? Clin. Microbiol. Rev. 15:155-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fergie, N., R. Bayston, J. P. Pearson, and J. P. Birchall. 2004. Is otitis media with effusion a biofilm infection? Clin. Otolaryngol. Allied Sci. 29:38-46. [DOI] [PubMed] [Google Scholar]
- 19a.Friedman, L., and R. Kolter. 2004. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol. Microbiol. 51:675-690. [DOI] [PubMed] [Google Scholar]
- 19b.Friedman, L., and R. Kolter. 2004. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J. Bacteriol. 186:4457-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gallagher, L. A., and C. Manoil. 2001. Pseudomonas aeruginosa PAO1 kills Caenorhabditis elegans by cyanide poisoning. J. Bacteriol. 183:6207-6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galloway, D. R. 1991. Pseudomonas aeruginosa elastase and elastolysis revisited: recent developments. Mol. Microbiol. 5:2315-2321. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh, P. 2004. Process of protein transport by the type III secretion system. Microbiol. Mol. Biol. Rev. 68:771-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodman, A. L., B. Kulasekara, A. Rietsch, D. Boyd, R. S. Smith, and S. Lory. 2004. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7:745-754. [DOI] [PubMed] [Google Scholar]
- 24.Hall-Stoodley, L., J. W. Costerton, and P. Stoodley. 2004. Bacterial biofilms: from the natural environment to infectious diseases. Nat. Rev. Microbiol. 2:95-108. [DOI] [PubMed] [Google Scholar]
- 25.Heurlier, K., F. Williams, S. Heeb, C. Dormond, G. Pessi, D. Singer, M. Camara, P. Williams, and D. Haas. 2004. Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 186:2936-2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huo, A., B. Xu, M. A. Chowdhury, M. S. Islam, R. Montilla, and R. R. Colwell. 1996. A simple filtration method to remove plankton-associated Vibrio cholerae in raw water supplies in developing countries. Appl. Environ. Microbiol. 62:2508-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huq, A., P. A. West, E. B. Small, M. I. Huq, and R. R. Colwell. 1984. Influence of water temperature, salinity, and pH on survival and growth of toxigenic Vibrio cholerae serovar O1 associated with live copepods in laboratory microcosms. Appl. Environ. Microbiol. 48:420-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irie, Y., S. Mattoo, and M. H. Yuk. 2004. The Bvg virulence control system regulates biofilm formation in Bordetella bronchiseptica. J. Bacteriol. 186:5692-5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28a.Jackson, K. D., M. Starkey, S. Kremer, M. R. Parsek, and D. J. Wozniak. 2004. Identification of psl, a locus encoding a potential exopolysaccharide that is essential for Pseudomonas aeruginosa PAO1 biofilm formation. J. Bacteriol. 186:4466-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuchma, S. L., J. P. Connolly, and G. A. O'Toole. 2005. A three-component regulatory system regulates biofilm maturation and type III secretion in Pseudomonas aeruginosa. J. Bacteriol. 187:1441-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulasekara, H. D., I. Ventre, B. R. Kulasekara, A. Lazdunski, A. Filloux, and S. Lory. 2005. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol. Microbiol. 55:368-380. [DOI] [PubMed] [Google Scholar]
- 31.Laskowski, M. A., E. Osborn, and B. I. Kazmierczak. 2004. A novel sensor kinase-response regulator hybrid regulates type III secretion and is required for virulence in Pseudomonas aeruginosa. Mol. Microbiol. 54:1090-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, E. J., B. A. Cowell, D. J. Evans, and S. M. Fleiszig. 2003. Contribution of ExsA-regulated factors to corneal infection by cytotoxic and invasive Pseudomonas aeruginosa in a murine scarification model. Investig. Ophthalmol. Vis. Sci. 44:3892-3898. [DOI] [PubMed] [Google Scholar]
- 33.Locht, C., R. Antoine, and F. Jacob-Dubuisson. 2001. Bordetella pertussis, molecular pathogenesis under multiple aspects. Curr. Opin. Microbiol. 4:82-89. [DOI] [PubMed] [Google Scholar]
- 34.Mah, T. F., and G. A. O'Toole. 2001. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 9:34-39. [DOI] [PubMed] [Google Scholar]
- 34a.Matsukawa, M., and E. P. Greenberg. 2004. Putative exopolysaccharide synthesis genes influence Pseudomonas aeruginosa biofilm development. J. Bacteriol. 186:4449-4456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsumoto, K. 2004. Role of bacterial proteases in pseudomonal and serratial keratitis. Biol. Chem. 385:1007-1016. [DOI] [PubMed] [Google Scholar]
- 36.Mattoo, S., A. K. Foreman-Wykert, P. A. Cotter, and J. F. Miller. 2001. Mechanisms of Bordetella pathogenesis. Front. Biosci. 6:E168-E186. [DOI] [PubMed] [Google Scholar]
- 37.Parkins, M. D., H. Ceri, and D. G. Storey. 2001. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol. 40:1215-1226. [DOI] [PubMed] [Google Scholar]
- 38.Parsek, M. R., and P. K. Singh. 2003. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 57:677-701. [DOI] [PubMed] [Google Scholar]
- 39.Pessi, G., F. Williams, Z. Hindle, K. Heurlier, M. T. Holden, M. Camara, D. Haas, and P. Williams. 2001. The global posttranscriptional regulator RsmA modulates production of virulence determinants and N-acylhomoserine lactones in Pseudomonas aeruginosa. J. Bacteriol. 183:6676-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Post, J. C. 2001. Direct evidence of bacterial biofilms in otitis media. Laryngoscope 111:2083-2094. [DOI] [PubMed] [Google Scholar]
- 41.Post, J. C., P. Stoodley, L. Hall-Stoodley, and G. D. Ehrlich. 2004. The role of biofilms in otolaryngologic infections. Curr. Opin. Otolaryngol. Head Neck Surg. 12:185-190. [DOI] [PubMed] [Google Scholar]
- 42.Priebe, G. P., C. R. Dean, T. Zaidi, G. J. Meluleni, F. T. Coleman, Y. S. Coutinho, M. J. Noto, T. A. Urban, G. B. Pier, and J. B. Goldberg. 2004. The galU gene of Pseudomonas aeruginosa is required for corneal infection and efficient systemic spread following pneumonia but not for infection confined to the lung. Infect. Immun. 72:4224-4232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 44.Rahme, L. G., M.-W. Tan, L. Le, S. M. Wong, R. G. Tompkins, S. B. Calderwood, and F. M. Ausubel. 1997. Use of model plant hosts to identify Pseudomonas aeruginosa virulence factors. Proc. Natl. Acad. Sci. USA 94:13245-13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reimmann, C., M. Beyeler, A. Latifi, H. Winteler, M. Foglino, A. Lazdunski, and D. Hass. 1997. The global activator GacA of Pseudomonas aeruginosa PAO1 positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24:309-319. [DOI] [PubMed] [Google Scholar]
- 46.Resch, A., R. Rosenstein, C. Nerz, and F. Gotz. 2005. Differential gene expression profiling of Staphylococcus aureus cultivated under biofilm and planktonic conditions. Appl. Environ. Microbiol. 71:2663-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roy-Burman, A., R. H. Savel, S. Racine, B. L. Swanson, N. S. Revadigar, J. Fujimoto, T. Sawa, D. W. Frank, and J. P. Wiener-Kronish. 2001. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183:1767-1774. [DOI] [PubMed] [Google Scholar]
- 48.Safdar, N., and D. G. Maki. 2002. The commonality of risk factors for nosocomial colonization and infection with antimicrobial-resistant Staphylococcus aureus, enterococcus, gram-negative bacilli, Clostridium difficile, and Candida. Ann. Intern. Med. 136:834-844. [DOI] [PubMed] [Google Scholar]
- 49.Schaber, J. A., N. L. Carty, N. A. McDonald, E. D. Graham, R. Cheluvappa, J. A. Griswold, and A. N. Hamood. 2004. Analysis of quorum sensing-deficient clinical isolates of Pseudomonas aeruginosa. J. Med. Microbiol. 53:841-853. [DOI] [PubMed] [Google Scholar]
- 50.Singh, P. K., M. R. Parsek, E. P. Greenberg, and M. J. Welsh. 2002. A component of innate immunity prevents bacterial biofilm development. Nature 417:552-555. [DOI] [PubMed] [Google Scholar]
- 51.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 52.Smith, R. S., S. G. Harris, R. Phipps, and B. Iglewski. 2002. The Pseudomonas aeruginosa quorum-sensing molecule N-(3-oxododecanoyl)homoserine lactone contributes to virulence and induces inflammation in vivo. J. Bacteriol. 184:1132-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith, R. S., and B. H. Iglewski. 2003. P. aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 6:56-60. [DOI] [PubMed] [Google Scholar]
- 54.Smith, R. S., M. C. Wolfgang, and S. Lory. 2004. An adenylate cyclase-controlled signaling network regulates Pseudomonas aeruginosa virulence in a mouse model of acute pneumonia. Infect. Immun. 72:1677-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tiscler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vallet, I., J. W. Olson, S. Lory, A. Lazdunski, and A. Filloux. 2001. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. USA 98:6911-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vance, R. E., A. Rietsch, and J. J. Mekalanos. 2005. Role of the type III secreted exoenzymes S, T, and Y in systemic spread of Pseudomonas aeruginosa PAO1 in vivo. Infect. Immun. 73:1706-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vuong, C., C. Gerke, G. A. Somerville, E. R. Fischer, and M. Otto. 2003. Quorum-sensing control of biofilm factors in Staphylococcus epidermidis. J. Infect. Dis. 188:706-718. [DOI] [PubMed] [Google Scholar]
- 59.Vuong, C., S. Kocianova, Y. Yao, A. B. Carmody, and M. Otto. 2004. Increased colonization of indwelling medical devices by quorum-sensing mutants of Staphylococcus epidermidis in vivo. J. Infect. Dis. 190:1498-1505. [DOI] [PubMed] [Google Scholar]
- 60.Vuong, C., H. L. Saenz, F. Gotz, and M. Otto. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182:1688-1693. [DOI] [PubMed] [Google Scholar]
- 61.West, S. E. H., A. K. Sample, and L. J. Runyen-Janecky. 1994. The vfr gene product, required for Pseudomonas aeruginosa exotoxin A and protease production, belongs to the cyclic AMP receptor protein family. J. Bacteriol. 176:7532-7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Whitchurch, C. B., S. A. Beatson, J. C. Comolli, T. Jakobsen, J. L. Sargent, J. J. Bertrand, J. West, M. Klausen, L. L. Waite, P. J. Kang, T. Tolker-Nielsen, J. S. Mattick, and J. N. Engel. 2005. Pseudomonas aeruginosa fimL regulates multiple virulence functions by intersecting with Vfr-modulated pathways. Mol. Microbiol. 55:1357-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whitchurch, C. B., A. J. Leech, M. D. Young, D. Kennedy, J. L. Sargent, J. J. Bertrand, A. B. Semmler, A. S. Mellick, P. R. Martin, R. A. Alm, M. Hobbs, S. A. Beatson, B. Huang, L. Nguyen, J. C. Commolli, J. N. Engel, A. Darzins, and J. S. Mattick. 2004. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 52:873-893. [DOI] [PubMed] [Google Scholar]
- 64.Wolfgang, M. C., V. T. Lee, M. E. Gilmore, and S. Lory. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 4:253-263. [DOI] [PubMed] [Google Scholar]
- 65.Worlitzsch, D., R. Tarran, M. Ulrich, U. Schwab, A. Cekici, K. C. Meyer, P. Birrer, G. Bellon, J. Berger, T. Weiss, K. Botzenhart, J. R. Yankaskas, S. Randell, R. C. Boucher, and G. Doring. 2002. Effects of reduced mucus oxygen concentration in airway Pseudomonas infections of cystic fibrosis patients. J. Clin. Investig. 109:317-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yap, M. N., C. H. Yang, J. D. Barak, C. E. Jahn, and A. O. Charkowski. 2005. The Erwinia chrysanthemi type III secretion system is required for multicellular behavior. J. Bacteriol. 187:639-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zaidi, T. S., S. M. J. Fleiszig, M. J. Preston, J. B. Goldberg, and G. B. Pier. 1996. Lipopolysaccharide outer core is a ligand for corneal cell binding and ingestion of Pseudomonas aeruginosa. Investig. Ophthalmol. Vis. Sci. 37:976-986. [PubMed] [Google Scholar]
- 68.Zaretzky, F. R., M. C. Gray, and E. L. Hewlett. 2002. Mechanism of association of adenylate cyclase toxin with the surface of Bordetella pertussis: a role for toxin-filamentous haemagglutinin interaction. Mol. Microbiol. 45:1589-1598. [DOI] [PubMed] [Google Scholar]
- 69.Zegans, M. E., H. I. Becker, J. Budzik, and G. O'Toole. 2002. The role of bacterial biofilms in ocular infections. DNA Cell Biol. 21:415-420. [DOI] [PubMed] [Google Scholar]
- 70.Zhu, H., R. Bandara, T. C. Conibear, S. J. Thuruthyil, S. A. Rice, S. Kjelleberg, M. Givskov, and M. D. Willcox. 2004. Pseudomonas aeruginosa with lasI quorum-sensing deficiency during corneal infection. Investig. Ophthalmol. Vis. Sci. 45:1897-1903. [DOI] [PubMed] [Google Scholar]
- 71.Zolfaghar, I., D. J. Evans, and S. M. Fleiszig. 2003. Twitching motility contributes to the role of pili in corneal infection caused by Pseudomonas aeruginosa. Infect. Immun. 71:5389-5393. [DOI] [PMC free article] [PubMed] [Google Scholar]