Abstract
Expression of several major histocompatibility complex (MHC) class I alleles is associated with a protective effect against disease progression in both human immunodeficiency virus type 1 and simian immunodeficiency virus infection. To understand the mechanism underlying this effect, we investigated the expression of the MHC class I allele Mamu-A*01 in simian-human immunodeficiency virus (SHIV) infection, one of the major models for evaluation of AIDS vaccine candidates. We found that disease progression was significantly delayed in Mamu-A∗01-positive rhesus monkeys infected with the highly pathogenic SHIV 89.6P. The delay corresponded not only to a noted Mamu-A∗01-restricted dominant cytotoxic T-lymphocyte (CTL) response but also to a lower viral load in lymph nodes (LN) and, importantly, to minimal destruction of LN structure during early infection. In contrast, Mamu-A∗01-negative monkeys exhibited massive destruction of LN structure with accompanying rapid disease progression. These data indicate that MHC class I allele-restricted CTL responses may play an important role in preservation of lymphoid tissue structure, thereby resulting in attenuation of disease progression in immunodeficiency virus infection.
Numerous observations have shown that host genetic factors play important roles in disease progression in human immunodeficiency virus type 1 (HIV-1) infection (24, 28). The genetics of the major histocompatibility complex (MHC) appears to be a particularly critical factor (5). Rapid disease progression has been demonstrated to be associated with certain class I HLA molecules, such as the HLA-B35-Cw∗04 haplotype (3, 11). Slow disease progression is linked with several class I HLA molecules, notably HLA-B∗27 and HLA-B∗57 (13, 17, 23). Recently, this protective effect was demonstrated in a mother-to-child HIV-1 transmission setting, where it was found that HLA-B∗27-restricted immunodominant cytotoxic T-lymphocyte (CTL) responses play a crucial role in delaying disease progression (12).
Similar protective effects were also observed in simian immunodeficiency virus (SIV) infection, particularly in association with expression of the Mamu-A∗01 MHC allele. For instance, expression of Mamu-A∗01 significantly delayed disease progression in SIVmac 251 infection (8, 9, 25). Since this allele has been consistently associated with the CTL response to an immunodominant epitope derived from the SIV Gag region (SIV Gag residues 181 to 189, designated p11CM), detectable by sensitive tetramer and ELISPOT assays, Mamu-A∗01 monkeys have been favored for evaluation of AIDS vaccine candidates (1, 2, 29).
Infection with a pathogenic isolate of simian-human immunodeficiency virus (SHIV 89.6P), causes severe CD4 lymphopenia and rapid disease progression in rhesus monkeys (16, 27). Because of the highly pathogenic nature of this virus and its consistent replication kinetics in rhesus monkeys, this challenge model is widely adopted for evaluation of potential AIDS vaccines (1, 2, 29). To further understand the pathogenesis of SHIV 89.6P infection, particularly the impact of expression of the Mamu-A∗01 allele on disease progression, we monitored a cohort of intravenously infected rhesus monkeys with and without expression of the Mamu-A∗01 allele. Here we report that expression of the Mamu-A∗01 allele was associated with delayed disease progression to AIDS in SHIV 89.6P infection. Early virus-specific CTL responses, especially Mamu-A∗01-restricted CTL responses, likely contributed to the significant reduction of initial viral replication observed in lymphoid tissue. Reduced viral replication in lymphoid tissue allowed lymphoid tissue structure to remain relatively intact, thereby resulting in attenuation of disease progression.
MATERIALS AND METHODS
Animals, virus, infections, and collection of LN biopsy specimens.
Rhesus monkeys (Macaca mulatta) were maintained in accordance with the institutional animal care protocols of Merck Research Laboratories (West Point, Pa.), New Iberia Research Center (New Iberia, La.), and Primedica. Monkeys were typed for Mamu-A*01 allele expression by PCR with sequence-specific primers as previously described (18). Animals that were positive by PCR were subsequently confirmed by DNA sequencing of the amplified region. Eight Mamu-A∗01+ monkeys and eight Mamu-A∗01− monkeys were challenged intravenously with 50 50% monkey infectious doses (50 MID50) of cell-free SHIV 89.6P (27). Sequential lymph node (LN) biopsies were performed on three Mamu-A∗01+ and three Mamu-A∗01− monkeys at days 12, 20, 40, and 100 postchallenge. LN tissues were collected and fixed in 4% paraformaldehyde and Molecular Biology Fixative (Streck Laboratories, Omaha, Nebr.). Animal maintenance and procedures were carried out in accordance with the guidelines established by the National Institutes of Health “Guide for the Care and Use of Laboratory Animals.” In brief, the animals were euthanized under institutional guidelines that were established prior to initiation of the studies, in order to provide a basis for protecting the welfare of the experimental animals. The moribund monkeys were euthanized at the discretion of the veterinarians overseeing the study, and the decisions were based on clinical evaluations of laboratory data and clinical manifestations: clinical blood chemistry and hematology parameters, CD4 counts, viral load levels, prolonged opportunistic infections, weight loss, and most importantly, the clinical condition of the animal. The veterinarians were not biased in their decisions by whether the subject to be euthanized expressed the Mamu-A∗01 allele, and the final euthanasia decisions were usually made after consultations with other veterinarians on staff.
ISH of vRNA in LNs.
Viral RNA (vRNA) was detected by in situ hybridization (ISH) with 35S-labeled RNA probes as described previously(35). Briefly, after fixed tissues were embedded in paraffin, 5-μm-thick sections were cut, adhered to silanized slides, and deparaffinized. Sections were then immersed in 0.2 N HCl for 30 min, 0.15 M triethanolamine (pH 7.4) for 15 min, and 0.005% digitonin for 5 min, all at room temperature. The slides were then incubated for 15 min at 37°C in a buffered solution containing 2 mM CaCl2 and proteinase K (5 μg/ml). After the slides were removed and washed, they were acetylated (0.25% acetic anhydride) for 10 min and then dehydrated through graded solutions of ethanol. The sections were then hybridized to SIV-specific RNA riboprobes complementary to about 10% of the sequences at the 5′ end of the SIVmac251 RNA genome labeled to specific activities of about 2 × 109 dpm/μg by incorporation of 35S-labeled UTP. The hybridization solution with the probe was sealed under a coverslip with rubber cement, and the slides were incubated for 24 h at 45°C. After the coverslips were removed, the slides were immersed in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-10 mM dithiothreitol at 42°C for 30 min, in 2× SSC-50% formamide-10 mM dithiothreitol at 60°C for 20 min, and in 1× riboprobe wash solution (RWS, comprising 0.1 M Tris-HCl [pH 7.5], 0.4 M NaCl, and 50 mM EDTA) for 10 min. The sections were then digested at 37°C with RNases A (25 μg/ml) and T1 (25 U/ml) for 30 min, washed at 37°C in RWS for 15 min, washed in 2× SSC in RWS at 37°C for 15 min, washed in 0.1× SSC at 37°C for 15 min, dehydrated through graded alcohols containing 0.3 M ammonium acetate, dried, and coated with Kodak NTB-2 emulsion. After autoradiographic exposure for 10 days at 4°C, the slides were developed and stained for 1 min in hematoxylin. Nonspecific background was determined as previously described (35). Cells with 8 grains over background were regarded as SHIV RNA+ cells.
IHC staining.
CD4+ cells and follicular dendritic cells (FDCs) were identified by immunohistochemical (IHC) staining with an anti-CD4 monoclonal antibody (NCL-CD4-IF6; Novocastra Laboratories, Newcastle, United Kingdom) and an anti-human CD35 monoclonal antibody at a 1:10 dilution (Dako, Carpinteria, Calif.), respectively. At least six 5-μm-thick sections were cut at different levels from the tissue blocks, attached to silane-treated slides, and then deparaffinized, rehydrated, immersed in 10 mM sodium citrate buffer (pH 6.0), and microwaved for 5 min. The Envision kit (Dako) was used to visualize positively stained cells according to the manufacturer's instructions. To investigate B-cell proliferation, tissue sections were costained with anti-Ki67 and anti-CD20 monoclonal antibodies at a 1:10 dilution (Novocastra Laboratories). The Envision double stain kit (Dako) was used according to the manufacturer's instructions.
QIA.
Silver grain counts were determined by quantitative image analysis (QIA) of autoradiographs illuminated by epipolarized light (14). Video images were captured with a low-light cooled charge-coupled device camera (Optronics TEC-470) and MetaMorph software (Universal Imaging, West Chester, Pa.). Silver grains were distinguished from the background and measured with the “threshold” and “measure object” tools of the MetaMorph software. The frequency of cells with vRNA in a defined tissue area (in square millimeters) was determined with the calibration tool. The amount of vRNA on the FDC network was determined as the silver grain counts of vRNA per germinal center (GC) (also in square millimeters). The number of CD4+ cells and the fraction of FDC network staining in LNs were also determined by QIA (36, 37).
Tetramer staining for the SIV Gag p11CM epitope.
Preparation of the p11CM tetramer reagent and staining of lymphocytes within whole blood have been described previously (29). Briefly, phycoerythrin-labeled tetrameric Mamu-A∗01-peptide complexes were used in conjunction with peridinin chlorophyll protein-labeled anti-human CD8α (SK1; Becton Dickinson, San Jose, Calif.) and allophycocyanin-labeled anti-rhesus monkey CD3+ (FN18) monoclonal antibodies to stain peptide-specific CD8+ T cells. A 100-μl sample of whole blood from each monkey was directly stained with these reagents, lysed, washed, and fixed. Samples were analyzed by flow cytometry on a FACScalibur (Becton Dickinson). Gated CD3+ CD8+ T cells were examined for staining with the tetrameric Mamu-A∗01-p11CM complex.
IFN-γ ELISPOT assay.
Sterile 96-well microtiter plates (polyvinylidene difluoride, MAIP S45; Millipore) were coated overnight at 4°C with 100 μl of a mouse anti-human gamma interferon (IFN-γ) monoclonal antibody/well, diluted at 10 μg/ml in sterile phosphate-buffered saline (PBS) (R&D Systems, Minneapolis, Minn.). Coated plates were washed with sterile PBS and blocked for 2 h at 37°C with 200 μl of R10 complete medium (RPMI-1640 plus 10% heat-inactivated fetal bovine serum)/well. Rhesus peripheral blood mononuclear cells (PBMCs) diluted in R10 were added with synthetic peptides to represent the SIV Gag antigen at a final concentration of 2.5 μg/ml per peptide. A peptide-free dimethyl sulfoxide diluent matching the dimethyl sulfoxide concentration in the peptide solutions was used as a negative control (medium control). Plates were incubated for 24 h in a humidified CO2 incubator at 37°C and then washed seven times with PBS containing 0.05% Tween 20 (Sigma). A biotinylated goat anti-human IFN-γ monoclonal antibody (catalog no. BAF285; R&D Systems) was diluted to 0.1 μg/ml in assay diluent consisting of PBS, 5% heat inactivated fetal bovine serum, and 0.005% Tween 20. The diluted antibody was added to the plates at 100 μl/well and incubated overnight at 4°C. After seven washes with PBS-Tween, 100 μl of alkaline phosphatase-conjugated streptavidin (BD PharMingen, San Diego, Calif.)/well at 1:2,500 in assay diluent was added to each well. After a wash, 100 μl of the precipitating alkaline phosphatase substrate NBT/BCIP (Nitro Blue Tetrazolium-5-bromo-4-chloro-3-indolylphosphate; Pierce)/well was added to each well for spot development.
RESULTS
Delayed disease progression in Mamu-A∗01+ rhesus monkeys following intravenous inoculation with SHIV 89.6P.
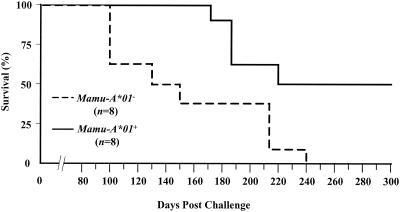
Sixteen rhesus monkeys were inoculated intravenously with 50 MID50 of SHIV 89.6P (2, 29). Survival analysis showed that the median times to death after challenge were 221 days for Mamu-A∗01+ monkeys and 131 days for Mamu-A∗01− monkeys (Fig. 1). The difference in survival rates between these two groups of monkeys was highly significant (P = 0.0137 by the log rank test). Evidently, expression of the Mamu-A∗01 allele corresponded to delayed disease progression in SHIV 89.6P infection.
FIG. 1.
Survival curves of Mamu-A∗01+ and Mamu-A∗01− monkeys following intravenous challenge with SHIV 89.6P.
Plasma viral load and peripheral CD4 counts.
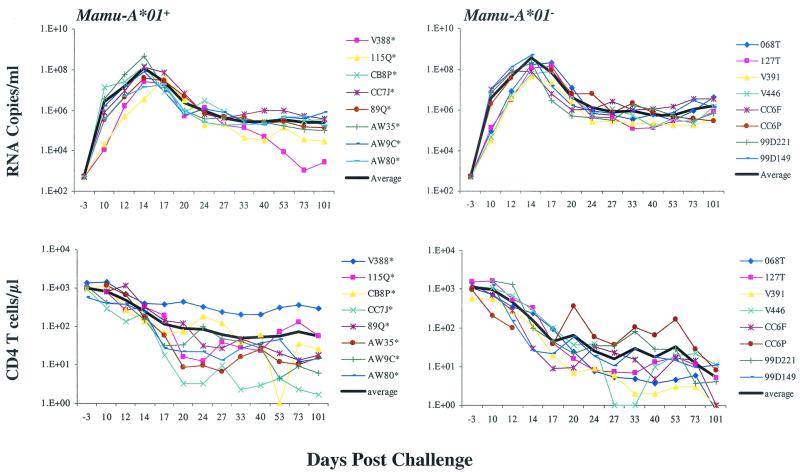
Plasma viral load is a proven surrogate for prognosis of disease progression in HIV-1-infected individuals (21). We therefore sought to determine whether slower disease progression in Mamu-A∗01+ monkeys was associated with lower plasma viral loads. We measured plasma viral loads in the monkeys by a sensitive branched-DNA amplification assay with a detection limit of 500 copies/ml (Bayer Diagnostics, San Jose, Calif.). We observed that although the average plasma viral loads in Mamu-A∗01− monkeys were approximately threefold higher at days 12, 14, and 17 than those in Mamu-A∗01+ monkeys, there was no statistically significant difference in plasma viral load at either the acute viremia peak level or the persistent set point level at day 40 (P = 0.1949 by the Wilcoxon rank sum test) (Fig. 2). The acute peripheral CD4+ T-cell lymphopenia associated with the infection was also observed, but no statistically significant differences at day 40 between Mamu-A∗01+ and Mamu-A∗01− monkeys could be documented (P = 0.1049 by the Wilcoxon rank sum test) (Fig. 2).
FIG. 2.
Postchallenge viremia and CD4+ cell count profiles. Plasma viral levels were determined by use of a branched-DNA amplification assay with a detection limit of 500 vRNA copies/ml (Bayer Diagnostics). For CD4+ cell counts, total lymphocyte numbers were determined by the percent CD3+/CD4+ lymphocyte staining based on flow cytometry and reported as the number of cells per microliter of whole blood. The thick black line in each figure represents the average curve.
Viral load in LNs.
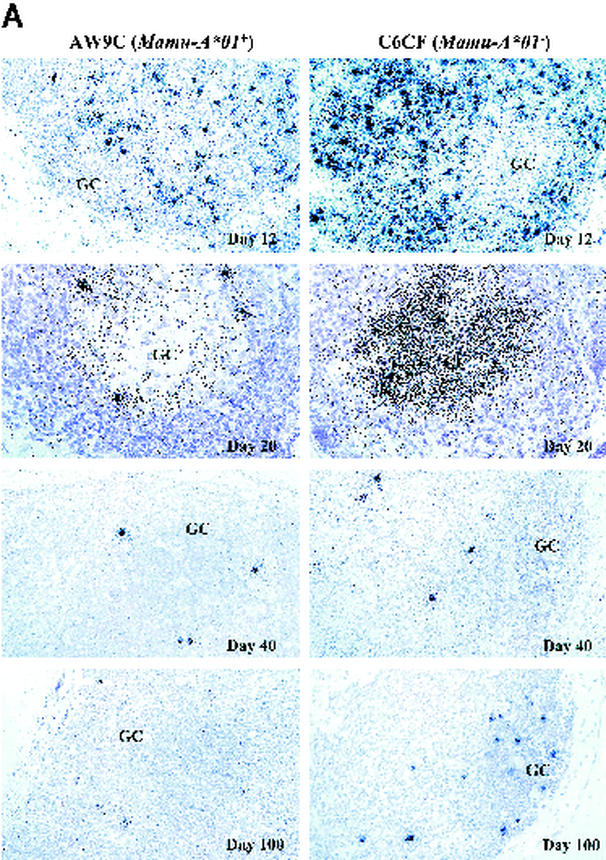
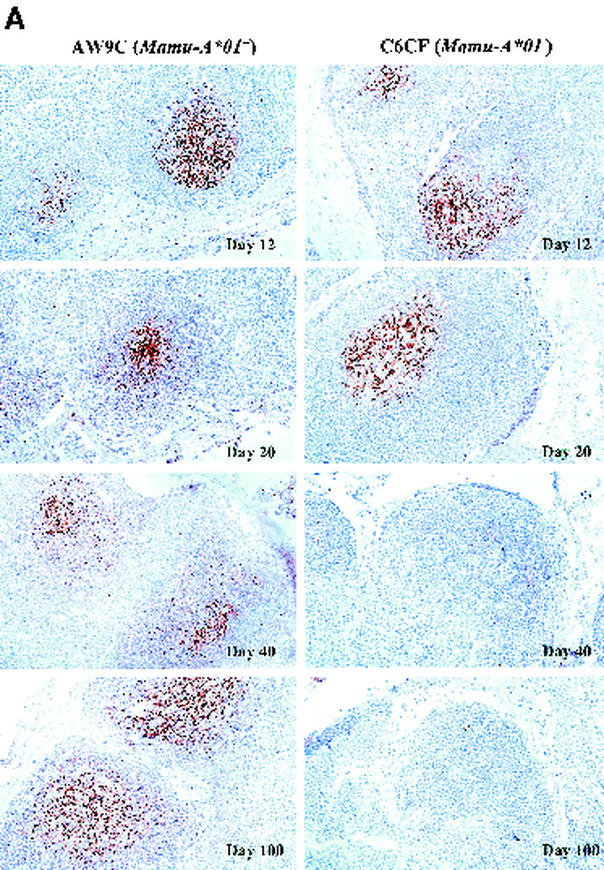
Lymphoid tissue is the major reservoir of HIV-1 replication in vivo (7, 26, 34). To determine whether there were any differences in tissue viral load between Mamu-A∗01+ and Mamu-A∗01− monkeys, we analyzed LN biopsy samples collected sequentially from six monkeys (three Mamu-A∗01+ and three Mamu-A∗01−) at 12, 20, 40, and 100 days after SHIV 89.6P challenge. We examined tissue sections by ISH for cells with detectable SHIV RNA (12). Strikingly, the frequency of vRNA+ cells was significantly lower in Mamu-A∗01+ monkeys than in Mamu-A∗01− monkeys (Fig. 3A). QIA revealed that at day 12, the average frequency of vRNA+ cells was threefold lower in Mamu-A∗01+ monkeys than in Mamu-A∗01− monkeys (Table 1). Moreover, the average level of viral gene expression per infected cell was twofold lower in Mamu-A∗01+ monkeys (data not shown). However, no difference was observed at day 20 or at succeeding time points (Table 1; Fig. 3A). These data demonstrate that during the acute phase of infection, peak levels of viral replication in LNs are markedly lower in Mamu-A∗01+ monkeys than in Mamu-A∗01− monkeys.
FIG. 3.
ISH of SHIV RNA (A) and staining of CD4+ cells (B) in LNs. The frequency of vRNA+ cells and the amount of vRNA deposited on the FDC network in the GC are shown for two representative monkeys, AW9C (Mamu-A∗01+) and C6CF (Mamu-A∗01−), at days 12, 20, 40, and 100 postchallenge. In the day 12 panels, the frequency of vRNA+ cells is significantly lower in AW9C than in C6CF (magnification, ×160; original magnification, ×200). In the day 20 panels, at a higher magnification (×320; original magnification, ×400), the level of vRNA in GCs is also significantly lower in AW9C than in C6CF. By days 40 and 100, there is no significant difference in either the frequency of vRNA+ cells or the amount of vRNA in GCs between these two monkeys. CD4 T-cell staining shows a higher density of CD4+ cells in the paracortical region (T-cell zone) in AW9C than in C6CF (magnification, ×160; original magnification, ×200). From day 20 to day 100, most CD4+ cells were depleted in both animals.
TABLE 1.
Changes in viral load and architecture in LNs following SHIV 89.6P challengea
| Monkey | Time of biopsy | Frequency of SHIV RNA+ cells per 1-mm2 tissue section | Silver grain counts per GC | No. of CD4+ cells per 1-mm2 tissue section | Fraction of FDC network (%) | Fraction of Ki67+ CD20+ GCs (%) |
|---|---|---|---|---|---|---|
| Mamu-A*01+ | ||||||
| 89Q | Day 12 | 128.0 | 0 | 11,100 | 0.01 | 100 |
| Day 20 | 12.0 | 792 | 275 | 0.01 | NT | |
| Day 40 | 1.7 | 0 | 125 | NT | NT | |
| Day 100 | 0.1 | 0 | 75 | 0.07 | 91 | |
| AW9C | Day 12 | 279.0 | 0 | 13,225 | 0.34 | 100 |
| Day 20 | 0.7 | 987 | 225 | 0.42 | 44 | |
| Day 40 | 5.4 | 0 | 250 | 0.15 | 72 | |
| Day 100 | 0.3 | 0 | 475 | 0.22 | 84 | |
| CB8P | Day 12 | 482.0 | 0 | 9,650 | 0.31 | 100 |
| Day 20 | 6.0 | 815 | 650 | 0.11 | 80 | |
| Day 40 | 1.1 | 944 | 575 | 0.3 | 72 | |
| Day 100 | 1.8 | 0 | 750 | 0.6 | 64 | |
| Mamu-A*01− | ||||||
| 99D221 | Day 12 | 1,206.0 | 0 | 4,675 | 0.22 | 100 |
| Day 20 | 3.0 | 1,626 | 175 | 0.06 | 13 | |
| Day 40 | 3.3 | 0 | 300 | 0 | 0 | |
| Day 100 | 0.6 | 0 | 375 | 0 | 0 | |
| C6CF | Day 12 | 990.0 | 0 | 6,200 | 0.14 | 96 |
| Day 20 | 2.0 | 6,409 | 1,025 | 0.19 | 4 | |
| Day 40 | 7.0 | 0 | 275 | 0 | 0 | |
| Day 100 | 24.0 | 0 | 325 | 0 | 0 | |
| CC6P | Day 12 | 421.0 | 0 | 5,925 | 0.2 | 100 |
| Day 20 | 5.0 | 7,276 | 1,875 | 0.09 | 61 | |
| Day 40 | 0.6 | 1,351 | 2,350 | 0.42 | 83 | |
| Day 100 | 0.9 | 1,620 | 2,450 | 0.08 | 14 |
The frequency of SHIV RNA+ cells, levels of SHIV RNA in GCs, and number of CD4+ cells in LNs were quantitated by QIA (35, 36). The cutoff value for silver grain counts per GC is 500. The fraction of the FDC network is calculated as the area staining positive for CD35 divided by the total LN area (37). The fraction of Ki67+ CD20+ GCs is calculated as the total number of Ki67+ CD20+ GCs divided by the total number of GCs in LN sections. NT, not tested.
A significant proportion of the viral load in the LNs is associated with FDCs in HIV-1 infection (10, 14). Virions are trapped on the FDC surfaces in the form of virus-antibody complexes, and these complexes may serve as continuing sources of infectious virus (15). Using QIA, at day 12 we observed no detectable vRNA in the FDC compartments of either Mamu-A∗01+ and Mamu-A∗01− monkeys (Table 1; Fig. 3A). As previously reported (4), this could be due to insufficient levels of anti-SHIV antibodies at this time point. By day 20, vRNA was detectable in the FDC compartment in all monkeys. Interestingly, the amounts of vRNA in the FDC compartments were significantly lower in Mamu-A∗01+ than in Mamu-A∗01− monkeys (Table 1; Fig. 3A). From day 40 to day 100, vRNA in the FDC compartment became undetectable in most of the Mamu-A∗01+ and Mamu-A∗01− animals. Hence, at day 20, the amount of vRNA trapped on FDCs reflects the level of viral replication in the LNs at day 12.
CD4+ cells in LNs.
We next analyzed the kinetics of depletion of LN CD4+ cells following challenge by using IHC detection with an anti-CD4 monoclonal antibody (36). At day 12, the average number of CD4+ cells was twofold higher in Mamu-A∗01+ monkeys than in Mamu-A∗01− monkeys (Table 1; Fig. 3B). The average number of CD4+ cells decreased more than 10-fold by day 20 and stayed at similar levels at days 40 and 100 in both monkey groups. Reduced depletion of LN CD4+ cells in Mamu-A∗01+ monkeys was observed at day 12, corresponding to the lower levels of viral replication at the same time point.
Pathological changes in the FDC network.
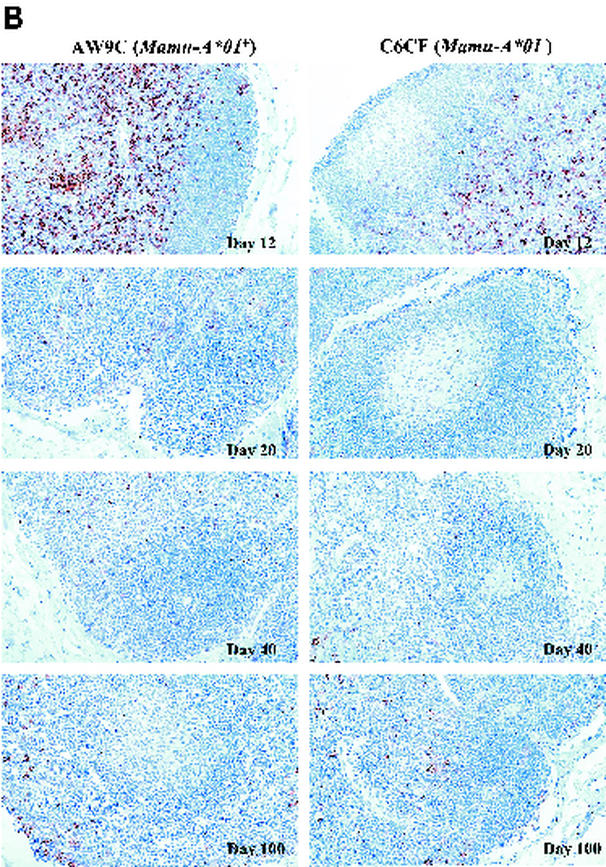
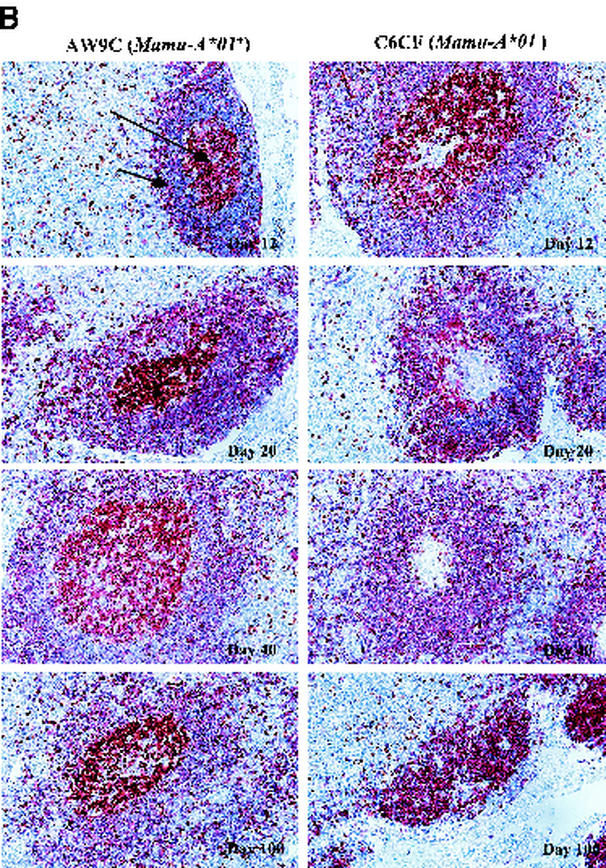
We then evaluated morphological changes of the FDC network, one of the key physiological elements in LNs, which serves as an antigen repository and provides critical anatomical structure for the interactions between B cells and T cells (30, 31). In HIV-1 infection, the FDC network is gradually disrupted and eventually destroyed by undefined mechanisms (32). We performed IHC staining on sequential LN biopsy specimens by using an anti-CD35 monoclonal antibody (37). We observed relatively intact morphology of the FDC network at day 12 in both the Mamu-A∗01+ and Mamu-A∗01− groups (Table 1; Fig. 4A). However, morphological changes, including fragmentation and thickening of the FDC network processes, became evident at day 20 in two of three MamuA∗01− monkeys (Fig. 4A). Furthermore, the FDC network processes could not be detected in entire tissue sections from these two Mamu-A∗01− monkeys at days 40 and 100, indicating complete destruction of the FDC network (Fig. 4A). The third MamuA∗01− monkey showed slightly delayed degeneration of the FDC network, and this corresponded to lower levels of initial viral replication observed at day 12 (Table 1). In contrast, all three Mamu-A∗01+ monkeys consistently exhibited relatively intact morphology of FDC network processes at these time points (Fig. 4A and Table 1). To quantitatively document the degree of destruction of the FDC network, we used QIA to measure the fraction of the LN that was occupied by the FDC network (37), and these data were consistent with the morphological observations (Table 1). The kinetics of destruction of the FDC network, therefore, is associated with the amounts of vRNA in the FDC compartment and the levels of initial viral replication in the LNs during the acute phase of infection.
FIG. 4.
Staining of FDC network (A) and proliferating B cells in GCs (B) in LNs. (A) The normal morphology of the FDC network is shown in day 12 panels for both monkeys, AW9C (Mamu-A∗01+) and C6CF (Mamu-A∗01−). In day 20 panels, the processes appear to be fragmented in C6CF but not in AW9C. At days 40 and 100, staining of the FDC network is completely absent in C6CF but obvious in AW9C. (B) Ki67+ cells and CD20+ cells are shown in brown and purple, respectively. The long arrow points to proliferating B cells (Ki67+ CD20+) locating in GCs. The short arrow points to the follicular mantle area, which is packed with nonproliferating CD20+ B cells. Proliferating B cells in GCs were observed in both monkeys at day 12. However, from day 20, there were no proliferating B cells in GCs in C6CF (shown as a blank, nonstaining GC area). GCs gradually became smaller and were almost invisible at day 100 in C6CF, indicating complete destruction of GCs. Magnification, ×160; original magnification, ×200.
Pathological changes of proliferating B cells in GCs.
FDCs not only serve as the major antigen-presenting cell network for B cells in the GC but also are responsible for the initiation of B-cell responses and maintenance of B-cell memory (33). To determine whether the destruction of the FDC network affected B-cell proliferation in the GC, we double-stained LN tissue sections with the cellular proliferation marker Ki67 and the B-cell surface marker CD20. At day 12, B-cell proliferation was evident in almost all GCs in LN samples from both monkey groups (Table 1; Fig. 4B), and the results were comparable to those for non-SHIV-infected monkeys (data not shown). Higher levels of GC hyperplasia were observed in Mamu-A∗01− monkeys at this time point (Fig. 4B and unpublished data). The GCs were surrounded by a follicular mantle consisting of nonproliferating B cells. Strikingly, at day 20, loss of proliferating B cells in most of the GCs was evident in two of three Mamu-A∗01− monkeys, and these two monkeys also started showing pathological changes in the FDC network at the same time point (Fig. 4A). Moreover, we could not detect any proliferating B cells in the GCs of these two Mamu-A∗01− monkeys at day 40. This observation coincided with complete destruction of the FDC network in these animals (Fig. 4A). By day 100, all three Mamu-A∗01− animals essentially had no proliferating B cells in the GCs. In contrast, proliferating B cells were consistently observed in most of the GCs of all three Mamu-A∗01+ animals, although a minor loss of proliferating B cells also occurred in these animals at the later time points (Table 1). No obvious depletion of nonproliferating B cells in the follicular mantle area was observed for either group (Fig. 4B). Noticeable disruption of the FDC network, therefore, occurred simultaneously with a significant loss of GC proliferating B cells during the acute phase of infection in Mamu-A∗01− monkeys. This histopathological change progressed rapidly and apparently led to the complete destruction of the FDC network and finally to a complete loss of proliferating B cells in GCs. However, destruction of the FDC network and loss of GC proliferating B cells were much slower in Mamu-A∗01+ monkeys.
Kinetics of tetramer staining and ELISPOT assay for the SIV Gag p11CM epitope.
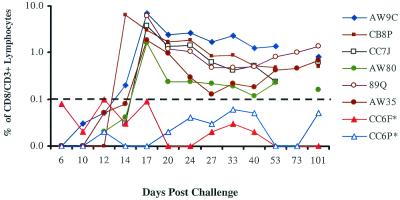
Mamu-A∗01-restricted immunodominant CTL responses against the SIV Gag p11CM epitope have been shown to be associated with reduction of early viral replication in vaccine studies (1, 2, 29). In this study, sequential tetramer staining on whole-blood samples showed an active anti-p11CM CD8+ T-cell response in all Mamu-A∗01+ monkeys, as expected. Tetramer staining levels reached a peak at day 17, ranging from 2 to 7% of circulating CD3+ CD8+ T cells, and gradually declined to around 1% of CD3+ CD8+ T cells (Fig. 5). We also conducted an IFN-γ ELISPOT assay with PBMCs at two selected time points postinfection to confirm the functional nature of the tetramer-positive CD3+ CD8+ T cells. PBMCs from eight monkeys (four Mamu-A∗01+ and four Mamu-A∗01− monkeys) were incubated with either the p11CM peptide or a pool of overlapping peptides (designated the KD pool) covering the SIV gag open reading frame. The KD peptide pool spans the length of SIV Gag but deletes the two overlapping peptides which contain the p11CM sequence and was designed to measure non-p11CM-directed T-cell responses in the ELISPOT assay. All Mamu-A∗01+ monkeys developed strong T-cell responses primarily directed against the p11CM epitope (Table 2). Conversely, very low or undetected levels of T-cell responses were noted at both time points in Mamu-A∗01− monkeys.
FIG. 5.
Kinetics of CD8+ T-cell responses detected by tetramer staining following intravenous challenge with SHIV 89.6P. The percentage of CD3+ CD8+ cells indicates the levels of circulating p11CM-specific CD8+ T cells for Mamu-A∗01-expressing monkeys. The cutoff value was 0.1%. Two animals (designations marked with asterisks) were Mamu-A∗01 negative (CC6F and CC6P).
TABLE 2.
IFN-γ ELISPOT response in monkeys infected with SHIV 89.6P
| Monkey | No. of spot-forming cells/106 PBMC in response to the indicated antigen at the following day postchallenge:
|
|||||
|---|---|---|---|---|---|---|
| Day 24
|
Day 73
|
|||||
| Medium | KDa | p11CM | Medium | KD | p11CM | |
| Mamu-A*01+ | ||||||
| AW80 | 0 | 22 | 352 | 0 | 3 | 175 |
| AW9C | 0 | 110 | 2,912 | 3 | 218 | 1,755 |
| CB8P | 2 | 25 | 2,425 | 0 | 48 | 1,083 |
| CC7J | 2 | 10 | 2,048 | 0 | 5 | 348 |
| Mamu-A*01− | ||||||
| CC6F | 0 | 10 | 2 | 5 | 5 | 5 |
| CC6P | 2 | 32 | 2 | 13 | 8 | 0 |
| 99D221 | 0 | 8 | 0 | NTb | NT | NT |
| 99D149 | 0 | 58 | 10 | NT | NT | NT |
KD, a pool of overlapping peptides covering the SIV gag open reading frame. The peptides containing the p11CM epitope sequence are omitted from the pool.
NT, not tested.
DISCUSSION
Our data demonstrate that expression of the Mamu-A∗01 allele has a profound positive impact on disease progression in SHIV 89.6P-infected rhesus monkeys. This immunodominant MHC class I allele mediates a substantial cellular immune response against the virus that is associated with a noted preservation of LN architecture. This preservation appears to be the underlying basis for the delay in disease progression. It has been suggested that in HIV-1 infection, the attenuation of disease progression associated with the expression of several MHC class I alleles, such as HLA-B∗27 and HLA-B∗57, may result from early induction of the allele-restricted immunodominant CTL responses (12, 17). Based on our observations with the SHIV infection model, we hypothesize that MHC class I allele-restricted CTL responses may significantly blunt initial viral replication and thus limit the destruction of lymphoid tissue, the major structural component of the host immune system. Preservation of lymphoid tissue structure enables the host to maintain a relatively intact global immune function, which plays an important role in controlling HIV-1 replication and other opportunistic infections, thus resulting in slower disease progression.
The mechanisms by which the FDC networks were disrupted and proliferating B cells in the GCs were lost following SHIV infection are unclear. For the Mamu-A∗01− monkeys in which these effects were most profound, the severe depletion of CD4+ T cells observed during the acute phase of infection was likely a contributing factor, given the essential role of CD4+ cells in the formation of the FDC network and in providing the B-cell helper function. It is also possible that the presence of large amounts of virus-antibody complexes in the FDC compartment may have played a role, as has been suggested for HIV-1 infection (32).
Under normal physiological conditions, GC B cells are constantly undergoing vigorous proliferation in response to foreign antigens and therefore play an important role in host immune surveillance. GCs have been shown to be occupied by virus-specific and nonspecific B cells in SHIV 89.6P infection (20). Thus, the severe loss of proliferating B cells in GCs observed in the Mamu-A∗01− monkeys may have caused the substantial loss of general B-cell immunity that has been documented for HIV-1 and SIV infection (6, 22). Along with T-cell dysfunction, loss of B-cell immunity likely contributes to the rapid development of the immunodeficiency and clinical deterioration seen in these animals.
Our findings have important implications for HIV-1 vaccine development and for strategies for antiretroviral therapy. Intervention should be targeted at slowing down the process of destruction of lymphoid tissues by inhibiting early viral replication. It has been demonstrated that the CTL-based AIDS vaccine candidates and early antiretroviral regimens that significantly reduce the initial level of viral replication are notably beneficial to the hosts (1, 2, 19, 29). Therefore, early preservation of lymphoid tissue from HIV-mediated destruction is likely to be a requirement for an effective AIDS vaccine as well as an important goal for intervention by antiretroviral therapy.
Acknowledgments
We are grateful to Ashley Haase for critical review of the manuscript, to Norman Letvin for the SHIV 89.6P challenge stock, and to Robert Druilhet and Rachel Colligan of New Iberia Research Center for contributions to this research. We also thank Susan Alderfer, Carey Hauer, and Jill Williams for assistance with preparation of the figures.
REFERENCES
- 1.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 2.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, and M. G. Lewis. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 3.Carrington, M., G. W. Nelson, M. P. Martin, T. Kissner, D. Vlahov, J. J. Goedert, R. Kaslow, S. Buchbinder, K. Hoots, and S. J. O'Brien. 1999. HLA and HIV-1: heterozygote advantage and B∗35-Cw∗04 disadvantage. Science 283:1748-1752. [DOI] [PubMed] [Google Scholar]
- 4.Chakrabarti, L., M. C. Cumont, L. Montagnier, and B. Hurtrel. 1994. Variable course of primary simian immunodeficiency virus infection in lymph nodes: relation to disease progression. J. Virol. 68:6634-6643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Sorrentino, A. H., K. Marinic, P. Motta, A. Sorrentino, R. Lopez, and E. Illiovich. 2000. HLA class I alleles associated with susceptibility or resistance to human immunodeficiency virus type 1 infection among a population in Chaco Province, Argentina. J. Infect. Dis. 182:1523-1526. [DOI] [PubMed] [Google Scholar]
- 6.Dykhuizen, M., J. L. Mitchen, D. C. Montefiori, J. Thomson, L. Acker, H. Lardy, and C. D. Pauza. 1998. Determinants of disease in the simian immunodeficiency virus-infected rhesus macaque: characterizing animals with low antibody responses and rapid progression. J. Gen. Virol. 79:2461-2467. [DOI] [PubMed] [Google Scholar]
- 7.Embretson, J., M. Zupancic, J. Beneke, M. Till, S. Wolinsky, J. L. Ribas, A. Burke, and A. T. Haase. 1993. Analysis of human immunodeficiency virus-infected tissues by amplification and in situ hybridization reveals latent and permissive infections at single-cell resolution. Proc. Natl. Acad. Sci. USA 90:357-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evans, D. T., P. Jing, T. M. Allen, D. H. O'Connor, H. Horton, J. E. Venham, M. Piekarczyk, J. Dzuris, M. Dykhuizen, J. Mitchen, R. A. Rudersdorf, C. D. Pauza, A. Sette, R. E. Bontrop, R. DeMars, and D. I. Watkins. 2000. Definition of five new simian immunodeficiency virus cytotoxic T-lymphocyte epitopes and their restricting major histocompatibility complex class I molecules: evidence for an influence on disease progression. J. Virol. 74:7400-7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Evans, D. T., L. A. Knapp, P. Jing, J. L. Mitchen, M. Dykhuizen, D. C. Montefiori, C. D. Pauza, and D. I. Watkins. 1999. Rapid and slow progressors differ by a single MHC class I haplotype in a family of MHC-defined rhesus macaques infected with SIV. Immunol. Lett. 66:53-59. [DOI] [PubMed] [Google Scholar]
- 10.Fauci, A. S. 1993. Multifactorial nature of human immunodeficiency virus disease: implications for therapy. Science 262:1011-1018. [DOI] [PubMed] [Google Scholar]
- 11.Gao, X., G. W. Nelson, P. Karacki, M. P. Martin, J. Phair, R. Kaslow, J. J. Goedert, S. Buchbinder, K. Hoots, D. Vlahov, S. J. O'Brien, and M. Carrington. 2001. Effect of a single amino acid change in MHC class I molecules on the rate of progression to AIDS. N. Engl. J. Med. 344:1668-1675. [DOI] [PubMed] [Google Scholar]
- 12.Goulder, P. J., C. Brander, Y. Tang, C. Tremblay, R. A. Colbert, M. M. Addo, E. S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, M. Altfeld, S. He, M. Bunce, R. Funkhouser, S. I. Pelton, S. K. Burchett, K. McIntosh, B. T. Korber, and B. D. Walker. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412:334-338. [DOI] [PubMed] [Google Scholar]
- 13.Goulder, P. J., R. E. Phillips, R. A. Colbert, S. McAdam, G. Ogg, M. A. Nowak, P. Giangrande, G. Luzzi, B. Morgan, A. Edwards, A. J. McMichael, and S. Rowland Jones. 1997. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat. Med. 3:212-217. [DOI] [PubMed] [Google Scholar]
- 14.Haase, A. T., K. Henry, M. Zupancic, G. Sedgewick, R. A. Faust, H. Melroe, W. Cavert, K. Gebhard, K. Staskus, Z. Q. Zhang, P. J. Dailey, H. H. Balfour, A. Erice, and A. S. Perelson. 1996. Quantitative image analysis of HIV-1 infection in lymphoid tissue. Science 274:985-989. [DOI] [PubMed] [Google Scholar]
- 15.Heath, S. L., J. G. Tew, A. K. Szakal, and G. F. Burton. 1995. Follicular dendritic cells and human immunodeficiency virus infectivity. Nature 377:740-744. [DOI] [PubMed] [Google Scholar]
- 16.Igarashi, T., C. R. Brown, Y. Endo, A. Buckler White, R. Plishka, N. Bischofberger, V. Hirsch, and M. A. Martin. 2001. Macrophages are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): implications for HIV-1 infections of humans. Proc. Natl. Acad. Sci. USA 98:658-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaslow, R. A., M. Carrington, R. Apple, L. Park, A. Munoz, A. J. Saah, J. J. Goedert, C. Winkler, S. J. O'Brien, C. Rinaldo, R. Detels, W. Blattner, J. Phair, H. Erlich, and D. L. Mann. 1996. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat. Med. 2:405-411. [DOI] [PubMed] [Google Scholar]
- 18.Knapp, L. A., E. Lehmann, M. S. Piekarczyk, J. A. Urvater, and D. I. Watkins. 1997. A high frequency of Mamu-A∗01 in the rhesus macaque detected by polymerase chain reaction with sequence-specific primers and direct sequencing. Tissue Antigens 50:657-661. [DOI] [PubMed] [Google Scholar]
- 19.Lifson, J. D., J. L. Rossio, M. Piatak, Jr., T. Parks, L. Li, R. Kiser, V. Coalter, B. Fisher, B. M. Flynn, S. Czajak, V. M. Hirsch, K. A. Reimann, J. E. Schmitz, J. Ghrayeb, N. Bischofberger, M. A. Nowak, R. C. Desrosiers, and D. Wodarz. 2001. Role of CD8+ lymphocytes in control of simian immunodeficiency virus infection and resistance to rechallenge after transient early antiretroviral treatment. J. Virol. 75:10187-10199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Margolin, D. H., E. F. H. Saunders, B. Bronfin, N. de Rosa, M. K. Axthelm, X. Alvarez, and N. L. Letvin. 2002. High frequency of virus-specific B lymphocytes in germinal centers of simian-human immunodeficiency virus-infected rhesus monkeys. J. Virol. 76:3965-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mellors, J. W., C. R. Rinaldo, P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 22.Michael, N. L., A. E. Brown, R. F. Voigt, S. S. Frankel, J. R. Mascola, K. S. Brothers, M. Louder, D. L. Birx, and S. A. Cassol. 1997. Rapid disease progression without seroconversion following primary human immunodeficiency virus type 1 infection—evidence for highly susceptible human hosts. J. Infect. Dis. 175:1352-1359. [DOI] [PubMed] [Google Scholar]
- 23.Migueles, S. A., M. S. Sabbaghian, W. L. Shupert, M. P. Bettinotti, F. M. Marincola, L. Martino, C. W. Hallahan, S. M. Selig, D. Schwartz, J. Sullivan, and M. Connors. 2000. HLA B∗5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA 97:2709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Brien, S. J., and J. P. Moore. 2000. The effect of genetic variation in chemokines and their receptors on HIV transmission and progression to AIDS. Immunol. Rev. 177:99-111. [DOI] [PubMed] [Google Scholar]
- 25.Pal, R., D. Venzon, N. L. Letvin, S. Santra, D. C. Montefiori, N. R. Miller, E. Tryniszewska, M. G. Lewis, T. C. VanCott, V. Hirsch, R. Woodward, A. Gibson, M. Grace, E. Dobratz, P. D. Markham, Z. Hel, J. Nacsa, M. Klein, J. Tartaglia, and G. Franchini. 2002. ALVAC-SIV-gag-pol-env-based vaccination and macaque major histocompatibility complex class I (A∗01) delay simian immunodeficiency virus SIVmac-induced immunodeficiency. J. Virol. 76:292-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pantaleo, G., C. Graziosi, J. F. Demarest, L. Butini, M. Montroni, C. H. Fox, J. M. Orenstein, D. P. Kotler, and A. S. Fauci. 1993. HIV infection is active and progressive in lymphoid tissue during the clinically latent stage of disease. Nature 362:355-358. [DOI] [PubMed] [Google Scholar]
- 27.Reimann, K. A., J. T. Li, G. Voss, C. Lekutis, K. Tenner Racz, P. Racz, W. Lin, D. C. Montefiori, D. E. Lee Parritz, Y. Lu, R. G. Collman, J. Sodroski, and N. L. Letvin. 1996. An env gene derived from a primary human immunodeficiency virus type 1 isolate confers high in vivo replicative capacity to a chimeric simian/human immunodeficiency virus in rhesus monkeys. J. Virol. 70:3198-3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rowland Jones, S., S. Pinheiro, and R. Kaul. 2001. New insights into host factors in HIV-1 pathogenesis. Cell 104:473-476. [DOI] [PubMed] [Google Scholar]
- 29.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 30.Szakal, A. K., M. H. Kosco, and J. G. Tew. 1989. Microanatomy of lymphoid tissue during humoral immune responses: structure function relationships. Annu. Rev. Immunol. 7:91-109. [DOI] [PubMed] [Google Scholar]
- 31.Szakal, A. K., and J. G. Tew. 1992. Follicular dendritic cells: B-cell proliferation and maturation. Cancer Res. 52(19 Suppl.):5554s-5556s. [PubMed] [Google Scholar]
- 32.Tenner Racz, K., and P. Racz. 1995. Follicular dendritic cells initiate and maintain infection of the germinal centers by human immunodeficiency virus. Curr. Top. Microbiol. Immunol. 201:141-159. [DOI] [PubMed] [Google Scholar]
- 33.Tew, J. G., J. Wu, D. Qin, S. Helm, G. F. Burton, and A. K. Szakal. 1997. Follicular dendritic cells and presentation of antigen and costimulatory signals to B cells. Immunol. Rev. 156:39-52. [DOI] [PubMed] [Google Scholar]
- 34.Veazey, R. S., M. DeMaria, L. V. Chalifoux, D. E. Shvetz, D. R. Pauley, H. L. Knight, M. Rosenzweig, R. P. Johnson, R. C. Desrosiers, and A. A. Lackner. 1998. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science 280:427-431. [DOI] [PubMed] [Google Scholar]
- 35.Zhang, Z. Q., T. Schuler, M. Zupancic, S. Wietgrefe, K. A. Staskus, K. A. Reimann, T. A. Reinhart, M. Rogan, W. Cavert, C. J. Miller, R. S. Veazey, D. Notermans, S. Little, S. A. Danner, D. D. Richman, D. Havlir, J. Wong, H. L. Jordan, T. W. Schacker, P. Racz, K. Tenner Racz, N. L. Letvin, S. Wolinsky, and A. T. Haase. 1999. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science 286:1353-1357. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, Z. Q., D. W. Notermans, G. Sedgewick, W. Cavert, S. Wietgrefe, M. Zupancic, K. Gebhard, K. Henry, L. Boies, Z. Chen, M. Jenkins, R. Mills, H. McDade, C. Goodwin, C. M. Schuwirth, S. A. Danner, and A. T. Haase. 1998. Kinetics of CD4+ T cell repopulation of lymphoid tissues after treatment of HIV-1 infection. Proc. Natl. Acad. Sci. USA 95:1154-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, Z. Q., T. Schuler, W. Cavert, D. W. Notermans, K. Gebhard, K. Henry, D. V. Havlir, H. F. Gunthard, J. K. Wong, S. Little, M. B. Feinberg, M. A. Polis, L. K. Schrager, T. W. Schacker, D. D. Richman, L. Corey, S. A. Danner, and A. T. Haase. 1999. Reversibility of the pathological changes in the follicular dendritic cell network with treatment of HIV-1 infection. Proc. Natl. Acad. Sci. USA 96:5169-5172. [DOI] [PMC free article] [PubMed] [Google Scholar]