Abstract
The molecular typing methods used so far for Mycobacterium ulcerans isolates have not been able to identify genetic differences among isolates from Africa. This apparent lack of genetic diversity among M. ulcerans isolates is indicative of a clonal population structure. We analyzed the genetic diversity of 72 African isolates, including 57 strains from Ghana, by variable number of tandem repeat (VNTR) typing based on a newly identified polymorphic locus designated ST1 and the previously described locus MIRU 1. Three different genotypes were found in Ghana, demonstrating for the first time the genetic diversity of M. ulcerans in an African country. While the ST1/MIRU 1 allele combination BD/BAA seems to dominate in Africa, it was only rarely found in isolates from Ghana, where the combination BD/B was dominant and observed in all districts studied. A third variant genotype (C/BAA) was found only in the Amansie-West district. The results indicate that new genetic variants of M. ulcerans emerged and spread within Ghana and support the potential of VNTR-based typing for genotyping of M. ulcerans.
Mycobacterium ulcerans, the causative agent of Buruli ulcer (BU), is an emerging pathogen, particularly in Sub-Saharan African countries; it is also found in tropical and subtropical regions of Asia, the western Pacific, and Latin America (4). BU is characterized by chronic necrotic lesions of subcutaneous tissues. Due to the lack of an established effective antimicrobial therapy, surgical excision and skin grafting is currently the recommended treatment (27).
While it is known that proximity to slowly flowing or stagnant bodies of water is a risk factor for M. ulcerans infection, the exact mode of transmission is unknown (4). This is partly because there is no molecular typing method that has sufficiently high resolution for microepidemiological analyses. The apparent lack of genetic diversity of M. ulcerans in individual geographical regions (7, 8, 16, 19-21) is indicative of a clonal population structure. Genetic analyses have suggested that M. ulcerans recently diverged from M. marinum (5, 21, 26), which is a well-known fish pathogen and can cause limited granulomatous skin infections in humans (10, 11, 13). One of the hallmarks of the emergence of M. ulcerans as a more severe pathogen is the acquisition of a 174-kb plasmid bearing a cluster of genes necessary for the synthesis of the macrolide toxin mycolactone, which is responsible for the massive tissue destruction seen in BU (22).
Variable number of tandem repeat (VNTR) typing is a PCR-based technique for identifying alleles of defined regions of DNA that contain variable numbers of copies of short sequence stretches. The resolution of the method is cumulative and can be increased by inclusion of additional loci. Tandem repeats are easily identified from genome sequence data, measurement of PCR fragment sizes is relatively straightforward, and VNTR typing data from different laboratories can be digitalized and compared. The availability of complete genomic sequences has facilitated identification of repetitive genetic elements of M. tuberculosis (12, 15, 24, 25), M. bovis (14, 18), and M. avium (6), including short tandem repeats designated exact tandem repeats and mycobacterial interspersed repetitive units (MIRUs). Strain typing with these sets of polymorphic loci is becoming an important tool in the epidemiological analysis of tuberculosis (9), and ordinary agarose gel electrophoretic separation of PCR products is usually sufficient to estimate the number of repeat units in an allele. In a study of M. bovis strains from Chad, VNTR typing of a distinct number of loci was most discriminative for strains of the same clone (14).
More recently, MIRUs and other VNTRs have also been described for M. ulcerans and M. marinum (3, 23) typing. Most of the described sequences are orthologues of sequences in the M. tuberculosis genome database, and the resolution seems to be comparable to that of the currently most discriminatory methods, 2426 PCR analysis (19) and IS2404-Mtb2 PCR (2), which discriminate among isolates having different geographical origins but not among strains from different regions of Africa where the organisms are endemic.
We describe in this report a new M. ulcerans-specific VNTR locus (ST1) which together with the previously described MIRU 1 locus (23) differentiated clinical isolates from Ghana into groups with three VNTR allele combinations.
MATERIALS AND METHODS
Identification of the VNTR locus ST1.
Tandem repeat finder software (http://www.c3.biomath.mssm.edu/trf.thml) was used to screen the M. marinum sequence data bank (www.sanger.ac.uk/projects/M_marinum) and to identify a tandem repeat-containing locus which was designated ST1. This locus is present both in M. marinum and in M. ulcerans (http://genopole.pasteur.fr/Mulc/BuruList.html), but it is not present in M. tuberculosis (http://www.sanger.ac.uk/Projects/M_tuberculosis). A forward primer (CTGAGGGGATTTCACGACCAG) and a reverse primer (CGCCACCCGCGGACACAGTCG) located in the sequences flanking the identified locus that yielded a 423-bp PCR product were designed. Genomic sequences corresponding to the primers were 100% identical for M. marinum and M. ulcerans.
Bacterial strains.
A panel consisting of 11 M. ulcerans clinical isolates of human origin from diverse geographical areas and six M. marinum clinical isolates of human origin from Switzerland was used to assess the polymorphism of ST1 (Table 1). Most of the M. marinum isolates were from patients living in the vicinity of Zurich; the only exception was M. marinum N119, which was isolated from a patient living in Biel. The years of isolation were 1995 for strains 853 and 894, 1997 for strains 8972 and 946, and 1998 for strains N119 and 3023. In order to analyze the diversity of African M. ulcerans strains, 66 additional clinical isolates (12 isolates from Benin and 54 isolates from Ghana) were included in this study. The Ghanaian strains were isolated (28) between 2001 and 2003 from patients being treated at the Amasaman Health Centre in the Greater Accra region of Ghana (48 isolates) or at Saint Martin Hospital Agroyesum in the Ashanti region of Ghana (six isolates).
TABLE 1.
ST1 alleles of M. ulcerans and M. marinum disease isolates
| Species | Isolate | Country of origin | No. of repeats | Arrangement of the variant repeat DNA sequences |
|---|---|---|---|---|
| M. ulcerans | ITM 8756 | Japan | 2 | CF |
| ITM 980912 | China | 2 | CF | |
| ITM 941328 | Malaysia | 2 | BD | |
| ITM 884 | Australia | 2 | BD | |
| ITM 9357 | Papua-New Guinea | 2 | BD | |
| ITM 7922 | French Guyana | 1 | C | |
| ITM 970359a | Ghana | 1 | C | |
| ITM 970321 | Ghana | 2 | BD | |
| ITM 940886 | Benin | 2 | BD | |
| ITM 940662 | Côte d'Ivoire | 2 | BD | |
| ITM 960658 | Angola | 2 | BD | |
| M. marinum | 894/1995 | Switzerland | 3 | ACE |
| 853/1995 | Switzerland | 3 | NDc | |
| 8972/1997 | Switzerland | 3 | ND | |
| 946/1997 | Switzerland | 3 | ND | |
| 3023/1998 | Switzerland | 3 | ND | |
| N119/1998 | Switzerland | 2 | ND | |
| M. marinumb | Unknown | 2 | AC |
Isolated in 1997 from a patient living in the Amansie-West district.
Isolate used for the M. marinum genome sequencing project.
ND, not determined.
Further information on the origins of the isolates is presented in Table S1 in the supplemental material.
TABLE 2.
ST1 and MIRU 1 allele combinations for M. ulcerans strains from Africaa
| Country of origin | District | No. of strains | ST1 allele | MIRU 1 allele | Allele combi- nation |
|---|---|---|---|---|---|
| Ghana | Ga | 47 | 2 (BD) | 1 (B) | 1 |
| Gae | 1 | 2 (BD) | 3 (BAA) | 3 | |
| Akwapim South | 1 | 2 (BD) | 1 (B) | 1 | |
| East Akim | 1 | 2 (BD) | 1 (B) | 1 | |
| Ahafo-Ano | 1 | 2 (BD) | 1 (B) | 1 | |
| Amansie-West | 1 | 2 (BD) | 1 (B) | 1 | |
| Amansie-West | 4 | 1 (C) | 3 (BAA) | 2 | |
| Unknownb | 1 | 2 (BD) | 3 (BAA) | 3 | |
| Benin | 13 | 2 (BD)c | 3 (BAA)c, d | 3 | |
| Côte d'Ivoire | 1 | 2 (BD) | 3d | 3 | |
| Angola | 1 | 2 (BD) | 3d | 3 |
VNTR copy numbers for ST1 and MIRU 1 were determined, and allele combinations (combinations 1 to 3) were assigned. The sequence profiles of M. ulcerans strains are indicated in parentheses.
Isolated at Saint Martin's Hospital in the Ashanti region.
Only one strain (ITM 94-0886) was analyzed by sequencing (23).
MIRU 1 copy number as previously described.
Agy99 used for the M. ulcerans genome sequencing project.
DNA extraction.
DNA was extracted as described previously (17). Briefly, small bacterial pellets were heated for 1 h at 95°C in 500 μl of an extraction mixture (50 mM Tris-HCl, 25 mM EDTA, 5% monosodium glutamate). One hundred microliters of a 50-mg/ml lysozyme solution was then added and incubated for 2 h at 37°C. Seventy microliters of proteinase K-10× buffer (100 mM Tris-HCl, 50 mM EDTA, 5% sodium dodecyl sulfate [pH 7.8]) and 10 μl of a 20-mg/ml proteinase K solution were added. After incubation at 45°C overnight, 300 μl of 0.1-mm-diameter zirconia beads (BioSpec Products) was added to each sample and vortexed at full speed for 4 min. The beads and large debris were removed by brief centrifugation, and the supernatants were transferred to fresh tubes for phenol-chloroform (Fluka) extraction. The DNA in the upper phase was precipitated with ethanol and resuspended in 100 μl of water.
PCR analysis and sequencing of PCR products.
PCR mixtures (final volume, 20 μl) contained 1× Taq PCR buffer, deoxynucleoside triphosphates (0.2 mM each), 1 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer Applied Biosystems), each primer at a concentration of 0.5 μM, and mycobacterial DNA. Addition of 5% dimethyl sulfoxide to the reaction mixture improved the yield of PCR products. The reaction was carried out using a Perkin-Elmer 9600 cycler starting with denaturation for 10 min at 95°C. After denaturation, the PCR was performed for 40 cycles of 0.5 min at 94°C, 0.5 min at 65°C, and 1 min at 72°C. The reactions were terminated by incubation for 10 min at 72°C. PCR fragments were analyzed by agarose gel electrophoresis using 2% NuSieve agarose. The sizes of the amplicons were estimated by comparison with Size Marker VIII (Roche). PCR products were directly sequenced with an ABI Prism 310 genetic analysis system. For ST1 423-bp and 369-bp PCR products corresponded to copy numbers of 2 and 1, respectively. For MIRU 1, copy numbers were assigned as described previously (23).
RESULTS
Identification and characterization of a new VNTR locus found in M. ulcerans and M. marinum.
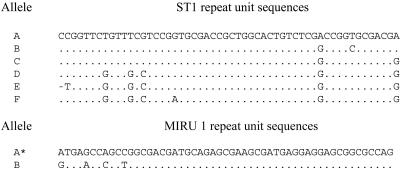
A new VNTR locus designated ST1 was identified by screening the M. marinum sequence data bank with tandem repeat finder software. Orthologues of ST1 were found in M. ulcerans but not in M. tuberculosis. The repeat length (54 bp) is suitable for size analysis by standard agarose gel electrophoresis. In contrast to MIRUs (23), ST1 is not intergenic but is part of a pseudogene and therefore of no known functional interest (T. Stinear, personal communication). When 11 M. ulcerans isolates that had geographically diverse origins were typed by agarose gel electrophoresis of PCR products, two different alleles of ST1 were identified (Table 1). While eight strains had two repeats, two isolates, one from French Guyana and one of the two isolates from Ghana analyzed, had only one repeat. Seven M. marinum clinical isolates were tested, and all five strains from patients in the vicinity of Zurich (strains 8972, 946, 3023, 853, and 894) had three repeats, whereas strain N119 isolated in a different part of Switzerland and the reference strain used for the M. marinum genome sequencing project both had two repeats. Sequence analysis of PCR products confirmed the size differences observed by agarose gel electrophoresis and identified six sequence variants of the repeat unit (designated A to F) (Fig. 1). Unlike many other VNTRs (1), ST1 showed no microdeletions but only single-nucleotide polymorphisms in the sequence variants. The M. ulcerans strains from China and Japan had a different allele (CF) (Table 1) than the other M. ulcerans strains with two repeats (BD). A third allele with two repeats (AC) and an allele with three repeats (ACE) were found in M. marinum (Table 1). Thus, sequencing of ST1 improved the discriminatory power for M. ulcerans and M. marinum strains compared to gel electrophoresis analysis alone and revealed distinctive genotypes for M. marinum and M. ulcerans.
FIG. 1.
Sequence variation of ST1 and MIRU 1 tandem repeat units. The dash indicates a base deletion; dots indicate identical sequence positions; and the asterisk indicates that allele A of MIRU 1 corresponds to variant A2 (23).
Diversity of M. ulcerans isolates from Ghana.
Evidence for diversity of the ST1 locus in African isolates (Table 1) prompted us to analyze additional collections consisting of 12 disease isolates from Benin and 54 isolates from Ghana (Table 2). All of the strains from Benin and the majority of the Ghanaian strains had an ST1 allele (BD) with two repeats. However, in most strains from the Amansie-West district (including ITM-970359) (Table 1), a second allele with only one repeat (C) was identified (Table 2). When the Ghanaian isolates were tested for diversity in the MIRU 1 (23) and VNTR 8, 9, and 19 loci, which were previously described as polymorphic in M. ulcerans strains having different geographical origins (3), diversity was also found in the MIRU 1 locus. Sequence analysis of PCR products of selected strains confirmed the size differences observed by agarose gel electrophoresis and identified two sequence variants of the MIRU 1 repeat unit (designated A and B) (Fig. 1).
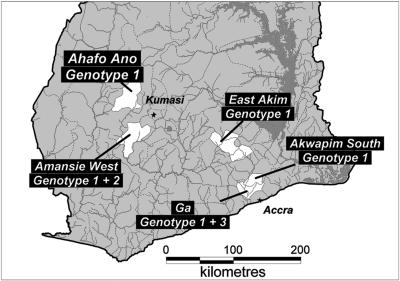
Altogether, three VNTR allele combinations were found in the clinical isolates from Ghana (Table 2 and Fig. 2 and 3). While all isolates from the Ga, Akwapim South, Ahafo-Ano North, and Akim Abuakwa districts had ST1/MIRU 1 allele combination 1 (BD/B), two allele combinations, allele combinations 1 and 2 (C/BAA), were found in the five isolates from the Amansie-West district. A third allele combination (BD/BAA) was found in two strains (Agy99 from the Ga district and ITM 97-0359 from the Ashanti region) isolated before 2000 in Ghana and in all other African isolates.
FIG. 2.
Map of southern Ghana, showing the residential districts of patients from whom the Ghanaian isolates analyzed in this study were obtained. The ST1/MIRU 1 allele combinations are genotype 1 (BD/B), genotype 2 (C/BAA), and genotype 3 (BD/BAA).
FIG. 3.
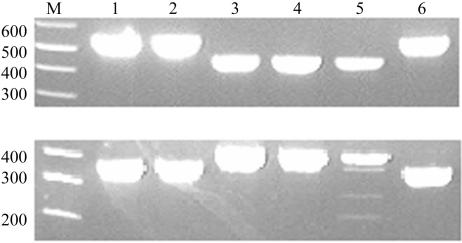
Agarose gel electrophoretic analysis of PCR products from amplifications with MIRU 1 primers (upper panel) and ST1 primers (lower panel). The results for selected isolates are shown. Lanes 1, 2, and 6, strains from the Amansie-West district; lanes 3, 4, and 5, strains from the Ga district.
DISCUSSION
Molecular typing methods, such as multilocus sequence typing, 16S rRNA sequencing, restriction fragment length polymorphism analysis, and variable number of tandem repeat typing, have revealed a remarkable lack of genetic diversity of M. ulcerans and a clonal population structure in certain geographical regions. In particular, the discriminatory power of all these methods is insufficient to differentiate between African isolates. Innovative molecular genetic fingerprinting methods are therefore required for local epidemiological studies that aim to reveal transmission pathways and environmental reservoirs of M. ulcerans. The first attempts to use VNTR typing for M. ulcerans (3, 23) identified variable loci suitable for discrimination of disease isolates at the continent level. In this study we used a newly identified VNTR (ST1) and four previously described VNTRs to analyze the genetic diversity in a collection of 71 M. ulcerans strains from Africa, including 57 isolates from Ghana. Three of the previously described VNTRs, VNTRs 8, 9, and 19 (3), were not able to discriminate among the African strains. However, MIRU 1 (23) and the newly identified locus (ST1) defined three subgroups for the Ghanaian strains.
The fact that two allele combinations (BD/B and C/BAA) that differ from the common African combination (BD/BAA) were found in a recent (2001 to 2003) collection of Ghanaian isolates is indicative of the ongoing microevolution of M. ulcerans and of the spread of new variants within Ghana. It is tempting to hypothesize that allele combination 3 (BD/BAA) represents an ancestor-like genotype and that the other combinations evolved by reduction of the number of the repeat unit in the ST1 locus (from BD to C) or in the MIRU 1 locus (from BAA to B). While conversion of the MIRU 1 locus from BAA to B could be explained by deletion of the two A repeat units, conversion of the ST1 locus from BD to C by a deletion mechanism would require that a central sequence stretch of the BD repeat region comprising portions of both the B and D repeat units was lost, yielding the hybrid repeat unit C.
While allele combination 3 seems to be the most common combination in Africa, most of the M. ulcerans strains from Ghana analyzed had allele combination 1. This genotype was found in all Ghanaian districts included in this study. Allele combination 2 dominated in the Amansie-West district, but it was found exclusively there. In the future a follow-up study of the temporal and spatial patterns of emergence and spread of genotypes may contribute to our understanding of the transmission and epidemiology of Buruli ulcer. From the present data, we cannot draw any conclusions concerning why certain variants appear to be the more successful than others. The fact that we were not able to subgroup the 47 isolates from the Ga district by VNTR (with the exception of “older” isolate Agy99) confirms that M. ulcerans has a clonal population structure that is associated with a low rate of genomic drift. The availability of the fully assembled and annotated genome sequence of M. ulcerans in the near future should facilitate identification of further polymorphic VNTR loci and potentially contribute to further refinement of genetic fingerprinting of M. ulcerans isolates.
Supplementary Material
Acknowledgments
We acknowledge Edwin Ampadu of the Ghana National Buruli Ulcer Control program for his assistance with collection of clinical samples and Anthony Ablordey for a critical review of the manuscript. We also thank Franca Baggi of the Swiss Centre of Mycobacteria for providing M. marinum strains and the heads of the M. ulcerans and M. marinum genome sequencing projects for permission to use the sequence data.
NCCR North-South IP-4 is acknowledged for financial support.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ablordey, A., M. Hilty, P. Stragier, J. Swings, and F. Portaels. 2005. Comparative nucleotide sequence analysis of polymorphic variable-number tandem-repeat loci in Mycobacterium ulcerans. J. Clin. Microbiol. 43:5281-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ablordey, A., R. Kotlowski, J. Swings, and F. Portaels. 2005. PCR amplification with primers based on IS2404 and GC-rich repeated sequence reveals polymorphism in Mycobacterium ulcerans. J. Clin. Microbiol. 43:448-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ablordey, A., J. Swings, C. Hubans, K. Chemlal, C. Locht, F. Portaels, and P. Supply. 2005. Multilocus variable-number tandem repeat typing of Mycobacterium ulcerans. J. Clin. Microbiol. 43:1546-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asiedu, K., R. Scherpbier, and M. Raviglione. 2000. Executive summary, p. 1-4. In Buruli ulcer—Mycobacterium ulcerans infection. WHO document WHO/CDS/CPE/GBUI/2000.1. World Health Organization, Geneva, Switzerland.
- 5.Boddinghaus, B., T. Rogall, T. Flohr, H. Blocker, and E. C. Bottger. 1990. Detection and identification of mycobacteria by amplification of ribosomal RNA. J. Clin. Microbiol. 28:1751-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bull, T. J., K. Sidi-Boumedine, E. J. Mcminn, K. Stevenson, R. Pickup, and J. Hermon-Taylor. 2003. Mycobacterial interspersed repetitive units (MIRU) differentiate Mycobacterium avium subspecies paratuberculosis from other species of the Mycobacterium avium complex. Mol. Cell. Probes 17:157-164. [DOI] [PubMed] [Google Scholar]
- 7.Chemlal, K., K. De Ridder, P. A. Fonteyne, W. M. Meyers, J. Swings, and E. Portaels. 2001. The use of IS2404 restriction fragment length polymorphisms suggests the diversity of Mycobacterium ulcerans from different geographical areas. Am. J. Trop. Med. Hyg. 64:270-273. [DOI] [PubMed] [Google Scholar]
- 8.Chemlal, K., G. Huys, P. A. Fonteyne, V. Vincent, A. G. Lopez, L. Rigouts, J. Swings, W. M. Meyers, and F. Portaels. 2001. Evaluation of PCR-restriction profile analysis and IS2404 restriction fragment length polymorphism and amplified fragment length polymorphism fingerprinting for identification and typing of Mycobacterium ulcerans and M. marinum. J. Clin. Microbiol. 39:3272-3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crawford, J. T. 2003. Genotyping in contact investigations: a CDC perspective. Int. J. Tuberc. Lung D 7:S453-S457. [PubMed] [Google Scholar]
- 10.Dailloux, M., C. Laurain, R. Weber, and P. Hartemann. 1999. Water and nontuberculous mycobacteria. Water Res. 33:2219-2228. [Google Scholar]
- 11.Dobos, K. M., F. D. Quinn, D. A. Ashford, C. R. Horsburgh, and C. H. King. 1999. Emergence of a unique group of necrotizing mycobacterial diseases. Emerg. Infect. Dis. 5:367-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frothingham, R., and W. A. Meeker-O'Connell. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189-1196. [DOI] [PubMed] [Google Scholar]
- 13.Gluckman, S. J. 1995. Mycobacterium marinum. Clin. Dermatol. 13:273-276. [DOI] [PubMed] [Google Scholar]
- 14.Hilty, M., C. Diguimbaye, E. Schelling, F. Baggi, M. Tanner, and J. Zinsstag. 2005. Evaluation of the discriminatory power of variable number tandem repeat (VNTR) typing of Mycobacterium bovis strains. Vet. Microbiol. 109:217-222. [DOI] [PubMed] [Google Scholar]
- 15.Mazars, E., S. Lesjean, A. L. Banuls, M. Gilbert, V. Vincent, B. Gicquel, M. Tibayrenc, C. Locht, and P. Supply. 2001. High-resolution minisatellite-based typing as a portable approach to global analysis of Mycobacterium tuberculosis molecular epidemiology. Proc. Natl. Acad. Sci. USA 98:1901-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Portaels, F., P. A. Fonteyne, H. DeBeenhouwer, P. DeRijk, A. Guedenon, J. Hayman, and W. M. Meyers. 1996. Variability in 3′ end of 16S rRNA sequence of Mycobacterium ulcerans is related to geographic origin of isolates. J. Clin. Microbiol. 34:962-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rondini, S., E. Mensah-Quainoo, H. Troll, T. Bodmer, and G. Pluschke. 2003. Development and application of real-time PCR assay for quantification of Mycobacterium ulcerans DNA. J. Clin. Microbiol. 41:4231-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skuce, R. A., T. P. McCorry, J. F. McCarroll, S. M. M. Roring, A. N. Scott, D. Brittain, S. L. Hughes, R. G. Hewinson, and S. D. Neill. 2002. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 148:519-528. [DOI] [PubMed] [Google Scholar]
- 19.Stinear, T., J. K. Davies, G. A. Jenkin, F. Portaels, B. C. Ross, F. Oppedisano, M. Purcell, J. A. Hayman, and P. D. R. Johnson. 2000. A simple PCR method for rapid genotype analysis of Mycobacterium ulcerans. J. Clin. Microbiol. 38:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stinear, T., B. C. Ross, J. K. Davies, L. Marino, R. M. Robins-Browne, F. Oppedisano, A. Sievers, and P. D. R. Johnson. 1999. Identification and characterization of IS2404 and IS2606: two distinct repeated sequences for detection of Mycobacterium ulcerans by PCR. J. Clin. Microbiol. 37:1018-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stinear, T. P., G. A. Jenkin, P. D. R. Johnson, and J. K. Davies. 2000. Comparative genetic analysis of Mycobacterium ulcerans and Mycobacterium marinum reveals evidence of recent divergence. J. Bacteriol. 182:6322-6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stinear, T. P., A. Mve-Obiang, P. L. C. Small, W. Frigui, M. J. Pryor, R. Brosch, G. A. Jenkin, P. D. R. Johnson, J. K. Davies, R. E. Lee, S. Adusumilli, T. Garnier, S. F. Haydock, P. F. Leadlay, and S. T. Cole. 2004. Giant plasmid-encoded polyketide synthases produce the macrolide toxin of Mycobacterium ulcerans. Proc. Natl. Acad. Sci. USA 101:1345-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stragier, P., A. Ablordey, W. M. Meyers, and F. Portaels. 2005. Genotyping Mycobacterium ulcerans and Mycobacterium marinum by using mycobacterial interspersed repetitive units. J. Bacteriol. 187:1639-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Supply, P., J. Magdalena, S. Himpens, and C. Locht. 1997. Identification of novel intergenic repetitive units in a mycobacterial two-component system operon. Mol. Microbiol. 26:991-1003. [DOI] [PubMed] [Google Scholar]
- 25.Supply, P., E. Mazars, S. Lesjean, V. Vincent, B. Gicquel, and C. Locht. 2000. Variable human minisatellite-like regions in the Mycobacterium tuberculosis genome. Mol. Microbiol. 36:762-771. [DOI] [PubMed] [Google Scholar]
- 26.Tonjum, T., D. B. Welty, E. Jantzen, and P. L. Small. 1998. Differentiation of Mycobacterium ulcerans, M. marinum, and M. haemophilum: mapping of their relationships to M. tuberculosis by fatty acid profile analysis, DNA-DNA hybridization, and 16S rRNA gene sequence analysis. J. Clin. Microbiol. 36:918-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Werf, T. S., T. Stinear, Y. Stienstra, W. T. A. van der Graaf, and P. L. Small. 2003. Mycolactones and Mycobacterium ulcerans disease. Lancet 362:1062-1064. [DOI] [PubMed] [Google Scholar]
- 28.Yeboah-Manu, D., T. Bodmer, E. Mensah-Quainoo, S. Owusu, D. Ofori-Adjei, and G. Pluschke. 2004. Evaluation of decontamination methods and growth media for primary isolation of Mycobacterium ulcerans from surgical specimens. J. Clin. Microbiol. 42:5875-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.