Abstract
We identified two regulators of denitrification genes in Brucella melitensis 16M: NarR, which regulates the nitrate reductase (nar) operon, and NnrA, which is involved in the expression of the last three reductases of the denitrification pathway (nirK, norB, and nosZ). NnrA is required for virulence in mice and for intracellular resistance to nitric oxide.
Brucella species are facultative intracellular pathogens that infect animals and occasionally humans (10). Recently, analysis of the complete Brucella melitensis sequence suggested the presence of an anaerobic electron transfer system and the enzymes allowing the reduction of nitrate into dinitrogen gas (nitrate, nitrite, NO [1] and nitrous oxide reductases) (3). These reactions are collectively named denitrification (17) and could allow Brucella to grow under low-oxygen conditions by respiration of nitrate (11).
Since the replicative compartment of Brucella suis is characterized by low oxygen tension, it was postulated that, during the infectious process, Brucella could use denitrification to survive using nitrogen oxides as terminal electron acceptors (6). Another potential role of this system is limiting the production of reactive nitrogen intermediates by the host during infection. Indeed, Brucella abortus was shown to counteract the effect of NO after 24 h of infection in activated macrophages (14).
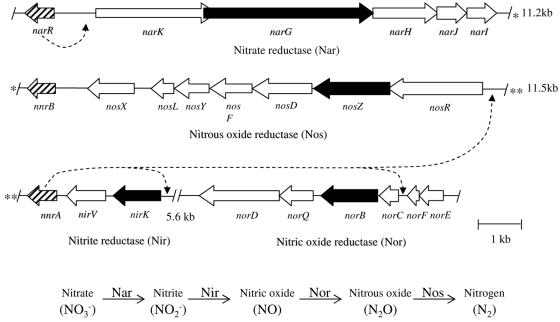
When analyzing the B. melitensis genome, we observed that three predicted coding sequences for Crp/Fnr regulators (narR, nnrA, and nnrB) are located near the genes putatively involved in denitrification (Fig. 1). NarR belongs to the NarR subfamily of the Dnr cluster, while NnrA and NnrB belong to the NnrR cluster, according to the Korner classification (7). Three other Crp/Fnr genes are predicted from the genomic sequence (5); two of them belong to the A cluster while the last belongs to the FnrN family (7). The B. melitensis genome contains the narKGHJI, nirKV, norEFCBQD, and nosRZDFYSLX gene clusters that are coding for a respiratory membrane-bound nitrate reductase (Nar, with the nitrite extrusion protein NarK), a copper nitrite reductase (Nir), a NO reductase (Nor), and a nitrous oxide reductase (Nos), respectively. The narR gene is found next to genes encoding the nitrate reductase, nnrA is next to genes encoding the nitrite and NO reductases, and nnrB is located downstream of genes encoding the nitrous oxide reductase. Furthermore, the regulators encoded by narR, nnrA, and nnrB genes are homologous to regulators involved in denitrification in other bacteria. Indeed, NarR from B. melitensis is homologous to the Paracoccus pantotrophus NarR, a regulator of the nitrate reductase genes (16). NnrB and NnrA belong to the NnrR family, comprising the NnrR regulator from Rhodobacter sphaeroides 2.4.3, which controls the expression of the nitrite and NO reductases (12).
FIG. 1.
Organization of the denitrifying reductase gene clusters in the B. melitensis 16M genome. The data presented here support a model (depicted as discontinuous lines) in which nnrA is a regulator of nir, nor, and nos genes, while narR is required for full nar expression. Each predicted coding sequence is indicated by an arrow. The arrows in contact and in the same orientation are expected to be coding sequences organized in operons. The genes coding for the catalytic subunit of the reductases are colored in black, and regulator genes are hatched. The three regions are located on the same portion of the small chromosome of B. melitensis 16M, as indicated by the short distances separating the nar, nos, nir, and nor loci. The overlap of narK and narG coding sequences is 86 bp long. The narR coding sequence (BMEII0947) is found close to genes encoding nitrate reductase homologs (nar genes and narK, encoding a putative nitrite extrusion protein), nnrA (BMEII0986) is next to genes encoding the nitrite and nitric oxide reductase homologs (nir and nor genes, respectively), and nnrB (BMEII0966) is downstream of nos genes, encoding the nitrous oxide reductase homolog. At the bottom, a summary of the denitrification pathway is shown. *, region located between narI and nnrB; **, region located between nosR and nnrA.
As a starting hypothesis, we postulated that the genomic colocalization between the denitrification genes and narR, nnrA, and nnrB could indicate that these Crp/Fnr regulators are involved in the transcriptional control of the denitrification genes. Since homology analyses support this hypothesis, we constructed mutants for NarR, NnrA, and NnrB regulators and for the catalytic subunits of the four reductases (narG, nirK, norB, and nosZ) of the denitrification process by using an integrative disruption strategy (5). Mutant genotypes were confirmed by Southern blotting as previously reported (5). We also constructed plasmidic promoter-lacZ fusions using the pBBRMCS1lacZ vector (S. Léonard, unpublished data), a derivative of pBBR1MCS (9), in order to follow the activity of narK, nirK, norC, and nosR promoters in the narR, nnrA, and nnrB mutants. These promoters, here defined as a 500-bp fragment upstream and including the predicted ATG, were amplified by PCR and inserted in frame with the lacZ coding sequence. The primer sequences are available upon request.
An evaluation of nitrite concentration in anaerobic conditions and in the presence of nitrate (15) showed that strains with nnrA and nirK mutants accumulate more nitrite than the wild-type (wt) strain, while narR and narG mutants accumulate a lower concentration of nitrite and no detectable nitrite, respectively (data not shown). These mutant phenotypes are complemented by the presence of an nnrA, nirK, narR, or narG coding sequence on a pBBRMCS4GW plasmid (data not shown). One interpretation for the nitrite accumulation data is that narR could activate the nar operon, probably through the promoter located upstream of narK, encoding a nitrite extrusion protein, and that nnrA could activate nirK. We therefore tested the activity of PnarK in the narR mutant and the activity of PnirK in the nnrA mutant by using the lacZ fusions described in the previous paragraph. Overnight cultures were diluted to an optical density at 600 nm of 0.1 in 2YT medium supplemented with 10 mM NaNO3 and incubated for 3 days in microaerobic conditions (using the CampyGen system, Oxoid) which allow a slow growth of B. melitensis 16M. We observed that the transcription from the PnarK promoter is lower in the narR mutant than in the wild-type strain (Table 1). A stronger effect was reported for NarR in the narK gene in Paracoccus (16), although the target promoter is also not completely switched off in the narR mutant of Paracoccus. In the nnrA mutant, the transcription from the PnirK promoter is much lower than that from the wild-type control (Table 1). We also tested the effect of nnrA and nnrB mutations on norC and nosR promoters that may control the expression of the norB and nosZ coding sequences. We observed that in the nnrA mutant, the transcription from both promoters is strongly reduced, while nnrB mutation does not affect PnorC or PnosR (Table 1).
TABLE 1.
β-Galactosidase activities from the reductase promoter-lacZ fusions in wt or mutant B. melitensis strains
| Fusion | Strain | β-Galactosidase activitya |
|---|---|---|
| PnarK-lacZ | wt | 1,091 ± 159 |
| narR | 457 ± 51 | |
| PnirK-lacZ | wt | 49 ± 12 |
| nnrA | 1 ± 0.6 | |
| PnorC-lacZ | wt | 640 ± 25 |
| nnrA | 9 ± 5 | |
| nnrB | 698 ± 46 | |
| PnosR-lacZ | wt | 304 ± 81 |
| nnrA | 4 ± 2 | |
| nnrB | 395 ± 61 |
Bacteria were grown microaerobically in rich medium (2YT) supplemented with 10 mM of NaNO3 before β-galactosidase activity was determined. Activity (in Miller units) (8) is the average of three independent clones.
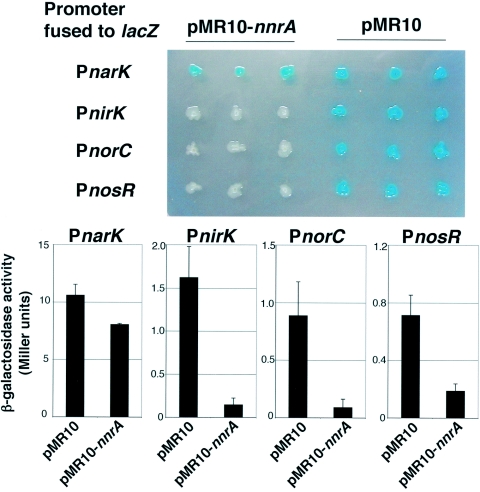
NnrA probably interacts with the Escherichia coli transcription machinery to modulate B. melitensis promoters nirK, norC, and nosR because when nnrA is used in a heterologous transcription interference assay with E. coli strain DH10B, transcription from these promoters is significantly reduced relative to a control in which nnrA is absent (Fig. 2). For the heterologous transcription interference assay, a plasmid encoding the regulator of interest is cotransformed in a host without close homologs to this regulator, with another plasmid bearing a promoter-reporter fusion. If the regulator is able to interfere with the basal transcription of the reporter, it is likely that the regulator binds to the promoter fused to the reporter. In this study, the regulator is encoded by pMR-nnrA, the empty vector pMR10 is used as a control, the host is E. coli strain DH10B, and each promoter tested is fused to the lacZ reporter as indicated in Fig. 2. The narK promoter, which was not affected by nnrA mutation in B. melitensis, is much less sensitive to the presence of nnrA than the other promoters tested. Moreover, sequence analysis of the nirK, norC, and nosR promoters reveals that each contains a putative NnrA binding site. Indeed, since NnrA is homologous to NnrR of R. sphaeroides, it is expected to recognize similar sites on DNA close to the TTGCG(N)4CACAA sequence found upstream of nnrS in R. sphaeroides (2). Similar sites are found in B. melitensis promoters nirK (AGGCGTGAA CACAA, at 198 nucleotides [nt] upstream of ATG), norC (TTGCCTATTCGCAA, at 129 nt upstream of ATG), and nosR (TTGCGTCATAT CAA, at 221 nt upstream of ATG) but not in the narK promoter, which also suggests that NnrA modulates transcription from the nirK, norC, and nosR promoters in B. melitensis.
FIG. 2.
Heterologous transcriptional interference test using nnrA of B. melitensis with the narK, nirK, norC, and nosR promoters. E. coli strains contain a pMR10 plasmid bearing nnrA (or a pMR10 without nnrA) and a pBBR-MCS1lacZ derivative, in which a promoter is fused to the lacZ reporter. It is expected that nnrA expression from pMR10-nnrA will interfere with a promoter-lacZ fusion if NnrA is able to modulate the promoter tested, probably by direct binding. In the upper panel, E. coli strains were grown overnight (in aerobic conditions, without the addition of nitrate) on LB medium containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (40 μg/ml), chloramphenicol (20 μg/ml), and kanamycin (50 μg/ml) in order to select for strains harboring a pMR10-derived plasmid and pBBRMCS carrying a fusion to the lacZ reporter. In these experiments, nnrA is expressed under the control of an E. coli lac promoter. The strains having the pMR10-nnrA plasmid and the PnirK-lacZ, PnorC-lacZ, and PnosR-lacZ fusions are less stained than the controls (pMR10-containing strains and the strain containing pMR10-nnrA and the PnarK-lacZ fusion). Three independent clones are shown. These interferences of nnrA with nirK, norC, and nosR promoters that were observed with X-Gal staining are confirmed by a standard β-galactosidase (β-gal) assay (lower panel) made with clones grown overnight in liquid LB medium supplemented with chloramphenicol (20 μg/ml) and kanamycin (50 μg/ml) (8). The indicated values are the averages of measurements of three independent clones, and each error bar corresponds to one standard deviation.
In order to test the importance of denitrification control in Brucella's virulence, we tested the residual virulence of the narR, nnrA, and nnrB mutants. The reductase mutants were also tested as a reference. We evaluated the residual virulence of those mutants in a mouse model of infection by using the intraperitoneal route (Table 2). Briefly, 5 × 105 CFU was injected into 7-week-old BALB/c mice and, 4 weeks later, spleens were recovered, homogenized in phosphate-buffered saline with 0.1% Triton X-100, and plated on 2YT medium for CFU counting. The nnrA mutant is the only mutant strongly attenuated (about a 100-fold reduction in the number of CFU per spleen) after 4 weeks of infection relative to the wild-type strain. The mutants in genes coding for the reductases NnrB and NarR are slightly attenuated (0.5 to 0.9 log units of attenuation) (Table 2). From these data, we propose that nnrA function in mice is not restricted in its capacity to activate nir, nor, and nos genes in the appropriate conditions.
TABLE 2.
Virulence of the mutants in BALB/c mice after intraperitoneal infectiona
| Strain | Attenuation in mice |
|---|---|
| narR | 0.9 ± 0.1 |
| nnrA | 2.4 ± 0.5 |
| nnrB | 0.8 ± 0.1 |
| narG | 0.5 ± 0.1 |
| nirK | 0.6 ± 0.1 |
| norB | 0.6 ± 0.1 |
| nosZ | 0.7 ± 0.1 |
Virulence is expressed as the log of attenuation of the mutant compared to the wild-type strain. Each group tested contained four mice at 4 weeks postinfection. Differences larger than 0.25 log units are statistically significant (P was <0.001 using standard ANOVA test). In each experiment, close to 4.5 log CFU was recovered from the spleen of mice infected with the wild-type strain.
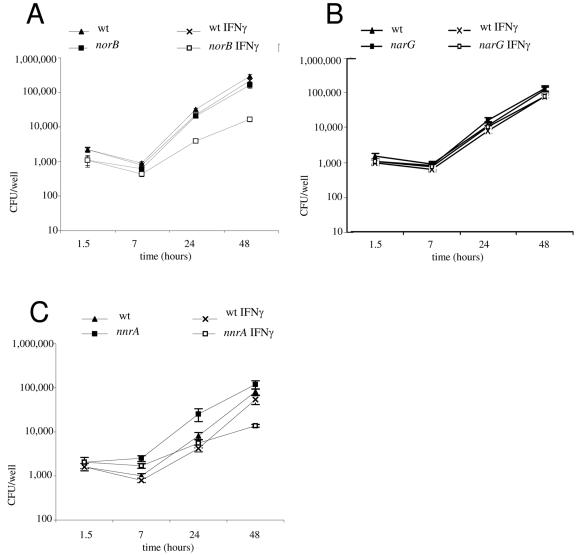
We also compared the phenotypes of nnrA and norB reductase mutants in activated macrophages. Indeed, it has been proposed that nitric oxide reductase eliminates the NO produced by the host cells and thus prevents the killing of intracellular Brucella by reactive nitrogen intermediates like peroxynitrite generated from NO (14). If NnrA is also an activator of nor genes ex vivo, the nnrA and norB mutants are expected to share similar phenotypes during a cellular infection. In order to test this hypothesis, J774A.1 macrophages were cultured in Iscove medium supplemented with 5 mM glutamine (Gibco BRL) and 10% fetal calf serum (Sigma Chimie) at 37°C in 5% CO2 and were treated or not treated with gamma interferon (IFN-γ) (10 U/ml of mrIFN-γ; Pharmingen, San Diego, CA) before being infected with the norB mutant as well as with the nnrA mutant (Fig. 3). The narG mutant was used as a negative control in this experiment. Unstimulated macrophages did not release any detectable NO (4) (data not shown), and in those conditions, the mutants appeared to be as virulent as was the wild type (Fig. 3). In IFN-γ-activated cells, which release the nitrogen radical upon infection (4), the growth of the narG mutant was not affected. On the contrary, we observed a clear difference between the wild type and the norB or nnrA mutants at 48 h of infection. Indeed, the number of wild-type bacteria strongly increased between 7 and 48 h of infection for the wild-type strain, while the multiplication of the norB and nnrA mutants was clearly affected (about 10-fold reductions at 48 h relative to unactivated macrophage condition). Measurements at 48 h postinfection (p.i.) revealed a concentration of nitrite (expected to result from NO oxidation) ranging around 15 ± 5 μM in supernatants of cells infected with the different Brucella strains (mutants or wild type), while nitrite was undetectable from the supernatants of cells infected in the absence of IFN-γ (data not shown). In order to check whether NO production was indeed involved in the lower resistance of norB and nnrA mutants in activated macrophages, we performed the same infections but in the presence of 2 mM L-NAME (Nω-nitro-l-arginine-methyl-ester), an inhibitor of NO synthase. In the presence of L-NAME, the survival of norB and nnrA mutants at 48 h p.i. was almost restored, confirming that the reduction of CFU of norB and nnrA mutants in activated macrophages is NO-dependent (Table 3). The norB and nnrA mutants are therefore sensitive to NO and its derivatives, which was not observed with the wild-type strain and with the narG mutant. Sensitivity of nnrA and norB mutants to NO and its derivatives was confirmed using sin-1, a generator of NO and peroxynitrite (data not shown). These results are in accordance with an nnrA role as activator of norB.
FIG. 3.
Growth of wt and norB (A), narG (B), and nnrA (C) mutants of B. melitensis in J774A.1 macrophages with or without IFN-γ stimulation. Cells treated or not treated with IFN-γ (10 U/ml) were infected under the conditions indicated in Materials and Methods. At the indicated time p.i., the number of intracellular bacteria was measured and expressed in CFU/well. Measurements performed (six replicates) ± standard deviations are presented. At 48 h p.i., the difference between activated and inactivated samples is statistically significant (P < 0.001 in a standard t test) for the norB and nnrA mutants (panels A and C).
TABLE 3.
L-NAME, an inhibitor of NO synthase, is able to suppress the survival defect of norB and nnrA mutants in activated macrophagesa
| Mutant strain | Suppression activity with (CFU [104/well]):
|
||
|---|---|---|---|
| Control | IFN-γ | IFN-γ and L-NAME | |
| norB | 12.5 ± 1.0 | 2.4 ± 0.2 | 7.8 ± 1.1 |
| nnrA | 10.4 ± 0.5 | 2.4 ± 0.3 | 7.8 ± 1.9 |
Murine macrophages are activated using IFN-γ, and L-NAME (2 mM) was added to inhibit the NO synthase activity. Six replicates were performed for each condition. Both treatments, activation with IFN-γ and inhibition with L-NAME, give a statistically significant test (P was <0.001 using standard ANOVA test).
It is noticeable that the norB mutant is attenuated in activated macrophages (Fig. 3) but only slightly attenuated in mice at 4 weeks postinfection (Table 2). Many reasons could explain this apparent discrepancy, including a minor or redundant role of bacterial destruction by NO and its derivatives in this mouse infection model.
In conclusion, our data demonstrate that NnrA-regulated genes, probably different from denitrification genes, are of importance for Brucella virulence. Further experiments will be required to identify new NnrA target genes, since more than 80 putative NnrA binding sites were found in a sequence analysis of the B. melitensis genome using the DNA pattern search at the RSA Tools website (http://rsat.ulb.ac.be/rsat/) (13), using default parameters and the NnrR predicted binding site [TTGCG(1)4CACAA] with the possibility of one substitution. It will also be interesting to discover the signal(s) perceived by or transmitted to NnrA; NO could be one of the obvious candidates.
Acknowledgments
Valérie Haine and Marie Dozot hold a specialization grant from the “Fonds pour la Formation à la Recherche dans l'Industrie et dans l'Agriculture” (FRIA).
We thank the “Fonds de la Recherche Fondamentale Collective” (FRFC, convention no. 2.4521.04). We thank URPhyM (University of Namur) for DNA sequencing facilities. We also thank Régis Hallez and Sandrine Léonard for the construction of the pBBRMCS4GW and pBBRMCS1lacZ plasmids, respectively.
REFERENCES
- 1.Anjum, M. F., T. M. Stevanin, R. C. Read, and J. W. Moir. 2002. Nitric oxide metabolism in Neisseria meningitidis. J. Bacteriol. 184:2987-2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartnikas, T. B., Y. Wang, T. Bobo, A. Veselov, C. P. Scholes, and J. P. Shapleigh. 2002. Characterization of a member of the NnrR regulon in Rhodobacter sphaeroides 2.4.3 encoding a haem-copper protein. Microbiology 148:825-833. [DOI] [PubMed] [Google Scholar]
- 3.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H. Elzer, S. Hagius, D. O'Callaghan, J. J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gross, A., S. Spiesser, A. Terraza, B. Rouot, E. Caron, and J. Dornand. 1998. Expression and bactericidal activity of nitric oxide synthase in Brucella suis-infected murine macrophages. Infect. Immun. 66:1309-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haine, V., A. Sinon, F. Van Steen, S. Rousseau, M. Dozot, P. Lestrate, C. Lambert, J.-J. Letesson, and X. De Bolle. 2005. Systematic targeted mutagenesis of Brucella melitensis 16M reveals a major role for GntR regulators in the control of virulence. Infect. Immun. 73:5578-5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kohler, S., V. Foulongne, S. Ouahrani-Bettache, G. Bourg, J. Teyssier, M. Ramuz, and J. P. Liautard. 2002. The analysis of the intramacrophagic virulome of Brucella suis deciphers the environment encountered by the pathogen inside the macrophage host cell. Proc. Natl. Acad. Sci. USA 99:15711-15716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Korner, H., H. J. Sofia, and W. G. Zumft. 2003. Phylogeny of the bacterial superfamily of Crp-Fnr transcription regulators: exploiting the metabolic spectrum by controlling alternative gene programs. FEMS Microbiol. Rev. 27:559-592. [DOI] [PubMed] [Google Scholar]
- 8.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 9.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 10:783-791. [Google Scholar]
- 10.Smith, L. D., and T. A. Ficht. 1990. Pathogenesis of Brucella. Crit. Rev. Microbiol. 17:209-230. [DOI] [PubMed] [Google Scholar]
- 11.Sperry, J. F., and D. C. Robertson. 1975. Erythritol catabolism by Brucella abortus. J. Bacteriol. 121:619-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tosques, I. E., J. Shi, and J. P. Shapleigh. 1996. Cloning and characterization of nnrR, whose product is required for the expression of proteins involved in nitric oxide metabolism in Rhodobacter sphaeroides 2.4.3. J. Bacteriol. 178:4958-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Helden, J. 2003. Regulatory sequence analysis tools. Nucleic Acids Res. 31:3593-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang, M., N. Qureshi, N. Soeurt, and G. Splitter. 2001. High levels of nitric oxide production decrease early but increase late survival of Brucella abortus in macrophages. Microb. Pathog. 31:221-230. [DOI] [PubMed] [Google Scholar]
- 15.Weber, I., C. Fritz, S. Ruttkowski, A. Kreft, and F. C. Bange. 2000. Anaerobic nitrate reductase (narGHJI) activity of Mycobacterium bovis BCG in vitro and its contribution to virulence in immunodeficient mice. Mol. Microbiol. 35:1017-1025. [DOI] [PubMed] [Google Scholar]
- 16.Wood, N. J., T. Alizadeh, S. Bennett, J. Pearce, S. J. Ferguson, D. J. Richardson, and J. W. Moir. 2001. Maximal expression of membrane-bound nitrate reductase in Paracoccus is induced by nitrate via a third FNR-like regulator named NarR. J. Bacteriol. 183:3606-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]