Abstract
The complete genome sequence of Thiobacillus denitrificans ATCC 25259 is the first to become available for an obligately chemolithoautotrophic, sulfur-compound-oxidizing, β-proteobacterium. Analysis of the 2,909,809-bp genome will facilitate our molecular and biochemical understanding of the unusual metabolic repertoire of this bacterium, including its ability to couple denitrification to sulfur-compound oxidation, to catalyze anaerobic, nitrate-dependent oxidation of Fe(II) and U(IV), and to oxidize mineral electron donors. Notable genomic features include (i) genes encoding c-type cytochromes totaling 1 to 2 percent of the genome, which is a proportion greater than for almost all bacterial and archaeal species sequenced to date, (ii) genes encoding two [NiFe]hydrogenases, which is particularly significant because no information on hydrogenases has previously been reported for T. denitrificans and hydrogen oxidation appears to be critical for anaerobic U(IV) oxidation by this species, (iii) a diverse complement of more than 50 genes associated with sulfur-compound oxidation (including sox genes, dsr genes, and genes associated with the AMP-dependent oxidation of sulfite to sulfate), some of which occur in multiple (up to eight) copies, (iv) a relatively large number of genes associated with inorganic ion transport and heavy metal resistance, and (v) a paucity of genes encoding organic-compound transporters, commensurate with obligate chemolithoautotrophy. Ultimately, the genome sequence of T. denitrificans will enable elucidation of the mechanisms of aerobic and anaerobic sulfur-compound oxidation by β-proteobacteria and will help reveal the molecular basis of this organism's role in major biogeochemical cycles (i.e., those involving sulfur, nitrogen, and carbon) and groundwater restoration.
Thiobacillus denitrificans, first isolated by Beijerinck over a century ago (4), was one of the first nonfilamentous bacteria ever described to be capable of growth on inorganic sulfur compounds as sole energy sources (47, 49). Characterized by its ability to conserve energy from the oxidation of inorganic sulfur compounds under either aerobic or denitrifying conditions, T. denitrificans is the best studied of the very few obligate chemolithoautotrophic species known to couple denitrification tosulfur-compound oxidation (Thiomicrospira denitrificans and Thioalkalivibrio thiocyanodenitrificans also have this ability [76, 85]). Despite many years of work on the biochemistry of inorganic sulfur-compound oxidation by Thiobacillus thioparus and T. denitrificans, the mechanisms of oxidation and how they are coupled to energy conservation are still not well understood in these β-proteobacteria, relative to the advances made with facultatively chemolithotrophic α-proteobacterial genera, such as Paracoccus and Starkeya (28, 39, 45, 50). The availability of the complete genome sequence should enable elucidation of the sulfur-oxidation pathway(s) and lead to specifically focused biochemical investigations to resolve these knowledge gaps.
Recent studies have revealed that, in addition to sulfur-compound oxidation, T. denitrificans has broader oxidative capabilities that may not contribute to energy conservation, including anaerobic, nitrate-dependent oxidation of certain metals, such as iron (78). The metabolic repertoire of this widely distributed bacterium can influence the carbon, nitrogen, sulfur, and iron cycles in many soil, aquifer, and sediment environments and is particularly relevant to in situ bioremediation of contaminated groundwater. Environmentally relevant capabilities of T. denitrificans include intrinsic biodegradation of nitrate, one of the most problematic groundwater contaminants worldwide, by anaerobic, nitrate-dependent oxidation of minerals such as FeS and pyrite (FeS2) (see, e.g., references 6, 56, and 78). T. denitrificans is the first and onlyautotrophic bacterium reported to carry out anaerobic (nitrate-dependent) oxidation of U(IV) oxide minerals (5), which could partially counteract efforts to remediate uranium-contaminated aquifers by in situ reductive immobilization [i.e., microbially mediated conversion of relatively soluble U(VI) species to poorly soluble U(IV) minerals]. The intriguing mechanism by which this species can oxidize mineral electron donors that cannot be taken into the cell is currently unknown, but its elucidation will be facilitated by the availability of the genome.
In this article, we present the complete genome sequence of T.denitrificans strain ATCC 25259, the first obligately chemolithoautotrophic, sulfur-oxidizing, β-proteobacterium to be sequenced. We describe some general features of the T. denitrificans genome, including recent gene acquisition, as well as genetic components involved in sulfur-compound oxidation, hydrogen metabolism, aerobic respiration, denitrification, autotrophy, central carbon metabolism, and heavy metal resistance.
MATERIALS AND METHODS
Organism source.
T. denitrificans strain ATCC 25259 was obtained from the American Type Culture Collection (ATCC). This strain was originally isolated by B. F. Taylor in the 1960s (83, 84) and was deposited with the ATCC in 1969, where it was freeze-dried for storage and distribution. For this study, unless noted otherwise, T. denitrificans was grown with thiosulfate under denitrifying conditions, as described elsewhere (5).
Sequencing, coding sequence prediction, and annotation.
Genomic DNA was isolated from T. denitrificans and the complete genome was sequenced as described previously (15). Briefly, small-insert (2- to 3-kb), medium-insert (6.5- to 8.5-kb), and large-insert (35- to 45-kb) libraries were generated by random mechanical shearing of genomic DNA. In the initial random sequencing phase, approximately ninefold sequence coverage was achieved. The sequences from all libraries were assembled together and viewed using the Phred/Phrap/Consed suite (P. Green, University of Washington) (22, 23, 31). Physical gaps were closed by PCR and sequencing. Open reading frames likely to encode proteins (coding sequences [CDS]) were identified and annotated by automated and manual curation as previously described (15).
Comparative genomics.
The Integrated Microbial Genomes system of the Joint Genome Institute (http://img.jgi.doe.gov/) was used for identification of orthologs and to identify CDS unique to T. denitrificans on the basis of BLASTP results; the cutoff values applied were E < 10−5 and 30% identity and E < 10−2 and 20% identity, respectively.
Generation of phylogenetic trees and other analyses.
Phylogenetic trees were generated by identifying potential homologs of translated T. denitrificans CDS by use of BLASTP (1) searches against the nonredundant (nr) GenBank database from the National Center for Biotechnology Information. Typically, trees included the top 50 BLASTP matches. However, sequences were excluded when their BLASTP E values fell below a cutoff of 10−5. For the phylogenetic tree results presented in this article, the lowest ranking sequences shared 24 to 54% identity and 43 to 71% similarity with the query sequences. Sequences were aligned and alignments refined using ClustalX version 1.8 (38) and manual adjustments. Phylogenetic trees were generated by using the protdist program and the neighbor program of the PHYLIP package (24, 25) to calculate distances (using the Jones-Taylor-Thorton matrix) and for clustering (neighbor-joining method), respectively. Membrane-spanning domains of proteins were identified using TmHMM (52, 75). The SignalP program (7) was used to identify putative signal peptides.
RT quantitative PCR analysis.
For 18 selected genes, mRNA levels under exposure conditions of interest were determined by reverse transcription-quantitative PCR (RT-qPCR) analysis. The target genes included Tbd0210, Tbd0561, Tbd0562, Tbd0822, Tbd0823, Tbd0872, Tbd0873, Tbd0874, Tbd1365, Tbd1408, Tbd1926, Tbd2282, Tbd2283, Tbd2326, Tbd2327, Tbd2480, Tbd2488, and Tbd2658. Cultures of T. denitrificans were grown anaerobically with thiosulfate and nitrate and were then anaerobically harvested by centrifugation and washed using techniques and conditions described elsewhere (5). Washed cells (∼6.5 mg protein) were resuspended in stoppered serum bottles under strictly anaerobic conditions in 5 or 10 ml of bicarbonate-buffered medium. Cells were resuspended with either thiosulfate (20 mM) and nitrate (20 mM) or FeCO3 (3.5 mmol/liter) and nitrate (3.5 mM). Chemical analyses (ion chromatography, spectrophotometric determination of ferrozine-iron complexes) of the resuspended cultures were performed to confirm that nitrate reduction and either thiosulfate or Fe(II) oxidation were occurring when the cultures were harvested for RNA. RNA extraction wascarried out using a MasterPure Complete DNA and RNA Purification Kit (EpiCentre) and a modified protocol. Total RNA was reverse transcribed and amplified using a QuantiTect SYBR Green RT-PCR kit (QIAGEN) with gene-specific primers. Each gene-specific PCR was performed in triplicate using 25-μl reactions containing ∼20 ng of template on a Prism 7000 cycler (ABI). Calibration curve determinations were performed with genomic DNA serially diluted over a range of 4 to 5 orders of magnitude. The PCR conditions were optimized to be performed as follows for all transcripts: 30 to 35 cycles at 50°C for 30 min; 95°C for 15 min; 94°C for 15 s; 58°C for 30 s; 72°C for 30 s.
Nucleotide sequence accession number.
The annotated genome sequence has been deposited in the GenBank/EMBL sequence database under accession no. CP000116.
RESULTS AND DISCUSSION
General genome features.
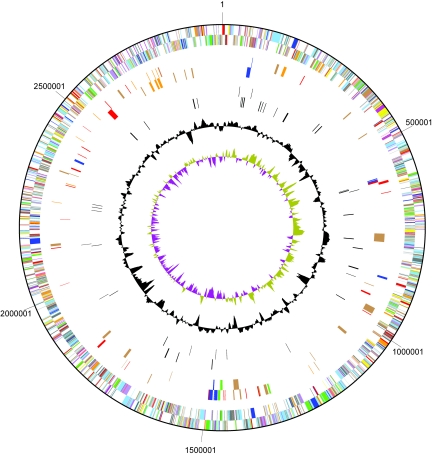
The genome of T. denitrificans strain ATCC 25259 consists of a single circular chromosome 2,909,809 bp in length with an average G+C content of 66.1% (Fig. 1 and Table 1). GC skew analysis does not reveal the origin of replication. Nucleotide position 1 of the chromosome is assigned to the predicted origin of replication, flanked on one side by the dnaA (Tbd0001), dnaN (Tbd0002), and gyrB (Tbd0003) genes and on the other by rpmH (Tbd2827) and rnpA (Tbd2826). Two copies of the 16S rRNA operon are located in regions of the genome separated by more than 1,380 kb. Few other repeated sequences were discovered in the genome, with the exception of a duplicated IS4-like insertion sequence. Thus, the numbers of repeated elements, insertion sequence elements, and transposons are low compared with those typically found in bacterial genomes sequenced to date.
FIG. 1.
Schematic circular diagram of the T. denitrificans ATCC 25259 genome. Outer circle, predicted coding regions on the forward strand, color-coded by role categories (dark gray, hypothetical proteins; light gray, conserved hypothetical and unknown function; brown, general function; red, DNA replication and repair; green, energy metabolism; blue, carbon and carbohydrate metabolism; cyan, lipid metabolism; magenta, transcription; yellow, translation; orange, amino acid metabolism; pink, metabolism of cofactors and vitamins; light red, purine and pyrimidine metabolism; lavender, signal transduction; sky blue, cellular processes; pale green, structural RNAs); second circle, predicted coding regions on the reverse strand, color coded as for the outer circle; third and fourth circles, coding regions (on forward and reverse strands) predicted to be involved in denitrification (blue), sulfur-compound oxidation (red), hydrogen oxidation (green), autotrophic carbon assimilation (orange), and metal ion transport-resistance (brown); fifth and sixth circles, coding regions found to have a CXXCH heme-binding motif and therefore potentially encoding c-type cytochromes; seventh circle, deviation from the average G+C; eighth circle, GC skew (olive, positive; purple, negative).
TABLE 1.
General features of the T. denitrificans strain ATCC 25259 genome
| Characteristic | Value |
|---|---|
| Chromosome size (bp) | 2,909,809 |
| G+C ratio (%) | 66.07 |
| Coding density (%) | 92.5 |
| No. of predicted protein coding genes | 2,827 |
| Average CDS length (bp) | 952 |
| No. of predicted proteins unique to T. denitrificans (%) | 89 (3.1) |
| No. of rRNA operons | 2 |
| No. of tRNA genes | 43 |
| No. of other small RNAs | 3 |
| No. of predicted proteins with putative function (%) | 2,183 (77.2) |
| No. of predicted proteins with unknown function (%) | 644 (22.8) |
| Protein categories (%) | |
| Energy production | 6.6 |
| Inorganic ion transport | 5.7 |
| Cell envelope biogenesis | 6.0 |
| BLASTP comparison against the KEGG completed microbial genomes database (no. of top KEGG hits) | |
| β-Proteobacteria | 1,624 |
| γ-Proteobacteria | 455 |
| α-Proteobacteria | 119 |
| δ-Proteobacteria | 92 |
| Archaea | 22 |
A total of 2,827 protein-encoding genes were identified, with an average length of 952 bp and accounting for 92.5% of thesequence. Among the predicted genes, 2,183 (77.2%) have been assigned a putative function, with the remainder designated as encoding a protein with unknown function, a conserved hypothetical protein, or a hypothetical protein. Of these, 89 predicted genes had no match in comparisons performed using BLAST versus the nr database with an E value cutoff of 10−6 or better. When distributed into biological role categories based on the COG database (82), the largest number of predicted proteins fell into the categories of energy production (6.6%), cell envelope biogenesis (6.0%), and inorganic ion transport (5.7%).
When searches using the KEGG database of complete genomes were performed, more than half (1,624) of the predicted proteins revealed top BLAST hits to one of the 12 β-proteobacteria available in this database, including Azoarcus sp. strain EbN1 (626 hits), the obligate chemolithoautotrophic bacterium Nitrosomonas europaea (346 hits), Chromobacterium violaceum (236 hits), Ralstonia solanacearum GMI1000 (189 hits), Burkholderia pseudomallei (118 hits), and Bordetella bronchiseptica (61 hits). Aside from β-proteobacteria, the organisms most frequently associated with the top BLAST hits were Methylococcus capsulatus (107 hits), Pseudomonas aeruginosa (75 hits), an environmentally versatile γ-proteobacterium, Pseudomonas putida (42 hits), and Geobacter sulfurreducens (42 hits), a δ-proteobacterium best known for its versatility in dissimilatory metal reduction.
Recent gene acquisition.
There is ample evidence of horizontal transfer in the T. denitrificans genome, inferred from local base composition to have been imported from an evolutionarily distant source. At least 13 regions, up to 25 kb in size, have been identified (on the basis of anomalies in observed G+C content and trinucleotide signature [40]) as likely recent integration events into the T. denitrificans genome that have not had time to drift toward the genome average (Table 2). Almost all of these putative regions of horizontal gene transfer carry phage integrases or other phage remnants, and a few of these regions are also flanked by tRNA genes, which are used as integration sites for many bacteriophages. Most of these regions are found to harbor many hypothetical or conserved hypothetical genes, and several carry restriction modification systems, which are known to be associated with mobile elements and indeed act as selfish mobile genetic elements themselves (51), whereas another region carries a cluster of genes encoding part of a type IV pilus. Although these likely recent insertions in the T. denitrificans chromosome are consistent with the concept of a fluid genome, it remains to be shown what role these regions may play in the metabolic or defense repertoire of T. denitrificans.
TABLE 2.
Regions with uncharacteristic G+C content and Karlin signaturesa
| Location in genome (nt)b | % G+C | CDS | BLAST hits |
|---|---|---|---|
| 394058-402433 | 58.87 | Tbd0363-Tbd0373 | Phage integrase, regulatory protein, hypothetical proteins |
| 979890-1004773 | 57.39 | Tbd0925-Tbd0943 | Methylase, transposase, regulatory protein, helicase (Snf2/Rad54 family), type III restriction-modification system (methylase, restriction enzyme), phage integrases, and several hypothetical proteins |
| 1264595-1271704 | 56.99 | Tbd1207-Tbd1215 | Phage integrase, conserved imported protein, and hypothetical proteins |
| 1395068-1400129 | 52.43 | Tbd1314-Tbd1318 | Phage integrase and hypothetical and conserved hypothetical proteins |
| 1424272-1431083 | 60.88 | Tbd1342-Tbd1354 | Mostly hypothetical proteins and a conserved imported protein |
| 1575347-1583813 | 57.71 | Tbd1487-Tbd1492 | Type II/III restriction-modification system (helicase, methylase), conserved hypothetical and hypothetical proteins |
| 1767440-1787406c | 60.46 | Tbd1679-Tbd1695 | Phage integrase, conjugal transfer region (TraWBAY), a number of hypothetical proteins, and imported AAA superfamily ATPase and conserved hypothetical protein |
| 1955614-1961331 | 61.96 | Tbd1861-Tbd1865 | Type IV pilus proteins (PilEWXV and FimT) |
| 2052544-2061073 | 59.64 | Tbd1958-Tbd1969 | Transposases, conserved imported hypothetical proteins, and hypothetical proteins |
| 2089032-2093604c | 57.82 | Tbd2000-Tbd2005 | Phage integrase, plasmid recombination protein, and several hypothetical proteins |
| 2160137-2166983c | 62.36 | Tbd2066-Tbd2073 | Phage integrase, phage replication protein, DNA helicase, and several hypothetical proteins |
| 2750091-2764591 | 57.46 | Tbd2675-Tbd2685 | Phage integrase, prophage regulatory protein, type I restriction-modification system (HsdMS, HsdR) |
| 2900270-2905890 | 61.02 | Tbd2817-Tbd2823 | Phage integrase, phage primase and phage regulatory protein |
All regions are also supported by Karlin signature difference.
nt, nucleotide.
Flanked by a tRNA gene.
Taxonomy of T. denitrificans strain ATCC 25259.
Following the reclassification of numerous α-, β-, and γ-proteobacterial species previously classified as Thiobacillus spp., only three β-proteobacteria are now securely placed in that genus: T. thioparus, T. denitrificans, and T. aquaesulis (47, 49). Of these, the last is a moderate thermophile and only T. denitrificans is capable of strictly chemolithotrophic anaerobic growth with denitrification using inorganic sulfur-compound oxidation as the sole source of energy (46, 49). Apart from this feature, T. denitrificans and T. thioparus are physiologically very similar mesophilic obligate chemolithoautotrophs. The G+C content of different strains ranges between 62 and 67% for T. thioparus and 63 and 68% for T. denitrificans (46, 79, 88). DNA hybridization distinguished the two species, which show only 22 to 29% cross-hybridization (46). Comparison of 16S rRNA gene sequences in the GenBank nr database shows that identities between different strains of T. thioparus range between 95.2 and 99.6% and that the type strains of T. thioparus (ATCC 8158) and T. denitrificans (NCIMB 9548) show 96.3% identity to each other (D. P. Kelly, unpublished data). Consequently, the ability to denitrify is a key physiological criterion in distinguishing between the species, given the similarity of their 16S rRNA gene sequences. T. denitrificans strain ATCC 25259 was originally isolated from Texas soil (84; ATCC catalog) and has been shown to have physiological properties characteristic of this species. T. denitrificans ATCC 25259 shows 97.6% identity (with respect to 16S rRNA genes) to the type strain T. denitrificans NCIMB 9548.
Sulfur-compound oxidation.
A number of enzymes involved in inorganic sulfur-compound oxidation and energy conservation have previously been studied in T. denitrificans and the closely related β-proteobacterium T. thioparus; some of these enzymes have been purified and characterized biochemically. The T. denitrificans enzymes include APS (adenylylsulfate) reductase, an AMP-independent sulfite oxidase, a siroheme sulfite reductase, and APS:phosphate adenylyltransferase (or APAT) (2, 12, 13, 29, 69-71). Biochemical studies, as well as the high growth yields and the entry into the electron transport chain of electrons from sulfur-compound oxidation at the level of quinone-cytochrome b, indicate highly efficient energy-conserving mechanisms of sulfur-compound oxidation in T. denitrificans (42-45). The mechanisms of inorganic-sulfur oxidation by T. denitrificans, which oxidizes polythionates as well as thiosulfate, sulfide, and, for some strains, thiocyanate, appear more similar to those of γ-proteobacteria such as Halothiobacillus spp. than to the well-defined thiosulfate-oxidizing, multienzyme system of α-proteobacterial chemolithotrophs (45, 50, 68).
As would be expected from previous biochemical work with T. denitrificans, its genome contains a diverse complement of genes encoding enzymes that catalyze inorganic sulfur-compound oxidation and energy conservation (by both substrate-level and electron transport-linked phosphorylation). The importance of sulfur-compound oxidation to T. denitrificans is underscored by the occurrence of multiple oxidation pathways for certain sulfur compounds and multiple copies of a number of genes associated with sulfur-compound oxidation (see overview in Fig. 2). The unusual ability of this bacterium to oxidize inorganic sulfur compounds under both aerobic and denitrifying conditions raises the question of whether different sulfur-oxidizing enzymes are involved under aerobic versus anaerobic conditions. Although such questions cannot be resolved by genome analysis alone, they can be addressed by transcriptional studies using whole-genome oligonucleotide microarrays, which are now enabled by the availability of the T. denitrificans genome. Included among the genes that are likely to be critical to sulfur-compound oxidation by T. denitrificans are sox (sulfur-oxidation) genes, dsr (dissimilatory sulfite reductase) genes, and genes associated with the AMP-dependent oxidation of sulfite to sulfate (Fig. 2). These genes serve as the focus of this section.
FIG. 2.
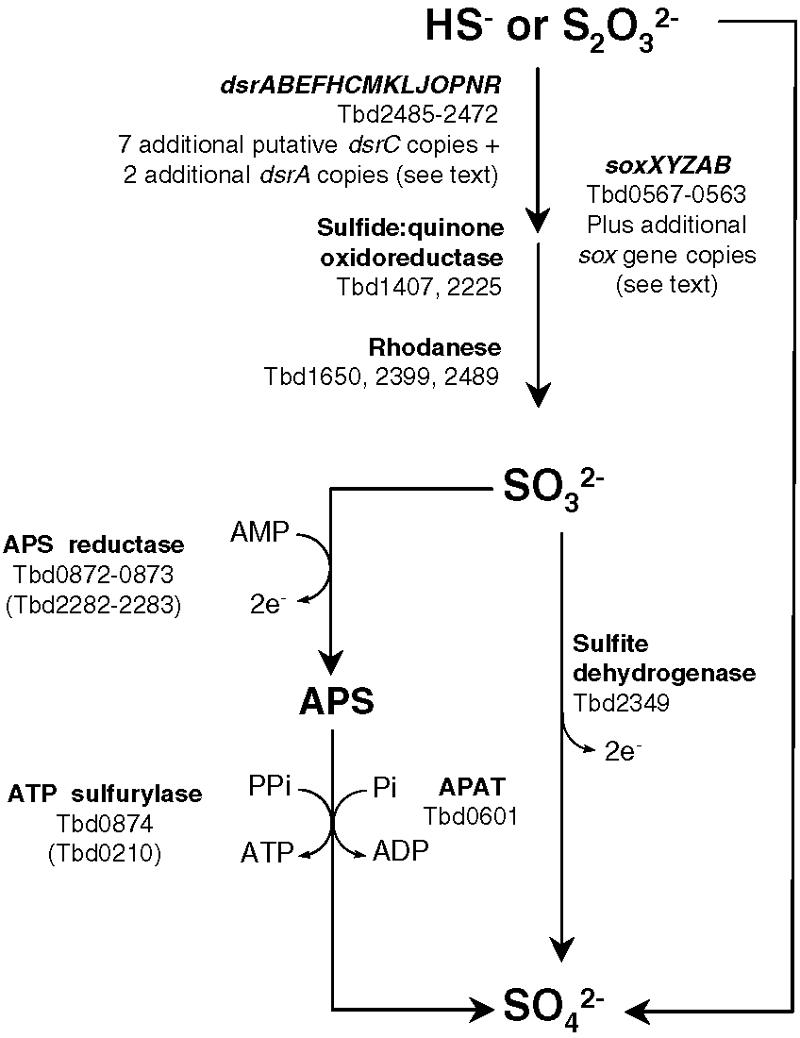
Schematic overview of key genes/enzymes putatively associated with sulfur-compound oxidation in T. denitrificans. Genes in parenthesis have been shown to be lesser expressed paralogs (this study). The biochemical roles of a number of gene products represented in this figure have not been experimentally demonstrated in T. denitrificans and are uncertain. Sulfide:quinone oxidoreductase is not proposed to catalyze the direct oxidation of sulfide to sulfite but rather may participate in an indirect pathway (20). The arrow between thiosulfate and sulfate (right side) represents the possibility that SoxB catalyzes a sulfate thiohydrolase reaction (28) in T. denitrificans. APAT, APS:phosphate adenylyltransferase.
Little molecular genetic work on the sulfur-oxidation (sox) systems of T. denitrificans has been conducted to date. The model for genes encoding sulfur-oxidation enzymes has been derived primarily from three α-proteobacteria: Paracoccus pantotrophus, Starkeya novella, and Pseudaminobacter salicylatoxidans. In T. denitrificans, genes showing various levels of sequence identity to sox genes of these α-proteobacteria have been detected, but gene clusters of the length found in facultatively chemolithotrophic, aerobic, thiosulfate-oxidizing bacteria do not occur. Thus, extensive sox clusters have been observed in P. pantotrophus, S. novella, and P. salicylatoxidans: soxRSVWXYZABCDEFGH, soxFDCBZYAXWV, and soxGTRSVWXYZABCD, respectively (GenBank X79242, AF139113, and AJ404005) (27, 28, 91). In contrast, the largest cluster in T. denitrificans consists of soxXYZAB (Tbd0567-Tbd0563), using the P. pantotrophus naming scheme. Comparison of deduced SoxX, -Y, -Z, -A, and -B sequences of T. denitrificans to those of P. pantotrophus, Chlorobium tepidum, P. salicylatoxidans, and S. novella revealed sequence identity in the range of 28 to 55%. This indicated that while the genes encoding these Sox functions had been putatively identified, they differed significantly from those of the reference organisms. Interestingly, the translated soxXYZA genes showed higher identities to those of the green sulfur bacterium C. tepidum (42 to 55%) than to those of Paracoccus, Starkeya, and Pseudaminobacter spp. (28 to 38%). Additional copies of soxXA were identified outside of the soxXYZAB cluster in T. denitrificans (Tbd0917-Tbd0918). A noteworthy difference between SoxA encoded in the T. denitrificans genome and those of P. pantotrophus and P. salicylatoxidans is that the latter are diheme cytochromes (28) whereas SoxA copies in T. denitrificans (Tbd0564 and, putatively, Tbd0918) are monoheme cytochromes.
For soxB, the T. denitrificans sequence can be compared to that of another β-proteobacterium as well as to those of α-proteobacteria, because the sequence from T. thioparus is available (GenBank AJ294326; partial sequence, 344 translated amino acids) (59). In fact, of the sox genes, only soxB has thus far been the subject of intercomparison across the α-, β-, and γ-proteobacterial groups (59); this comparison revealed a distant phylogenetic relationship of the soxB sequence of T. thioparus to those of α- and γ-proteobacteria. Consistent with phylogenetic relationships based on 16S rRNA, the predicted SoxB sequence of T. denitrificans (Tbd0563) is much more similar to that of T. thioparus (88% identity) than to those of P. pantotrophus, S. novella, and P. salicylatoxidans (48 to 50% identity). The encoded SoxB sequences for T. thioparus and P. pantotrophus (GenBank CAC82470 and CAA55824) showed 50% identity to each other.
Other sox genes found in the contiguous cluster of P. pantotrophus were remote from each other in the T. denitrificans genome. These included tentatively identified copies of soxH (Tbd1041, Tbd1103), soxE (Tbd2027, Tbd2034), soxF (Tbd2035), and soxW (Tbd2117). Tbd2349 corresponded to the soxC (sulfite dehydrogenase) of P. salicylatoxidans and P. pantotrophus and the sorA of S. novella; thus, the encoded protein may be a sulfite dehydrogenase, catalyzing AMP-independent oxidation of sulfite to sulfate. BLAST probing of the genome with the nucleotide and deduced amino acid sequences of the soxD of P. pantotrophus and S. novella produced no hits, so a gene corresponding to the α-proteobacterial soxD appeared to be absent. As the proteins encoded by soxCD in Paracoccus spp. are believed to catalyze the oxidation of sulfane-sulfur to the oxidation level of sulfite (28), an alternative system is likely present in T. denitrificans.
Coding functions have been ascribed to many of the α-proteobacterial sox genes discussed above (27, 28, 63-65, 67, 68). soxX and soxA encode SoxXA, a heterodimeric c-type cytochrome. soxY and soxZ encode SoxYZ, the “thiosulfate-binding” enzyme A of P. pantotrophus. soxC and soxD encode the molybdoprotein SoxC and diheme c-type cytochrome SoxD of an α2β2-heterodimeric “sulfur dehydrogenase”; SoxCD has also been shown to function as a sulfite dehydrogenase. soxB encodes SoxB, a sulfur-thiol esterase, identified in P. pantotrophus as enzyme B, a protein with a dinuclear manganese center that catalyzes thiosulfate cleavage and sulfate production. soxE encodes a c-type cytochrome. soxF encodes a sulfide dehydrogenase/flavocytochrome-c oxidoreductase protein. soxRS are reported to have a regulatory function. soxGH are currently of uncertain function. Finally, the SoxFGH proteins (which are all periplasmic) were reportedly not required for lithotrophic growth of P. pantotrophus on thiosulfate, although they were induced by thiosulfate. The proteins SoxXA, SoxYZ, SoxB, and SoxCD can be reconstituted into a system catalyzing thiosulfate-, sulfite-, sulfur-, and hydrogen sulfide-dependent cytochrome c reduction in P. pantotrophus and P. versutus, although this multienzyme system may not operate in the facultative chemolithotroph S. novella. The soxB gene has also been identified in T. thioparus, some γ-proteobacteria (including Halothiobacillus and Thiomicrospira spp.), and some phototrophic sulfur bacteria (59).
In T. denitrificans, central roles for the putative SoxXA, SoxB, and SoxYZ gene products are suggested by the clustering of the genes encoding them, but the low polypeptide sequence identity of these to the corresponding sequences in P. pantotrophus could mean that their biochemical functions might differ considerably from those of P. pantotrophus. The thiosulfate-oxidizing multienzyme system of P. pantotrophus (and of other α-proteobacteria) is located in the periplasm, but there is considerable evidence from studies of T. denitrificans (and other obligately chemolithotrophic sulfur oxidizers) that at least some reactions of thiosulfate, sulfite, and sulfide oxidation require membrane-associated processes (43). Deduction of the functions of the putative T. denitrificans sox genes solely by reference to the roles of the sox complexes in Paracoccus species must clearly be done with caution, because even if they were acquired primordially by lateral gene transfer (59), they could encode significantly modified enzyme functions in extant α- and β-proteobacteria.
In a few bacteria that oxidize inorganic sulfur compounds, namely T. denitrificans, Allochromatium vinosum, and C. tepidum, a siroheme-containing sulfite reductase has been proposed to catalyze the oxidation of certain inorganic sulfur species (e.g., hydrogen sulfide or sulfane-sulfur derived from thiosulfate) to sulfite (20, 71, 86). Thus, dissimilatory sulfite reductase, which is encoded by dsr genes and named for its catalytic role in sulfate-reducing bacteria and archaea, is apparently used in the reverse direction for dissimilatory oxidation of sulfur compounds. Siroheme-containing sulfite reductase (with an α2β2 structure encoded by dsrAB) was previously purified from T. denitrificans strain DSM 807 (86). A gene cluster (dsrABEFHCMKLJOPNR; Tbd2485-Tbd2472) occurs in T. denitrificans ATCC 25259 that is very similar in terms of gene sequence and organization to a dsr cluster in A. vinosum that was studied by Dahl et al. (17). A notable difference in gene organization is that dsrS in T. denitrificans (Tbd2558) is not part of the dsr cluster, as it is in A. vinosum, and that a cysG (siroheme synthase)-like gene is located adjacent to dsrR in T. denitrificans (Tbd2471). For the translations of most of these dsr genes, the degree of identity with the corresponding A. vinosum sequences is >55%. This is the only major cluster of dsr genes in the finished T. denitrificans sequence, which contrasts with the claim by Dahl et al. (17) that there are two dsr gene clusters (on the basis of their examination of shotgun clone sequences in GenBank).
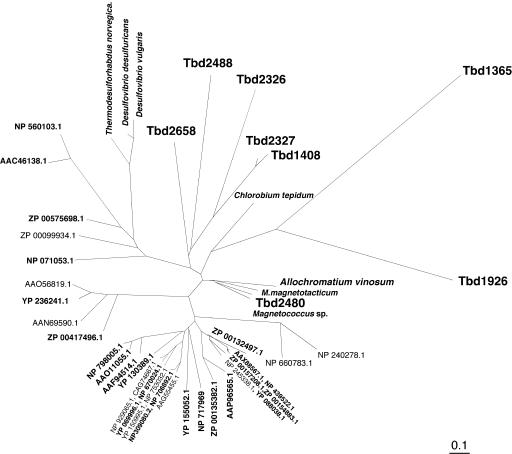
A noteworthy finding with regard to dsr genes in T. denitrificans was that two genes, dsrC and dsrA, are replicated multiple times in the genome, typically with no more than one or two adjacent genes that are putatively associated with sulfur-compound oxidation. Eight putative copies of dsrC were identified (Fig. 3): Tbd2480 (located in the large dsr gene cluster), Tbd2488 (located next to Tbd2489, which encodes a rhodanese-related sulfurtransferase), Tbd2658, Tbd2326 and Tbd2327 (which are adjacent to each other), Tbd1365 (located near a dsrA copy, Tbd1369), Tbd1408 (located next to Tbd1407, which encodes sulfide:quinone oxidoreductase), and Tbd1926. The degree of sequence identity among the corresponding predicted DsrC copies ranges from 26 to 88%. DsrC is a soluble, cytoplasmic protein whose function is not currently known (62). A cysteine residue at the C terminus of DsrC that is highly conserved in a range of bacteria including A. vinosum and various sulfate-reducing bacteria (62) is present in only two of the eight copies encoded in the T. denitrificans genome, including the copy located in the dsr cluster (Tbd2480) and Tbd2658. In fact, overall, the translated DsrC from Tbd2480 was more similar to DsrC in A. vinosum than to the other seven copies in T. denitrificans (Fig. 3). Inasmuch as Tbd2480 is the dsrC copy in T. denitrificans most likely to code for functional DsrC, it is noteworthy that, like dsrC in A. vinosum (17, 62) and unlike all other dsrC copies in T. denitrificans, Tbd2480 appears to be constitutively expressed [as indicated by RT-qPCR results for T. denitrificans carrying out thiosulfate or Fe(II) oxidation under denitrifying conditions; Table 3 ]. As shown in Table 3, Tbd2480 is relatively highly expressed when oxidizing either thiosulfate or Fe(II), whereas none of the other seven putative dsrC copies is highly expressed under both conditions.
FIG. 3.
Phylogenetic relationships among the eight putative DsrC proteins encoded in T. denitrificans ATCC 25259 and the top BLASTP matches from the GenBank nr database for Tbd2480. Of the proteins represented in this figure that are not from T. denitrificans, more than 70% are known or predicted to be DsrC or more broadly related to sulfite reductases (indicated in boldface type). For limbs that show species names rather than GenBank accession numbers, the corresponding accession numbers are as follows: A. vinosum (AAC35399.1), M. magnetotacticum (ZP 00052645.1), Magnetococcus sp. (ZP 00287929.1), C. tepidum (NP 663123.1), T. norvegica (CAC36215.1), D. desulfuricans (ZP 00130056.2), and D. vulgaris (YP 011988).
TABLE 3.
Differential transcription of selected genes in T. denitrificans that occur in multiple copies
| Genes compared | Thiosulfate-induced transcription (fold difference)a | FeCO3-induced transcription (fold difference)b |
|---|---|---|
| dsrC (putative) | ||
| Tbd2480/Tbd2480c | 1d | 0.38 |
| Tbd2488/Tbd2480c | 1.4 | 0.025 |
| Tbd2658/Tbd2480c | 0.069 | 0.006 |
| Tbd2326/Tbd2480c | 0.015 | 0.002 |
| Tbd2327/Tbd2480c | 0.009 | 0.002 |
| Tbd1365/Tbd2480c | 0.039 | 0.009 |
| Tbd1408/Tbd2480c | 0.82 | 0.031 |
| Tbd1926/Tbd2480c | 0.001 | 0.002 |
| APS reductase | ||
| α subunit | ||
| Tbd0872/Tbd2282 | 80 | NAe |
| β subunit | ||
| Tbd0873/Tbd2283 | 740 | NA |
| ATP sulfurylase | ||
| Tbd0874/Tbd0210 | 5 | NA |
| Nitric oxide reductase | ||
| norC | ||
| Tbd0562/Tbd0822 | 63 | NA |
| norB | ||
| Tbd0561/Tbd0823 | 61 | NA |
Cells harvested for RNA while carrying out thiosulfate oxidation and denitrification (see Materials and Methods).
Cells harvested for RNA while carrying out oxidation of Fe(II) (in FeCO3) and denitrification (see Materials and Methods).
Comparison made to the number of transcripts for Tbd2480 under thiosulfate-induced conditions. The transcript copy number for Tbd2480 under these conditions was relatively high (in the copy number range of Tbd0562 and Tbd0561, which encode subunits of nitric oxide reductase, a key enzyme involved in denitrification).
By definition.
NA, not analyzed.
Three putative copies of dsrA were identified in the T. denitrificans genome: Tbd2485 (located in the large dsr gene cluster), Tbd1309, and Tbd1369 (located near a dsrC copy). The degree of sequence identity among the corresponding predicted DsrA copies was high (from 78 to 83%). As dsrA encodes the α subunits of the α2β2-structured siroheme sulfite reductase, it is curious that two copies of dsrA in the T. denitrificans genome are not located near copies of dsrB, which encodes the β subunits; indeed, only one dsrB copy was identified in the genome (Tbd2484). Thus, the nature of dsr gene duplication in T. denitrificans differs from that observed in C. tepidum, whose genome includes two copies of the dsrCABL cluster (17, 20).
T. denitrificans ATCC 25259 contains genes encoding all the enzymes necessary to catalyze the AMP-dependent oxidation of sulfite to sulfate (Fig. 2). All but one of these enzymes is used in the reverse direction by sulfate-reducing bacteria for the activation of sulfate to APS and the subsequent AMP-yielding reduction of APS to sulfite. APS reductase, an αβ-heterodimeric iron-sulfur flavoenzyme, is encoded by Tbd0872-Tbd0873 and catalyzes the AMP-dependent oxidation of sulfite to APS. Another pair of genes, Tbd2282-Tbd2283, also putatively encodes APS reductase, but RT-qPCR results suggest that the Tbd0872-Tbd0873 genes are much more highly expressed during thiosulfate oxidation (Table 3). ATP sulfurylase, which catalyzes an ATP-yielding substrate level phosphorylation that converts APS to sulfate, is encoded by Tbd0874 and also by Tbd0210 (with the former being more highly expressed; Table 3). APAT, which is encoded by Tbd0601 and catalyzes an alternative substrate-level phosphorylation that converts APS to sulfate (yielding ADP rather than ATP), is not reversible (although it was formerly misnamed ADP sulfurylase) (13).
The T. denitrificans genome includes other genes that are likelyto play a role in sulfur-compound oxidation, including sulfide:quinone oxidoreductase (Tbd1407, Tbd2225) and rhodanese (thiosulfate-sulfurtransferase; Tbd1650, Tbd2399, Tbd2489). Genes for dimethyl sulfoxide (DMSO) reductase (Tbd0570-Tbd0572) and tetrathionate reductase (Tbd1739-Tbd1741) are also present in the T. denitrificans genome, but these may have a role as anaerobic electron acceptors rather than in sulfur-compound oxidation. No genes for the α- and β-subunits of DMSO dehydrogenase were detected. The type strain of T. denitrificans (NCIMB 9548) and some strains of T. thioparus can oxidize and grow on thiocyanate as a sole electron donor (47), and three genes of T. thioparus encode the thiocyanate hydrolase enzyme that initiates thiocyanate degradation (GenBank AB007989; scnBAC). In contrast, T. denitrificans strain ATCC 25259 does not oxidize thiocyanate, and its genome lacks the genes encoding thiocyanate hydrolase.
Hydrogen metabolism.
Analysis of the genome of T. denitrificans ATCC 25259 has revealed the presence of genes encoding two [NiFe]hydrogenases. Hydrogenases are metalloenzymes that catalyze the reversible oxidation of H2 to protons and are vital components of the energy metabolism of many microbes. Notably, hydrogenases have not previously been reported in T. denitrificans, and the sequenced strain does not appear to be able to grow on hydrogen as a sole electron donor under denitrifying conditions (H. Beller, unpublished data); however, hydrogen oxidation appears to be required for nitrate-dependent U(IV) oxidation by T. denitrificans (5).
One of the hydrogenases encoded in the T. denitrificans genome is putatively a cytoplasmic, heterotetrameric, group 3b [NiFe]hydrogenase (following the classification system described by Vignais et al. [87]). The four-gene cluster (Tbd1260-Tbd1263) does not appear to be near any accessory or maturation genes encoding proteins necessary for assembly of the functional holoenzyme. Although group 3b hydrogenases have primarily been found in hyperthermophilic archaea (87), BLASTP analysis revealed that a group of similar predicted proteins (43 to 53% identity for the four subunits) occur in Azotobacter vinelandii, a mesophilic, δ-proteobacterium; gene organization in T. denitrificans and A. vinelandii was also similar. Group 3b hydrogenases in the hyperthermophile Pyrococcus furiosus are among the better characterized and are thought to play a role in disposing of excess reductant generated during fermentation (whereby NADPH can serve as the physiological electron donor for H2 evolution) (87). The role of a group 3b hydrogenase in T. denitrificans, if indeed this hydrogenase is expressed in functional form, is not currently known.
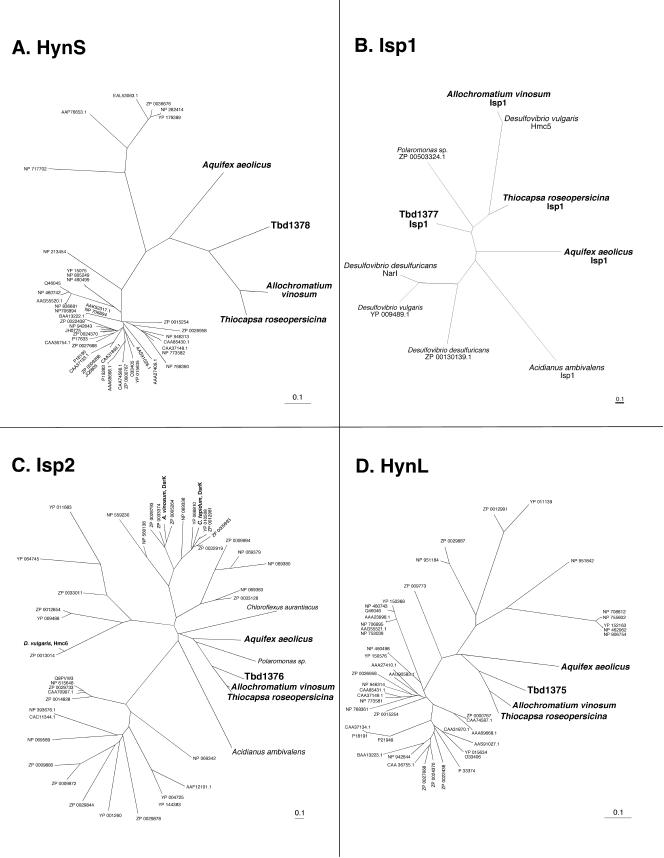
The other hydrogenase encoded in the T. denitrificans genome is putatively a periplasmic, group 1 [NiFe]hydrogenase (following the classification system described by Vignais et al. [87]). Although the sequences of the small (HynS) and large (HynL) subunits of this hydrogenase are similar to those in many bacteria and archaea, they occur in an unusual gene cluster (hynS-isp1-isp2-hynL; Tbd1378-Tbd1375) that has only been observed in four other microbes to date (none of which is a mesophilic, chemolithoautotrophic bacterium like T. denitrificans): the phototrophic sulfur bacteria Thiocapsa roseopersicina (66) and A. vinosum (16), the hyperthermophilic bacterium Aquifex aeolicus (GenBank NP 213658.1-213655.1), and the archaeon Acidianus ambivalens (53). Typically, hynS and hynL are adjacent to one another, but they are separated by two intervening genes in these species. Not only is the organization of the hynS-isp1-isp2-hynL gene cluster in T. denitrificans identical to that of the other four species, the sequence similarity of the four predicted proteins is also relatively high among these species (Fig. 4). The degree of sequence identity for HynS, Isp1, Isp2, and HynL in T. denitrificans compared to T. roseopersicina, A. aeolicus, and A. vinosum ranges from 26 to 68%. A phylogenetic tree of Isp1 in T. denitrificans ATCC 25259 and the small complement of related proteins from the GenBank nr database (BLASTP E values < 10−5) reveals similarity not only to Isp1 copies of other species but also to NarI (the γ subunit of cytoplasmic nitrate reductase) and Hmc5 (part of the high-molecular-weight, transmembrane, electron transport protein complex found in Desulfovibrio vulgaris) (Fig. 4B). Alignment of the predicted amino acid sequence of Isp1 from T. denitrificans with other sequences represented in Fig. 4B shows that four histidine residues (located in two of the five predicted transmembrane helices in Isp1, NarI, and Hmc5) are highly conserved. Similar observations have been made for other Isp1 sequences (16, 53, 66), and Berks et al. (8) elucidated how the conserved histidines (and the two b-hemes that they putatively bind, one in each half of the membrane bilayer) play a role in mediating transport of electrons across the cytoplasmic membrane as part of an electron-carrying arm of a redox loop in the integral membrane proteins NarI and HyaC (a b-type cytochrome associated with periplasmic [NiFe]hydrogenases). On the basis of these and other observations, it is likely that Isp1 serves two functions: (i) to anchor HynSL on the periplasmic side of the cytoplasmic membrane, where H2 can be converted to 2H+ and 2 e− by HynSL, and (ii) to mediate transmembrane electron transfer from HynSL to the quinone pool of electron transport chains and thereby participate in a chemiosmotic mechanism of energy conservation. Notably, in serving these functions, Isp1 may be substituting for the b-type cytochrome HupC/HoxZ/HupZ/HyaC that is typically found in group 1 [NiFe]hydrogenases but which is apparently absent in the T. denitrificans genome. A phylogenetic tree of Isp2 in T. denitrificans (Fig. 4C) reveals a similarity not only to Isp2 sequences of other species but also to sequences of various iron-sulfur-containing proteins such as heterodisulfide reductases, DsrK (from other sulfur-compound oxidizers A. vinosum and C. tepidum), and Hmc6 from D. vulgaris; such similarities have been noted previously for other Isp2 copies (16, 66).
FIG.4.
Phylogenetic relationships among predicted amino acid sequences for HynS (A), Isp1 (B), Isp2 (C), and HynL (D) in T. denitrificans and the best BLASTP matches from the GenBank nr database. For limbs that show species names rather than GenBank accession numbers, the corresponding accession numbers are as follows: (HynS) A. vinosum (AAU93828.1), T. roseopersicina (AAC38281.1), A. aeolicus (NP 213658.1); (Isp1) A. vinosum (AAU93829.2), T. roseopersicina (AAC38283.1), A. aeolicus (NP 213657.1), D. vulgaris Hmc5 (YP 009755), A. ambivalens (CAC86885.1), D. desulfuricans NarI (ZP 00128546.1); (Isp2) A. vinosum (AAY89333.1), T. roseopersicina (AAC38284.1), A. aeolicus (NP213656.1), A. ambivalens (CAC86886.1), Polaromonas sp. (ZP 00503323.1), C. aurantiacus (ZP 00356812), A. vinosum DsrK (AAC35401.2), C. tepidum DsrK (NP 663117.1), D. vulgaris Hmc6 (YP 009754.1); (HynL) A. vinosum (AAY89334.1), T. roseopersicina (AAC38282.1), A. aeolicus (NP213655.1).
The two adjacent gene clusters (Tbd1380-Tbd1374 and Tbd1381-Tbd1386 [on opposite DNA strands]) that code for the group 1 [NiFe]hydrogenase of T. denitrificans include a number of genes putatively involved in biosynthesis and maturation of the hydrogenase. This clustering of maturation genes along with the hynS-isp1-isp2-hynL cluster in T. denitrificans distinguishes its gene organization from that of T. roseopersicina and A. aeolicus, which do not have accessory genes in the immediate vicinity of the hynS-isp1-isp2-hynL cluster. In the Tbd1380-Tbd1374 gene cluster, Tbd1374 codes for a homolog of UreJ and is probably involved in Ni transport; hynD (Tbd1380) codes for a putative maturation protease. Most or all of the genes in the hypCABDFE gene cluster (Tbd1381-Tbd1386) encode proteins that are putatively involved with insertion of Ni, Fe, CO, and CN in the active site of the hydrogenase (87).
Aerobic respiration.
The genome of T. denitrificans, a facultative anaerobe, encodes all the necessary machinery for aerobic respiration, including NADH:ubiquinone oxidoreductase (complex I; Tbd1142-Tbd1155), succinate dehydrogenase (complex II; Tbd1182-Tbd1185), and cytochrome bc1-type ubiquinol oxidoreductase (complex III; Tbd1831-Tbd1833) (9). These components can provide the reducing equivalents needed for terminal respiration with nitrate or oxygen, the latter in conjunction with one of three terminal cytochrome c oxidases encoded by two gene clusters on the T. denitrificans genome. The first cluster encodes a cytochrome aa3-type cytochrome c oxidase (Tbd0325, Tbd0326, Tbd0328, Tbd0330) and a cbb3-type cytochrome c oxidase (Tbd0338-Tbd0341), whereas the second cluster (Tbd0640-Tbd0643) encodes a second cbb3-type cytochrome c oxidase. The presence of both aa3- and cbb3-type oxidases, in addition to the denitrification machinery, allows T. denitrificans to survive under a wide range of redox conditions; presumably, the aa3 oxidase operates under high oxygen tension, cbb3 oxidases operate under microaerophilic conditions, and the denitrification complex operates under anaerobic conditions (58, 61).
Denitrification.
T. denitrificans has all necessary genes encoding the four essential enzymes that catalyze denitrification (reduction of nitrate to nitrogen gas): nitrate reductase, nitrite reductase, nitric oxide reductase, and nitrous oxide reductase (60, 94). The membrane-bound, dissimilatory nitrate reductase is encoded in a narKK2GHJI cluster (Tbd1401-Tbd1406), whereas the NarXL two-component regulatory system is encoded on the reverse DNA strand (Tbd1400-Tbd1399). This gene organization is similar to that described for Pseudomonas aeruginosa PAO1 and P. fluorescens C7R12 (60). A nir operon including the CDS for cytochrome cd1-nitrite reductase (nirS; Tbd0077) is present in T. denitrificans, and this protein was in fact purified from a different strain of T. denitrificans (strain DSM 807) (36). T. denitrificans ATCC 25259 contains two nor gene clusters that include the norCB structural genes encoding nitric oxide reductase, a membrane-anchored protein complex. RT-qPCR analyses carried out with thiosulfate-oxidizing, denitrifying cells (Table 3) indicated that the norCB genes Tbd0562-Tbd0561 had >50-fold-higher expression levels than norCB genes Tbd0822-Tbd0823, strongly indicating that the former genes are of greater functional importance. The predicted NorC amino acid sequence for the less-expressed copy (Tbd0822) contains >120 more residues at the C terminus than the highly expressed copy (Tbd0562) or a similar copy in Azoarcus sp. strain EbN1 (GenBank YP 157125); these additional amino acid residues for Tbd0822 include a second CXXCH heme-binding motif. The enzyme catalyzing the final step of denitrification, nitrous oxide reductase, is associated with the structural gene nosZ (Tbd1389) and, like NirS, was purified from T. denitrificans strain DSM 807 (36). Organization of nos genes in T. denitrificans is similar, but not identical, to that described for Ralstonia metallidurans CH34 (60). As has been observed for some other β-proteobacteria (60), nosR (Tbd1390) is located downstream from nosZ in T. denitrificans.
c-type cytochromes.
T. denitrificans strain ATCC 25259 has a higher number of c-type cytochromes than most bacteria with finished genomes, as suggested by analysis of the characteristic CXXCH heme-binding motif throughout the genome. Fifty-six genes, or approximately 2% of the total CDS, contained at least one CXXCH motif (some encoded proteins, such as DnaJ and ribosomal protein L31, are not truly c-type cytochromes; Table 4). In the context of bacteria reported to have relatively high numbers of c-type cytochromes, the number in T. denitrificans is less than that cited for Geobacter sulfurreducens (111, or 3.2% of the total CDS) (54) but more than for Shewanella oneidensis (39, or 0.8% of the total CDS) (34) and Pseudomonas aeruginosa (35, or 0.6% of the total CDS) (77). Overall, c-type cytochromes in T. denitrificans range in predicted molecular mass from 9.5 to 138 kDa and in number of heme groups from one to three (with only one triheme c-type cytochrome and the vast majority as monoheme proteins) (Table 4). Although the functions of some c-type cytochromes in T. denitrificans can be confidently predicted, such as NirS and NorC, some are of less certain or unknown function (Table 4). The ability of T. denitrificans to catalyze anaerobic, nitrate-dependent oxidation of metals with high reduction potentials, such as Fe(II) or U(IV) (5, 78), may be mediated by c-type cytochromes, as few other electron carriers in a bacterial cell would have sufficiently high reduction potentials to accept electrons from compounds such as uraninite, or UO2 (the UO22+/UO2 couple has an E0′ value of +0.26V) (5).
TABLE 4.
CDS potentially encoding c-type cytochromes in the T. denitrificans genomea
| CDS | Annotationb | Molecular mass (Da)c | No. of hemes |
|---|---|---|---|
| Tbd0055 | Cytochrome c family protein | 20,116 | 1 |
| Tbd0064 | Cytochrome c-553 | 23,727 | 2 |
| Tbd0070 | Probable nirN | 63,589 | 1 |
| Tbd0076 | Probable nirC | 10,547 | 1 |
| Tbd0077 | nirS (cytochrome cd1) | 62,992 | 1 |
| Tbd0094 | Hypothetical protein | 17,109 | 1 |
| Tbd0128 | Cytochrome c | 38,000 | 2 |
| Tbd0129 | Cytochrome c | 21,784 | 2 |
| Tbd0137 | Diheme cytochrome c | 19,427 | 2 |
| Tbd0138 | Cytochrome c-type protein | 14,395 | 1 |
| Tbd0146 | Probable cytochrome c5 | 26,734 | 2 |
| Tbd0187 | Cytochrome c | 21,387 | 2 |
| Tbd0219 | Flavin adenine dinucleotide-flavin mononucleotide-containing dehydrogenase | 138,251 | 1 |
| Tbd0325 | aa3-type cytochrome c oxidase, subunit II | 41,279 | 1 |
| Tbd0339 | cbb3-type cytochrome c oxidase, subunit II | 28,037 | 1 |
| Tbd0341 | cbb3-type cytochrome c oxidase, subunit III | 33,124 | 2 |
| Tbd0436 | Excinuclease ATPase subunit | 103,145 | 1 |
| Tbd0562 | norC | 15,845 | 1 |
| Tbd0564 | soxA | 30,960 | 1 |
| Tbd0567 | soxX | 12,712 | 1 |
| Tbd0571 | DMSO reductase chain B | 25,909 | 1 |
| Tbd0640 | cbb3-type cytochrome c oxidase, subunit III | 33,873 | 2 |
| Tbd0642 | cbb3-type cytochrome c oxidase, subunit II | 22,314 | 1 |
| Tbd0723 | Possible high-affinity Fe2+/Pb2+ permease | 69,511 | 1 |
| Tbd0752 | Mannose-sensitive hemagglutinin pilin biogenesis ATPase protein MshE | 62,481 | 1 |
| Tbd0820 | Cytochrome c (in or near nonfunctional nor cluster) | 57,261 | 2 |
| Tbd0822 | norC-related (potentially not functional) | 30,200 | 2 |
| Tbd0840 | Probable cytochrome c5 | 16,718 | 1 |
| Tbd0917 | soxX | 13,246 | 1 |
| Tbd0918 | soxA | 30,875 | 1 |
| Tbd1169 | Ferredoxin, 2Fe-2S | 12,259 | 1 |
| Tbd1357 | Unknown | 16,635 | 1 |
| Tbd1398 | Putative cytochrome c-type protein | 15,842 | 1 |
| Tbd1404 | narH | 59,191 | 1 |
| Tbd1484 | Cytochrome c | 9,544 | 1 |
| Tbd1520 | Putative Fe-S protein | 48,430 | 1 |
| Tbd1542 | ATPase involved in DNA replication | 61,297 | 1 |
| Tbd1564 | Probable ribonuclease E | 95,354 | 1 |
| Tbd1585 | Putative pyruvate formate-lyase-activating enzyme | 40,800 | 1 |
| Tbd1831 | Putative cytochrome c1 | 27,211 | 1 |
| Tbd1840 | Unknown | 11,061 | 1 |
| Tbd2026 | Possible cytochrome c4 or c-553 | 11,253 | 1 |
| Tbd2027 | Cytochrome c, class IC | 11,521 | 1 |
| Tbd2034 | Possible cytochrome subunit of sulfide dehydrogenase | 10,395 | 1 |
| Tbd2060 | Possible alpha-mannosidase | 64,835 | 1 |
| Tbd2157 | Cytochrome c | 18,742 | 1 |
| Tbd2170 | Activase of anaerobic class III ribonucleotide reductase | 24,585 | 1 |
| Tbd2181 | Unknown | 20,904 | 1 |
| Tbd2476 | dsrJ | 17,948 | 3 |
| Tbd2477 | dsrL | 71,328 | 1 |
| Tbd2545 | Diheme cytochrome c | 37,973 | 2 |
| Tbd2726 | Cytochrome c | 11,096 | 1 |
| Tbd2727 | Conserved protein of unknown function | 74,616 | 1 |
| Tbd2738 | Zinc-dependent hydrolase | 26,287 | 1 |
As defined by the presence of at least one CXXCH heme-binding motif. Tbd0039 (which encodes ribosomal protein L31) and Tbd1539 (which encodes DnaJ) include the CXXCH motif but were excluded from this table.
Best attempt at annotation based on examination of best BLASTP hits, top 10 PSI-BLAST hits, and genomic context.
Molecular mass predicted for the unprocessed gene product without cofactors.
Autotrophy.
The genome of this obligate chemolithoautotroph encodes both form I and form II ribulose 1,5-bisphosphate carboxylase/oxygenase (RubisCO) enzymes for CO2 fixation (21, 35). The form I genes (cbbL and cbbS) occur in an operon with cbbQ and cbbO genes (cbbLSQO; Tbd2624-Tbd2621). The form II gene, cbbM, is in a separate operon that also includes cbbQ and cbbO genes; the operon has the form cbbMQO (Tbd2638-Tbd2636). Both operons are preceded by divergently transcribed cbbR genes encoding LysR-type transcriptional regulators (Tbd2625 and Tbd2639). The cbbQ and cbbO genes encode proteins involved in the posttranslational activation of RubisCO (33). The two copies of each gene are quite distinct: the two CbbQ proteins have 71% identical residues, whereas the two CbbO proteins only share 34% identity. The two RubisCO operons, although separate, fall within a 33-kb region that also includes an operon of 10 genes (Tbd2641-Tbd2650) encoding the carboxysome shell proteins (14). The carboxysome operon does not begin with cbbLS, unlike other known examples (14). These three operons, all located on the reverse strand, are oriented to be transcribed in the same direction. The carboxysome operon includes the gene (Tbd2649) for carbonic anhydrase (CA) epsilon (74). The carboxysome operon is also preceded by a divergently transcribed cbbR gene (Tbd2651) encoding a LysR-type transcriptional regulator. In addition to the epsilon-type CA, a eukaryotic-type CA (Tbd2167) is encoded elsewhere in the genome. Since there is no evidence that the carboxysome operon is functional (14, 72), this alternative CA may be the primary source of this activity.
Inasmuch as T. denitrificans can grow under both aerobic and denitrifying conditions, and form I and form II RubisCO in this species were shown to have markedly different abilities to discriminate between CO2 and O2 (35), it is possible that form I and II RubisCO are differentially expressed in T. denitrificans as a function of O2 concentration. Molecular oxygen competes with CO2 for the active site of RubisCO and thereby decreases its efficiency for carbon fixation. Hence, form I, which has a higher CO2/O2 specificity, should be more highly expressed under aerobic conditions, whereas form II should be more highly expressed under anaerobic conditions.
Genes for all enzymes to complete the Calvin-Benson-Bassham cycle are present. Transketolase, NAD-dependent glyceraldehyde 3-phosphate dehydrogenase (EC 1.2.1.12), phosphoglycerate kinase, pyuvate kinase, and fructose 1,6-bisphosphate aldolase (Tbd0159-Tbd0163) are encoded by an operon, whereas fructose 1,6-/sedoheptulose-1,7-bisphosphatase (Tbd2577), ribose 5-phosphate isomerase (Tbd2364), and phosphoribulokinase (Tbd2447) are encoded by isolated genes. Genes encoding ribulose 5-phosphate 3-epimerase and phosphoglycolate phosphatase (Tbd2230-Tbd2229) are adjacent but may not be cotranscribed.
Central intermediary metabolism.
Organic storage materials in T. denitrificans have not been reported, but the presence of genes encoding glycogen synthase (Tbd2057), maltooligosyl trehalose synthase (Tbd1174), and various glucan branching enzymes (e.g., an α-1,4-d-glucan branching enzyme; Tbd1173, Tbd2058) and glucano- and glycosyl-transferases suggests that the bacterium synthesizes a polyglucose storage product (cf. Halothiobacillus neapolitanus [10]). The gene for the key enzyme of the Embden-Meyerhof-Parnas (EMP) pathway necessary for it to effect gluconeogenesis (fructose 1,6-bisphosphatase) is present in the genome (Tbd2577). The synthesis of such a storage product would provide a rationale for the presence of genes for glucokinase (Tbd2062, Tbd2216), which could thus assist in the endogenous catabolism of stored polyglucose. Several genes encoding enzymes for polysaccharide hydrolysis also appear to be present: β-galactosidase (Tbd2429), several glycosidases (Tbd0727, Tbd0923, Tbd1172) and glycosyl transferases (e.g., Tbd0289, Tbd0293, Tbd0294, Tbd0301, Tbd2139), an α-mannosidase (Tbd2060), and an α-arabinofuranosidase (Tbd1789).
Genes encoding all the enzymes of the EMP pathway necessary for it to function in the forward direction for the conversion of glucose to pyruvate are present in the genome, including the gene for phosphofructokinase (Tbd1502), which is unique to the degradative EMP pathway. Genes for all the enzymes of the oxidative pentose phosphate pathway for the oxidation of glucose to carbon dioxide are also present in thegenome. Some of the enzymes of the EMP and oxidative pentose phosphate pathways are also common to the Calvin-Benson-Bassham reductive pentose phosphate cycle for carbon dioxide fixation, and ribose 5-phosphate for nucleic acid synthesis can be produced from glucose 6-phosphate by glucose 6-phosphate dehydrogenase, phosphogluconolactonase, and 6-phosphogluconate dehydrogenase (encoded by Tbd2121-Tbd2123). In addition, genes encoding alcohol dehydrogenase (EC 1.1.1.1; Tbd1767) and other short-chain alcohol dehydrogenases (Tbd0924, Tbd1469, Tbd1549, Tbd1699, Tbd1886, Tbd2701, Tbd2756), lactate dehydrogenase (Tbd1998), and phosphoketolase (EC 4.1.2.9) (Tbd0049, Tbd0831) suggest that T. denitrificans might be able to produce ethanol and lactate (by homo- or hetero-fermentative metabolism) from endogenous glucose under anoxic conditions. This would parallel the heterolactic fermentation of stored polyglucose carried out anaerobically by Halothiobacillus neapolitanus (11). Glucose 6-phosphatase and glycerol phosphatase are absent from the T. denitrificans genome, reflecting the lack of need to produce free glucose or glycerol.
Genes for some enzymes of the Entner-Doudoroff pathway are present, but the gene encoding 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase is notably absent, meaning that T. denitrificans cannot express a functional Entner-Doudoroff pathway. We failed to detect KDPG aldolase in the genome in BLAST searches conducted using database polypeptide sequences from Escherichia coli and Zymomonas, Gluconobacter, and Neisseria spp. The gene putatively encoding 6-phosphogluconate (6-PG) dehydratase (Tbd2730), whose product is KDPG, may be a false identification, as a TBLASTN search of the T. denitrificans genome with the 6-PG dehydratase protein sequence of Xanthomonas axonopodis (GenBank NP_642389) failed to detect any matching sequences, although direct BLAST2(P) comparison of the polypeptide encoded by Tbd2730 did show low (27 to 29%) identity to the 6-PG dehydratases of Xanthomonas sp. and Helicobacter pylori (GenBank NP_223740). The highest identities of Tbd2730 indicated by BLAST analyses were to dihydroxyacid dehydratases, and comparisons by BLASTP of the polypeptide sequences of the dihydroxyacid dehydratase of Sinorhizobium meliloti (GenBank AL591792) with the 6-PG dehydratase of Xanthomonas sp. also showed 29% identity. Assigning putative function must thus be done with caution.
Genes for all the enzymes of the Krebs tricarboxylic acid cycle were identified in the genome. The E1 and E2 subunits of 2-oxoglutarate dehydrogenase are encoded by Tbd1188 and Tbd1189, whereas the E3 subunit (common also to pyruvate dehydrogenase) is encoded by Tbd0652. The genes encoding the E1, E2, and E3 subunits of pyruvate dehydrogenase are Tbd0652, Tbd0654, and Tbd0655, whereas Tbd1847 putatively encodes all three subunits, with highest identity to the pdhA, pdhB, and pdhL genes of Ralstonia eutropha. It was surprising to find genes for the E1 and E2 subunits of 2-oxoglutarate dehydrogenase, as T. denitrificans strains (including ATCC 25259) do not express an active 2-oxoglutarate dehydrogenase enzyme when growing autotrophically (57, 83). This inability is shared with other obligate chemolithotrophs and methanotrophs, including Nitrosomonas europaea and Methylococcus capsulatus, and has been proposed as a contributory factor in the obligate growth modes of these bacteria (15, 41, 48, 73, 89, 90, 92). The E3 component of the pyruvate and 2-oxoglutarate dehydrogenases is also known to be controlled by multiple regulatory mechanisms (93), so it may be that failure of obligate chemolithotrophs to express active 2-oxoglutarate dehydrogenase results from regulation at the E3 gene expression level.
While identifying genes encoding some Krebs cycle enzymes, it was found that a gene (Tbd2119) showed significant identity to both fumarase (fumarate hydratase; EC 4.2.1.2) and aspartate ammonia-lyase (EC 4.3.1.1). BLASTP analysis showed the translated sequence of Tbd2119 to share 55% sequence identity with the aspartate ammonia-lyase of Geobacter sulfurreducens (GenBank NP_951538.1), 55% identity with the fumarase sequence of Pelobacter propionicus (GenBank ZP_00677090.1), and 41% with the fumarase C of E. coli. We suggest that the role of the Tbd2119 gene product is as a fumarase in the Krebs cycle. This similarity of genes for fumarase and aspartate ammonia-lyase is common across the database sequences for numerous organisms, possibly indicating multifunctional roles for the genes or their encoded proteins.
Genes encoding isocitrate lyase and malate synthase have not been detected using BLASTP searches with the translated polypeptide sequences of the genes from E. coli, meaning that the glyoxylate cycle cannot be present and hence exogenous acetate could not be metabolized as a sole source of carbon by that route.
Transport systems for organic nutrients.
Permease systems for inorganic ions and numerous ABC transporter components are encoded within the genome, but relatively few specific systems have been detected for the uptake of sugars or organic acids. Those detected include various components constituting a tripartite ATP-independent periplasmic (TRAP)-type C4-dicarboxylate transporter system (Tbd0466, Tbd0467, Tbd0468, Tbd2151, Tbd2164) and phosphotransferase system components including enzyme I (Tbd2414), HPr (Tbd2413), two enzyme IIA subunits specific for mannose/fructose (Tbd2412, Tbd0531), and Hpr(ser) kinase/phosphorylase (Tbd0530). The absence from the T. denitrificans genome of a gene encoding enzyme IIC (the sugar permease component) suggests that the role of this PTS system may be regulatory rather than for sugar transport (30). The genome of T. denitrificans also encodes two sodium:solute symporter family proteins that may be involved in acetate uptake (Tbd0088, Tbd0212). A sodium:solute symporter protein, ActP, has been shown to be involved in acetate uptake in E. coli (GenBank P32705). A functional transport system for acetate in T. denitrificans was indicated by the uptake of 14C-labeled acetate into bacteria growing chemolithotrophically (acetate provided 6 to 11% of the cell carbon of strain ATCC 25259) (84). As well as being used for lipid biosynthesis, [14C]acetate was incorporated only into the protein-amino acids glutamate, proline, and arginine, as reported for other obligate chemolithotrophs and methanotrophs that lack 2-oxoglutarate dehydrogenase (19, 41, 73, 92). Incorporation of acetate-carbon into glutamate by T. denitrificans was unaffected by exogenous glutamic acid, which was presumably not taken up significantly by the bacteria. The possibility remains to be tested that chemolithoorganotrophic growth by T. denitrificans might be possible if the organism were presented with compounds such as acetate, or a suitable C4-dicarboxylic acid, in the presence of thiosulfate and nitrate as the energy source. This would be akin to the chemolithoorganotrophic growth of Nitrosomonas europaea on fructose or pyruvate and ammonia (15, 37).
Inorganic ion transport and heavy metal resistance.
A number of genes were identified in T. denitrificans that putatively encode transporters that can mediate either the uptake or efflux of a range of inorganic ions. In all, at least 18 complete ABC (ATP Binding Cassette) transporters predominantly for inorganic molecules are present in the genome, allowing for the uptake of Fe3+, thiosulfate, nitrate, nitrite, and many other ions (18). Numerous non-ABC-type transporters allowing for uptake of other ions, such as various sulfur-containing compounds and bicarbonate, are also present. The genome of T. denitrificans also contains a surprisingly large number of metal resistance systems, particularly considering its relatively small genome size (Table 5). In total, the T. denitrificans genome encodes as many as 17 possible metal resistance systems (described by Nies [55]), including five heavy metal efflux (HME) systems from the resistance-nodulation-cell division (RND) family of transporters; three cation diffusion facilitators (CDF); three CPx-type ATPases (heavy metal-specific P-type ATPases); and five additional gene clusters encoding possible resistance systems specific for metals such as Ni2+ (nreB), Pb2+ (pbrT), Hg2+ (3), chromium (as chromate; chrA), and Cu2+/Ag2+ (26, 32). Although the T. denitrificans genome has fewer systems than the model metal-resistant bacterium Ralstonia metallidurans (55), it has more than most other bacteria characterized to date (Table 5). Notably, T. denitrificans (or species with >98% 16S rRNA gene sequence similarity) was found to be prevalent in an inactive uranium mine with relatively high concentrations of uranium, nickel, cobalt, and zinc (80).
TABLE 5.
Comparison of encoded efflux-mediated heavy metal resistance systems among the genomes of T. denitrificans and other selected bacteria
| Bacterial species | Genome size (Mbp) | No. of HME RNDs | No. of CDF | No. of CPx-type ATPases |
|---|---|---|---|---|
| Thiobacillus denitrificansa | 2.9 | 5 | 3 | 3 |
| Geobacter sulfurreducens PCAa | 3.8 | 3 | 2 | 2 |
| Ralstonia metalliduransb | 6.9 | 12 | 3 | 5 |
| Pseudomonas aeruginosab | 6.3 | 1 | 3 | 4 |
| Escherichia colib | 4.6 | 1 | 2 | 2 |
Putative heavy metal-exporting protein families for T. denitrificans and G. sulfurreducens PCA (GenBank AE01780) were identified as described by Nies (55). Confirmatory data on metal transport in G. sulfurreducens was obtained from an unpublished source (H. A. Vrionis and D. R. Lovley, 105th Gen. Meet. Am. Soc. Microbiol., poster I-054, 2005).
From Nies (55).
Several of the efflux-mediated heavy metal resistance systems, along with various systems involved in metal uptake and storage, are found in large gene clusters on the T. denitrificans genome (Fig. 1). The largest of these metal transport clusters, Tbd0704-Tbd0726, encodes proteins allowing for high-affinity Fe3+ acquisition, Fe3+ storage, Pb2+ resistance, and heavy metal resistance. Genes involved in high-affinity Fe3+ acquisition include homologs of a portion of the Vibrio parahaemolyticus polyhydroxycarboxylate-type siderophore biosynthesis, secretion, and uptake gene cluster pvuApvsABCDE psuA (Tbd0722-Tbd0717, Tbd0715) (81) and tonBexbBD (Tbd0713-Tbd0711), which allows for active transport across the outer membrane of Fe3+-bound siderophore. Also found are genes encoding the Fe3+ storage proteins bacterioferritin and bacterioferritin-associated ferredoxin (Tbd0704-Tbd0705), Pb2+ resistance (Tbd0723), and CDF family heavy metal resistance (Tbd0726). A second large gene cluster (encompassing genes on both the forward and reverse strands) encodes proteins primarily associated with metal resistance (Tbd1324-Tbd1341). This cluster includes a multi-copper oxidase (Tbd1324) (32), a periplasmic Cu2+-binding protein (Tbd1326) (26), two HME-RND systems (Tbd1327-Tbd1329, Tbd1333-Tbd1335), and a Hg2+ resistance system, merRTPA (Tbd1338-Tbd1341) (3).
Future prospects.
The availability of the T. denitrificans genome has fostered new insights into this bacterium and will effectively focus further investigations into its biochemistry and physiology. Among the new insights reported here are the following genomic characteristics: an unusually large number of genes encoding c-type cytochromes, a relatively large complement of genes associated with inorganic ion transport and heavy metal resistance, and the presence of genes encoding two [NiFe]hydrogenases, which is particularly significant because no physiological, biochemical, or genetic information on hydrogenases has previously been reported for this species. The genome also provides much more information on the genetic basis of sulfur-compound oxidation in chemolithotrophic β-proteobacteria (particularly with respect to sox and dsr genes). Much more work will be required to understand the genetic and biochemical basis of unusual and enigmatic metabolic capabilities of this bacterium, including the coupling of denitrification with sulfur-compound oxidation, the use of mineral electron donors, and the anaerobic, nitrate-dependent oxidation of metals. Whole-genome transcriptional studies with cDNA microarrays are currently under way to address such questions.
Acknowledgments
We thank A. Lapidus and C. Detter (Department of Energy Joint Genome Institute), M. Land (Oak Ridge National Laboratory), and M. Shin, T. Legler, F. Bourguet, W. Marrs, and A. Krefft (Lawrence Livermore National Laboratory) for their valuable contributions.
This work was performed under the auspices of the U.S. Department of Energy by the University of California, Lawrence Livermore National Laboratory, under contract W-7405-Eng-48.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aminuddin, M., and D. J. D. Nicholas. 1974. An AMP-independent sulphite oxidase from Thiobacillus denitrificans: purification and properties. J. Gen. Microbiol. 82:103-113. [Google Scholar]
- 3.Barkay, T., S. M. Miller, and A. O. Summers. 2003. Bacterial mercury resistance from atoms to ecosystems. FEMS Microbiol. Rev. 27:355-384. [DOI] [PubMed] [Google Scholar]
- 4.Beijerinck, M. W. 1904. Phénomènes de réduction produits par les microbes (Conférence avec demonstrations faite—Delft, le 16 avril 1903). Arch. Neerl. Sci. Ser. 2 9:131-157. [Google Scholar]
- 5.Beller, H. R. 2005. Anaerobic, nitrate-dependent oxidation of U(IV) oxide minerals by the chemolithoautotrophic bacterium Thiobacillus denitrificans. Appl. Environ. Microbiol. 71:2170-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beller, H. R., V. Madrid, G. B. Hudson, W. W. McNab, and T. Carlsen. 2004. Biogeochemistry and natural attenuation of nitrate in groundwater at an explosives test facility. Appl. Geochem. 19:1483-1494. [Google Scholar]
- 7.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 8.Berks, B. C., M. D. Page, D. J. Richardson, A. Reilly, A. Cavill, F. Outen, and S. J. Ferguson. 1995. Sequence analysis of subunits of the membrane-bound nitrate reductase from a denitrifying bacterium: the integral membrane subunit provides a prototype for the dihaem electron-carrying arm of a redox loop. Mol. Microbiol. 15:319-331. [DOI] [PubMed] [Google Scholar]
- 9.Berlyn, M. K. B., K. B. Low, K. E. Rudd, and M. Singer. 1996. Linkage map of Escherichia coli K12, edition 9, p. 1715-1902. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 2. ASM Press, Washington, D.C. [Google Scholar]
- 10.Beudeker, R. F., J. W. M. Kerver, and J. G. Kuenen. 1981. Occurrence, structure and function of intracellular polyglucose in the obligate chemolithotroph Thiobacillus neapolitanus. Arch. Microbiol. 129:221-226. [Google Scholar]
- 11.Beudeker, R. F., W. de Boer, and J. G. Kuenen. 1981. Heterolactic fermentation of intracellular polyglucose by the obligate chemolithotroph Thiobacillus neapolitanus under anaerobic conditions. FEMS Microbiol. Lett. 12:337-342. [Google Scholar]
- 12.Bowen, T. J., F. C. Happold, and B. F. Taylor. 1966. Studies on the adenosine-5′-phosphosulfate reductase from Thiobacillus denitrificans. Biochim. Biophys. Acta 118:566-576. [DOI] [PubMed] [Google Scholar]
- 13.Brüser, T., T. Selmer, and C. Dahl. 2000. “ADP sulfurylase” from Thiobacillus denitrificans is an adenylylsulfate:phosphate adenylyltransferase and belongs to a new family of nucleotidyltransferases. J. Biol. Chem. 275:1691-1698. [DOI] [PubMed] [Google Scholar]
- 14.Cannon, G. C., S. H. Baker, F. Soyer, D. R. Johnson, C. E. Bradburne, J. L. Mehlman, P. S. Davies, Q. L. Jiang, S. Heinhorst, and J. M. Shively. 2003. Organization of carboxysome genes in the thiobacilli. Curr. Microbiol. 46:115-119. [DOI] [PubMed] [Google Scholar]
- 15.Chain, P., J. Lamerdin, F. Larimer, W. Regala, V. Lao, M. Land, L. Hauser, A. Hooper, M. Klotz, J. Norton, L. Sayavedra-Soto, D. Arciero, N. Hommes, M. Whittaker, and D. Arp. 2003. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J. Bacteriol. 185:2759-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahl, C., G. Rakhely, A. S. Pott-Sperling, B. Fodor, M. Takacs, A. Toth, M. Kraeling, K. Gyorfi, A. Kovacs, J. Tusz, and K. L. Kovacs. 1999. Genes involved in hydrogen and sulfur metabolism in phototrophic sulfur bacteria. FEMS Microbiol. Lett. 180:317-324. [DOI] [PubMed] [Google Scholar]
- 17.Dahl, C., S. Engels, A. S. Pott-Sperling, A. Schulte, J. Sander, Y. Lübbe, O. Deuster, and D. C. Brune. 2005. Novel genes of the dsr gene cluster and evidence for close interaction of Dsr proteins during sulfur oxidation in the phototrophic sulfur bacterium Allochromatium vinosum. J. Bacteriol. 187:1392-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dassa, E., and P. Bouige. 2001. The ABC of ABCs: a phylogenetic and functional classification of ABC systems in living organisms. Res. Microbiol. 152:211-229. [DOI] [PubMed] [Google Scholar]
- 19.Eccleston, M., and D. P. Kelly. 1973. Assimilation and toxicity of some exogenous C-1 compounds, alcohols, sugars and acetate in the methane-oxidizing bacterium Methylococcus capsulatus. J. Gen. Microbiol. 75:211-221. [DOI] [PubMed] [Google Scholar]
- 20.Eisen, J. A., K. E. Nelson, I. T. Paulsen, J. F. Heidelberg, M. Wu, R. J. Dodson, R. Deboy, M. L. Gwinn, W. C. Nelson, D. H. Haft, E. K. Hickey, J. D. Peterson, A. S. Durkin, J. L. Kolonay, F. Yang, I. Holt, L. A. Umayam, T. Mason, M. Brenner, T. P. Shea, D. Parksey, W. C. Nierman, T. V. Feldblyum, C. L. Hansen, M. B. Craven, D. Radune, J. Vamathevan, H. Khouri, O. White, T. M. Gruber, K. A. Ketchum, J. C. Venter, H. Tettelin, D. A. Bryant, and C. M. Fraser. 2002. The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl. Acad. Sci. USA 99:9509-9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.English, R. S., C. A. Williams, S. C. Lorbach, and J. M. Shively. 1992. Two forms of ribulose 1,5-bisphosphate carboxylase/oxygenase from Thiobacillus denitrificans. FEMS Microbiol. Lett. 73:111-119. [DOI] [PubMed] [Google Scholar]
- 22.Ewing, B., and P. Green. 1998. Basecalling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 23.Ewing, B., L. Hillier, M. Wendl, and P. Green. 1998. Basecalling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 24.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 25.Felsenstein, J. 2002. PHYLIP (Phylogeny Inference Package) version 3.6a3. Department of Genome Sciences, University of Washington, Seattle, Wash.
- 26.Franke, S., G. Grass, C. Rensing, and D. H. Nies. 2003. Molecular analysis of the copper-transporting efflux system CusCFBA of Escherichia coli. J. Bacteriol. 185:3804-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Friedrich, C. G., A. Quentmeier, F. Bardichewsky, D. Rother, R. Kraft, S. Kostka, and H. Prinz. 2000. Novel genes coding for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17. J. Bacteriol. 182:4677-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Friedrich, C. G., D. Rother, F. Bardichewsky, A. Quentmeier, and J. Fischer. 2001. Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl. Environ. Microbiol. 67:2873-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fritz, G., T. Büchert, H. Huber, K. O. Stetter, and P. M. H. Kroneck. 2000. Adenylylsulfate reductases from archaea and bacteria are 1:1 αβ-heterodimeric iron-sulfur flavoenzymes—high similarity of molecular properties emphasizes their central role in sulfur metabolism. FEBS Lett. 473:63-66. [DOI] [PubMed] [Google Scholar]
- 30.Gonzalez, C. F., A. J. Stonestrom, G. L. Lorca, and M. H. Saier. 2005. Biochemical characterization of phosphoryl transfer involving HPr of the phosphoenolpyruvate-dependent phosphotransferase system in Treponema denticola, an organism that lacks PTS permeases. Biochemistry 44:598-608. [DOI] [PubMed] [Google Scholar]
- 31.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 32.Grass, G., and C. Rensing. 2001. CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem. Biophys. Res. Commun. 286:902-908. [DOI] [PubMed] [Google Scholar]
- 33.Hayashi, N. R., H. Arai, T. Kodama, and Y. Igarashi. 1997. The novel genes, cbbQ and cbbO, located downstream from the RubisCO genes of Pseudomonas hydrogenothermophila, affect the conformational states and activity of RubisCO. Biochem. Biophys. Res. Commun. 241:565-569. [DOI] [PubMed] [Google Scholar]
- 34.Heidelberg, J. F., I. T. Paulsen, K. E. Nelson, E. J. Gaidos, W. C. Nelson, T. D. Read, J. A. Eisen, R. Seshadri, N. Ward, B. Methe, R. A. Clayton, T. Meyer, A. Tsapin, J. Scott, M. Beanan, L. Brinkac, S. Daugherty, R. T. DeBoy, R. J. Dodson, A. S. Durkin, D. H. Haft, J. F. Kolonay, R. Madupu, J. D. Peterson, L. A. Umayam, O. White, A. M. Wolf, J. Vamathevan, J. Weidman, M. Impraim, K. Lee, K. Berry, C. Lee, J. Mueller, H. Khouri, J. Gill, T. R. Utterback, L. A. McDonald, T. V. Feldblyum, H. O. Smith, J. C. Venter, K. H. Nealson, and C. M. Fraser. 2002. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat. Biotechnol. 20:1118-1123. [DOI] [PubMed] [Google Scholar]
- 35.Hernandez, J. M., S. H. Baker, S. C. Lorbach, J. M. Shively, and F. R. Tabita. 1996. Deduced amino acid sequence, functional expression, and unique enzymatic properties of the form I and form II ribulose bisphosphate carboxylase/oxygenase from the chemoautotrophic bacterium Thiobacillus denitrificans. J. Bacteriol. 178:347-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hole, U. H., K.-U. Vollack, W. G. Zumft, E. Eisenmann, R. A. Siddiqui, B. Friedrich, and P. M. H. Kroneck. 1996. Characterization of the membranous denitrification enzymes nitrite reductase (cytochrome cd1) and copper-containing nitrous oxide reductase from Thiobacillus denitrificans. Arch. Microbiol. 165:55-61. [DOI] [PubMed] [Google Scholar]
- 37.Hommes, N. G., L. A. Sayavedra-Soto, and D. J. Arp. 2003. Chemolithoorganotrophic growth of Nitrosomonas europaea on fructose. J. Bacteriol. 185:6809-6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 39.Kappler, U., C. G. Friedrich, H. G. Trüper, and C. Dahl. 2001. Evidence for two pathways of thiosulfate oxidation in Starkeya novella (formerly Thiobacillus novellus). Arch. Microbiol. 175:102-111. [DOI] [PubMed] [Google Scholar]
- 40.Karlin, S., J. Mrazek, and A. M. Campbell. 1997. Compositional biases of bacterial genomes and evolutionary implications. J. Bacteriol. 179:3899-3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kelly, D. P. 1967. The incorporation of acetate by the chemoautotroph Thiobacillus neapolitanus strain C. Arch. Mikrobiol. 58:99-116. [DOI] [PubMed] [Google Scholar]
- 42.Kelly, D. P. 1982. Biochemistry of the chemolithotrophic oxidation of inorganic sulphur. Philos. Trans. R. Soc. Lond. B Biol. Sci. 298:499-528. [DOI] [PubMed] [Google Scholar]
- 43.Kelly, D. P. 1989. Physiology and biochemistry of unicellular sulfur bacteria, p. 193-217. In H. G. Schlegel and B. Bowien (ed.), Biology of autotrophic bacteria. Science Tech Publishers, Madison, Wis.
- 44.Kelly, D. P. 1990. Energetics of chemolithotrophic bacteria, p. 479-503. In T. A. Krulwich (ed.), Bacterial energetics, Academic Press, San Diego, Calif.
- 45.Kelly, D. P. 1999. Thermodynamic aspects of energy conservation by chemolithotrophic sulfur bacteria in relation to the sulfur oxidation pathways. Arch. Microbiol. 171:219-229. [Google Scholar]
- 46.Kelly, D. P., and A. P. Harrison. 1989. Genus Thiobacillus, p. 1842-1858. In J. T. Staley, M. P. Bryant, N. Pfennig, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, 1st ed., vol. 3. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 47.Kelly, D. P., and A. P. Wood. 2000. Confirmation of Thiobacillus denitrificans as a species of the genus Thiobacillus, in the beta-subclass of the Proteobacteria, with strain NCIMB 9548 as the type strain. Int. J. Syst. Evol. Microbiol. 50:547-550. [DOI] [PubMed] [Google Scholar]
- 48.Kelly, D. P., and A. P. Wood. 2000. The chemolithotrophic prokaryotes. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes, an evolving electronic resource for the microbiological community (PKIIIE). Springer-Verlag, New York, N.Y. [Online.] http://www.prokaryotes.com.
- 49.Kelly, D. P., A. P. Wood, and E. Stackebrandt. 2005. Genus II. Thiobacillus Beijerinck, p. 764-769. In G. M. Garrity (eds.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 2, part C. Springer, New York, N.Y. [Google Scholar]
- 50.Kelly, D. P., J. K. Shergill, W. P. Lu, and A. P. Wood. 1997. Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Leeuwenhoek 71:95-107. [DOI] [PubMed] [Google Scholar]
- 51.Kobayashi, I. 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29:3742-3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krogh, A., B. Larsson, G. von Heijne, and E. L. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 53.Laska, S., F. Lottspeich, and A. Kletzin. 2003. Membrane-bound hydrogenase and sulfur reductase of the hyperthermophilic and acidophilic archaeon Acidianus ambivalens. Microbiology 149:2357-2371. [DOI] [PubMed] [Google Scholar]
- 54.Methé, B. A., K. E. Nelson, J. A. Eisen, I. T. Paulsen, W. Nelson, J. F. Heidelberg, D. Wu, M. Wu, N. Ward, M. J. Beanan, R. J. Dodson, R. Madupu, L. M. Brinkac, S. C. Daugherty, R. T. DeBoy, A. S. Durkin, M. Gwinn, J. F. Kolonay, S. A. Sullivan, D. H. Haft, J. Selengut, T. M. Davidsen, N. Zafar, O. White, B. Tran, C. Romero, H. A. Forberger, J. Weidman, H. Khouri, T. V. Feldblyum, T. R. Utterback, S. E. Van Aken, D. R. Lovley, and C. M. Fraser. 2003. Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science 302:1967-1969. [DOI] [PubMed] [Google Scholar]
- 55.Nies, D. H. 2003. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27:313-339. [DOI] [PubMed] [Google Scholar]
- 56.Pauwels, H., W. Kloppmann, J.-C. Foucher, A. Martelat, and V. Fritsche. 1998. Field tracer test for denitrification in a pyrite-bearing schist aquifer. Appl. Geochem. 13:767-778. [Google Scholar]
- 57.Peeters, T. L., M. S. Liu, and M. I. H. Aleem. 1970. The tricarboxylic acid cycle in Thiobacillus denitrificans and Thiobacillus-A2. J. Gen. Microbiol. 64:29-35. [DOI] [PubMed] [Google Scholar]
- 58.Pereira, M. M., and M. Teixeira. 2004. Proton pathways, ligand binding and dynamics of the catalytic site in haem-copper oxygen reductases: a comparison between the three families. Biochim. Biophys. Acta 1655:340-346. [DOI] [PubMed] [Google Scholar]
- 59.Petri, R., L. Podgorsek, and J. F. Imhoff. 2001. Phylogeny and distribution of the soxB gene among thiosulfate-oxidizing bacteria. FEMS Microbiol. Lett. 197:171-178. [DOI] [PubMed] [Google Scholar]
- 60.Philippot, L. 2002. Denitrifying genes in bacterial and Archaeal genomes. Biochim. Biophys. Acta 1577:355-376. [DOI] [PubMed] [Google Scholar]
- 61.Pitcher, R. S., and N. J. Watmough. 2004. The bacterial cytochrome cbb3 oxidases. Biochim. Biophys. Acta 1655:388-399. [DOI] [PubMed] [Google Scholar]
- 62.Pott, A. S., and C. Dahl. 1998. Sirohaem sulfite reductase and other proteins encoded by genes at the dsr locus of Chromatium vinosum are involved in the oxidation of intracellular sulfur. Microbiology (Reading) 144:1881-1894. [DOI] [PubMed] [Google Scholar]
- 63.Quentmeier, A., P. Hellwig, F. Bardichewsky, G. Grolle, R. Kraft, and C. G. Friedrich. 2003. Sulfur oxidation in Paracoccus pantotrophus: interaction of the sulfur-binding protein SoxYZ with the dimanganese SoxB protein. Biochem. Biophys. Res. Commun. 312:1011-1018. [DOI] [PubMed] [Google Scholar]
- 64.Quentmeier, A., P. Hellwig, F. Bardichewsky, R. Wichmann, and C. G. Friedrich. 2004. Sulfide dehydrogenase activity of the monomeric flavoprotein SoxF of Paracoccus pantotrophus. Biochemistry 43:14696-14703. [DOI] [PubMed] [Google Scholar]
- 65.Quentmeier, A., R. Kraft, S. Kostka, R. Klockemkaper, and C. G. Friedrich. 2000. Characterization of a new type of sulfite dehydrogenase from Paracoccus pantotrophus GB17. Arch. Microbiol. 173:117-125. [DOI] [PubMed] [Google Scholar]
- 66.Rakhely, G., A. Colbeau, J. Garin, P. M. Vignais, and K. L. Kovacs. 1998. Unusual organization of the genes coding for HydSL, the stable [NiFe]hydrogenase in the photosynthetic bacterium Thiocapsa roseopersicina BBS. J. Bacteriol. 180:1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rother, D., and C. G. Friedrich. 2002. The cytochrome complex SoxXA of Paracoccus pantotrophus is produced in Escherichia coli and functional in the reconstituted sulfur-oxidizing enzyme system. Biochim. Biophys. Acta 1598:65-73. [DOI] [PubMed] [Google Scholar]
- 68.Rother, D., H. J. Henrich, A. Quentmeier, F. Bardichewsky, and C. G. Friedrich. 2001. Novel genes of the sox gene cluster, mutagenesis of the flavoprotein SoxF, and evidence for a general sulfur-oxidizing system in Paracoccus pantotrophus GB17. J. Bacteriol. 183:4499-4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sawnhey, V., and D. J. D. Nicholas. 1977. Sulphite-dependent nitrate reductase and NADH-dependent nitrate reductase from Thiobacillus denitrificans. J. Gen. Microbiol. 100:49-58. [Google Scholar]
- 70.Sawnhey, V., and D. J. D. Nicholas. 1978. Sulphide-linked nitrite reductase from Thiobacillus denitrificans with cytochrome oxidase activity: purification and properties. J. Gen. Microbiol. 106:119-128. [Google Scholar]
- 71.Schedel, M., and H. G. Trüper. 1979. Purification of Thiobacillus-denitrificans siroheme sulfite reductase and investigation of some molecular and catalytic properties. Biochim. Biophys. Acta 568:454-466. [DOI] [PubMed] [Google Scholar]
- 72.Shively, J. M., G. L. Decker, and J. W. Greenawalt. 1970. Comparative ultrastructure of the thiobacilli. J. Bacteriol. 101:618-627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Smith, A. J., and D. S. Hoare. 1977. Specialist phototrophs, lithotrophs, and methylotrophs: a unity among a diversity of prokaryotes? Bacteriol. Rev. 41:419-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.So, A. K.-C., G. S. Espie, E. B. Williams, J. M. Shively, S. Heinhorst, and G. C. Cannon. 2004. A novel evolutionary lineage of carbonic anhydrase (ɛclass) is a component of the carboxysome shell. J. Bacteriol. 186:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sonnhammer, E. L. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences, p. 175-182. In J. Glasgow, T. Littlejohn, F. Major, R. Lathrop, D. Sankoff, and C. Sensen (ed.), Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, CA. [PubMed]
- 76.Sorokin, D. Y., T. P. Tourova, A. N. Antipov, G. Muyzer, and J. G. Kuenen. 2004. Anaerobic growth of the haloalkaliphilic denitrifying sulfur-oxidizing bacterium Thialkalivibrio thiocyanodenitrificans sp. nov. with thiocyanate. Microbiology (Reading) 150:2435-2442. [DOI] [PubMed] [Google Scholar]
- 77.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D.Spencer, G. K.-S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 78.Straub, K. L., M. Benz, B. Schink, and F. Widdel. 1996. Anaerobic, nitrate-dependent microbial oxidation of ferrous iron. Appl. Environ. Microbiol. 62:1458-1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stubner, S., T. Wind, and R. Conrad. 1998. Sulfur oxidation in rice field soil: activity, enumeration, isolation and characterization of thiosulfate-oxidizing bacteria. Syst. Appl. Microbiol. 21:569-578. [DOI] [PubMed] [Google Scholar]
- 80.Suzuki, Y., S. D. Kelly, K. M. Kemner, and J. F. Banfield. 2003. Microbial populations stimulated for hexavalent uranium reduction in uranium mine sediment. Appl. Environ. Microbiol. 69:1337-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tanabe, T., T. Funahashi, H. Nakao, S. Miyoshi, S. Shinoda, and S. Yamamoto. 2003. Identification and characterization of genes required for biosynthesis and transport of the siderophore vibrioferrin in Vibrio parahaemolyticus. J. Bacteriol. 185:6938-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Taylor, B. F., and D. S. Hoare. 1971. Thiobacillus denitrificans as an obligate chemolithoautotroph. Cell suspension and enzymic studies. Arch. Mikrobiol. 80:262-276. [DOI] [PubMed] [Google Scholar]
- 84.Taylor, B. F., D. S. Hoare, and S. L. Hoare. 1971. Thiobacillus denitrificans as an obligate chemolithoautotroph. Isolation and growth studies. Arch. Mikrobiol. 78:193-204. [DOI] [PubMed] [Google Scholar]
- 85.Timmer-Ten Hoor, A. 1975. A new type of thiosulfate oxidizing, nitrate reducing microorganism: Thiomicrospira denitrificans sp. nov. Neth. J. Sea Res. 9:344-350. [Google Scholar]
- 86.Trüper, H. G. 1994. Reverse siroheme sulfite reductase from Thiobacillus denitrificans. Meth. Enzymol. 243:422-426. [Google Scholar]
- 87.Vignais, P. M., B. Billoud, and J. Meyer. 2001. Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25:455-501. [DOI] [PubMed] [Google Scholar]
- 88.Vlasceanu, L., R. Popa, and B. K. Kinkle. 1997. Characterization of Thiobacillus thioparus LV43 and its distribution in a chemoautotrophically based groundwater ecosystem. Appl. Environ. Microbiol. 63:3123-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ward, N., O. Larsen, J. Sakwa, L. Bruseth, H. Khouri, A. S. Durkin, G. Dimitrov, L. X. Jiang, D. Scanlan, K. H. Kang, M. Lewis, K. E. Nelson, B. Methé, M. Wu, J. F. Heidelberg, I. T. Paulsen, D. Fouts, J. Ravel, H. Tettelin, Q. Ren, T. Read, R. T. DeBoy, R. Seshadri, S. L. Salzberg, H. B. Jensen, N. K. Birkeland, W. C. Nelson, R. J. Dodson, S. H. Grindhaug, I. Holt, I. Eidhammer, I. Jonasen, S. Vanaken, T. Utterback, T. V. Feldblyum, C. M. Fraser, J. R. Lillehaug, and J. A. Eisen. 2004. Genomic insights into methanotrophy: the complete genome sequence of Methylococcus capsulatus (Bath). PLoS Biol. 2:e303. [Online.] http://biology.plosjournals.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Whittenbury, R., and D. P. Kelly. 1977. Autotrophy: a conceptual phoenix. Symp. Soc. Gen. Microbiol. 27:121-149. [Google Scholar]
- 91.Wodara, C., S. Kostka, M. Egert, D. P. Kelly, and C. G. Friedrich. 1994. Identification and sequence analysis of the soxB gene essential for sulfur oxidation of Paracoccus denitrificans GB17. J. Bacteriol. 176:6188-6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wood, A. P., J. P. Aurikko, and D. P. Kelly. 2004. A challenge for 21st century molecular biology and biochemistry: what are the causes of obligate autotrophy and methanotrophy? FEMS Microbiol. Rev. 28:335-352. [DOI] [PubMed] [Google Scholar]
- 93.Zaman, Z., S. B. Bowman, G. D. Kornfeld, A. J. Brown, and I. W. Dawes. 1999. Transcription factor GCN4 for control of amino acid biosynthesis also regulates the expression of the gene for lipoamide dehydrogenase. Biochem. J. 340:855-862. [PMC free article] [PubMed] [Google Scholar]
- 94.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]