Abstract
Methanococcus maripaludis is a mesophilic archaeon that reduces CO2 to methane with H2 or formate as an energy source. It contains two membrane-bound energy-conserving hydrogenases, Eha and Ehb. To determine the role of Ehb, a deletion in the ehb operon was constructed to yield the mutant, strain S40. Growth of S40 was severely impaired in minimal medium. Both acetate and yeast extract were necessary to restore growth to nearly wild-type levels, suggesting that Ehb was involved in multiple steps in carbon assimilation. However, no differences in the total hydrogenase specific activities were found between the wild type and mutant in either cell extracts or membrane-purified fractions. Methanogenesis by resting cells with pyruvate as the electron donor was also reduced by 30% in S40, suggesting a defect in pyruvate oxidation. CO dehydrogenase/acetyl coenzyme A (CoA) synthase and pyruvate oxidoreductase had higher specific activities in the mutant, and genes encoding these enzymes, as well as AMP-forming acetyl-CoA synthetase, were expressed at increased levels. These observations support a role for Ehb in anabolic CO2 assimilation in methanococci.
Methanogens are strictly anaerobic archaea that produce methane as the major product of their energy metabolism. They play an important role in the global carbon cycle, processing 1 to 2% of the carbon fixed per year and producing most of the earth's atmospheric methane (10, 21). Methanococcus maripaludis is a mesophile that reduces carbon dioxide to methane with H2 or formate as electron donor (10). In addition, M. maripaludis assimilates acetate and some amino acids as carbon sources when they are present (24, 25, 33). Progress in genetics tools (30), relatively fast growth (11), suitability for chemostats (7), and a complete genomic sequence (9) make M. maripaludis an excellent model for the physiology of hydrogenotrophic methanogens.
Hydrogenases catalyze the reaction H2 → 2 H+ + 2 e−. These enzymes are indispensable for the growth of hydrogenotrophic methanogens, which use H2 as an electron donor. M. maripaludis contains six nickel-iron hydrogenases, including two coenzyme F420-reducing hydrogenases and two non-F420-reducing hydrogenases (9). Of each of these pairs of enzymes, one contains a selenocysteinyl residue and the other contains a cysteinyl residue at the active site (3). In addition, M. maripaludis contains genes for two separate multisubunit energy-conserving hydrogenases, Eha and Ehb (9). These open reading frames (ORFs) include subunits that are homologous to the NADH-ubiquinone oxidoreductase or complex I of mitochondria (1). In the methanogens, the energy-converting [NiFe] hydrogenase (Ech) has been purified and characterized from Methanosarcina barkeri (12, 16). M. barkeri is only distantly related to the methanococci. Although it can reduce CO2 to CH4, it also utilizes acetate and methanol as substrates for methanogenesis. In this organism, the complex Ech enzyme contains six subunits, two predicted integral membrane-spanning proteins and four subunits expected to extrude in the cytoplasm. Two of the hydrophilic proteins are homologous to the large and small subunits of the soluble [NiFe] hydrogenases, and a third hydrophilic subunit contains two [4Fe-4S] cluster binding motifs. A mutant of M. barkeri containing a deletion of the genes encoding Ech is unable to reduce CO2 to methane, grow with acetate as a substrate for methanogenesis, or biosynthesize pyruvate (17). These properties suggest that Ech catalyzes the reduction of low-potential ferredoxins by H2. Because this reaction is unfavorable at the low partial pressures of H2 typical of the habitats of methanogens, the reduction is probably driven by the proton motive force.
The hydrogenotrophic methanogens, such as M. maripaludis and Methanothermobacter marburgensis, contain two Ech homologs, Eha and Ehb. In M. marburgensis (28), the eha operon (12.5 kb) and ehb operon (9.6 kb) are composed of 20 and 17 ORFs, respectively. These operons include homologs to the large and small subunits of [NiFe] hydrogenases and the two integral membrane proteins found in the M. barkeri enzyme. In addition, these operons encode a number of polyferredoxins and other integral membrane and hydrophilic subunits. Like the enzyme from M. barkeri, these enzymes may be necessary to reduce low-potential ferredoxins during growth with low concentrations of H2 (8, 28).
Although the eha operon of M. maripaludis is similar to that of M. marburgensis, the homologs to the genes encoding Ehb are scattered in several loci around the genome (9), with only 9 of the 16 homologs to the M. marburgensis ehb genes in one cluster in M. maripaludis. A mutant with an insertion in one gene of this cluster was an acetate auxotroph, suggesting that Ehb was involved in carbon assimilation (36). This paper describes the isolation and characterization of a deletion mutant in the ehb operon of M. maripaludis. Comparison of the transcriptome and proteome between the mutant and the wild type provides further insight into the role of the Ehb hydrogenase in the methanococci and reveals a global regulatory response to the loss of its activity (Q. Xia et al., submitted for publication).
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and culture conditions.
The Methanococcus maripaludis strains used in this work were two wild-type strains, S2 (32) and JJ (10), and the ΔehbF::pac mutant S40, which was derived from S2 (see below). The Escherichia coli TOP10F′ competent cells (Invitrogen, Carlsbad, CA) were used for construction of the plasmids. The plasmids used in this work were the puromycin-resistant integration vector pIJA03 (27) and pWDK40, pIJA10, pIJA175, and pIJA176 (this work; see below). E. coli was grown in Luria-Bertani medium with ampicillin (100 μg/ml) when needed. M. maripaludis was grown with 276 kPa H2-CO2 gas (80:20 [vol/vol]) at 37°C in the mineral media McN, McNA (McN plus 10 mM sodium acetate), McYA (McNA plus 0.2% [wt/vol] yeast extract), or McCA (McNA plus 0.2% [wt/vol] Casamino Acids) as described previously (32). Puromycin (2.5 μg/ml) was added when needed. For preparation of the cell extracts, M. maripaludis was grown in bottles with 100 ml McNA medium and 138 kPa H2-CO2 gas (80:20 [vol/vol]) at 37°C.
Construction of the M. maripaludis mutants.
The ΔehbF:pac mutant was made by transformation with the suicide vector pWDK40 based upon pIJA03 (27). For the construction of pWDK40, the upstream and downstream regions of the ehbF gene were PCR amplified from genomic DNA using the primers U1 and U2 and D1 and D2, respectively (Fig. 1 and Table 1). pWDK40 was constructed in E. coli TOP10F′ by cloning the U1-U2 PCR product into the BamHI-XbaI sites and the D1-D2 PCR product into the KpnI and NheI sites of pIJA03. pWDK40 was transformed into M. maripaludis S2 by the polyethylene glycol method (29), and transformants were plated on McYA medium plus puromycin and restreaked on the same medium. The colonies were picked into McYA broth medium plus puromycin, and glycerol stocks were prepared when the cultures reached early stationary phase (29).
FIG. 1.
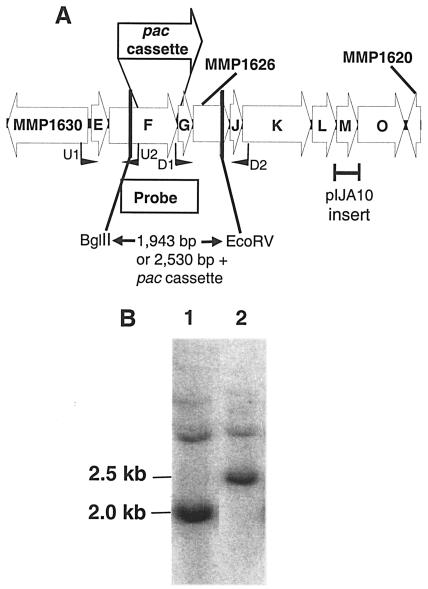
Construction of the ΔehbF:pac mutation. A. M. maripaludis S2 ehb operon (subunits E to O; Mmp1629 to Mmp1621). The ORFs Mmp1630, Mmp1626, and Mmp1620 were annotated as ATP/GTP-binding site motif A (P-loop):ABC transporter:AAA ATPase, conserved hypothetical protein, and hypothetical protein (9), respectively. The location of the primers U1, U2, D1, and D2 used to clone the upstream and downstream regions flanking ehbF are shown. The homologous portion of the DNA from M. maripaludis JJ cloned in pIJA10 is shown. B. Confirmation of the genotypes of the wild-type S2 and mutant S40 by Southern hybridization. The genomic DNA (1 μg) was digested with BglII and EcoRV prior to hybridization with the probe indicated in panel A. Lanes 1 and 2, digested genomic DNA of S2 and S40.
TABLE 1.
List of primers
| Purpose and primer name | Sequence | Details |
|---|---|---|
| Construction of plasmids | ||
| U1 | 5′-CGCGGATCCACCTTTTCTCCATACCGTTTTGTT | pWDK40 |
| U2 | 5′-CTAGTCTAGACCATAGCAAAGCCCAATAATAAGC | ΔebhF::pac mutant |
| D1 | 5′-CGGGGTACCAAACGAAATTGGAAGGGTATGGAC | |
| D2 | 5′-CTAGCTAGCACAGGTTCCGCAGGTAATACATGA | |
| BM1 | 5′-CCGAATTCCTGAAGAACCGCTATC | pIJA10, ehbM mutant |
| BM2 | 5′-GTACGCGTGGTCTTGGTGGGCATC | |
| HA1 | 5′-CGAAGATCTCAGAATGCGGTTGGTG | pIJ175, cdhA mutant |
| HA2 | 5′-GGTCTAGAGCATCAAAAATTCCTTCA | |
| HBC1 | 5′-GGACGCGTTTATCGGGGTTACTTACT | pIJ176, cdhBC mutant |
| HBC2 | 5′-GGTCTAGAGTGCATAAATCCCTGAA | |
| Probe for Southern hybridization | ||
| SH1 | 5′-GCAGTAGTTATGGCAGATGACC | ehbFG probe |
| SH2 | 5′-GAGTGTCAAAACCTCTCCAATCGAA | |
| RT-PCR | ||
| EH1 | 5′-GGGAATTAATTCGAAGCTGCTGG | Terminator pac |
| EH2 | 5′-GTGTCCGCCCAAAATTATGG | cassette, Mmp1626 |
pIJA10, an integration vector for gene disruption of ehbO expression, was constructed by inserting a 0.5-kb PCR product into EcoRI-MluI sites of pIJA03. The primers for the PCR were BM1 and BM2 (Table 1). The PCR product was cloned into EcoRI-MluI sites of pZErO-2 (Invitrogen, Carlsbad, CA) and then into pIJA03. In a similar way, pIJA175 and pIJA176 were constructed for the disruption of cdhA and cdhBC, respectively. For pIJA175, the primers used were HA1 and HA2 (Table 1), and the PCR product was finally cloned into BglII-XbaI sites of pIJA03. For pIJ176 the primers used were HBC1 and HBC2 (Table 1), and the PCR product was finally cloned into the MluI-XbaI sites of pIJA03. For each plasmid, 1 μg of supercoiled DNA was transformed into M. maripaludis JJ by the polyethylene glycol method (29).
Southern hybridization.
Genomic DNA (1 μg) was treated with 20 units of EcoRV and BglII for 16 h at 37°C. The restricted DNA was transferred to a positively charged nylon membrane (Boehringer, Mannheim, Germany) after separation on a 1.0% agarose gel. The probe for the hybridization was amplified and labeled by PCR using genomic DNA of M. maripaludis S2 as the template and 25 μCi of [α-32P]dATP (ICN, Aurora, OH). The primers used were SH1 and SH2 (Table 1). Standard techniques were used for hybridization and washing the membrane (22). The membrane was exposed in a phosphorimager for 3 h, and subsequent analysis was performed with the ImageQuant 1.1 software (Amersham Bioscience, Buckinghamshire, England).
Methanogenesis.
Methane production from pyruvate was determined as described previously (14, 35) except that the cells were grown in McCA medium, after which resting cells were assayed by incubation under N2-CO2 (80:20 [vol/vol]) with 100 mM pyruvate overnight at 37°C. For positive controls, 0.1 ml of the resting cells was incubated for 1 h under 70 kPa of H2-CO2. Methane was measured with a Shimazu GC-8A gas chromatograph (Shimadzu Scientific Instruments Inc., Columbia, MD) by flame ionization detection on a DB-624 column run at 60°C (J & W Scientific, Folsom, CA). The carrier gas was N2, and the injector temperature was 200°C.
Preparation of cell extracts and purification of membrane fractions.
Early-linear-phase cultures (optical density at 600 nm, 0.45 to 0.6) of S2 or S40 were grown in McNA medium in 1-liter bottles, each containing 100 ml of medium. Cultures were harvested by centrifugation at 10,000 × g for 30 min at 4°C. The cells (from a total of 400 ml of culture) were resuspended in 7.5 ml of buffer A [25 mM piperazine-N,N′-bis(2-ethanesulfonic acid)-KOH, pH 7.0, 10 mM Mg-acetate, 30 mM KCl, and 2 mM dithiothreitol] and immediately passed through a chilled French pressure cell at 110 MPa. DNase, 10 U, was added, and the suspension was incubated for 15 min at 37°C. The cell extract was obtained by separation of the cell debris and the unbroken cells by centrifugation at 8,000 × g for 30 min at 4°C. The membranes were further purified by centrifugation of the cell extract at 171,500 × g (50,000 rpm; Beckman 70.1 Ti rotor; Beckman Coulter, Inc., Fullerton, CA) for 50 min at 4°C. The pellet was resuspended in 0.2 ml buffer A and loaded onto a 3.9-ml 10-to-60% sucrose gradient prepared in buffer A (5, 6). Centrifugation was performed at 32,000 × g (20,000 rpm; Beckman MLS 50 rotor) for 16 to 18 h at 20°C. The gradient fractions were collected (∼0.3 ml each), and those with the highest hydrogenase activities were pooled. All the purification steps were performed in an anaerobic chamber, and the solutions were flushed with N2 to remove traces of O2.
Enzymatic assays.
The hydrogenase activity was assayed anaerobically, using the cell extract and the purified membrane fractions, based on H2-dependent reduction of methyl viologen as described previously (20), except that the reaction mixture contained 20 mM methyl viologen and 25 mM phosphate buffer (pH 7.5) in a final volume of 1 ml. In cell extract, the activity of the carbon monoxide dehydrogenase-acetyl coenzyme A (CoA) synthase (CODH-ACS) was measured similarly as the CO-dependent reduction of methyl viologen (24). Pyruvate oxidoreductase (POR) activity was assayed anaerobically as pyruvate- and CoA-dependent methyl viologen reduction (15). Because of the instability of the POR (15), this activity was assayed within 1 day of preparation of the cell extracts.
Proteomics, arrays, and real-time PCR.
Metabolic 15N/14N labeling and preparation of proteins, quantitative proteomic analysis by liquid chromatography and tandem mass spectrometry, array analysis, and real-time reverse transcriptase PCR (RT-PCR) were performed as described elsewhere (Xia et al., submitted). Briefly, S2 and the mutant S40 were grown in McNA medium for 13 and 21 h, respectively, when an absorbance of 0.56 was achieved. Four biological replicates were analyzed for the arrays, each with duplicate arrays on each slide and with flip-dye hybridizations, making a total of 16 measurements for each gene. Expression ratios (S40/S2) were calculated, as were standard deviations of the ratios and P values. Genes with P values less than 0.01 were regarded as differentially expressed. For the proteome, two independent cultures each of the S40 mutant and the wild-type S2 were grown in minimal medium with stable isotope (“light” 14N and “heavy” 15N) labeling. Soluble and insoluble protein fractions were collected, and differentially labeled fractions were combined. Thus, four samples were analyzed: soluble proteins with S2 14N-labeled and S40 15N-labeled, soluble proteins with the nitrogen labeling reversed, and the same with the insoluble proteins. Tryptic peptides derived from each sample were analyzed in duplicate by two-dimensional capillary high-performance liquid chromatography coupled with tandem mass spectrometry. Mass spectrometric identification and quantification of peptides were used to generate 14N/15N ratios. Expression ratios for each protein were derived by averaging the ratios from all peptide pairs measured for that protein. Data from all four samples were combined to produce a quantitative S40/S2 proteome summary data set (Xia et al., submitted). Differential protein levels were regarded as significant if n1 (the number of peptide pairs measured) was equal to or greater than 3 and the average ratio (S40/S2) differed from 1 by an amount greater than the standard deviation.
RT-PCR.
RNA was purified as described elsewhere (Xia et al., submitted). RT-PCR was performed on an Eppendorf Mastercycler gradient thermocycler (Eppendorf AG, Hamburg, Germany), using a OneStep RT-PCR kit from QIAGEN according to the manufacturer's protocol. Primers EH1 and EH2 were used at concentrations of 600 nM (Table 1), and RNA was added to 500 ng per 50-μl reaction mixture. Cycling parameters were initial RT incubation at 50°C for 30 min, initial denaturing at 95°C for 15 min, and 35 cycles of 94°C for 45 s, 55°C for 45 s, and 72°C for 2 min, followed by a final extension at 72°C for 10 min. The product was analyzed on a 1% agarose gel.
GEO accession numbers.
The GEO series accession numbers are GSE2744 for the proteomics data sets and GSE2745 for the spotted cDNA arrays (http://www.ncbi.nlm.nih.gov/geo/).
RESULTS
Mutations in Ehb affect the biosynthesis of acetate.
In previous studies, an acetate auxotroph of M. maripaludis JJ was isolated by transformation with integration plasmids containing degenerate PCR products targeted to metalloenzyme clusters (36). Subsequent analysis found that this mutant was formed by integration of a plasmid vector into the ehbM gene, which encodes the small subunit of the Ehb hydrogenase (Fig. 1A and data not shown). In order to confirm the role of ehbM in acetate biosynthesis, the plasmid pIJA10 was constructed. This plasmid contained only a 479-bp portion of the ehb operon, including the entire ehbM gene, and was sufficient to allow a single recombination event inside ehbM (Fig. 1A). Thus, it was expected to disrupt expression of the downstream gene ehbO. As controls, plasmids that disrupted cdhA and cdhBC (pIJA175 and pIJA176, respectively), which encode subunits of CODH-ACS, were also used. This enzyme is also required for acetate biosynthesis (13). Following transformation with these plasmids, no transformants were observed on minimal medium (McN). In contrast, 131, 88, and 150 transformants were obtained in medium with acetate (McNA), and 200, 25, and 400 transformants appeared in plates with yeast extract plus acetate (McYA) following transformation with 1 μg of DNA of pIJA175, pIJA176, and pIJA10, respectively. The transformation frequencies in McNA or in McYA medium were at the levels expected for plasmids containing cloned regions of this size (488 to 495 bp). The failure to observe transformants in mineral medium without acetate supported a role for Ehb in acetate biosynthesis. However, these mutants were unstable and readily reverted back to the wild-type phenotype (data not shown).
To examine more fully the role of the Ehb system, a deletion mutant of ehbF was constructed. In this case, a stable mutant, S40, was obtained by a double recombination event, exchanging a portion of the ehbF gene with the puromycin resistance marker in the pac cassette (Fig. 1A). The genotype of S40 was confirmed by Southern blotting. The replacement of most of ehbF with the pac cassette resulted in an increase in size of the BglII-EcoRV fragment from 2.0 kb in the wild type to 2.5 kb in the mutant (Fig. 1B). The absence of the 2.0-kb wild-type fragment in the mutant also confirmed that insertion occurred by a double recombination event that eliminated the wild-type copy of the gene.
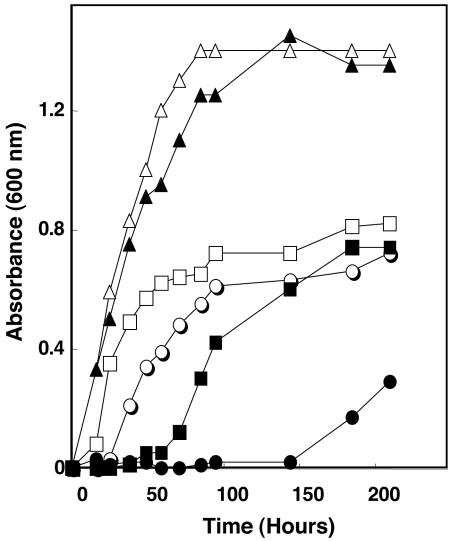
Although the growth of S40 closely resembled the wild type in rich medium, growth in minimal medium was severely impaired (Fig. 2). Growth on completely mineral McN medium without acetate was only observed after 150 h. Similarly, growth with acetate but in the absence of yeast extract was also severely delayed. Yeast extract provides a source of amino acids for methanococci, especially the branched-chain amino acids alanine, proline, and arginine (33). These results suggested that Ehb was involved in amino acid as well as acetyl-CoA biosynthesis during autotrophic growth with CO2 as a sole carbon source.
FIG. 2.
Stimulation of growth of M. maripaludis S40 by acetate and yeast extract. Growth of wild-type S2 in the minimal McN (open circles), McN plus acetate (open squares), and McN plus acetate and yeast extract (open triangles) media and growth of mutant S40 in McN (closed circles), McN plus acetate (closed squares), and McN plus acetate and yeast extract (closed triangles) media.
Although the growth phenotype of the mutation in S40 was severe, changes in the levels of hydrogenase activity in cell extracts and membranes were not found. The specific activities of the methyl viologen-linked hydrogenases were not significantly different in cell extracts of the wild type and S40 (Table 2). Similarly, the specific activities were the same in membranes collected from the extract by high-speed centrifugation, even after additional purification on a sucrose gradient. Therefore, the growth phenotype was not a result of a reduced level of hydrogenase activity per se.
TABLE 2.
Hydrogenase, POR, and CODH-ACS activities of M. maripaludis strains S2 and S40
| Fractiona | Sp act (U/mg)
|
|||||
|---|---|---|---|---|---|---|
| Hydrogenase
|
POR
|
CODH-ACS
|
||||
| S2b | S40b | S2c | S40b | S2c | S40b | |
| Low-speed centrifugation | ||||||
| Supernatant | 370 ± 46 | 350 ± 85 | 400 ± 130 | 1,230 ± 190 | 240 ± 110 | 950 ± 120 |
| High-speed centrifugation | ||||||
| Supernatant | 260 ± 82 | 280 ± 75 | ||||
| Pellet | 390 ± 110 | 330 ± 69 | ||||
| Sucrose gradientd | 830 ± 120 | 860 ± 110 | ||||
The fractions were obtained following cell lysis with a French press. After the low-speed centrifugation (8,000 × g, 30 min), the supernatant was further centrifuged at 171,500 × g for 50 min. The resulting pellet was loaded onto a 10-to-60% sucrose gradient.
Data are the averages of two to four assays from four independent cultures.
Data are the averages of two to four assays from three independent cultures.
Fraction with maximum specific activity in sucrose gradient.
Previously, Ehb was proposed to be coupled to POR in methanococci by two small ferredoxins associated with the POR genes (15). Consistent with this hypothesis, mutants with a deletion of the genes encoding these ferredoxins were unable to form methane with pyruvate as an electron donor. Thus, the POR-associated ferredoxins are believed to be the electron donors for the Ehb-catalyzed reduction of protons to H2, which is the proximal electron donor for methane biosynthesis with pyruvate. We used S40 to test the role of Ehb in methane production from pyruvate. Although S40 contained wild-type levels of hydrogenase activity, pyruvate-dependent methanogenesis was reduced by one-third, and the average rates for the three experiments were 2.0 ± 0.9 and 3.0 ± 1.9 nmol CH4 min−1 cell (dry weight)−1 for S40 and S2, respectively. These values were significantly different by analysis of variance, with a P value of <0.05. However, for H2-dependent methanogenesis the rates were 160.0 ± 120 and 160 ± 70 nmol CH4 min−1 cell (dry weight)−1 for S40 and S2, respectively, which were not significantly different. Thus, the mutation did not directly affect methane biosynthesis. These observations supported the hypothesis that S40 was deficient in coupling hydrogenase activity to biosynthetic enzymes such as POR.
Proteomics and arrays.
In order to further understand the function of the Ehb complex in M. maripaludis, array and proteome comparisons were performed between the ΔehbF:pac mutant S40 and the wild-type S2 (Xia et al., submitted). The results from the arrays and the proteome were highly correlated, and in many cases similar trends for multiple ORFs within an operon corroborated the results. The complete data set is available at the NCBI GEO database. The following presents the results that are of particular interest to the biology of the Ehb mutant.
Expression of carbon assimilation genes.
The expression of a number of genes involved in carbon assimilation increased in S40 relative to the wild type. In both the proteome and the transcriptome, the expression of the CODH-ACS (Mmp0979 to -0985) and POR (Mmp1502 to -1507) increased significantly in S40 relative to the wild type (Table 3). These trends were consistent for multiple subunits encoded in their respective operons. These enzymes carry out the first two steps of CO2 fixation. Both reactions require low-potential electrons which are proposed to originate from the Ehb hydrogenase. Expression of the AMP-forming acetyl-CoA synthetase (Mmp0148) also increased (Table 3). This enzyme is the major alternative route of acetyl-CoA formation in M. maripaludis in the absence of CO2 fixation by CODH-ACS (24). Lastly, three of the five proteins composing the 2-oxoisovalerate oxidoreductase (VOR; Mmp1271 to -1275) (Table 3) were overexpressed in the proteome by about 50%. Although small, these changes were significant. The VOR catalyzes the reductive carboxylation of branched-chain fatty acids to branched-chain amino acids (7). Like the POR, it is expected to utilize low-potential electron donors generated by Ehb hydrogenase. Thus, the increased expression of all these genes was consistent with a general pattern of limitation for fixed carbon.
TABLE 3.
Differential expression of some genes by proteomic and transcriptional array analyses
| ORF and function | Descriptiona | Proteomic datab
|
Array datac
|
||||
|---|---|---|---|---|---|---|---|
| Avg ratio | n1 | SD | Ratio | SD | P value | ||
| Mmp0148 | Acetyl-CoA synthetase | 8.94 | 62 | 5.28 | 3.23 | 1.17 | 1.70E-06 |
| CODH-ACS | |||||||
| Mmp0979 | Conserved archaeal protein | 5.75 | 6 | 1.93 | 1.63 | 0.27 | 1.04E-03 |
| Mmp0980 | Subunit gamma | 2.87 | 30 | 1.00 | 1.77 | 0.28 | 9.11E-04 |
| Mmp0981 | Subunit delta | 3.17 | 14 | 1.38 | 1.42 | 0.19 | 2.57E-03 |
| Mmp0982d | Conserved hypothetical protein | 1.47 | 0.20 | 1.04E-03 | |||
| Mmp0983 | Subunit beta | 3.33 | 21 | 1.51 | 1.47 | 0.18 | 1.36E-03 |
| Mmp0984 | Subunit epsilon | 3.10 | 9 | 1.50 | 1.37 | 0.16 | 1.16E-02 |
| Mmp0985 | Subunit alpha | 3.73 | 38 | 2.72 | 1.42 | 0.10 | 2.56E-04 |
| POR | |||||||
| Mmp1502 | Conserved archaeal protein | 1.58 | 20 | 0.70 | 3.05 | 0.64 | 1.41E-06 |
| Mmp1503 | Conserved archaeal protein | 2.00 | 29 | 0.90 | 2.79 | 0.63 | 1.46E-05 |
| Mmp1504 | Subunit beta | 2.68 | 76 | 0.77 | 2.43 | 0.62 | 1.23E-05 |
| Mmp1505 | Subunit alpha | 2.73 | 117 | 1.06 | 2.36 | 0.37 | 1.23E-05 |
| Mmp1506 | Subunit delta | 2.73 | 13 | 0.57 | 2.23 | 0.35 | 3.43E-04 |
| Mmp1507 | Subunit gamma | 2.68 | 39 | 1.30 | 1.95 | 0.35 | 1.64E-04 |
| VOR | |||||||
| Mmp1271 | VOR subunit alpha | 1.40 | 14 | 0.30 | 1.11 | 0.16 | 1.63E-01 |
| Mmp1272 | VOR subunit beta | 1.39 | 5 | 0.06 | 1.15 | 0.16 | 7.65E-02 |
| Mmp1273d | VOR subunit gamma | 1.23 | 0.38 | 1.32E-01 | |||
| Mmp1274 | Acetyl-CoA synthetase related | 1.33 | 54 | 0.58 | 1.22 | 0.24 | 1.22E-01 |
| Mmp1275 | Transcriptional regulator protein | 2.08 | 11 | 0.49 | 1.27 | 0.12 | 7.65E-02 |
| Ehb | |||||||
| Mmp1621 | Integral membrane subunit | 3.95 | 2 | 0.41 | 3.93 | 0.81 | 1.41E-06 |
| Mmp1622 | Small subunit | 3.63 | 9 | 1.43 | 3.84 | 0.97 | 1.41E-06 |
| Mmp1623 | Ferredoxin | 3.54 | 17 | 2.14 | 4.38 | 0.81 | 1.41E-06 |
| Mmp1624 | Polyferredoxin | 6.87 | 19 | 2.69 | 4.77 | 1.50 | 1.41E-06 |
| Mmp1625d | Conserved hypothetical protein | 6.73 | 1.81 | 1.41E-06 | |||
| Mmp1626d | Conserved hypothetical protein | 6.06 | 1.67 | 1.41E-06 | |||
| Mmp1627d | Conserved hypothetical protein | 7.25 | 2.01 | 2.05E-06 | |||
| Mmp1628d,e | Transmembrane subunit | 0.59 | 0.17 | 2.56E-04 | |||
| Mmp1629d | Conserved hypothetical protein | 1.42 | 0.25 | 2.57E-03 | |||
The ORF description is derived from the genome annotation (9).
Numerical data for proteomic analysis report average S40/S2 ratios, numbers of peptide pairs (n1) used to derive ratios, and standard deviations for the combination of four samples as described in the text and by Xia et al. (submitted for publication). An average ratio different from 1 by an amount greater than the standard deviation was taken to indicate differential protein expression. Boldface indicates significant differential expression.
Numerical data for array analysis report average S40/S2 ratios, standard deviations, and P values (values less than or equal to 0.01 were taken to indicate differential expression). The number of replicates for array analysis was 16. Boldface indicates significant differential expression.
Protein not detected.
Mmp1628 was deleted in the S40 mutant.
The increased expression of some of these genes was confirmed by alternative methods. In cell extracts, the specific activities of CODH-ACS and POR increased four- and threefold, respectively, in the mutant S40 relative to the wild type (Table 2). Similarly, the increased expression of Mmp1503, which encodes PorE, was confirmed by real-time RT-PCR (Xia et al., submitted). By this method, expression of Mmp1503 increased between 3- and 6-fold, compared to 2.8-fold in the arrays. In contrast, the levels of mRNA for Mmp1094 and Mmp1478, two genes whose expression was not changed in the microarrays, were also not elevated in the real-time RT-PCR experiments.
Expression of genes in Ehb and Eha complexes.
There was an increase in the expression of many of the genes encoding Ehb (Mmp1621 to -1629) in S40. One exception was Mmp1628, which contained the deletion and the pac cassette insertion and was not effectively probed by the arrays (Table 3). The high-level expression of the other genes was confirmed by real-time RT-PCR of Mmp1623, which encoded ehbL and was differentially expressed nearly fivefold in the mutant. In this case, high levels of expression for these genes may have been due to transcription from the pac promoter. Reverse transcriptase PCR demonstrated the presence of a transcript extending upstream of the terminator in the pac cassette into the downstream ehbG gene (data not shown). In contrast, Mmp1629, which encoded the first gene of the operon and was upstream of the pac cassette, was only moderately differentially expressed (1.4-fold) (Table 3). Presumably, the expression of this gene was under control of the native promoter. Some components of Ehb were encoded by genes found outside this operon, and their expression was also of interest (9). For ORFs Mmp0400, Mmp0940, Mmp1049, and Mmp1153, the expression in the mutant was not significantly different from that in the wild type (Xia et al., submitted). Mmp1074, which encoded the hydrogenase large subunit, was 1.3-fold more highly expressed in the mutant, which was similar to the expression of Mmp1629. The modest differential expression observed for Mmp1629 and Mmp1074 in the mutant was consistent with a small effect on the expression of these genes and the absence of a significant change in hydrogenase specific activity in cell extracts.
Little evidence was found for the differential expression of Eha. No significant differences were observed for the 18 ORFs in the arrays (Mmp1448 to -1467) (Xia et al., submitted). Six ORFs were detected in the proteome. Only the expression of Mmp1462, which encodes the large hydrogenase subunit, was moderately increased (Xia et al., submitted).
Expression of other genes.
Expression of most of the other genes detected in the microarrays and proteome was not significantly altered in the mutant (Xia et al., submitted). However, expression of a few genes was affected. The levels of mRNA for an operon encoding flagellar biosynthetic genes (Mmp1666 to -1673) decreased in S40 (Xia et al., submitted). Expression of four of these genes (Mmp1666 to -1669) decreased by 2.5-fold, and expression of two genes (Mmp1670 and Mmp1672) decreased by 1.5-fold. The flagellar genes of Methanocaldococcus jannaschii are down-regulated by high levels of H2 (2, 18). In addition, in wild-type M. maripaludis the expression of the flagellar genes is also decreased under high levels of H2 (E. L. Hendrickson and J. A. Leigh, unpublished data). Thus, the lower expression of the flagellar genes in S40 may be a response to high levels of H2. Presumably, the slower growth of the mutant reduces the H2 demand for methanogenesis and increases the H2 concentrations in the medium during the linear growth phase. Moreover, the levels of mRNA for three other multicistronic operons that may be affected by the hydrogen concentration were decreased by about 50% in the mutant. These operons encoded the ATPase (Mmp1038 to -1046), methyl coenzyme M reductase (Mmp1555 to -1559), and methyltetrahydromethanopterin:coenzyme M methyltransferase (Xia et al., submitted). Finally, the levels of mRNA for a potential operon encoding components of a transporter of uncertain function (Mmp0165 to -0168) increased in S40 by 1.8- to 4.5-fold (Xia et al., submitted).
DISCUSSION
The homology of subunits of the energy-coupling hydrogenases (Ech) of bacteria and archaea to subunits of the NADH-ubiquinone oxidoreductase or complex I of mitochondria suggests that these enzymes possess a fundamental role in energy metabolism (for reviews, see references 8 and 31). In the methanogenic archaea, two different physiological roles have been proposed (8). During aceticlastic growth of Methanosarcina, the conversion of CO to CO2 plus H2 is coupled to generation of a proton motive force (4). The Ech is believed to participate in this reaction by oxidizing the low-potential ferredoxin generated from CO by the CODH-ACS system. The Ech then reduces protons to generate H2 for methanogenesis as well as a proton motive force (17). This function is similar to that of the homologous complex in the hyperthermophilic archaeon Pyrococcus (23, 26). However, in this case, the low-potential ferredoxins are generated by the fermentation of sugars or amino acids and H2 is an end product. In the second physiological role, the Ech is believed to generate low-potential ferredoxins for CO2 reduction to methane as well biosynthetic reactions (8). At the low partial pressures of H2 common in natural environments, the E′ of the 2H+/H2 couple is near −286 mV. For methanogens utilizing H2 as an electron donor, the initial reduction of CO2 to methane is catalyzed by the formylmethanofuran dehydrogenase (FMD). This reaction, which has an E′ near −500 mV, as well as a number of biosynthetic reactions, becomes problematic under these conditions. During growth on H2, the Ech of Methanosarcina is also believed to generate low-potential ferredoxins for FMD and other biosynthetic reactions (17).
The obligately hydrogenotrophic methanogens, such as Methanococcus and Methanothermobacter, possess two homologs of the energy-coupling hydrogenases. While there is little direct evidence for their physiological functions, the presence of two enzymes makes it possible for the activities to be differentially regulated. Like Methanosarcina, these enzymes would be expected to be necessary for activity of the FMD as well as biosynthetic reactions, and there may be a physiological advantage for one enzyme system to specialize in the ATP-generating pathway of methanogenesis and the second enzyme system to specialize in the anabolic CO2 assimilation pathways. Unlike Methanosarcina, FMD activity is essential for these methanogens, and so mutations in an enzyme coupled specifically to this system would be lethal. Because mutations in ehb are not lethal, it is unlikely that Ehb is specifically coupled to the FMD. Instead, the properties of the ΔehbF:pac mutant S40 are consistent with a role of Ehb in carbon assimilation. First, the growth of the mutant in rich medium and the rate of methanogenesis from H2-CO2 are comparable to that of the wild type. Thus, the mutation does not affect central pathways of energy conservation. The slow growth in the absence of organic carbon sources implies a role in autotrophic CO2 assimilation. In methanococci, autotrophic CO2 assimilation proceeds from methyltetrahydromethanopterin, an intermediate in methanogenesis, to acetyl-CoA and pyruvate (13, 24). Sugars and amino acids are then formed by gluconeogenesis and the reductive incomplete tricarboxylic acid cycle, respectively (25, 37). Because acetate greatly stimulated growth of the mutant, Ehb is expected to play a role in autotrophic acetyl-CoA biosynthesis. However, because acetate alone was not sufficient to restore growth to wild-type levels, the mutation must have pleiotrophic affects on other steps of carbon assimilation. In the mutant, many of the genes involved in carbon assimilation were also differentially overexpressed. This response was also consistent with growth of the mutant during carbon limitation and a role of Ehb in carbon assimilation.
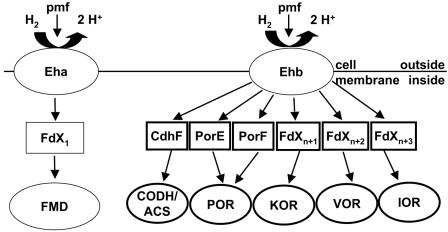
A working model for the role of Ehb in providing low-potential electrons for biosynthesis is shown in Fig. 3. Enzyme systems in M. maripaludis that are expected to require low-potential electron donors in addition to FMD include CODH-ACS, which catalyzes the biosynthesis of acetyl-CoA from methyltetrahydromethanopterin and CO2; POR, which catalyzes the reductive carboxylation of acetyl-CoA (14, 15); 2-ketoglutarate oxidoreductase, which catalyzes the reductive carboxylation of succinyl-CoA; and branched-chain VOR and two indole-pyruvate oxidoreductases, which are involved in biosynthesis of amino acids from the corresponding carboxylic acids (7, 19). In addition, the genome of M. maripaludis contains an oxidoreductase of unknown specificity (9) which could also be coupled to Ehb.
FIG. 3.
Working model for the role of Ehb in generation of low-potential electron donors in M. maripaludis. The cellular membrane contains two energy-conserving [NiFe] hydrogenases, Eha and Ehb. Eha is proposed to be coupled to FMD, a key enzyme in methanogenesis from CO2, via an unidentified ferredoxin, FdX1. Ehb is proposed to be coupled to CODH-ACS, a key enzyme in autotrophic CO2 fixation, and a number of ferredoxin-dependent oxidoreductases that catalyze important anabolic reactions, including POR, 2-ketoglutarate oxidoreductase (KOR), branched-chain VOR, and indole-pyruvate oxidoreductase (IOR). Ferredoxins that may be involved in coupling these enzymes to Ehb include CdhF, PorE, and PorF. FdX2, FdX3, and FdX4 are unidentified ferredoxins.
The proposal that Ehb is specifically involved in CO2 assimilation depends greatly upon the properties of the S40 mutant discussed above. In addition, genes for two possible electron carriers, named PorE and PorF, are found adjacent to the genes encoding the POR subunits (14, 15). One of these proteins copurifies with POR. Mutants with deletions in these genes grow poorly in mineral medium and are unable to use pyruvate as an electron donor for methanogenesis (14), similar to the phenotype of the ehbF mutant described here. In addition, a homolog of PorE, named CdhF, is adjacent to the genes encoding the subunits for CODH-ACS (15). Those electron carriers could transfer the electrons from the Ehb system to the POR and CODH-ACS reactions.
Although Ehb plays the primary role in providing electrons for carbon assimilation, it is not absolutely essential, since S40 still grew slowly in minimal medium. However, POR is required for pyruvate biosynthesis in methanococci even in the presence of abundant sources of organic carbon (34), and mutations in the structural genes encoding POR are apparently lethal (W. Lin and W. B. Whitman, unpublished data). Therefore, mutations in Ehb or the coupling ferredoxins would be lethal if an alternative source of low-potential electrons did not exist. For this reason, it is likely that Eha and the electron carriers for FMD possess partial activity with the Ehb-dependent enzyme systems and that leakage from Eha allows slow growth of the Ehb mutant.
Acknowledgments
This work is funded by a DOE Microbial Cell Project grant, DE-FG03-01ER15252, a grant from DOE Energy Biosciences to W.W., DE-FG02-97ER20269, and NIH grant GM60403 to J.A.L. and M. V. Olson.
We thank Michael R. Strand for the use of his ultracentrifuge, Fredrick Bohanon and Magdalena Sieprawska-Lupa for preparing media and for growing M. maripaludis strains, and Mark A. Schell for his advice on the purification of membrane proteins.
REFERENCES
- 1.Albracht, S. P., and R. Hedderich. 2000. Learning from hydrogenases: location of a proton pump and of a second FMN in bovine NADH-ubiquinone oxidoreductase (complex I). FEBS Lett. 485:1-6. [DOI] [PubMed] [Google Scholar]
- 2.Babnigg, G., and C. S. Giometti. 2004. GELBANK: a database of annotated two-dimensional gel electrophoresis patterns of biological systems with completed genomes. Nucleic Acids Res. 32(database issue):D582-D585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berghofer, Y., K. Agha-Amiri, and A. Klein. 1994. Selenium is involved in the negative regulation of the expression of selenium-free [NiFe] hydrogenases in Methanococcus voltae. Mol. Gen. Genet. 242:369-373. [DOI] [PubMed] [Google Scholar]
- 4.Bott, M., and R. K. Thauer. 1989. Proton translocation coupled to the oxidation of carbon monoxide to CO2 and H2 in Methanosarcina barkeri. Eur. J. Biochem. 179:469-472. [DOI] [PubMed] [Google Scholar]
- 5.Chen, W., and J. Konisky. 1993. Characterization of a membrane-associated ATPase from Methanococcus voltae, a methanogenic member of the Archaea. J. Bacteriol. 175:5677-5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dharmavaram, R. M., and J. Konisky. 1987. Identification of a vanadate-sensitive, membrane-bound ATPase in the archaebacterium Methanococcus voltae. J. Bacteriol. 169:3921-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haydock, A. K., I. Porat, W. B. Whitman, and J. A. Leigh. 2004. Continuous culture of Methanococcus maripaludis under defined nutrient conditions. FEMS Microbiol. Lett. 238:85-91. [DOI] [PubMed] [Google Scholar]
- 8.Hedderich, R. 2004. Energy-converting [NiFe] hydrogenases from archaea and extremophiles: ancestors of complex I. J. Bioenerg. Biomembr. 36:65-75. [DOI] [PubMed] [Google Scholar]
- 9.Hendrickson, E. L., R. Kaul, Y. Zhou, D. Bovee, P. Chapman, J. Chung, E. Conway de Macario, J. A. Dodsworth, W. Gillett, D. E. Graham, M. Hackett, A. K. Haydock, A. Kang, M. L. Land, R. Levy, T. J. Lie, T. A. Major, B. C. Moore, I. Porat, A. Palmeiri, G. Rouse, C. Saenphimmachak, D. Soll, S. Van Dien, T. Wang, W. B. Whitman, Q. Xia, Y. Zhang, F. W. Larimer, M. V. Olson, and J. A. Leigh. 2004. Complete genome sequence of the genetically tractable hydrogenotrophic methanogen Methanococcus maripaludis. J. Bacteriol. 186:6956-6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jones, W. J., M. J. B. Paynter, and R. Gupta. 1983. Characterization of Methanococcus maripaludis sp. nov., a new methanogen isolated from salt marsh sediment. Arch. Microbiol. 135:91-97. [Google Scholar]
- 11.Jones, W. J., W. B. Whitman, R. D. Fields, and R. S. Wolfe. 1983. Growth and plating efficiency of methanococci on agar media. Appl. Environ. Microbiol. 46:220-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunkel, A., J. A. Vorholt, R. K. Thauer, and R. Hedderich. 1998. An Escherichia coli hydrogenase-3-type hydrogenase in methanogenic archaea. Eur. J. Biochem. 252:467-476. [DOI] [PubMed] [Google Scholar]
- 13.Ladapo, J., and W. B. Whitman. 1990. Method for isolation of auxotrophs in the methanogenic archaebacteria: role of the acetyl-CoA pathway of autotrophic CO2 fixation in Methanococcus maripaludis. Proc. Natl. Acad. Sci. USA 87:5598-5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin, W., and W. B. Whitman. 2004. The importance of porE and porF in the anabolic pyruvate oxidoreductase of Methanococcus maripaludis. Arch. Microbiol. 181:68-73. [DOI] [PubMed] [Google Scholar]
- 15.Lin, W. C., Y. L. Yang, and W. B. Whitman. 2003. The anabolic pyruvate oxidoreductase from Methanococcus maripaludis. Arch. Microbiol. 179:444-456. [DOI] [PubMed] [Google Scholar]
- 16.Meuer, J., S. Bartoschek, J. Koch, A. Kunkel, and R. Hedderich. 1999. Purification and catalytic properties of Ech hydrogenase from Methanosarcina barkeri. Eur. J. Biochem. 265:325-335. [DOI] [PubMed] [Google Scholar]
- 17.Meuer, J., H. C. Kuettner, J. K. Zhang, R. Hedderich, and W. W. Metcalf. 2002. Genetic analysis of the archaeon Methanosarcina barkeri Fusaro reveals a central role for Ech hydrogenase and ferredoxin in methanogenesis and carbon fixation. Proc. Natl. Acad. Sci. USA 99:5632-5637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mukhopadhyay, B., E. F. Johnson, and R. S. Wolfe. 2000. A novel pH2 control on the expression of flagella in the hyperthermophilic strictly hydrogenotrophic methanarchaeaon Methanococcus jannaschii. Proc. Natl. Acad. Sci. USA 97:11522-11527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Porat, I., B. W. Waters, Q. Teng, and W. B. Whitman. 2004. Two biosynthetic pathways for aromatic amino acids in the archaeon Methanococcus maripaludis. J. Bacteriol. 186:4940-4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ragsdale, S. W., and L. G. Ljungdahl. 1984. Hydrogenase from Acetobacterium woodii. Arch. Microbiol. 139:361-365. [DOI] [PubMed] [Google Scholar]
- 21.Reeburgh, W. S. 2003. Global methane biogeochemistry, p. 65-89. In H. D. Holland and K. K. Turekian (ed.), The atmosphere, vol. 4, treatise on geochemistry. Elsevier-Pergamon, Oxford, England. [Google Scholar]
- 22.Sambrook, J., and D. W. Russell. 2003. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Sapra, R., K. Bagramyan, and M. W. W. Adams. 2003. A simple energy-conserving system: proton reduction coupled to proton translocation. Proc. Natl. Acad. Sci. USA 100:7545-7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shieh, J., and W. B. Whitman. 1988. Autotrophic acetyl coenzyme A biosynthesis in Methanococcus maripaludis. J. Bacteriol. 170:3072-3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shieh, J. S., and W. B. Whitman. 1987. Pathway of acetate assimilation in autotrophic and heterotrophic methanococci. J. Bacteriol. 169:5327-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silva, P. J., E. C. van den Ban, H. Wassink, H. Haaker, B. de Castro, F. T. Robb, and W. R. Hagen. 2000. Enzymes of hydrogen metabolism in Pyrococcus furiosus. Eur. J. Biochem. 267:6541-6551. [DOI] [PubMed] [Google Scholar]
- 27.Stathopoulos, C., W. Kim, T. Li, I. Anderson, B. Deutsch, S. Palioura, W. Whitman, and D. Soll. 2001. Cysteinyl-tRNA synthetase is not essential for viability of the archaeon Methanococcus maripaludis. Proc. Natl. Acad. Sci. USA 98:14292-14297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tersteegen, A., and R. Hedderich. 1999. Methanobacterium thermoautotrophicum encodes two multisubunit membrane-bound [NiFe] hydrogenases. Transcription of the operons and sequence analysis of the deduced proteins. Eur. J. Biochem. 264:930-943. [DOI] [PubMed] [Google Scholar]
- 29.Tumbula, D. L., R. A. Makula, and W. B. Whitman. 1994. Transformation of Methanococcus maripaludis and identification of a PstI-like restriction system. FEMS Microbiol. Lett. 121:309-314. [Google Scholar]
- 30.Tumbula, D. L., and W. B. Whitman. 1999. Genetics of Methanococcus: possibilities for functional genomics in Archaea. Mol. Microbiol. 33:1-7. [DOI] [PubMed] [Google Scholar]
- 31.Vignais, P. M., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25:455-501. [DOI] [PubMed] [Google Scholar]
- 32.Whitman, W. B., J. S. Shieh, S. H. Sohn, D. S. Caras, and U. Premachandran. 1986. Isolation and characterization of 22 mesophilic methanococci. Syst. Appl. Microbiol. 7:235-240. [Google Scholar]
- 33.Whitman, W. B., S. Sohn, and R. Y. Xing. 1987. Role of amino acids and vitamins in nutrition of mesophilic Methanococcus spp. Appl. Environ. Microbiol. 53:2373-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, Y. L., J. N. Glushka, and W. B. Whitman. 2002. Intracellular pyruvate flux in the methane-producing archaeon Methanococcus maripaludis. Arch. Microbiol. 178:493-498. [DOI] [PubMed] [Google Scholar]
- 35.Yang, Y. L., J. A. Lapado, and W. B. Whitman. 1992. Pyruvate oxidation by Methanococcus spp. Arch. Microbiol. 158:271-275. [Google Scholar]
- 36.Yu, J. P. 1997. Investigation of carbon metabolism in Methanococcus maripaludis. Ph.D. dissertation. University of Georgia, Athens.
- 37.Yu, J. P., J. Ladapo, and W. B. Whitman. 1994. Pathway of glycogen metabolism in Methanococcus maripaludis. J. Bacteriol. 176:325-332. [DOI] [PMC free article] [PubMed] [Google Scholar]