Abstract
Furaquinocin (FQ) A, produced by Streptomyces sp. strain KO-3988, is a natural polyketide-isoprenoid hybrid compound that exhibits a potent antitumor activity. As a first step toward understanding the biosynthetic machinery of this unique and pharmaceutically useful compound, we have cloned an FQ A biosynthetic gene cluster by taking advantage of the fact that an isoprenoid biosynthetic gene cluster generally exists in flanking regions of the mevalonate (MV) pathway gene cluster in actinomycetes. Interestingly, Streptomyces sp. strain KO-3988 was the first example of a microorganism equipped with two distinct mevalonate pathway gene clusters. We were able to localize a 25-kb DNA region that harbored FQ A biosynthetic genes (fur genes) in both the upstream and downstream regions of one of the MV pathway gene clusters (MV2) by using heterologous expression in Streptomyces lividans TK23. This was the first example of a gene cluster responsible for the biosynthesis of a polyketide-isoprenoid hybrid compound. We have also confirmed that four genes responsible for viguiepinol [3-hydroxypimara-9(11),15-diene] biosynthesis exist in the upstream region of the other MV pathway gene cluster (MV1), which had previously been cloned from strain KO-3988. This was the first example of prokaryotic enzymes with these biosynthetic functions. By phylogenetic analysis, these two MV pathway clusters were identified as probably being independently distributed in strain KO-3988 (orthologs), rather than one cluster being generated by the duplication of the other cluster (paralogs).
Isoprenoids are the largest single family of natural compounds, with more than 23,000 known examples, and include industrially useful compounds such as flavors, antibiotics, and plant hormones (6). All of these metabolites can be classified into several groups. This classification is based on the number of C5 units derived from isopentenyl diphosphate (IPP), which is biosynthesized via the 2-C-methyl-d-erythritol 4-phosphate (MEP) and/or mevalonate (MV) pathway (25); the groups include monoterpenes (C10), sesquiterpenes (C15), and diterpenes (C20). All these compounds are biosynthesized from the corresponding polyprenyl diphosphate. Geranyl diphosphate gives rise to monoterpenes (45), farnesyl diphosphate to sesquiterpenes (3), and geranylgeranyl diphosphate to diterpenes (28). In eukaryotes such as plants and fungi, which produce the vast majority of isoprenoids, the polyprenyl diphosphate is generally cyclized by an organism-specific isoprenoid cyclase to form a basic skeleton. This is followed by successive reactions, such as hydroxylation, methylation, and glycosylation, that give rise to many thousands of compounds (2, 5).
In contrast to eukaryotes, prokaryotes are known to produce a limited number of isoprenoids (7). Among prokaryotes, actinomycete strains have been known to produce several isoprenoid compounds, such as 2-methylisoborneol (10), pentalenene (4), geosmin (11), and squalene-hopene (35). Moreover, it was recently revealed that among prokaryotes, actinomycetes produce many types of isoprenoids in relatively large numbers, and the structures of these compounds are unique and different from those of eukaryotic origin (7). In particular, isoprenoid moieties of the compounds produced by actinomycetes are generally attached to other moieties, such as an aromatic ring, an amino acid, and a phenazine moiety. These moieties are biosynthesized via pathways independent of isoprenoids to further give rise to the so-called isoprenoid-hybrid compounds, such as novobiocin (27), clorobiocin (27), brasilicardin A (22), KS505a (31), and lavanducyanin (16). Moreover, several actinomycete strains are known to produce polyketide-isoprenoid hybrid compounds, such as furaquinocin (FQ) (23), naphterpin (38), napyradiomycin (40), and marinone (33), all of which were reported to show biological activities and could act as an antitumor drug, an antioxidative agent, a nonsteroidal estrogen receptor antagonist, and an anticancer drug, respectively. Considering that the structures of polyketide moieties, which are derived from 1,3,6,8-tetrahydroxynaphthalene (THN), are almost the same in these compounds, the prenyl moieties are suggested to play important roles in exhibiting diversity in the biological activities of these compounds. In contrast to studies on the biological activities of these compounds, there are few reports on the biosynthetic genes and enzymes of the compounds. Although an enzyme that catalyze the attachment of the C10 (geranyl) prenyl group to polyketide has been recently identified in the naphterpin biosynthetic gene cluster (24), to the best of our knowledge there are no reports on the entire biosynthetic gene cluster of this type of compound. Therefore, as a first step toward understanding the biosynthetic machinery of these unique compounds, we have cloned and identified an FQ biosynthetic gene cluster from Streptomyces sp. strain KO-3988 by using a recently developed methodology. An isoprenoid biosynthetic gene cluster generally exists in the flanking regions of the MV pathway gene cluster in actinomycetes possessing both the MEP and MV pathways for the formation of isopentenyl diphosphate (7). Moreover, we confirmed that strain KO-3988 is the first example of a microorganism equipped with two distinct MV pathway gene clusters, and we identified a viguiepinol [3-hydroxypimara-9(11),15-diene] biosynthetic gene cluster in the upstream region of the other cluster, MV1.
MATERIALS AND METHODS
Chemicals.
[α-32P]dCTP and (R)-[2-14C] mevalonolactone (CFA 660) were obtained from Amersham.
Bacterial strains.
Streptomyces sp. strain KO-3988 was used for the cloning experiment. Media and growth conditions for strain KO-3988 were as described by Komiyama et al. (23). Streptomyces lividans TK23 (15) and pWHM3 (44) were used for heterologous expression of the MV pathway gene clusters and their flanking regions. The Escherichia coli mutant DYM1, in which the dxr gene, encoding 1-deoxy-d-xylulose 5-phosphate reductoisomerase, was disrupted (26), was also used for heterologous expression of the MV pathway gene clusters. E. coli JM110 (Toyobo, Osaka, Japan) and plasmids pUC118 and pUC119 were used for sequencing analysis. A cosmid library of DNA of strain KO-3988 had been constructed previously (9). Ampicillin (100 μg/ml) and kanamycin (25 μg/ml) were added to the medium as required.
DNA isolation and manipulation.
Plasmids from E. coli were prepared using a QIAGEN plasmid kit. All restriction enzymes, T4 DNA ligase, and calf intestinal alkaline phosphatase were obtained from Toyobo and used in accordance with the manufacturer's protocols. Transformation of E. coli with plasmid DNA by electroporation was performed under standard conditions by using a BTX ECM 600 electroporation system (Biotechnologies and Experimental Research, Inc., San Diego, CA). Other general procedures were performed as described by Maniatis et al. (29).
Sequence analysis.
A 7.2-kb BamHI fragment that had been previously determined to carry regions homologous to the 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase gene by Southern blot analysis (20) was cloned from strain KO-3988 and used as a probe for colony hybridization. A cosmid containing the BamHI fragment, which was used as the probe, was selected and used for sequencing analysis. Sequencing was carried out with an automatic DNA sequencer (LiCor model 4000L).
Expression of the MV2 cluster in E. coli and S. lividans.
The entire MV pathway gene cluster (MV2) containing putative mevalonate kinase (MK), mevalonate diphosphate decarboxylase (MDPD), phosphomevalonate kinase (PMK), type 2 IPP isomerase, HMG-CoA reductase, and HMG-CoA synthase genes was amplified by PCR. The 5′ and 3′ primers had the respective sequences 5′-TGCTCTAGAGCGATCTCACCGTAGGAACGACAAGG-3′ and 5′-ACCAAGCTTCTAGCGCGCCGTGTAGATGCGCTTGT-3′. In order to facilitate cloning, an additional restriction site (underlined) was incorporated into both primers. After sequence confirmation, the XbaI-HindIII fragment was inserted into the same sites of pWHM3 (44) to yield pFQ-MEV2. The dxr defective strain DYM1 was transformed with pFQ-MEV2, and the growth of the transformants was examined on an LB agar plate supplemented or not with 2-C-methyl-d-erythritol.
The plasmid was also introduced into S. lividans TK23. Approximately 5 μg of cell extract of S. lividans TK23 harboring pFQ-MEV2 was incubated with (R)-[2-14C]mevalonolactone for 2 h at 30°C. The reaction products were spotted on cellulose thin-layer chromatography sheets (Merck, catalog no. 1.05628) and developed in ethanol, ammonia, and water (80:12.5:15) (Rf values: mevalonate, 0.90; mevalonate 5-phosphate, 0.51; IPP, 0.39; mevalonate diphosphate, 0.28). The sheets were exposed to an imaging plate (Fujifilm, Tokyo, Japan), and radiolabeled products were detected with BAS 1000 (Fujifilm, Tokyo, Japan).
Heterologous expression of the upstream region of the MV1 cluster in S. lividans.
In order to obtain the entire gene without the excess flanking region, PCR amplification was carried out. The 5′ and 3′ primers with an additional restriction site (underlined) had the respective 5′-TGCTCTAGATACTTGGAGCCATGCCGGAGCTACCT-3′ and 5′-ACCAAGCTTTCAGGTGGTCCTGAGCGTGGTGGTGG-3′. The amplified PCR product was digested with XbaI and HindIII, separated by agarose gel electrophoresis, and then purified with a gel extraction kit (QIAGEN). After sequence confirmation, the fragment was inserted into the same sites of pWHM3 to yield pWHM-Fura1. Metabolites from 5 liters of the culture broth of the transformant were purified as described below. The purity of the metabolite was examined by reverse-phase high-pressure liquid chromatography (HPLC). The analytical conditions were as follows: C18 reverse-phase column (Merck Mightysil RP-18 column; 250 by 4.6 mm), column temperature of 30°C, detection at 210 nm, a linear gradient from 85% to 100% acetonitrile for 0 to 10 min and 100% acetonitrile for an additional 50 min, and a flow rate of 1 ml/min.
Isolation of viguiepinol and (−)-pimara-9(11),15-diene.
S. lividans TK23 harboring pWHM-Fura1 was grown in several 300-ml Erlenmeyer flasks containing 30 ml of SK no. 2 medium (8) and thiostrepton (10 μg/ml). Fermentation was carried out for 7 days at 30°C with agitation (200 rpm). The culture broths (5 liters), in which approximately 1.3 μg/ml and 0.7 μg/ml of viguiepinol and (−)-pimara-9(11),15-diene, respectively, were accumulated, were centrifuged, and the precipitated mycelial cake was suspended in 1 liter of methanol. After vigorous shaking, the suspension was filtered and the methanol filtrate was concentrated to dryness in vacuo. The dried material was dissolved in 500 ml of ethyl acetate and water (1:1). After centrifugation, in order to separate the emulsion, the organic layer was recovered and evaporated to dryness under reduced pressure. The dried material was dissolved in a small volume of ethyl acetate and then fractionated by preparative HPLC (Merck Mightysil RP-18 column [250 by 20 mm]; mobile phase, 100% acetonitrile; flow rate, 5 ml/min; detection, 210 nm).
Heterologous expression of the upstream and downstream regions of the MV2 cluster in S. lividans.
Two DNA fragments carrying the regions that are upstream and downstream of MV2 were amplified by PCR with the following two sets of primers: (i) Fura-up5 (5′-TGCTCTAGAGTCCTCCGGGCAGCTGAAGCCCCTTC-3′) and Fura-up3 (5′-CCAATGCATCACCGCGGCTGGGGGACCAGGAGCAG-3′) and (ii) Fura-dn5 (5′-CCAATGCATGAAGTCGGGATTCTCGGCACGGGTGC-3′) and Fura-dn3 (5′-ACCAAGCTTTCAGTCCCGCTTCGTCGCGCAGATCA-3′). Other procedures were the same as those for the construction of pWHM-Fura1. This constructed plasmid was designated pWHM-Fura2. S. lividans TK23 harboring pWHM-Fura2 was grown in 300-ml Erlenmeyer flasks containing 30 ml of SK no. 2 medium (8) and thiostrepton (10 μg/ml). Fermentation was carried out for 7 days at 30°C with agitation (200 rpm). The culture broth (20 liters), containing approximately 0.2 μg/ml of FQ D, was collected and the compound was purified as described by Ishibashi et al. (17). The purity of the metabolite was examined by reverse-phase HPLC.
Analysis of metabolites.
1H- and 13C-nuclear magnetic resonance (NMR) spectra were recorded at 500 and 125 MHz, respectively, using a JEOL A500 spectrometer. One- and two-dimensional experiments were performed at ambient temperature. The sample (2 mg) was dissolved in 0.2 ml of CD3OD or CDCl3. Mass spectra were obtained using a JEOL JMS-AX500 mass spectrometer.
Structure determination.
The molecular formula of (−)-pimara-9(11),15-diene (21) was confirmed to be C20H32 by mass spectral data. The 13C-NMR δC values in CD3OD are 153.0, 151.5, 116.7, 109.6, 46.8, 43.6, 43.1, 42.3, 39.1, 38.8, 35.8, 34.4, 33.7, 30.6, 28.2, 25.7, 22.9, 22.2, 20.5, and 19.9. The structure shown in Fig. 4 was determined by comparing the 1H- and 13C-NMR spectral data with those for the 3-O-acyl derivative of pimara-9(11),15-diene, which had been recently isolated from culture broth of strain KO-3988 (30). The molecular formula of viguiepinol [3-hydroxypimara-9(11),15-diene] (41) was determined to be C20H32O by mass spectral data. The 13C-NMR δC values in CD3OD are 152.6, 151.4, 117.1, 109.6, 79.8, 46.2, 43.0, 40.3, 40.3, 38.8, 38.8, 35.9, 30.7, 28.8, 28.5, 28.1, 25.8, 23.0, 19.5, and 16.0. The structure was determined by comparing the 1H- and 13C-NMR spectral data with those for (−)-pimara-9(11),15-diene (see Fig. 4). The molecular formula of FQ D (17) was confirmed to be C22H26O6 by mass spectral data. The 13C-NMR δC values in CDCl3 are 183.7, 180.7, 160.4, 158.4, 156.9, 139.3, 134.1, 133.7, 124.4, 118.3, 110.8, 109.2, 88.7, 72.9, 60.7, 52.0, 31.9, 26.1, 18.9, 18.3, 16.1, and 9.3.
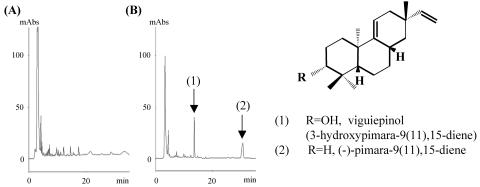
FIG. 4.
HPLC analysis of the products of S. lividans TK23 harboring pWHM-Fura1. Products of the culture broths of S. lividans TK23 harboring pWHM3 (A) and S. lividans TK23 harboring pWHM-Fura1 (B) were analyzed by HPLC. The arrows indicate peaks specifically detected in the culture broth of S. lividans TK23 harboring pWHM-Fura1. The structures of the metabolites revealed by NMR and mass spectral analyses are also shown.
Nucleotide sequence accession number.
The DNA sequences determined in this study (MV2) have been deposited in the DDBJ, EMBL, and GenBank databases with accession number AB212624.
RESULTS
Cloning of the MV2 cluster from an FQ A producer.
In order to clone an FQ biosynthetic gene cluster, we employed the following methodology, which has been recently developed. Most Streptomyces strains are equipped with only the MEP pathway for the formation of IPP, a common precursor of isoprenoids. In addition to this pathway, some Streptomyces strains possess the MV pathway, via which isoprenoid antibiotics are produced (7, 25). We have recently cloned and analyzed the MV pathway gene clusters and their flanking regions from terpentecin (12) and BE-40644 (19) producers. By analyzing the flanking regions of the MV pathway gene clusters, we could identify the biosynthetic genes of terpentecin (8, 13) and BE-40644 (19) in the regions just upstream and/or downstream of the MV pathway gene cluster. These facts suggested that all the actinomycete strains possessing both the MV and MEP pathways produce isoprenoid compounds, and the biosynthetic genes of one of these isoprenoids generally exist adjacent to the MV pathway gene cluster. Therefore, we first searched for an MV pathway gene cluster in strain KO-3988.
We had previously cloned the HMG-CoA reductase gene as a 1.6-kb BamHI fragment from Streptomyces sp. strain KO-3988 (20). By analyzing the flanking regions of the fragment, we identified an MV1 cluster comprising the MK, MDPD, PMK, type 2 IPP isomerase, HMG-CoA reductase, and HMG-CoA synthase genes arranged in a manner similar to those in actinomycetes possessing the MV and MEP pathways (20). Although we could not find any genes related to FQ biosynthesis, diterpene (viguiepinol) biosynthetic genes (see below) were observed in the flanking region of the MV1 cluster. Previously, we identified a second gene homologous to the HMG-CoA reductase gene that was used as a probe in the form of a 7.2-kb BamHI fragment (20). Therefore, we expected that strain KO-3988 had an additional MV pathway gene cluster and attempted to clone the cluster. A cosmid carrying the 7.2-kb BamHI fragment was selected by colony and Southern hybridizations, and nucleotide sequences were determined. Finally, we identified the second MV cluster, MV2, containing the same genes as those in MV1 (Fig. 1).
FIG. 1.
MV2 and its flanking regions in Streptomyces sp. strain KO-3988. ORFs deduced by sequencing analysis are shown. The bars with arrowheads show the DNA fragment used for the heterologous expression experiment. IPP iso, type 2 IPP isomerase; HMGR, HMG-CoA reductase; HMGS, HMG-CoA synthase; DAD, DNA Data Bank of Japan.
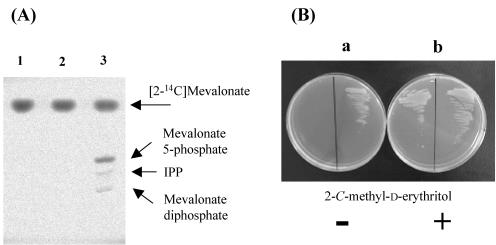
In order to examine whether the MV2 gene cluster cloned in this study would indeed encode the predicted enzymes, these genes were expressed in S. lividans and E. coli, which have only the MEP pathway for the formation of IPP. A shuttle plasmid, pFQ-MEV2, carrying the MV2 cluster was constructed and introduced into S. lividans. After cell extracts of S. lividans TK23 harboring pFQ-MEV2 were incubated with (R)-[2-14C]mevalonolactone, the products were analyzed by thin-layer chromatography. As shown in Fig. 2, spots of mevalonate-5-phosphate, mevalonate diphosphate, and IPP were clearly detected. These results showed that at least the genes that coded for MK, PMK, and MDPD were included in the DNA fragment employed.
FIG. 2.
Tracer and complementation experiments to confirm that the MV2 cluster encodes enzymes involved in the mevalonate pathway. (A) Tracer experiments using (R)-[2-14C]mevalonolactone. The enzyme reaction was performed in 100 mM potassium phosphate buffer (pH 7.3), 4 mM ATP, 6 mM MgCl2, 0.4 mM (R)-[2-14C]mevalonolactone (3.2 mCi/mmol), and a cell extract of S. lividans harboring pFQ-MEV2. Lanes: 1, reaction mixture without enzyme; 2, mixture with a cell extract of S. lividans harboring pWHM3 (vector); 3, mixture with a cell extract of S. lividans harboring pFQ-MEV2. (B) Phenotypes of the E. coli mutant DYM1 (yaeM) and its transformant harboring pFQ-MEV2. The mutant DYM1 harboring pWHM3 (vector) cannot grow on the LB plate (a, left); however, it can grow on the LB plate supplemented with 2-C-methyl-d-erythritol (0.01%) (b, left). The DYM1 strain transformed with pFQ-MEV2, which carries the gene cluster for the mevalonate pathway, was able to grow on the LB plate without 2-C-methyl-d-erythritol (with isopropyl-β-d-thiogalactopyranoside, 0.1 mM) (a, right). Since the DYM1 strain harboring pFQ-MEV2 grew poorly on the LB plate, the plate (a) shown is after incubation for 3 days at 37°C.
We have previously demonstrated that the E. coli mutant DYM1, in which the dxr gene (encoding 1-deoxy-d-xylulose 5-phosphate reductoisomerase, which is responsible for the MEP pathway and essential for cell growth) was disrupted, could be complemented by the MV pathway gene clusters because the IPP required for cell growth was supplied by the MV pathway (12, 19, 20). This strategy was also employed in this study. The DYM1 mutant was complemented by pFQ-MEV2 (Fig. 2), confirming that the DNA fragment cloned in this study contained a set of the MV pathway genes. These two heterologous expression experiments confirmed that the DNA fragment cloned in this study contained a set of the MV pathway genes.
FQ A biosynthetic genes are located at both upstream and downstream regions of the MV2 cluster.
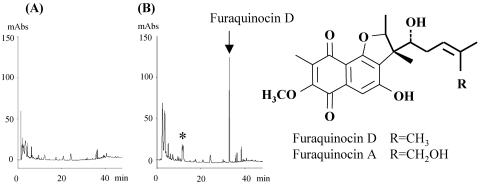
We next analyzed the flanking regions of the MV2 cluster. We identified a putative FQ A biosynthetic gene cluster in both the upstream and downstream regions of the MV2 cluster, as expected (Fig. 1). This cluster included putative genes coding for THN synthase (type III polyketide synthase), P450, methyltransferase, prenyl transferase (similar to CloQ), etc. To examine whether these genes would encode enzymes responsible for FQ A biosynthesis, a heterologous expression experiment was performed. Two DNA fragments containing the upstream region and the downstream region of the MV2 cluster were amplified by PCR, and these were then inserted into pWHM3 to yield pWHM-Fura2. S. lividans TK23 harboring pWHM-Fura2 was cultivated, and the production of a new compound was investigated by HPLC analysis. One major compound was specifically detected in the culture broth of the transformant (Fig. 3). The product was purified, and its structure was determined to be that of FQ D, a compound related to FQ A (Fig. 3), by mass spectral and NMR spectral analyses, suggesting that the genes specific to the biosynthesis of FQ A were contained in the DNA fragment that was used.
FIG. 3.
HPLC analysis of the products of S. lividans TK23 harboring pWHM-Fura2. Products of the culture broths of S. lividans TK23 harboring pWHM3 (A) and S. lividans TK23 harboring pWHM-Fura2 (B) were analyzed by HPLC. A major peak and a minor peak that were specifically detected in the culture broth of S. lividans TK23 harboring pWHM-Fura2 are indicated by the arrow and the asterisk, respectively. The structure corresponding to the former peak (FQ D) was determined by NMR and mass spectral analyses and is shown together with that of FQ A.
Viguiepinol [3-hydroxypimara-9(11),15-diene] biosynthetic genes are located in the upstream region of the MV1 cluster.
We have previously shown that genes for a putative cytochrome P450 (open reading frame 1 [ORF1]), a copalyl diphosphate synthase (ORF2), an unknown protein (ORF3), and a geranylgeranyl disphosphate (GGDP) synthase (ORF4) were located in the upstream region of the MV1 cluster (20). Using recombinant enzymes, it was confirmed that of these genes, ORF2 and ORF4 possessed the estimated enzymatic activities. However, we could not estimate the functions of ORF3, because it did not show any significant similarities with other proteins and the recombinant enzymes of ORF3 always formed inclusion bodies. Considering that copalyl diphosphate was known to be converted into several diterpene compounds in plants and fungi (28), ORF3 was expected to catalyze the conversion of copalyl diphosphate into a diterpene compound. In order to confirm this possibility and to elucidate the structure of a reaction product formed by the enzyme, a heterologous expression experiment was performed. We constructed a plasmid, pWHM-Fura1, carrying the four genes (ORF1 to ORF4) and transformed S. lividans TK23 with the plasmid. The transformant was cultivated in a liquid medium, and the metabolites were analyzed by HPLC. We specifically identified two new compounds in the culture broth of the transformant (Fig. 4). The products were purified, and their structures were determined by mass and NMR spectral analyses. Finally, one product was determined to be (−)-pimara-9(11),15-diene (21), and the other was its hydroxylated compound, viguiepinol [3-hydroxypimara-9(11),15-diene] (41). This result suggested that ORF1 and ORF3 probably encode enzymes that, respectively, catalyze a hydroxylation reaction at the 3 position of (−)-pimara-9(11),15-diene and the conversion of copalyl diphosphate into (−)-pimara-9(11),15-diene.
Phylogenetic analysis of the genes involved in the MV1 and MV2 clusters.
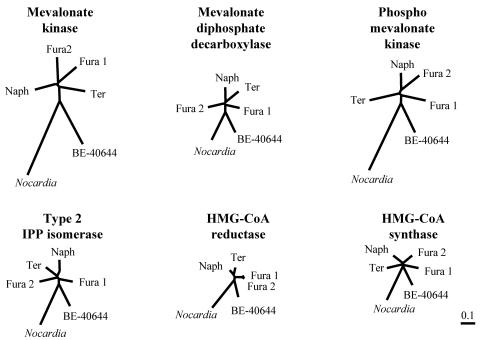
Streptomyces sp. strain KO-3988 was the first example of a microorganism equipped with two distinct MV pathway gene clusters. Therefore, a phylogenetic analysis of the genes involved in the MV1 and MV2 clusters was performed in order to examine whether the MV1 and MV2 clusters were independently distributed in strain KO-3988 or whether one cluster was generated by a duplication of the other cluster. To date, we have cloned five MV pathway gene clusters from actinomycetes. In addition, by performing whole-genome sequencing, Nocardia farcinica was recently shown to possess an MV pathway gene cluster (18). Therefore, these six clusters were used for the analyses. First, the amino acid similarities of each gene involved in these clusters were investigated by using the FASTA program (34). The MK, MDPD, PMK, type 2 IPP isomerase, HMG-CoA reductase, and HMG-CoA synthase encoded by the MV1 cluster were, respectively, 66%, 75%, 64%, 76%, 90%, and 79% identical to those encoded by the MV2 cluster. All values were almost the same as those obtained by comparing the genes involved in the MV1 cluster and those in the clusters cloned from the naphterpin producer (42) and the terpentecin producer (12). Subsequently, phylogenetic analysis was performed by using the ClustalW program (43), which provides both a multiple-sequence alignment and a phylogenetic tree based on the neighbor-joining method (36). The calculated results were visualized as unrooted trees by the TreeView program (32), as shown in Fig. 5. The genes that were cloned from the Streptomyces strains (including Kitasatospora griseola, which was formerly classified as a Streptomyces strain) were at almost the same distance from each other. On the other hand, each of the genes cloned from the genus Nocardia and the Actinoplanes strains were relatively distant from those of the Streptomyces strains. These results suggested that the MV1 and MV2 clusters are probably independently distributed in strain KO-3988.
FIG. 5.
Phylogenetic analysis of the MV pathway genes found in actinobacteria. Phylogenetic analysis was performed by using the ClustalW program, and the calculated results were visualized as unrooted trees by the TreeView program. Abbreviations: Ter, terpentecin producer Kitasatospora griseola; Naph, naphterpin producer Streptomyces sp. strain CL-190; Fura1, the MV1 cluster in strain KO-3988; Fura2, the MV2 cluster in strain KO-3988; BE-40644, BE-40644 producer Actinoplanes sp. strain A-40644; Nocardia, Nocardia farcinica IFM 10152.
DISCUSSION
In this study, we have identified the FQ D biosynthetic gene cluster by using heterologous expression in S. lividans TK23; this is the first example of a gene cluster responsible for the biosynthesis of polyketide-isoprenoid hybrid compounds. By HPLC analysis, we were able to detect one major peak of the transformant in the culture broth (Fig. 3). The compound was purified, and its structure was determined to be that of FQ D (17). Compared with FQ D, FQ A has one additional hydroxyl group at the isoprenoid moiety (Fig. 3). However, we could identify only one putative gene encoding the P450 enzyme, which generally catalyzes hydroxylation reactions in isoprenoid natural product biosynthesis, in the FQ D biosynthetic gene cluster. We therefore analyzed the upstream region of the fur1 gene and the downstream region of the fur21 gene. However, we could not identify any genes related to FQ biosynthesis, including an additional P450 gene. Subsequently, we carefully searched for FQ A, which is more hydrophilic than FQ D and would be eluted at an earlier retention time than FQ D by reverse-phase HPLC analysis, in the culture broth of the transformant, because a P450 enzyme responsible for the biosynthesis of gibberellin in fungi is known to catalyze multiple hydroxylation reactions (14). We detected one specific compound (Fig. 3) which was eluted with almost the same retention time as authentic FQ A and has almost the same UV spectrum as FQ A (23). We attempted to purify the compound; however, we could not prepare a sufficient quantity of the compound for structural analysis because of its low productivity. Although we could not obtain direct evidence that the cluster contained all the FQ biosynthetic genes, almost all genes responsible for the biosynthesis of FQs were confirmed to be involved in the cluster.
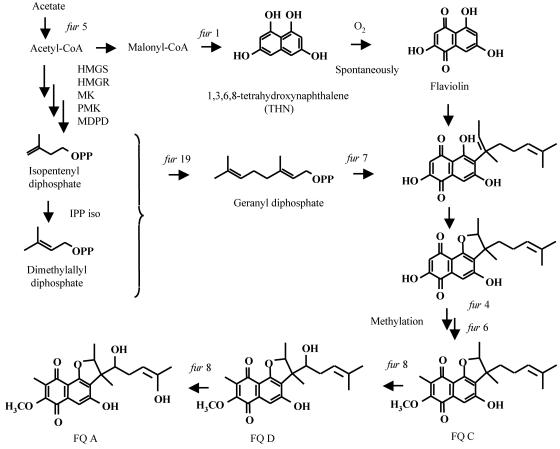
Among the genes involved in the cluster, we could estimate the functions of most of the genes which were located in the upstream region of the MV2 pathway gene cluster. These gene functions were estimated by searching databases with their translated products, such as THN synthase (fur1), C-methyltransferase (fur4), O-methyltransferase (fur6), prenylation enzyme (fur7), and P450 enzyme (fur8), by means of the sequence similarity search programs BLAST (1) and FASTA (34), as shown in Fig. 6. Moreover, a homologue of fur2 was also reported to exist in a flanking region of a type III polyketide synthase gene in Saccharopolyspora erythraea (accession number AY078067-7; 61.1% identity in a 175-amino-acid overlap), although its function remains unclear. The fur5 gene had a significant similarity to those for acyl-CoA ligases, suggesting that this gene might be responsible for the formation of acetyl-CoA from acetate. Therefore, these two genes might also participate in the biosynthesis of FQ. However, we could not estimate the function of fur3, which has significant similarity to the gene for an aminotransferase involved in streptomycin biosynthesis, because there are no amination steps in FQ biosynthesis.
FIG. 6.
Proposed biosynthetic pathway of FQ A.
In contrast, we could not annotate genes located in the downstream region of the MV2 cluster, with the exception of fur19. Considering that the fur19 gene is the only gene homologous to geranyl diphosphate synthase and that almost all genes responsible for the biosynthesis of secondary metabolites are generally clustered in a genomic region in actinomycetes, at least fur15 to fur18 were suggested to be involved in FQ biosynthesis. To examine this possibility, additional experiments are necessary. To date, some actinomycete strains are known to produce polyketide-isoprenoid hybrid compounds, such as FQ A (23), naphterpin (38), napyradiomycin A (40), and marinone (33). Among them, naphterpin (37, 42) and napyradiomycin A (39) producers have been shown to possess both the MEP and MV pathways. Therefore, biosynthetic genes of these compounds might also be present in the regions flanked by an MV pathway gene cluster, and cloning and sequencing of gene clusters of these compounds and comparison of the genes involved in these gene clusters will reveal the functions of each of the genes that we were unable to annotate in this study. Further, detailed enzymatic studies using recombinant enzymes will clarify the function of each of the genes and enable us to produce novel polyketide-isoprenoid hybrid compounds by using a combinatorial biosynthetic approach. These studies are now in progress and will be reported in the near future.
We have previously shown that ORF2, located in the upstream region of the MV1 cluster, catalyzed the conversion of GGDP into ent-copalyl diphosphate or its enantiomer, syn-copalyl diphosphate, although the absolute stereochemistry of the product remains unclear (20). However, recently we have succeeded in isolating a 3-O-acyl derivative of pimara-9(11),15-diene from the culture broth of strain KO-3988 (30). Mild alkaline treatment of this compound produced 3-hydroxypimara-9(11),15-diene, whose absolute stereochemistry was established by application of Mosher's method as shown in Fig. 4. Considering that strain KO-3988 has no additional copalyl diphosphate synthase gene other than the ORF2, the basic skeleton of the 3-O-acyl derivative of pimara-9(11),15-diene isolated from the culture broth would be synthesized by ORF2. Therefore, it is reasonable to assume that the 3-O-acyl derivative of pimara-9(11),15-diene and copalyl diphosphate formed in the in vitro experiment have the same absolute configuration. Based on this assumption, copalyl diphosphate generated by ORF2 in vitro would be ent-copalyl diphosphate, the first example of a prokaryotic enzyme with this biosynthetic function.
Previously, we did not identify any proteins homologous to ORF3 by use of the sequence similarity search programs BLAST (1) and FASTA (34). However, in this study, by performing the heterologous expression experiment, ORF3 was identified to be probably a diterpene cyclase catalyzing the conversion of ent-copalyl diphosphate to (−)-pimara-9(11),15-diene. Diterpene cyclases are classified into two major types based on their modes of cyclization (2, 28). One type of reaction is initiated by the ionization of GGDP to an allylic carbocation followed by cyclization and deprotonation to an olefin. The other type is initiated by protonation at the 14,15 double bond of GGDP. The former and the latter classes of enzymes are known to possess a DDXXD motif and a DXDD motif, respectively, which mediate substrate binding to a divalent metal ion by chelation. In eukaryotes, the former type of enzyme was known to catalyze the conversion of copalyl diphosphate to a variety of pimaradiene compounds (28). Therefore, we carefully compared ORF3 with the isoprenoid cyclases possessing the DDXXD motif by use of ClustalW (43). ORF3 was observed to possess a weak similarity to Cyc2, which was previously confirmed to convert terpentedienol diphosphate to terpentetriene in terpentecin biosynthesis (13) (15% identity) (Fig. 7). This result also suggested that ORF3 is probably a diterpene cyclase catalyzing the conversion of ent-copalyl diphosphate to (−)-pimara-9(11),15-diene, although an in vitro experiment is essential to draw a conclusion.
FIG. 7.
Alignment of (−)-pimara-9(11),15-diene synthase (ORF3) and Cyc2, which catalyzes the conversion of terpentedienol diphosphate into terpentetriene in terpentecin biosynthesis. Identical and similar amino acid residues are indicated by asterisks and colons, respectively. P, (−)-pimara-9(11),15-diene synthase; T, Cyc2. Amino acid residues that are known to be substrate binding sites are underlined.
Acknowledgments
We thank S. Omura of the Kitasato Institute for providing us with the FQ A-producing microorganism and B. Ikeda and K. Gankai for their excellent technical assistance.
This work was supported in part by a Grant-in-Aid for Scientific Research (B) and grants from the Naito Foundation, the Uehara Memorial Foundation, and the Novozymes Research Foundation to T. Dairi; a Grant-in-Aid for Scientific Research (B) to H. Seto; and a Grant-in-Aid for Scientific Research (A) to T. Sassa of Yamagata University.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Bohlmann, J., G. Meyer-Gauen, and R. Croteau. 1998. Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. USA 95:4126-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cane, D. E. 1999. Sesquiterpene biosynthesis, p. 155-200. In D. Barton, K. Nakanishi, and O. Meth-Cohn (ed.), Comprehensive natural products chemistry, vol. 2. Elsevier, Oxford, United Kingdom. [Google Scholar]
- 4.Cane, D. E., J. K. Sohng, C. R. Lamberson, S. M. Rudnicki, Z. Wu, M. D. Lloyd, J. S. Oliver, and B. R. Hubbard. 1994. Pentalenene synthase. Purification, molecular cloning, sequencing, and high-level expression in Escherichia coli of a terpenoid cyclase from Streptomyces UC5319. Biochemistry 33:5846-5857. [DOI] [PubMed] [Google Scholar]
- 5.Chappell, J. 2002. The genetics and molecular genetics of terpene and sterol origami. Curr. Opin. Plant Biol. 5:151-157. [DOI] [PubMed] [Google Scholar]
- 6.Connolly, J. D., and R. A. Hill. 1992. Dictionary of terpenoids. Chapman and Hall, New York, N.Y.
- 7.Dairi, T. 2005. Studies on biosynthetic genes and enzymes of isoprenoids produced by actinomycetes. J. Antibiot. 58:227-243. [DOI] [PubMed] [Google Scholar]
- 8.Dairi, T., Y. Hamano, T. Kuzuyama, N. Itoh, K. Furihata, and H. Seto. 2001. Eubacterial diterpene cyclase genes essential for production of the isoprenoid antibiotic terpentecin. J. Bacteriol. 183:6085-6094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dairi, T., Y. Motohira, T. Kuzuyama, S. Takahashi, N. Itoh, and H. Seto. 2000. Cloning of the gene encoding 3-hydroxy-3-methylglutaryl coenzyme A reductase from terpenoid antibiotic-producing Streptomyces strains. Mol. Gen. Genet. 262:957-964. [DOI] [PubMed] [Google Scholar]
- 10.Gerber, N. N. 1969. A volatile metabolite of actinomycetes, 2-methylisoborneol. J. Antibiot. 22:508-509. [DOI] [PubMed] [Google Scholar]
- 11.Gerber, N. N., and H. A. Lechevalier. 1965. Geosmin, an earthly-smelling substance isolated from actinomycetes. Appl. Microbiol. 13:935-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamano, Y., T. Dairi, M. Yamamoto, T. Kawasaki, K. Kaneda, T. Kuzuyama, N. Itoh, and H. Seto. 2001. Cloning of a gene cluster encoding enzymes responsible for the mevalonate pathway from a terpenoid-antibiotic-producing Streptomyces strain. Biosci. Biotechnol. Biochem. 65:1627-1635. [DOI] [PubMed] [Google Scholar]
- 13.Hamano, Y., T. Kuzuyama, N. Itoh, K. Furihata, H. Seto, and T. Dairi. 2002. Functional analysis of eubacterial diterpene cyclases responsible for biosynthesis of a diterpene antibiotic, terpentecin. J. Biol. Chem. 277:37098-37104. [DOI] [PubMed] [Google Scholar]
- 14.Hedden, P., A. L. Phillips, M. C. Rojas, E. Carrera, and B. Tudzynski. 2002. Gibberellin biosynthesis in plants and fungi: a case of convergent evolution? J. Plant Growth Regul. 20:319-331. [DOI] [PubMed] [Google Scholar]
- 15.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schremp. 1985. Gene manipulation of Streptomyces, a laboratory manual. The John Innes Foundation, Norwich, United Kingdom.
- 16.Imai, S., K. Furihata, Y. Hayakawa, T. Noguchi, and H. Seto. 1997. Lavanducyanin, a new antitumor substance produced by Streptomyces sp. J. Antibiot. 42:1196-1198. [DOI] [PubMed] [Google Scholar]
- 17.Ishibashi, M., S. Funayama, Y. Anraku, K. Komiyama, and S. Omura. 1991. Novel antibiotics, furaquinocins C, D, E, F, G and H. J. Antibiot. 44:390-395. [DOI] [PubMed] [Google Scholar]
- 18.Ishikawa, J., A. Yamashita, Y. Mikami, Y. Hoshino, H. Kurita, K. Hotta, T. Shiba, and M. Hattori. 2004. The complete genomic sequence of Nocardia farcinica IFM 10152. Proc. Natl. Acad. Sci. USA 101:14925-14930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawasaki, T., T. Kuzuyama, K. Furihata, N. Itoh, H. Seto, and T. Dairi. 2003. A relationship between the mevalonate pathway and isoprenoid production in actinomycetes. J. Antibiot. 56:957-966. [DOI] [PubMed] [Google Scholar]
- 20.Kawasaki, T., T. Kuzuyama, Y. Kuwamori, N. Matsuura, N. Itoh, K. Furihata, H. Seto, and T. Dairi. 2004. Presence of copalyl diphosphate synthase gene in an actinomycete possessing the mevalonate pathway. J. Antibiot. 57:739-747. [DOI] [PubMed] [Google Scholar]
- 21.Kim, Y. H., B. S. Chung, and U. Sankawa. 1988. Pimaradiene diterpenes from Acanthopanax koreanum. J. Nat. Prod. 51:1080-1083. [Google Scholar]
- 22.Komaki, H., A. Nemoto, Y. Tanaka, K. Yazawa, Y. Mikami, H. Shigemori, J. Kobayashi, A., Ando, and Y. Nagata. 1999. Brasilicardin A, a new terpenoid antibiotic from pathogenic Nocardia brasiliensis: fermentation, isolation and biological activity. J. Antibiot. 52:13-19. [DOI] [PubMed] [Google Scholar]
- 23.Komiyama, K., S. Funayama, Y. Anraku, M. Ishibashi, Y. Takahashi, and S. Omura. 1990. Novel antibiotics, furaquinocins A and B. Taxonomy, fermentation, isolation and physico-chemical and biological characteristics. J. Antibiot. 43:247-252. [DOI] [PubMed] [Google Scholar]
- 24.Kuzuyama, T., J. P. Noel, and S. B. Richard. 2005. Structural basis for the promiscuous biosynthetic prenylation of aromatic natural products. Nature 435:983-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuzuyama, T., and H. Seto. 2003. Diversity of the biosynthesis of the isoprene units. Nat. Prod. Rep. 20:171-183. [DOI] [PubMed] [Google Scholar]
- 26.Kuzuyama, T., S. Takahashi, and H. Seto. 1999. Construction and characterization of Escherichia coli disruptants defective in the yaeM gene. Biosci. Biotechnol. Biochem. 63:776-778. [DOI] [PubMed] [Google Scholar]
- 27.Li, S. M., and L. Heide. 2004. Functional analysis of biosynthetic genes of aminocoumarins and production of hybrid antibiotics. Curr. Med. Chem. Anti-Infect. Agents 3:279-295. [Google Scholar]
- 28.MacMillan, J., and M. H. Beale. 1999. Diterpene biosynthesis, p. 217-243. In D. Barton, K. Nakanishi, O. Meth-Cohn (ed.), Comprehensive natural products chemistry, vol. 2. Elsevier, Oxford, United Kingdom. [Google Scholar]
- 29.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Motohashi, K., R. Ueno, T. Miyamoto, K. Furihata, T. Dairi, and H. Seto. Submitted for publication.
- 31.Nakanishi, S., K. Osawa, Y. Saito, I. Kawamoto, K. Kuroda, and H. Kase. 1992. KS-505a, a novel inhibitor of bovine brain Ca2+ and calmodulin-dependent cyclic-nucleotide phosphodiesterase from Streptomyces argenteolus. J. Antibiot. 45:341-347. [DOI] [PubMed] [Google Scholar]
- 32.Page, R. D. M. 1996. TREEVIEW. An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 33.Pathirana, C., P. R. Jensen, and W. Fenical. 1992. Marinone and debromomarinone: antibiotic sesquiterpenoid naphthoquinones of a new structure class from a marine bacterium. Tetrahedron Lett. 33:7663-7666. [Google Scholar]
- 34.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence comparison. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poralla, K., G. Muth, and T. Hartner. 2000. Hopanoids are formed during transition from substrate to aerial hyphae in Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 189:93-95. [DOI] [PubMed] [Google Scholar]
- 36.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 37.Seto. H., H. Watanabe, and K. Furihata. 1996. Simultaneous operation of the mevalonate and non-mevalonate pathways in the biosynthesis of isopentenyl diphosphate in Streptomyces aeriouvifer. Tetrahedron Lett. 37:7979-7982. [Google Scholar]
- 38.Shin-ya, K., S. Imai, K. Furihata, Y. Hayakawa, Y. Kato, G. D. VanDuyne, J. Clardy, and H. Seto. 1990. Isolation and structural elucidation of an antioxidative agent, naphterpin. J. Antibiot. 43:444-447. [DOI] [PubMed] [Google Scholar]
- 39.Shiomi, K., H. Iinuma, H. Naganawa, K. Isshiki, T. Takeuchi, and H. Umezawa. 1987. Biosynthesis of napyradiomycins. J. Antibiot. 40:1740-1745. [DOI] [PubMed] [Google Scholar]
- 40.Shiomi, K., H. Nakamura, H. Iinuma, H. Naganawa, K. Isshiki, T. Takeuchi, H. Umezawa, and Y. Iitaka. 1986. Structures of new antibiotics napyradiomycins. J. Antibiot. 39:494-501. [DOI] [PubMed] [Google Scholar]
- 41.Soriano-Garcia, M., C. Guerrero, and R. A. Toscano. 1986. Structure and stereochemistry of (3R,5R,8S,10R,13R)-ent-pimera-9(11),15-dien-3p-bromobenzoate (viguiepinol). Acta Crystallogr. C 42:729-731. [Google Scholar]
- 42.Takagi, M., T. Kuzuyama, S. Takahashi, and H. Seto. 2000. A gene cluster for the mevalonate pathway from Streptomyces sp. strain CL190. J. Bacteriol. 182:4153-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTALW. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vara, J., M. Lewandowska-Skarbek, Y. G. Wang, S. Donadio, and C. R. Hutchinson. 1989. Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus). J. Bacteriol. 171:5872-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wise, M. L., and R. Croteau. 1999. Monoterpene biosynthesis, p. 97-153. In D. Barton, K. Nakanishi, O. Meth-Cohn (ed.), Comprehensive natural products chemistry, vol. 2. Elsevier, Oxford, United Kingdom. [Google Scholar]