Abstract
Under iron stress, Legionella pneumophila secretes legiobactin, a nonclassical siderophore that is reactive in the chrome azurol S (CAS) assay. Here, we have optimized conditions for legiobactin expression, shown its biological activity, and identified two genes, lbtA and lbtB, which are involved in legiobactin production. lbtA appears to be iron repressed and encodes a protein that has significant homology with siderophore synthetases, and FrgA, a previously described iron-regulated protein of L. pneumophila. lbtB encodes a protein homologous with members of the major facilitator superfamily of multidrug efflux pumps. Mutants lacking lbtA or lbtB were defective for legiobactin, producing 40 to 70% less CAS reactivity in deferrated chemically defined medium (CDM). In bioassays, mutant CDM culture supernatants, unlike those of the wild type, did not support growth of iron-limited wild-type bacteria in 2′,2′-dipyridyl-containing buffered charcoal yeast extract (BCYE) agar and a ferrous iron transport mutant on BCYE agar without added iron. The lbtA mutant was modestly defective for growth in deferrated CDM containing the iron chelator citrate, indicating that legiobactin is required in conditions of severe iron limitation. Complementation of the lbt mutants restored both siderophore expression, as measured by the CAS assay and bioassays, and bacterial growth in deferrated, citrate-containing media. The lbtA mutant replicated as the wild type did in macrophages, amoebae, and the lungs of mice. However, L. pneumophila expresses lbtA in the macrophage, suggesting that legiobactin, though not required, may play a dispensable role in intracellular growth. The discovery of lbtAB represents the first identification of genes required for L. pneumophila siderophore expression.
The gram-negative organism Legionella pneumophila is an inhabitant of natural and man-made aquatic environments (24). Importantly, it is also the primary agent of Legionnaires' disease, a serious form of pneumonia that often afflicts the immunocompromised (10, 24). In aquatic habitats, L. pneumophila survives free, in biofilms, and as an intracellular parasite of protozoa such as amoebae (32, 46, 71, 74, 79). In the lung, the organism replicates within alveolar macrophages (1, 66, 79, 83).
Iron has long been recognized as a key requirement for L. pneumophila replication, intracellular infection, and virulence (6, 13, 28, 34, 37, 45, 61). For many years, it was believed that L. pneumophila does not make siderophores, a conclusion based upon results obtained by Arnow and Csáky assays, which identify catecholate and hydroxamate structures, as well as the chrome azurol S (CAS) assay, which detects iron chelators independently of structure (36, 40, 62). The story of Legionella siderophores changed when we showed that L. pneumophila could produce a high-affinity iron chelator (39). When grown at 37°C in a low-iron chemically defined medium (CDM), L. pneumophila secretes a low-molecular-weight substance that is reactive in the CAS assay. The siderophore-like activity is iron repressed and is only observed when the CDM is inoculated with legionellae that had been grown to log or early stationary phase (39). Inocula from the late stationary phase, despite growing in the CDM, fail to result in CAS reactivity. CAS reactivity was seen with serogroup 1 strains 130b and Philadelphia-1, as well as isolates representing nine other L. pneumophila serogroups (39). We designated the iron-chelating activity in L. pneumophila supernatants as legiobactin (39), and later observed siderophore activity in the supernatants of 18 other Legionella species (75).
The first gene to be examined as a possible promoter of legiobactin production was frgA, a Fur-regulated gene whose predicted product had initially been shown to be homologous with IucA and IucC, two enzymes involved in aerobactin biosynthesis in Escherichia and Shigella species (35). Current BlastP results indicate that FrgA also shares homology with siderophore biosynthetic genes of Bordetella bronchiseptica, Erwinia chrysanthemi, Sinorhizobium meliloti, Staphylococcus aureus, and Vibrio parahaemolyticus (17, 26, 29, 43, 80). Although FrgA is homologous with hydroxamate synthetases and frgA promotes growth within the iron-limited intracellular niche in macrophages (35), frgA mutants proved not to be defective for CAS reactivity when grown in low-iron CDM (39). We also observed that the cytochrome c maturation operon, which promotes L. pneumophila growth in low-iron media as well as siderophore expression in several other types of bacteria, is not required for legiobactin production (47, 82). In another recent study, the feoB ferrous iron transporter showed no role in the generation of L. pneumophila CAS reactivity (63). Thus, the genetic basis of legiobactin production has remained unknown and served as the goal for the present study.
Here, we optimized conditions for legiobactin expression and developed a bioassay for the siderophore. Then, we identified two L. pneumophila genes, lbtA and lbtB, which are highly related to hydroxamate synthetases and permeases, respectively, of the major facilitator superfamily (MFS) class of proton motive force-dependent membrane efflux pumps. Mutational analysis revealed that lbtA and lbtB are required for optimal legiobactin production and as such represent our first insight into the genetics of Legionella siderophore expression.
MATERIALS AND METHODS
Bacterial strains.
L. pneumophila strain 130b (American Type Culture Collection strain BAA-74, also known as AA100) was used for mutagenesis of lbtA, lbtB, lbtC, pvcA, and pvcB and served as a wild-type control (22, 35). Other L. pneumophila strains and Legionella species tested for the presence of lbtA, frgA, and siderophore activity are listed in Table 1. The L. pneumophila frgA mutant NU229 as well as NU232 were previously isolated during a screen for iron-regulated genes (35). That NU232 has a mini-Tn10′lacZ fusion in frgA and is defective for macrophage infection was determined during the present study (data not shown). L. pneumophila ira mutants, 130b derivatives that appear impaired for iron acquisition, were described before (59, 81). The L. pneumophila feoB mutant NU269 was described in a previous study (63). Escherichia coli DH5α and DH5α λ pir were used as hosts for recombinant plasmids (Invitrogen, Carlsbad, Calif.).
TABLE 1.
lbtA, frgA, CAS reactivity, and feoB bioassay activity in Legionella strainsa
| Species | Strain | Hybridizationb
|
Growth in CDM lacking Fe | CAS reactivity | Bioassay activity | |
|---|---|---|---|---|---|---|
| lbtAb | frgAb | |||||
| L. pneumophila | Variousc | +++ | +++ | +++ | +++ | + |
| L. adelaidensis | 49625 | − | − | − | ? | − |
| L. anisa | 35292 | ++ | − | +++ | +++ | + |
| L. birminghamensis | 43702 | − | + | ++ | ++ | − |
| L. bozemanii | 33217 | ++ | − | ++ | ++ | + |
| L. brunensis | 43878 | ++ | − | ++ | ++ | − |
| L. cherrii | 35252 | + | − | − | ? | − |
| L. cincinnatiensis | 43753 | ++ | − | − | ? | − |
| L. erythra | 35303 | ++ | − | + | +/−d | + |
| L. fairfieldensis | 49588 | − | − | − | ? | − |
| L. feeleii | 35072 | ++ | − | +++ | ++ | + |
| L. gratiana | 49413 | − | ++ | − | ? | ND |
| L. hackeliae | 35250 | ++ | − | − | ? | − |
| L. jamestowniensis | 35298 | ++ | − | +++ | ++ | − |
| L. jordanis | 33623 | − | − | +/−d | ? | − |
| L. londiniensis | 49505 | − | − | +/−d | +/−d | − |
| L. longbeachae | 33462 | ++ | − | − | ? | ND |
| L. micdadei | 33218 | ++ | − | +++ | ++ | − |
| L. moravica | 43877 | ++ | − | ++ | + | + |
| L. oakridgensis | 33761 | ++ | − | − | ? | + |
| L. parisiensis | 35299 | ++ | − | ++ | ++ | ND |
| L. quinlivanii | 43830 | − | − | ++ | ++ | − |
| L. rubrilucens | 35304 | ++ | − | +++ | +++ | + |
| L. sainthelensi | 35248 | ++ | − | +++ | +++ | − |
| L. santicrusis | 35301 | ++ | − | + | ++ | + |
| L. spiritensis | 35249 | + | − | ++ | ++ | ND |
| L. steigerwaltii | 35302 | + | ND | ++ | +++ | − |
| L. wadsworthii | 33877 | ++ | − | ++ | ++ | ND |
For the origin of strains and the association of a strain with disease, see reference 75 and the references therein. The strain designations represent American Type Culture Collection strain numbers.
+++, hybridization to lbtA or frgA probe under high-stringency conditions; ++, moderate; +, weak; −, no hybridization to the probe under low-stringency conditions. In all cases, the lbtA-containing fragment was distinct from the frgA-containing fragment. ND, not determined.
L. pneumophila strains examined include strains BAA-74 (130b), 33217 (Philadelphia-1), 33154, 33155, 33156, 33216, 33823, 35096, 43736, and 43703.
The growth and/or CAS reactivity of this strain varied, for unknown reasons.
Bacteriological media and extracellular growth experiments.
Legionella strains were routinely grown at 37°C on buffered charcoal yeast extract (BCYE) agar or in buffered yeast extract (BYE) broth (41). These media contain an iron supplement consisting of 0.25 g of ferric pyrophosphate per liter. For selection of allelic exchange mutants, the BCYE agar was supplemented with 5% sucrose as well as kanamycin at 25 μg/ml, gentamicin at 2.5 μg/ml and chloramphenicol at 6 μg/ml. E. coli strains were grown in Luria-Bertani medium, with kanamycin (50 μg/ml), gentamicin (2.5 μg/ml) chloramphenicol (30 μg/ml), or ampicillin (100 μg/ml). Unless otherwise noted, chemicals were purchased from Sigma-Aldrich Chemical Co. (St. Louis, Mo.).
In order to assess siderophore production, legionellae were typically grown in CDM that lacked its iron component (39). The iron-deplete CDM was made using water that had been deferrated by passage through a Chelex-100 (Bio-Rad, Laboratories, Hercules, Calif.) column (15). The deferration of media was confirmed by the ferrozine assay (data not shown) (78). To further control the amount of iron in media, acid-washed glassware was used (15). To monitor the general extracellular growth capacity of L. pneumophila strains, bacteria grown on BCYE agar were inoculated into BYE broth, and the optical density of the resulting cultures was determined at 660 nm (OD660) (35, 63, 81). To assess extracellular growth under iron-limiting conditions, L. pneumophila was inoculated into either BYE broth that lacked its iron supplement, Chelex-treated BYE broth, deferrated CDM, or deferrated CDM with 10 μM to 2 mM citrate, 5 to 30 μM deferoxamine mesylate (DFX) or 5 to 30 μM ethylenediamine di(o-hydroxyphenylacetic acid) (EDDA) and then the OD660 of the cultures was monitored (63, 82). To examine growth and survival on iron-limited solid media, legionellae were tested for their ability to form colonies on BCYE agar that lacked its iron supplement (63). In addition, we tested L. pneumophila strains for their capacity to form colonies within nonsupplemented BCYE agar that had been made even more iron limited by the inclusion of 100 to 400 μM 2,2′-dipyridyl (DIP).
Siderophore assays.
Legionella culture supernatants were tested for siderophore activity using the CAS assay as previously described (39, 56, 75). Supernatants were also tested for catecholate and hydroxamate structures by the Arnow and Csáky assays (3, 16, 39, 56, 70). DFX was the standard for the CAS and Csáky assays, while 2,3-dihydroxybenzoic acid served that role for the Arnow procedure (39, 56). Low-molecular-weight fractions were obtained by passage of supernatants through Centricon filters (Millipore, Bedford, Mass.) having a 3-kDa size limit. The heat and protease susceptibility of the Legionella CAS reactivity were determined by either boiling for 5 min or incubation with proteinase K at 1 mg/ml for 3 h at 37°C (39).
Bioassays for L. pneumophila siderophores.
CAS-positive supernatants were tested for their ability to promote the growth of either wild-type 130b in non-iron-supplemented BCYE agar containing 100 to 600 μM of the iron chelator DIP or feoB mutant bacteria on non-iron-supplemented BCYE agar. To obtain the CAS-positive samples, strain 130b was grown in BYE to log phase (i.e., OD660 of 1.0), washed, and then inoculated into 20 ml of deferrated CDM to an OD660 of 0.25. After 20 h of incubation, a period of time sufficient to yield significant CAS reactivity (39), the culture was centrifuged at 3,000 rpm in a Beckman J2-21 for 10 min, and the supernatants were collected and passed through a 0.2-μm filter (Millipore, Bedford, Mass.).
To obtain a low-molecular-weight fraction, 2 ml of the sterile supernatant was passed through a 3-kDa-cutoff Centricon filter. To finally assess growth-stimulating activity, 50 μl of the sample was placed into a well cut out of the center of either the DIP-containing BCYE seeded with 104 CFU per ml of wild type or the non-iron-supplemented BCYE agar unto which had been spread 105 CFU of the feoB mutant. As a positive control, we tested the stimulatory activity of 50 μl of 10 mM FeCl3 (in water). For negative controls, we monitored growth around wells containing 50 μl of either deferrated CDM or a <3-kDa fraction obtained from a CAS-negative supernatant of strain 130b.
To obtain the CAS-negative fraction, 130b bacteria were grown in BYE to stationary phase (OD660 of 2.1), inoculated into deferrated CDM to an OD660 of 0.25, and then incubated for 20 h (39). The growth of the wild-type bacteria was observed upon incubation at room temperature, whereas the feoB mutant plates were examined at 37°C. To assess the direct growth-stimulating ability of L. pneumophila strains and other Legionella species, legionellae were grown on non-iron-supplemented BCYE agar at 37°C for 2 to 3 days and spotted with a sterile stick onto non-iron-supplemented BCYE agar with or without 2 mM isopropylthiogalactopyranoside (IPTG) onto which had been spread 105 CFU of the feoB mutant. The plates were then incubated and examined.
DNA isolation and sequencing analysis.
DNA was isolated from L. pneumophila as described previously (12). DNA sequencing was done by the Northwestern Biotech Laboratory. Primers were obtained from Integrated DNA Technologies (Coralville, IA). Nucleotide sequences were analyzed with Seqman (DNAStar; Madison, WI), and BLAST homology searches were conducted through GenBank at the National Center for Biotechnology Information. Protein alignments were performed by the BCM Search Launcher: Multiple Sequence Alignments (http://searchlauncher.bcm.tmc.edu/multi align/multi-align.html); and Boxshade 3.21 (http://www.ch.embnet.org/software/BOX_form.html). DNA motifs and structural analyses were conducted using the Prosite prediction model (http://us.expasy.org/prosite) and SOSUI program (http://sosui.proteome.bio.tuat.ac.jp).
RT-PCR analysis of L. pneumophila gene expression.
Reverse transcription (RT)-PCR was performed as before (42, 82). Legionella RNA was isolated from CDM-cultured bacteria or L. pneumophila-infected macrophages using RNA STAT-60 (TEL-TEST B, Inc, Friendswood, TX). Primers MIP1-F (5′-AAAGGCATGCAAGACGCTAA) and MIP1-R (5′-GTATCCGATTTTCCGGGTTT) were used to amplify a 260-bp internal fragment of mip (11) and LBTA-F (5′CATTTGATCGATGGCCTCTT) and LBTA-R (5′-GCGCGGAAATTAGGATGATA) were used to amplify a 226-bp fragment of lbtA. Control experiments in which the reverse transcriptase enzyme was omitted from the reaction were performed to rule out contributions of contaminating DNA in the DNase-treated RNA samples. These controls were performed with lbtA primers described above.
Mutation and complementation analysis.
To obtain a mutated lbtA gene with a kanamycin resistance (Kmr) or gentamicin resistance (Gmr) marker, an internal fragment of the L. pneumophila strain 130b lbtA gene was amplified by PCR using primers LBTA9-F (5′-ATACTGCCATAGCATCGGG) and LBTA8-R (5′-TTTTCAAAAACTGACCTGA). The resultant 2-kb DNA fragment was then cloned into pGemTeasy (Promega, Madison, WI), to give plasmid pVK120. The cloned L. pneumophila lbtA was mutated by deletion of a 51-bp fragment at the two MfeI sites, the first located 563 bp from the lbtA start site of pVK120 and following Klenow treatment, the insertion of either a 1.1-kb Kmr gene isolated from pMB2190 upon HincII digestion (31) to yield pVK121 or a Gmr gene isolated from pX1918G after HincII and PvuII digestion (69) to give pKA7.
The mutated Kmr lbtA gene was then cloned on a 3.3-kb SacI-SphI fragment from pVK121 into the counterselectable pBOC20, to yield pVK122. The chloramphenicol-resistant (Cmr) pBOC20 is a ColE1 replicon that facilitates allelic exchange in L. pneumophila by virtue of its sacB gene (49). The mutated Gmr lbtA gene was also cloned on a 3.3-kb SacI-SphI fragment into the counterselectable pRE112, to give pKA11. The pRE112 suicide vector has a conditional R6K ori that requires the π protein to replicate (21).
In addition, an unmarked, nonpolar lbtA deletion mutant was constructed. In this case, the lbtA gene was cloned on a 2.1-kb KpnI-XbaI fragment from pVK120 into pBluescriptII KS(+) (Stratagene, La Jolla, Calif.), to give plasmid pKA2. The cloned lbtA gene was mutated by removal of a 626-bp MfeI-BglII fragment, 563 bp from the lbtA start site, blunted by T4 polymerase and religated to give plasmid pKA16. The mutated lbtA gene was then cloned on a 1.4-kb KpnI-XbaI fragment into the counterselectable pRE112, to yield pKA17.
To obtain a mutated lbtB gene with a Gmr marker, an internal fragment of the L. pneumophila strain 130b with the lbtBC genes was amplified by PCR using primers LBTA7-F (5′-GCCACGAATTGAGTGATCT) and LBTC1-R (GACTGGAGGTGATACGGAAT). The resultant 2-kb DNA fragment was then cloned into pGemTeasy to give plasmid pKA4. The cloned L. pneumophila lbtB was mutated by insertion of a 1.1-kb Gmr gene on a HincII-PvuII fragment from PX1918G at the T4 polymerase-treated HindIII site of pKA4, 440 bp from the start site, resulting in pKA8. Mutated lbtB was then cloned on a 4.4-kb SacI-SphI fragment into pRE112, to yield pKA12. The mutated lbtC gene was obtained by inserting the same Gmr gene at the T4 polymerase-treated KpnI site of pKA4, 739 bp from the gene's start site, resulting in pKA9.
Multiple lbtC mutants were obtained through natural transformation of 130b with pKA9. Plasmid pKA11 was also used to construct lbtA frgA and lbtA pvcA double mutants. The lbtA frgA double mutant was constructed by introducing the lbtA::Gmr on pKA11 into the previously described Kmr NU229. Attempts to make an lbtA feoB double mutant were carried out by introducing pKA11 into the Kmr NU269 in the presence or absence of legiobactin-containing supernatant. To obtain a mutated pvcA and pvcB, a fragment containing pvcA and pvcB was amplified by PCR from 130b with primers PVCA9-F (5′-CGATAGTGACTCTGCTATGG) and PVCB6-R (5′-GGCAAGAGTCGTAAGACATC). The resultant 2.3-kb DNA fragment was then cloned into pGemTeasy to give plasmid pJSA1.
The cloned pvcA was mutated by the insertion of a Kmr gene PCR amplified from pVK3 (81) using primers USKanBam2 (5′-CGCGGATCCAAGCCACGTTGTGTCTCA) and DSKanBam2 (5′-CGCGGATCCAGAAGGTGTTGCTGACTCAT) at the first BglII-digested site of pJSA1 after a 34-bp deletion, to give pJS5. A mutated pvcB gene was generated by insertion of the PCR-amplified Kmr gene after BamHI digestion of pJSA1, to give plasmid pJA6. Multiple pvcA and pvcB mutants were obtained by natural transformation of 130b with pJS5 and pJS6, respectively (25, 77). The lbtA pvcA double mutant was constructed in the Kmr pvcA background by introducing pKA11 with lbtA::Gmr.
Production of competent 130b, NU229, lbtA, lbtB, and pvcA mutant cells and electroporation of pVK122, pKA17, pKA12, and pKA11 into L. pneumophila were achieved as previously described (12, 63). Potential mutants were selected based on Cm sensitivity, sucrose resistance, Km resistance (pVK122), and Gm resistance (pKA11 and pKA12) indicative of the introduction of the mutated gene into the 130b, NU229, or pvcA mutant chromosome by homologous recombination. Verification of the Kmr lbtA mutant genotype was carried out by PCR and Southern hybridization, using the same primers and DNA probe used to identify pVK122 (LBTA9-F and LBTA8-R). The lbtA deletion and double mutant phenotypes were verified by PCR using the same primers to identify lbtA on pVK122, pKA11, and pKA17. The lbtB and lbtC mutant phenotypes were verified by PCR with primers LBTA7-F and LBTB1-R (5′-ACTAATGATGGCAAGGCTGG) (lbtB) and LBTA7-F and LBTC1-R (lbtC). The pvcA and pvcB mutants were verified by PCR and Southern hybridization with the same primers used to identify pJSA1.
To facilitate complementation, a 2.1-kb KpnI-XbaI-digested fragment containing only the lbtA gene with its endogenous promoter was obtained from pKA2 and cloned under control of the tac promoter in pMMB2002 (65) to yield plbtA. For complementation of the lbtB mutants, a 1.6-kb fragment containing only the lbtB gene was amplified by PCR from L. pneumophila 130b DNA using primers LBTA7-F and LBTB1-R and cloned into pGemTeasy (Promega; Madison, WI) to yield pKA14. Finally, the wild-type lbtB gene was cloned on a 1.7-kb SacI-SphI fragment from pKA14 into pMMB2002 under the control of the tac promoter to yield plbtB. Complementation with plbtB was obtained when lbtB was under the control of the tac promoter and induced with 2 mM IPTG (68). Plasmids were electroporated into L. pneumophila strains as previously described (41).
Infection assays.
To examine the ability of L. pneumophila to grow intracellularly, Hartmannella vermiformis amoebae and human U937 cells were infected as previously described (2, 12, 63, 65). Infection of iron-depleted macrophages and amoebae was accomplished by the addition of 10 to 40 μM DIP or 10 to 160 μM DFX to the medium for 24 h prior to infection with L. pneumophila and/or during the incubation period (7, 28, 59, 63, 81, 82). We observed that as much as 160 μM DFX and 35 μM DIP in H. vermiformis cocultures and 5 μM DFX and 20 μM DIP in U937 macrophages had no effect on the growth of 130b (data not shown). To assess the virulence of bacteria, competition assays were done following intratracheal inoculation of A/J mice, as described previously (63-65).
Southern hybridization analysis.
Southern blots were carried out using EcoRI-restricted DNA from strains representing several L. pneumophila serogroups and a variety of Legionella spp. A digoxigenin nonradioactive labeling and detection system was used (Roche Molecular Biochemicals, Indianapolis, IN). The lbtA probe was produced by PCR incorporation according to the manufacturer's recommendations using primers LBTA1-F (5′-GCAGCACTTCGTGAAGGAT) and LBTA4-R (TAGGTACAGCAAGGCTTGC) and 130b DNA as a template. The frgA probe was generated by restriction digest of plasmid pEH44 and labeling as previously described (35). High-stringency washes (0 to 10% base pair mismatch) were employed for hybridization to L. pneumophila DNAs and low-stringency washes (∼30% base pair mismatch) were used for hybridization to genomic DNA from other Legionella spp (35).
Nucleotide sequence accession number.
The NCBI and GenBank accession number for the L. pneumophila lbtA gene is DQ118422.
RESULTS
Optimization and kinetics of siderophore expression by L. pneumophila 130b.
By acid-washing glassware and by deferrating the water used to make the CDM, we obtained culture supernatants that, based upon the CAS assay, consistently yielded ≥900 μM net DFX equivalents, with the highest values approaching 1,500 μM equivalents. The ability of strain 130b to elaborate CAS reactivity that was <3 kDa and heat- and protease-resistant still required the use of log-phase inocula. The increased level of siderophore activity observed in deferrated cultures was not associated with the expression of an Arnow- or Csáky-reactive material, suggesting that it is not due to turn-on of a “typical” catecholate or hydroxamate siderophore.
Mixtures of supernatants with siderophores DFX and DHB retained positivity in the structural assays, indicating L. pneumophila is not elaborating a substance that interferes with siderophore recognition. Since cysteine is reactive in the CAS assay (40), we optimized the detection of legiobactin by replacing the cysteine in deferrated CDM with cystine, a substance that is not CAS reactive (40). Supernatants obtained from cystine-containing cultures consistently displayed ≥900 μM DFX equivalents while having no background reactivity (data not shown). Thus, all subsequent legiobactin determinations were made using cystine-containing deferrated CDM and are presented as net DFX equivalents.
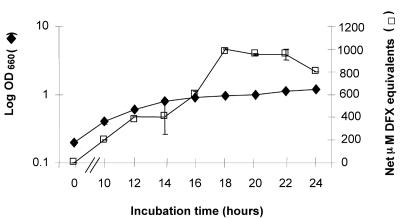
To better understand the kinetics of legiobactin production, we examined strain 130b cultures for CAS reactivity at multiple early time points (Fig. 1). Legiobactin expression was detectable as early as mid-log phase, and the level of siderophore increased within the culture until stationary phase was established and then it declined. The decline in CAS reactivity appeared to be due to the action of the bacteria; i.e., filter-sterilized supernatants obtained at 24 h of incubation maintained full CAS reactivity even when stored 37°C (data not shown). This suggests that stationary-phase L. pneumophila may degrade and/or not recycle legiobactin. Given the clear coincidence of CAS reactivity with the most active stage of bacterial growth, legiobactin is likely an enhancer of L. pneumophila replication in low-iron conditions.
FIG. 1.
Kinetics of siderophore production by L. pneumophila. Strain 130b bacteria were grown in BYE to an OD660 of 1.0, washed, and then inoculated into deferrated CDM to an OD660 of 0.2. Over the next day, the growth of the cultures was monitored spectrophotometrically (left y axis), and the CAS reactivity of culture supernatants, reported as net DFX equivalents, was examined (right y axis). The values presented are the means and standard deviations from triplicate cultures. The CAS reactivity of the cultures was significantly above the medium control at all times of incubation (P < 0.05; Student's t test). The results are characteristic of three independent experiments.
Effect of growth temperature on L. pneumophila siderophore expression.
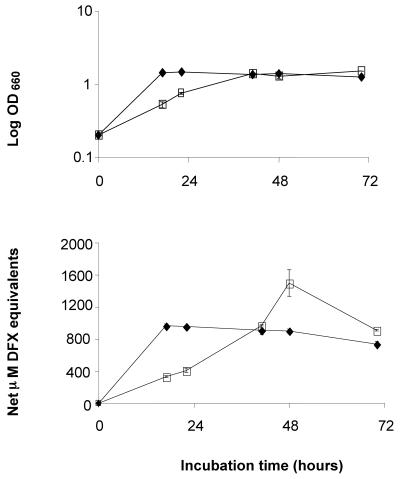
Since L. pneumophila survives and replicates at temperatures ranging from 4 to 63°C (33, 42, 73) and since siderophore expression is elevated at lower temperatures in some other aquatic bacteria (14), strain 130b was grown in deferrated CDM at 25°C and 37°C, and supernatants were tested in the CAS assay (Fig. 2). As before, the legionellae grew more slowly at room temperature than they did at 37°C (73), but the peak optical density for the 25°C cultures was always comparable to that seen with 37°C cultures. CAS reactivity was readily apparent in 25°C cultures, with the level of siderophore activity increasing along with the progression toward stationary phase and then declining afterward (Fig. 2, and data not shown). The maximal amount of CAS-reactive material in 25°C cultures was 2 to 3 times greater than in 37°C cultures. However, as was the case at 37°C, the CAS reactivity at 25°C was Arnow- and Csáky-negative and required that the CDM be inoculated with log-phase legionellae. The elevated levels of CAS reactivity at 25°C may be due to increased expression of legiobactin or another siderophore. Alternatively, it may mean that degradation of legiobactin is greater at higher temperatures.
FIG. 2.
Effect of temperature on L. pneumophila siderophore expression. Strain 130b bacteria were grown at 37°C in BYE to an OD660 of 1.0, washed, and then inoculated into deferrated CDM to an OD660 of 0.2. Over the next 3 days, one set of cultures was incubated at room temperature (□), and another set at 37°C (⧫). At various time points, the growth of the cultures was monitored spectrophotometrically (top), and the CAS reactivity of culture supernatants was examined (bottom). The values presented represent the means and standard deviations from duplicate cultures. The CAS reactivity of the room temperature cultures was significantly different from that of the 37°C cultures at all times of incubation, except at 40 h (P < 0.05; Student's t test). The results presented are representative of four independent experiments.
Biological activities associated with legiobactin.
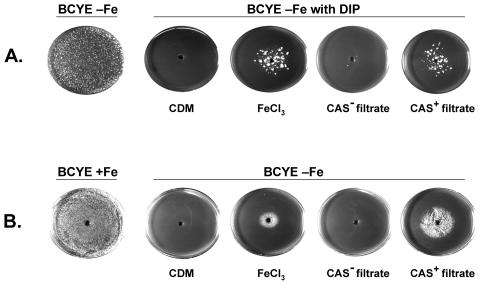
Next, we sought to develop a siderophore bioassay by testing CAS-positive supernatants for their ability to facilitate the growth of iron-starved wild-type legionellae. Toward that end, we first determined that wild-type 130b would not form colonies within 25°C BCYE agar that lacked its iron supplement and contained the ferrous iron chelator DIP at a concentration of ≥100 μM (Fig. 3A). As expected, bacteria did grow in the DIP-containing medium, if a solution of 10 mM ferric chloride was placed into a well cut out of the agar (Fig. 3A).
FIG. 3.
Biological activities associated with legiobactin. (A) 104 CFU of wild-type strain 130b were inoculated into non-iron-supplemented BCYE agar (leftmost plate) or non-iron-supplemented BCYE agar containing 600 μM DIP (four rightmost plates), and a center well was filled, as indicated, with 50 μl of either deferrated CDM, 10 mM FeCl3, or the <3-kDa fraction from a CAS-negative (i.e., 194 μM DFX equivalents) or CAS-positive (i.e., 1,100 μM DFX equivalents) supernatant of strain 130b. After 21 days of incubation at room temperature, the growth of the bacteria was recorded. (B) We plated 105 CFU of feoB mutant bacteria onto the surface of standard (i.e., iron-supplemented) BCYE agar (leftmost plate) or non-iron-supplemented BCYE agar (four rightmost plates), and a center well was filled, as indicated above, with either deferrated CDM, FeCl3, or a CAS-negative or CAS-positive supernatant fraction. After 5 days of incubation at 37°C, the growth of the bacteria was recorded. The results presented for each type of bioassay are representative of at least three independent experiments.
To ascertain the effect of legiobactin, sterile supernatants were obtained from 130b cultures that had been inoculated with either log-phase or stationary phase bacteria, and then the <3-kDa fraction from the resulting CAS-positive and CAS-negative samples were compared for their ability to promote colony formation in the DIP-BCYE agar. The CAS-positive supernatant fraction alone, even when diluted 5-fold, facilitated the growth of wild-type legionellae (Fig. 3A), suggesting that legiobactin has biological activity. In a previous study, we had isolated a L. pneumophila feoB mutant that is defective for ferrous iron transport and grows very poorly at 37°C on BCYE agar that lacks an iron supplement (63). This 130b derivative, however, forms colonies on the iron-deplete medium quicker when it is plated next to a lawn of wild-type bacteria, suggesting that a secreted factor can rescue the mutant from iron starvation. Thus, we compared CAS-positive and CAS-negative supernatants of strain 130b for their ability to stimulate the growth of the feoB mutant on unsupplemented BCYE agar (Fig. 3B). The CAS-positive sample, like ferric chloride, was able to enhance the growth of the iron-starved mutant, indicating once again that legiobactin can deliver iron to the L. pneumophila organism.
Identification of an L. pneumophila gene, lbtA, that is required for legiobactin production.
Since our frgA and ira mutants (59) continued to display wild-type levels of growth and CAS reactivity when cultured in deferrated, cystine-containing CDM (data not shown), it appeared that previously unrecognized genes encode legiobactin. While performing inverse PCR (42, 82) to characterize the nature of the mini-Tn10 insertions in the ira mutants, a primer (i.e., 5′-GGCTCACGATGGCACTTG-3′) that had been designed to facilitate the analysis of strain NU223 amplified, without the assistance of a second primer, a 1.5-kb fragment from 130b DNA. Sequence analysis of the PCR fragment identified an incomplete open reading frame whose predicted product appeared to have homology with the carboxyl end of FrgA.
Subsequent examination of what was, at the time, the unfinished genome database of L. pneumophila revealed the presence of a similar open reading frame within strain Philadelphia-1. Using PCR primers based upon sequences in the database, a ∼2-kb DNA fragment, predicted to contain the entire open reading frame, was amplified from strain 130b. Complete sequence analysis of the amplified fragment confirmed the existence of an L. pneumophila 1.74-kb gene, whose 63.8-kDa predicted product was 36% identical and 51% similar to the 63.3-kDa FrgA. On the basis of mutational analysis that is to be discussed below, we named this gene lbtA, for legiobactin gene A.
In addition to FrgA, the LbtA protein was related to several siderophore synthetases; e.g., it showed 23% identity and 40% similarity to IucA and 26% identity and 44% similarity to IucC (19). The significance of the homology between LbtA and other siderophore biosynthetic enzymes is similar to the 21% identity and 47% similarity between IucA and IucC (44). In addition, the 580-amino-acid LbtA aligned with and was comparable in size to the hydroxamate synthetases; e.g., it was predicted to have the same number of amino acids as IucC. The only known proteins to which the Legionella protein had significant homology were the enzymes involved in siderophore production. Further computer program analysis of the predicted protein showed an absence of transmembrane domains and secretion signals, suggesting that it is a cytosolic protein.
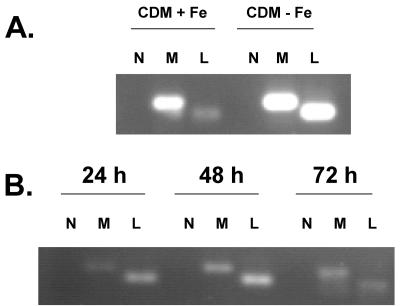
Given that L. pneumophila controls iron-regulated genes through Fur (34, 35), we looked for Fur boxes in the promoter region of lbtA. Two overlapping Fur boxes (5′-GCAAATGATAATCATTATC and 5′-GATAATCATTATCATTTAT) were identified that had only two mismatches to the L. pneumophila frgA Fur boxes (35). These data suggest that lbtA, like frgA and legiobactin, is subject to iron repression. Indeed, using RT-PCR, we saw that lbtA mRNA levels appeared greater in L. pneumophila 130b grown in deferrated CDM than in bacteria grown in iron-supplemented CDM (Fig. 4A).
FIG. 4.
Extra- and intracellular expression of L. pneumophila lbtA. (A) Wild-type 130b was grown on non-iron-supplemented BCYE agar for 3 days, subcultured into BYE lacking the iron supplement, and grown to an OD660 of 1.0. Bacteria were then inoculated into deferrated CDM (CDM-Fe) or CDM supplemented with 20 μM FeCl3 (CDM+Fe) at an OD660 of 0.3 and incubated at 37°C. Bacteria were harvested at mid-log phase and the RNA was isolated. RT-PCR was performed to detect mip (M) and lbtA (L) transcripts. Lanes N, negative controls for DNA contamination performed without reverse transcriptase. (B) Bacteria were grown as in A in BYE-Fe to an OD660 of 1.0. U937 cells were infected with log-phase bacteria and RNA was isolated from infected cells at 24, 48, and 72 h postinoculation. Between 0 and 24 h postinoculation, the numbers of legionellae increased 1,000-fold, and then between 24 and 72 h postinoculation, the numbers increased another 10-fold. Lane denotation is the same as in A. lbtA was also expressed intracellularly when a stationary-phase inoculum was used (data not shown). The results presented are representative of at least two independent experiments. RT-PCR products were electrophoresed through 1.5% agarose and stained with ethidium bromide.
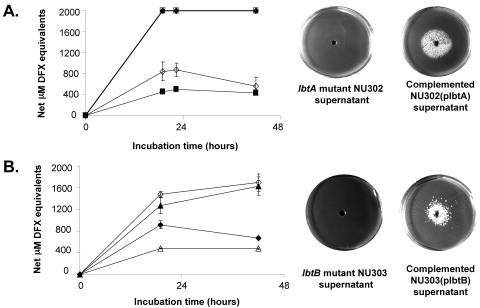
In order to determine if lbtA plays a role in siderophore expression, we constructed a nonpolar lbtA deletion mutant, NU302, which had a 626-bp fragment excised from lbtA. NU302 supernatants displayed 40 to 70% less CAS-reactive material than the wild type and did not promote the growth of the feoB mutant in iron-limiting BCYE agar (Fig. 5A). trans-Complementation with a plasmid (plbtA) containing lbtA as its only L. pneumophila gene restored both the CAS reactivity and bioactivity of NU302 to wild-type levels (Fig. 5A), indicating a role for lbtA in legiobactin production. By introducing plbtA into 130b, the amount of CAS reactivity was increased two- to threefold (Fig. 5A), suggesting that legiobactin production is increased if excess LbtA is available.
FIG. 5.
Siderophore production by L. pneumophila wild type and lbtA and lbtB mutants. (A) (Left panel) Wild-type130b with pMMB2002 (⋄) or plbtA (⧫) and lbtA deletion mutant NU302 with pMMB2002 (▪) or plbtA (□) were grown in BYE to an OD660 of 1.0, inoculated into deferrated CDM to an OD660 of 0.3, and then incubated at 37°C. At various times, the growth of the cultures was monitored spectrophotometrically (data not shown), and the CAS reactivity of supernatants was examined. The CAS values presented represent the means and standard deviations from duplicate cultures. The CAS reactivity of the mutant was significantly different from that of the complemented mutant at all time points and wild-type cultures up to 22 h postinoculation (P < 0.05; Student's t test). The results presented are representative of at least three independent experiments. (Right panel) We plated 105 CFU of feoB mutant bacteria onto the surface of non-iron-supplemented BCYE agar and a center well was filled with either a <3-kDa supernatant fraction from the lbtA mutant NU302 or a <3-kDa supernatant fraction from the complemented mutant NU302(plbtA). After 5 days of incubation at 37°C, the growth of the bacteria was recorded. The results presented are representative of at least three independent experiments. (B) (Left panel) Wild-type130b with pMMB2002 (⋄) or plbtB (⧫) and lbtB mutant NU303 with pMMB2002 (▵) or plbtB (▴) were inoculated into deferrated CDM containing 2 mM IPTG and then assayed for CAS reactivity as described in (A). The CAS reactivity of the mutant's cultures was significantly different from that of the wild-type and complemented mutant cultures, until 44 h (P < 0.05; Student's t test). The results presented are representative of at least four independent experiments. (Right panel) A <3-kDa supernatant fraction from the lbtB mutant and the complemented mutant NU303(plbtB) were tested in the feoB bioassay as indicated in A, with the results being representative of three independent experiments.
In support of the conclusion that lbtA is involved in siderophore expression, mutants NU300 and NU301 containing a kanamycin resistance (Kmr) cassette insertion in lbtA also displayed 40 to 70% less CAS-reactive material than the wild type and had no activity in the two siderophore bioassays (data not shown). The mutants showed a comparable reduction in siderophore expression when grown at room temperature (data not shown), suggesting that the increased CAS activity at room temperature is lbtA independent. The lbtA mutants grew normally on standard BCYE agar and in standard BYE broth (data not shown), indicating that lbtA is not generally required for extracellular growth. Based upon the lbtA mutant phenotypes, we believe that LbtA is involved in the expression of legiobactin. The homology of LbtA with siderophore synthetases and the protein's predicted cellular location suggests that LbtA is involved in the biosynthesis of the L. pneumophila siderophore.
Identification of a second gene, lbtB, involved in legiobactin expression.
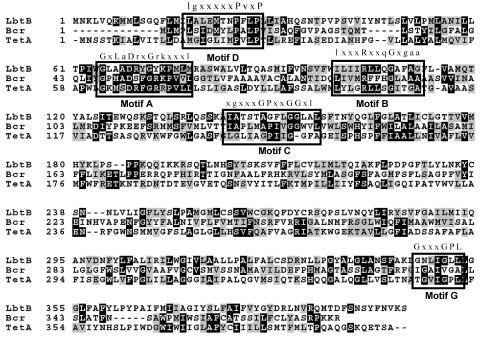
According to the completed genomes of L. pneumophila strains Philadelphia-1, Paris, and Lens (8, 9), lbtA is the first gene in a three-gene operon. The two genes downstream of lbtA are predicted to encode members of the MFS class of proton motive force-dependent membrane efflux pumps. The second gene in the operon, which we now designate lbtB, is a 1.2-kb gene that encodes a 44.4-kDa protein with 12 transmembrane (TMS) domains that is 23% identical and 44% similar to the E. coli bicyclomycin resistance protein Bcr and 21% identical and 39% similar to the E. coli tetracycline efflux pump TetA.
The homology between LbtB and these proteins is greatest in five of the amino acid motifs conserved among MFS transporters, i.e., motifs A, B, C, D, and G (Fig. 6). Recently, members of the MFS family have been shown to include transporters involved in the export of bacterial siderophores, such as the E. coli enterobactin exporter EntS (27). The last gene in the lbt operon, lbtC, encodes a 42.5-kDa protein with 12 predicted transmembrane domains that is related to LbtB and Bcr (data not shown). Given these data, we suspected that lbtB and lbtC are involved in legiobactin export.
FIG. 6.
Amino acid sequence alignments of L. pneumophila LbtB with E. coli Bcr and TetA. The consensus sequences for conserved motifs A, B, C, D, G are labeled above the boxed-in areas; x is any amino acid, upper case is a highly conserved amino acid, and lower case is a conserved, but variable amino acid. Motif A, conserved in both 12- and 14-TMS families, is located in the cytoplasmic loop between TMS 2 and TMS 3 and may be involved in substrate binding as well as opening and closing of the channel (54). Motif B, located in TMS 4, is predicted to be involved in proton transfer (55). Motif C, located in TMS 5, is implicated in the direction of transport and is only found in those transport proteins with efflux capacity (30, 55). The function of motif D, located in TMS 1 in 12- and 14- TMS families, has not been investigated. Motif G, located in TMS 11, is found only in 12-TMS families, although its function is unknown (55).
In order to assess the role of lbtB in legiobactin production, we constructed two lbtB insertion mutants, NU303 and NU304. Both mutants grew normally on standard BCYE agar and in standard BYE broth (data not shown), indicating that, like lbtA, lbtB is not generally required for extracellular growth. However, the lbtB mutants consistently produced 40 to 70% less CAS-reactive material than the wild type (Fig. 5B; NU303 with or without the vector pMMB2002 gave similar results, as did NU304). In addition, NU303 supernatants did not promote the growth of the feoB mutant in BCYE agar lacking the iron supplement (Fig. 5B). Thus, the lbtB mutant was defective for legiobactin production. Importantly, trans-complementation with lbtB under the control of the tac promoter in plasmid plbtB restored both siderophore expression of the mutant to wild-type levels and the ability of NU303 supernatants to promote growth of the feoB mutant on iron-limited BCYE agar (Fig. 5B).
Providing lbtB in trans to the wild type results in an almost twofold reduction in the amount of CAS reactivity detected in CDM supernatants. This does not occur in the lbtB mutant; reintroducing lbtB in this background only restores siderophore expression to normal wild-type levels. Since the lbtB mutant bears an insertion that may have downstream effects on lbtC expression, “excess” LbtB in the presence of “normal levels” of LbtC may have deleterious effects on siderophore excretion. Overall, these data confirm that LbtB is required for legiobactin expression.
Next, a role for lbtC in legiobactin production was assessed by introducing a mutation into the gene. Two mutants, NU305 and NU306, containing a 1.1-kb Gmr insertion in lbtC were obtained (data not shown). Both mutants grew normally on standard BCYE agar and in standard BYE broth (data not shown), indicating that lbtC is not generally required for extracellular growth. However, the lbtC mutants produced wild-type levels of legiobactin (data not shown), ruling out a required role for lbtC in legiobactin expression.
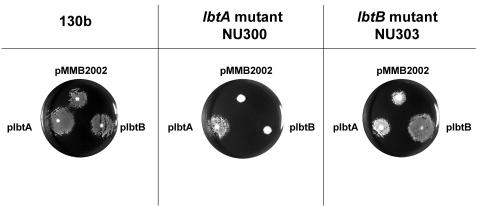
Since supernatants from lbtA and lbtB mutants had similar reductions in CAS reactivity and bioactivity, we believe that both lbtA and lbtB are required for optimal siderophore production by strain 130b. However, phenotypic differences between lbtA mutant NU300 and lbtB mutant NU303 were seen when the bacteria, as opposed to supernatants, were assessed in the feoB mutant bioassay (Fig. 7). When spotted atop the feoB mutant, 130b bacteria containing either vector pMMB2002, plbtA, or plbtB supported the growth of the iron-starved mutant. In keeping with the results obtained with supernatants, the lbtA mutant NU300 only stimulated growth if it contained plbtA (Fig. 7). In contrast, NU303 retained an ability to promote growth of the mutant regardless of recombinant plasmid content, suggesting that, unlike the lbtA mutant, the lbtB mutant still produces legiobactin. Indeed, the homology of lbtB with MFS exporters implies a role for LbtB in legiobactin export and not biosynthesis. We suspect that when the lbtB mutant is grown on the low-iron agar media, some cellular lysis occurs and released legiobactin stimulates the feoB mutant to grow, and the inability of NU303 supernatants to likewise promote growth is likely due to a dilution and/or breakdown of siderophore in the broth.
FIG. 7.
Phenotypes of lbt mutant bacteria in the feoB bioassay. 105 CFU of feoB mutant bacteria were plated unto the surface of non-iron-supplemented BCYE agar containing 2 mM IPTG, and then 130b, lbtA mutant NU300, or lbtB mutant NU303 containing either vector pMMB2002, plbtA, or plbtB was spotted on top of the agar as indicated. After 5 days of incubation at 37°C, the growth of the bacteria was recorded. The results presented are representative of three independent experiments.
Influence of L. pneumophila pvc-like genes and frgA on siderophore expression in the wild type and an lbtA mutant.
The fact that supernatants from the lbtA and lbtB mutants were not completely lacking CAS-reactive material suggested, for the first time, that the CAS reactivity produced by L. pneumophila is the result of multiple CAS-reactive substances and that strain 130b might secrete a second siderophore. An investigation of the L. pneumophila genome database revealed a gene (i.e., lpg0174 in Philadelphia-1, lpp0236 in Paris, and lpl0236 in Lens) that was homologous with the Pseudomonas aeruginosa pyoverdine biosynthetic gene pvcA as well as an adjacent gene homologous with pvcB (76). However, L. pneumophila pvcA mutants (i.e., NU307 and NU308) and pvcB mutants (i.e., NU309 and NU310) produced wild-type levels of CAS reactivity when grown in deferrated CDM (data not shown), indicating that pvcAB, like frgA, is a siderophore-like gene that is dispensable for normal CAS reactivity in strain 130b.
However, it remained possible that increases in CAS reactivity due to legiobactin could have been masking a loss of a siderophore activity associated with FrgA or PvcAB. Thus, we constructed and characterized lbtA frgA double mutants (NU311 and NU312) and lbtA pvcA double mutants (NU313 and NU314). Both types of double mutants were identical to the lbtA single mutants in the CAS assay and the feoB bioassay (data not shown), showing that frgA and pvcAB are not required for wild-type CAS reactivity or the residual CAS activity of lbtA mutants. Further BlastP searches of the L. pneumophila genome using known siderophore biosynthetic, transport, and receptor proteins, as well as an examination of the annotation of the L. pneumophila genome did not reveal any other candidate siderophore genes. Given these various data, we focused on determining the importance of lbtAB for L. pneumophila growth.
Role of lbtA in L. pneumophila extracellular growth.
lbt mutants NU300, NU302, and NU303 grew in deferrated CDM as well as did the wild type (data not shown), suggesting that lbtA and lbtB, though necessary for full siderophore expression, may not be required for extracellular growth in iron-deplete conditions, even though lbtA is expressed in low-iron CDM. Mutant NU302 also grew normally in BYE broth or on BCYE agar that lacked their iron supplements, on unsupplemented BCYE agar that contained 100 to 400 μM DIP, and in deferrated CDM with 5 to 30 μM EDDA or DFX (data not shown). These data suggested that the affinity of legiobactin for iron might be less than 1031, as the stability constants (Ks) for DFX-iron(III) and EDDA-iron(III) are 1031and 1034, respectively (48, 72).
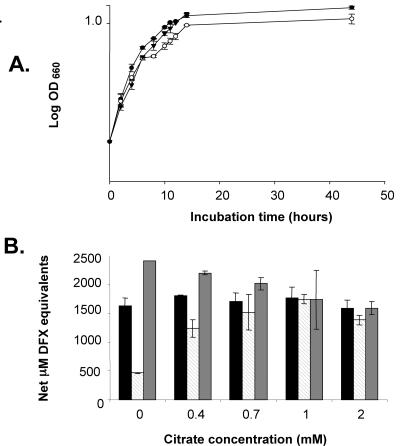
Therefore, to identify a requirement for legiobactin, we investigated bacterial growth in deferrated CDM containing citrate, a chelator with a notably lower affinity for iron(III), 1011 (72). Indeed, when grown in deferrated CDM supplemented with 1 mM citrate, NU302 showed modestly reduced growth at 8 to 44 h postinoculation, and this defect was fully complemented by reintroducing lbtA into the mutant on plbtA (Fig. 8A). When the lbtA mutant was grown in the presence of citrate, including chelator levels that were not inhibitory to bacterial growth, it produced increasing amounts of CAS reactivity, eventually achieving a degree of reactivity that rivaled the wild-type level (Fig. 8B). This could represent the upregulation of the residual CAS activity found in lbtA supernatants or the production of a new CAS reactive substance. However, the mutant supernatants continued to be inactive in the feoB bioassay and negative in the Csáky and Arnow assays, indicating that the heightened CAS reactivity was not due to the turning on of a typical hydroxamate or catecholate. In summary, L. pneumophila can replicate in a variety of low-iron conditions in the absence of lbtA, but legiobactin is beneficial for growth under conditions of severe iron limitation when residual iron is not sequestered by a chelator(s) of extraordinarily high affinity.
FIG. 8.
Growth of wild-type and lbtA mutant L. pneumophila in deferrated CDM containing citrate. (A) 130b with pMMB2002 (•) and NU302 with pMMB2002 (○) or plbtA (▾) were grown in BYE to an OD660 of 1.0, inoculated into deferrated CDM with 1 mM citrate to an OD660 of 0.3, and then incubated at 37°C. At various times, the growth of the cultures was monitored spectrophotometrically. The growth of NU302 was different from that of the wild type and complemented mutant from 8 to 44 h (P < 0.05; Student's t test). (B) L. pneumophila strains were grown in deferrated CDM containing the indicated amounts of citrate, and then, at 44 h, the CAS reactivity of supernatants from 130b (black bars), NU302 (striped bars), and NU302(plbtA) (gray bars) was examined. The values represent the means and standard deviations from duplicate cultures. The CAS reactivity of NU302 was significantly different from that of the wild type and complemented mutant in 0 and 0.4 mM citrate-containing cultures (P < 0.05; Student's t test). When the citrate-associated CAS activity was normalized across cultures, more CAS reactivity was detected in citrate-containing NU302 supernatants than in non-citrate-containing NU302 supernatants (P < 0.05; Student's t test). The results presented in this figure are representative of two independent experiments.
Role of lbtA in intracellular infection and virulence.
We next determined the ability of an lbtA mutant to infect human U937 cell macrophages and H. vermiformis amoebae. Under standard conditions, NU300 behaved as 130b did in both host cells (data not shown). Likewise, NU300 did not differ from 130b when grown in amoebal cultures treated with 0, 25, and 50 μM DIP or in U937 cultures treated with either 10 to 15 μM DFX or 20 to 40 μM DIP. NU300 behaved like the wild type did whether the inoculum was derived from BCYE agar or log-phase BYE cultures (data not shown). Together, these data indicate that lbtA and legiobactin are not required for optimal intracellular infection. The lbtA frgA double mutant NU311 was no more defective in U937 cells than the single frgA mutant, and pvcA and pvcB single mutants and an lbtA pvcA double mutant all grew normally in the macrophage cell line (data not shown), again indicating that lbtA is not necessary for intracellular growth.
However, RT-PCR experiments demonstrated that lbtA is expressed by L. pneumophila when growing within U937 cells (Fig. 4B). We were unable to obtain an lbtA feoB double mutant of strain 130b, whose isolation might have uncovered an intracellular role for a siderophore as it has in studies of Shigella (67). Next, a competition assay was performed in A/J mice (5, 63-65). However, the ratio of wild type to mutant in the mouse lung did not change significantly during the 3-day course of the experiment (data not shown), suggesting that lbtA and legiobactin are not required for L. pneumophila growth in the lungs of A/J mice.
Distribution of lbtA in L. pneumophila serogroups and in other Legionella species.
The L. pneumophila species consists of 15 serogroups (24). Our analysis of strain 130b as well as the data contained within the Philadelphia-1, Paris, and Lens genomic databases indicates that lbtA is present within L. pneumophila serogroup 1. Southern hybridization analysis determined that lbtA is also in strains representing serogroups 2 to 5, 7, 8, 13, and 14 (Table 1). All strains that were found to contain lbtA produce CAS reactivity when grown in iron-deplete CDM (39), supporting a correlation between the presence of lbtA and legiobactin production in L. pneumophila.
The Legionella genus contains 49 species, in addition to L. pneumophila (24, 53). DNAs from most (i.e., 20 out of 27) species tested hybridized with the lbtA probe (Table 1). In agreement with the results of our earlier study (35), frgA was nearly absent from Legionella species other than L. pneumophila (Table 1). Thus, lbtA, unlike frgA, appears to be present within most species of Legionella, including strains isolated from clinical and environmental sources. Most Legionella species tested secrete a siderophore-like activity (75) (Table 1). Fifteen of these 18 CAS-positive species contained lbtA, suggesting that they may produce legiobactin or a related siderophore (Table 1). However, only L. adelaidensis, L. anisa, L. erythra, L. feeleii, L. moravica, L. rubrilucens, and L. santicrusis stimulated L. pneumophila feoB mutant growth when tested in the bioassay, suggesting that they, more so than the others, express legiobactin.
Among the CAS-positive species, L. birminghamensis, L. londiniensis, and L. quinlivanii did not contain lbtA and were negative in the feoB bioassay (Table 1), indicating that lbtA-dependent legiobactin is not the only Legionella siderophore. Although not previously believed to have a siderophore activity (75), L. oakridgensis rescued feoB mutant growth in the bioassay (Table 1).
In summary, lbtA sequences were broadly distributed within the L. pneumophila genus. In all L. pneumophila strains and most other Legionella species tested, the presence of lbtA correlated with CAS reactivity. However, there were examples of both siderophore activity in the absence of lbtA and lack of siderophore expression despite the presence of lbtA.
DISCUSSION
To begin, we optimized the conditions for siderophore production by virulent L. pneumophila strain 130b and demonstrated that CAS reactive supernatants can stimulate growth of iron-starved legionellae. Thus, the CAS reactive material elaborated by L. pneumophila behaves as a bona fide siderophore. It is not surprising that the legionellae produce a siderophore(s), since many other aquatic bacteria, when examined, are found to produce this type of iron scavenger. The other main feature of this study is the first identification of genes (lbtA and lbtB) involved in L. pneumophila siderophore production. The behavior of the lbtA mutant in defined media containing chelators has given an initial impression of the affinity of legiobactin for iron. Since the lbtA mutant and wild type were similarly sensitive to DFX, we suspect that the iron(III) Ks for legiobactin is roughly between 1011 and 1031, the Kss for citrate and DFX, respectively (48, 72). Such a situation would not be incompatible with legiobactin being a siderophore, since the iron(III) stability constants of most siderophores range from 1022 to 1050 (60). In the case of a pathogen like L. pneumophila, such a siderophore is sufficiently strong to chelate iron away from host transferrin and lactoferrin, both with a Ks of ∼1020 (60).
Formally, the reduction in CAS reactivity displayed by the lbtA mutants could be due to alterations in siderophore biosynthesis or secretion. However, since LbtA is related to biosynthetic enzymes and does not contain any transmembrane domains or secretion signals, we suspect that LbtA is involved in the biosynthesis of legiobactin rather than siderophore export. The reactions catalyzed by the LbtA-related enzymes can give clues to the possible LbtA-mediated reaction. In E. coli, IucA catalyzes the addition of N′-acetyl-N′-hydroxylysine to citrate by formation of an amide bond to yield the intermediate Nα-citryl-′-acetyl-N′-hydroxylysine, and subsequently, IucC adds another N′-acetyl-N′-hydroxylysine moiety to the intermediate to form aerobactin (19).
In S. meliloti, it is believed that RhbC catalyzes the addition of N4-acetyl-N4-hydroxy-1-aminopropane to citrate to yield an intermediate to which RhbF then adds N4-acetyl-N4-hydroxy-1-aminopropane to form the immediate precursor to rhizobactin 1021 (43). Although these enzymes catalyze the formation of hydroxamate siderophores, and L. pneumophila is negative in the Csáky assay that detects hydroxamates, recent work in bacteria that produce polyhydroxycarboxylate siderophores has elucidated biosynthesis genes homologous with aerobactin iuc genes (18, 80). For example, although the structure of staphylobactin is unknown, the SbeE protein, like LbtA, is essential for siderophore production and is homologous with IucA (17). Similarly, vibrioferrin proteins PvsB and PvsD are homologous with IucC and IucA; these enzymes catalyze the formation of the two amide bonds contained in vibrioferrin (80). Given the relatedness of LbtA with synthetases of diverse siderophores, a simplest hypothesis is that legiobactin assembly involves an LbtA-catalyzed amide bond formation between precursors.
Despite the extensive understanding of siderophore import, only recently have determinants of siderophore export been identified (4, 26, 27, 38, 50, 58, 85). For example, ExiT, an ATP-binding cassette-like transporter, has been found in Mycobacterium smegmatis, and in Pseudomonas aeruginosa the RND efflux pump, OprM, is implicated in siderophore export (38, 58, 85). However, most of the identified exporters are members of the MFS family, a group of proteins historically viewed as transporting small solutes, such as antibiotics (51). The known MFS members involved in siderophore export include proteins that are required for export of enterobactin in E. coli (EntS), protochelin in Azotobacter vinelandii (CbsX), achromabactin in E. chrysanthemi (YhcA), and alcaligin in Bordetella species (AlcS) (4, 26, 27, 50). These siderophore export proteins constitute the inner membrane channels that facilitate export of the siderophore out of the cytoplasm and into the periplasm. Following the example of antibiotic export in gram-negative bacteria (84), it is likely that the MFS siderophore exporters recruit outer membrane protein channels to excrete the siderophore out of the cell (4, 26, 27, 50). Due to the relatedness of LbtB with MFS permeases, its predicted inner membrane localization, and the partial growth-promoting ability of lbtB mutants, we suspect LbtB to be the latest MFS protein involved in siderophore export.
While many bacteria organize their siderophore-encoding genes into large operons containing multiple biosynthetic genes and often a ferrisiderophore receptor gene, the lbt system may encode only three genes, and perhaps only one required biosynthetic and one required transport gene. The wild-type phenotype of the L. pneumophila mutants lacking lbtC, the last gene in the lbt operon, is similar to that of Yersinia pestis ybtX mutants that secrete siderophore despite their loss of a gene encoding an MFS family member (23). Given that we have thus far not identified other candidate legiobactin genes linked or unlinked to lbtABC, the biosynthesis of legiobactin may be uniquely simple, perhaps involving only LbtA and one or two precursor molecules. Alternatively, other L. pneumophila legiobactin genes exist but they would appear to be unusual in content and location. Interestingly, the gene directly upstream of the lbt locus is predicted to encode a 40-kDa outer membrane protein that, because of an iron box, seems to be Fur regulated. Thus, it is tempting to speculate that this protein is a receptor for ferrilegiobactin.
The fact that mutations in lbtAB do not completely abolish CAS reactivity suggests that there may be more than one L. pneumophila iron chelator produced in low iron environments. The residual activity was found to be the temperature-regulated component(s) of the CAS reactivity in CDM supernatants, since the lbtA mutants, like the wild type, showed an increase in CAS activity at room temperature. This activity might represent one or multiple molecules, including a legiobactin precursor(s), other low affinity siderophore(s), or nonsiderophore CAS-reactive specie(s). If the residual activity represents a new siderophore, it is still a member of the complexone class as it is not detected in the Arnow and Csáky assays.
The biological (i.e., growth-promoting) activity of this residual CAS activity is presently unclear. On the one hand, it was not active in the bioassays used in this study. On the other hand, since the lbtA and lbtB mutants grew normally in deferrated CDM, this residual CAS activity may promote bacterial growth so long as an additional iron chelator, such as citrate, is not present. When we investigated candidate siderophore genes, we found that neither frgA nor the pvc locus was involved in production of the activity, or legiobactin for that matter.
Numerous assays and the use of single and double mutants indicate that lbtA and legiobactin are not required for optimal intracellular infection or virulence. These data, however, do not demonstrate that legiobactin has no relevance for intracellular growth or in vivo persistence. Indeed, lbtA is expressed by L. pneumophila within the macrophage, suggesting a dispensable role for legiobactin in this intracellular environment that can be compensated for by another siderophore or another iron uptake system. It is also conceivable that lbtAB and legiobactin are only expressed and required for extracellular growth or persistence in aquatic environments. Further, legiobactin may be made during the log phase of growth but stored and only utilized during the planktonic phase and perhaps within biofilms.
In contrast to lbtAB, frgA is required for optimal intracellular growth in macrophages (35). These data and the sequence homology of FrgA raise the possibility that L. pneumophila encodes a siderophore that, unlike legiobactin, is necessary for optimal intracellular replication. Presently, there are few data concerning the role of siderophores in macrophage or amoebal intracellular infection, although mycobactin of Mycobacterium tuberculosis and 2,5-dihydroxybenzoic acid of Brucella abortus have been shown to promote infection of macrophages (20, 52). The hypothesis that L. pneumophila has evolved multiple siderophores in order to flourish in distinct intra- and extracellular niches is reasonable and worthy of future investigation.
Our understanding of L. pneumophila iron acquisition is still, relatively speaking, in its infancy. But LbtAB now join FrgA, FeoB, IraAB, Ccm proteins, Fur, and ferric reductases as L. pneumophila proteins required for growth in low iron and presumably in iron acquisition (34, 35, 47, 57, 59, 63, 81, 82). It remains to be determined whether these factors operate in common or distinct iron uptake pathways. However, we suspect that FeoB and LbtA are components of two critical pathways in iron uptake since simultaneous inactivation of feoB and lbtA was incompatible with growth under standard conditions. By utilizing various types of lbtAB mutations and mutants, we can design new genetic screens for identifying other components of L. pneumophila iron uptake.
Acknowledgments
We thank Tracy Aber Scheel, Erin Hickey, Joseph Salerno, Shawn Starkenburg, Ombeline Rossier, and Christopher Sampson for technical assistance. We acknowledge Marianne Robey for first determining that L. pneumophila supernatants can rescue a feoB mutant from iron starvation.
K.A. was partly supported by NIH Training Grant GM08061. This work was funded by NIH grant AI34937 awarded to N.P.C.
REFERENCES
- 1.Abu Kwaik, Y. 1998. Fatal attraction of mammalian cells to Legionella pneumophila. Mol. Microbiol. 30:689-695. [DOI] [PubMed] [Google Scholar]
- 2.Aragon, V., S. Kurtz, A. Flieger, B. Neumeister, and N. P. Cianciotto. 2000. Secreted enzymatic activities of wild-type and pilD-deficient Legionella pneumophila. Infect. Immun. 68:1855-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnow, L. E. 1937. Colorimetric determination of the components of 3,4-dihyrdoxyphenylalanine tyrosine mixtures. J. Biol. Chem. 118:531-537. [Google Scholar]
- 4.Brickman, T. J., and S. K. Armstrong. 2005. Bordetella AlcS transporter functions in alcaligin siderophore export and is central to inducer sensing in positive regulation of alcaligin system gene expression. J. Bacteriol. 187:3650-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brieland, J., P. Freeman, R. Kunkel, C. Chrisp, M. Hurley, J. Fantone, and C. Engleberg. 1994. Replicative Legionella pneumophila lung infection in intratracheally inoculated A/J mice. A murine model of human Legionnaires' disease. Am. J. Pathol. 145:1537-1546. [PMC free article] [PubMed] [Google Scholar]
- 6.Byrd, T. F., and M. A. Horwitz. 2000. Aberrantly low transferrin receptor expression on human monocytes is associated with nonpermissiveness for Legionella pneumophila growth. J. Infect. Dis. 181:1394-1400. [DOI] [PubMed] [Google Scholar]
- 7.Byrd, T. F., and M. A. Horwitz. 1991. Lactoferrin inhibits or promotes Legionella pneumophila intracellular multiplication in nonactivated and interferon gamma-activated human monocytes depending upon its degree of iron saturation. Iron-lactoferrin and nonphysiologic iron chelates reverse monocyte activation against Legionella pneumophila. J. Clin. Investig. 88:1103-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cazalet, C., C. Rusniok, H. Bruggemann, N. Zidane, A. Magnier, L. Ma, M. Tichit, S. Jarraud, C. Bouchier, F. Vandenesch, F. Kunst, J. Etienne, P. Glaser, and C. Buchrieser. 2004. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat. Genet. 36:1165-1173. [DOI] [PubMed] [Google Scholar]
- 9.Chien, M., I. Morozova, S. Shi, H. Sheng, J. Chen, S. M. Gomez, G. Asamani, K. Hill, J. Nuara, M. Feder, J. Rineer, J. J. Greenberg, V. Steshenko, S. H. Park, B. Zhao, E. Teplitskaya, J. R. Edwards, S. Pampou, A. Georghiou, I. C. Chou, W. Iannuccilli, M. E. Ulz, D. H. Kim, A. Geringer-Sameth, C. Goldsberry, P. Morozov, S. G. Fischer, G. Segal, X. Qu, A. Rzhetsky, P. Zhang, E. Cayanis, P. J. De Jong, J. Ju, S. Kalachikov, H. A. Shuman, and J. J. Russo. 2004. The genomic sequence of the accidental pathogen Legionella pneumophila. Science 305:1966-1968. [DOI] [PubMed] [Google Scholar]
- 10.Cianciotto, N. P. 2001. Pathogenicity of Legionella pneumophila. Int. J. Med. Microbiol. 291:331-343. [DOI] [PubMed] [Google Scholar]
- 11.Cianciotto, N. P., B. I. Eisenstein, C. H. Mody, G. B. Toews, and N. C. Engleberg. 1989. A Legionella pneumophila gene encoding a species-specific surface protein potentiates initiation of intracellular infection. Infect. Immun. 57:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cianciotto, N. P., and B. S. Fields. 1992. Legionella pneumophila mip gene potentiates intracellular infection of protozoa and human macrophages. Proc. Natl. Acad. Sci. USA 89:5188-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cianciotto, N. P., S. Kurtz, K. Krcmarik, S. Mody, U. Prasad, M. Robey, J. Salerno, and V. K. Viswanathan. 2001. Iron requirements of and acquisition of iron by Legionella pneumophila, p. 31-37. In R. Marre, Y. Abu Kwaik, C. Bartlett, N. P. Cianciotto, B. S. Fields, M. Frosch, J. Hacker, and P. C. Luck (ed.), Legionella. ASM Press, Washington, D.C.
- 14.Colquhoun, D. J., and H. Sorum. 2001. Temperature dependent siderophore production in Vibrio salmonicida. Microb. Pathog. 31:213-219. [DOI] [PubMed] [Google Scholar]
- 15.Cox, C. D. 1994. Deferration of laboratory media and assays for ferric and ferrous ions. Methods Enzymol. 235:315-329. [DOI] [PubMed] [Google Scholar]
- 16.Csáky, T. Z. 1948. On the estimation of bound hydroxylamines in biological materials. Acta Chem. Scand. 2:450-454. [Google Scholar]
- 17.Dale, S. E., A. Doherty-Kirby, G. Lajoie, and D. E. Heinrichs. 2004. Role of siderophore biosynthesis in virulence of Staphylococcus aureus: identification and characterization of genes involved in production of a siderophore. Infect Immun. 72:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dale, S. E., M. T. Sebulsky, and D. E. Heinrichs. 2004. Involvement of SirABC in iron-siderophore import in Staphylococcus aureus. J. Bacteriol. 186:8356-8362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Lorenzo, V., and J. B. Neilands. 1986. Characterization of iucA and iucC genes of the aerobactin system of plasmid ColV-K30 in Escherichia coli. J. Bacteriol. 167:350-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Voss, J. J., K. Rutter, B. G. Schroeder, H. Su, Y. Zhu, and C. E. Barry, 3rd. 2000. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. USA 97:1252-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Edwards, R. A., L. H. Keller, and D. M. Schifferli. 1998. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 207:149-157. [DOI] [PubMed] [Google Scholar]
- 22.Engleberg, N. C., D. J. Drutz, and B. I. Eisenstein. 1984. Cloning and expression of Legionella pneumophila antigens in Escherichia coli. Infect. Immun. 44:222-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fetherston, J. D., V. J. Bertolino, and R. D. Perry. 1999. YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol. Microbiol. 32:289-299. [DOI] [PubMed] [Google Scholar]
- 24.Fields, B. S., R. F. Benson, and R. E. Besser. 2002. Legionella and Legionnaires' disease: 25 years of investigation. Clin. Microbiol. Rev. 15:506-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flieger, A., B. Neumeister, and N. P. Cianciotto. 2002. Characterization of the gene encoding the major secreted lysophospholipase A of Legionella pneumophila and its role in detoxification of lysophosphatidylcholine. Infect. Immun. 70:6094-6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franza, T., B. Mahe, and D. Expert. 2005. Erwinia chrysanthemi requires a second iron transport route dependent of the siderophore achromobactin for extracellular growth and plant infection. Mol. Microbiol. 55:261-275. [DOI] [PubMed] [Google Scholar]
- 27.Furrer, J. L., D. N. Sanders, I. G. Hook-Barnard, and M. A. McIntosh. 2002. Export of the siderophore enterobactin in Escherichia coli: involvement of a 43 kDa membrane exporter. Mol. Microbiol. 44:1225-1234. [DOI] [PubMed] [Google Scholar]
- 28.Gebran, S. J., C. Newton, Y. Yamamoto, R. Widen, T. W. Klein, and H. Friedman. 1994. Macrophage permissiveness for Legionella pneumophila growth modulated by iron. Infect. Immun. 62:564-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giardina, P. C., L. A. Foster, S. I. Toth, B. A. Roe, and D. W. Dyer. 1997. Analysis of the alcABC operon encoding alcaligin biosynthesis enzymes in Bordetella bronchiseptica. Gene 194:19-24. [DOI] [PubMed] [Google Scholar]
- 30.Griffith, J. K., M. E. Baker, D. A. Rouch, M. G. Page, R. A. Skurray, I. T. Paulsen, K. F. Chater, S. A. Baldwin, and P. J. Henderson. 1992. Membrane transport proteins: implications of sequence comparisons. Curr. Opin. Cell Biol. 4:684-695. [DOI] [PubMed] [Google Scholar]
- 31.Grindley, N. D., and C. M. Joyce. 1980. Genetic and DNA sequence analysis of the kanamycin resistance transposon Tn903. Proc. Natl. Acad. Sci. USA 77:7176-7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harb, O. S., L.-Y. Gao, and Y. Abu Kwaik. 2000. From protozoa to mammalian cells: a new paradigm in the life cycle of intracellular bacterial pathogens. Environ. Microbiol. 2:251-265. [DOI] [PubMed] [Google Scholar]
- 33.Heuner, K., C. Dietrich, C. Skriwan, M. Steinert, and J. Hacker. 2002. Influence of the alternative sigma-28 factor on virulence and flagellum expression of Legionella pneumophila. Infect. Immun. 70:1604-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hickey, E. K., and N. P. Cianciotto. 1994. Cloning and sequencing of the Legionella pneumophila fur gene. Gene 143:117-121. [DOI] [PubMed] [Google Scholar]
- 35.Hickey, E. K., and N. P. Cianciotto. 1997. An iron- and fur-repressed Legionella pneumophila gene that promotes intracellular infection and encodes a protein with similarity to the Escherichia coli aerobactin synthetases. Infect. Immun. 65:133-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.James, B. W., W. S. Mauchline, P. J. Dennis, and C. W. Keevil. 1997. A study of iron acquisition mechanisms of Legionella pneumophila grown in chemostat culture. Curr. Microbiol. 34:238-243. [DOI] [PubMed] [Google Scholar]
- 37.James, B. W., W. S. Mauchline, R. B. Fitzgeorge, P. J. Dennis, and C. W. Keevil. 1995. Influence of iron-limited continuous culture on physiology and virulence of Legionella pneumophila. Infect. Immun. 63:4224-4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li, X. Z., H. Nikaido, and K. Poole. 1995. Role of mexA-mexB-oprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 39:1948-1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liles, M. R., T. Aber Scheel, and N. P. Cianciotto. 2000. Discovery of a nonclassical siderophore, legiobactin, produced by strains of Legionella pneumophila. J. Bacteriol. 182:749-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liles, M. R., and N. P. Cianciotto. 1996. Absence of siderophore-like activity in Legionella pneumophila supernatants. Infect. Immun. 64:1873-1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liles, M. R., P. H. Edelstein, and N. P. Cianciotto. 1999. The prepilin peptidase is required for protein secretion by and the virulence of the intracellular pathogen Legionella pneumophila. Mol. Microbiol. 31:959-970. [DOI] [PubMed] [Google Scholar]
- 42.Liles, M. R., V. K. Viswanathan, and N. P. Cianciotto. 1998. Identification and temperature regulation of Legionella pneumophila genes involved in type IV pilus biogenesis and type II protein secretion. Infect. Immun. 66:1776-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lynch, D., J. O'Brien, T. Welch, P. Clarke, P. O. Cuiv, J. H. Crosa, and M. O'Connell. 2001. Genetic organization of the region encoding regulation, biosynthesis, and transport of rhizobactin 1021, a siderophore produced by Sinorhizobium meliloti. J. Bacteriol. 183:2576-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martinez, J. L., M. Herrero, and V. de Lorenzo. 1994. The organization of intercistronic regions of the aerobactin operon of pColV-K30 may account for the differential expression of the iucABCD iutA genes. J. Mol. Biol. 238:288-293. [DOI] [PubMed] [Google Scholar]
- 45.Mengaud, J. M., and M. A. Horwitz. 1993. The major iron-containing protein of Legionella pneumophila is an aconitase homologous with the human iron-responsive element-binding protein. J. Bacteriol. 175:5666-5676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murga, R., T. S. Forster, E. Brown, J. M. Pruckler, B. S. Fields, and R. M. Donlan. 2001. Role of biofilms in the survival of Legionella pneumophila in a model potable-water system. Microbiology 147:3121-3126. [DOI] [PubMed] [Google Scholar]
- 47.Naylor, J., and N. P. Cianciotto. 2004. Cytochrome c maturation proteins are critical for in vivo growth of Legionella pneumophila. FEMS Microbiol. Lett. 241:249-256. [DOI] [PubMed] [Google Scholar]
- 48.Neilands, J. B. 1981. Microbial iron compounds. Annu. Rev. Biochem. 50:715-731. [DOI] [PubMed] [Google Scholar]
- 49.O'Connell, W. A., J. M. Bangsborg, and N. P. Cianciotto. 1995. Characterization of a Legionella micdadei mip mutant. Infect. Immun. 63:2840-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Page, W. J., E. Kwon, A. S. Cornish, and A. E. Tindale. 2003. The csbX gene of Azotobacter vinelandii encodes an MFS efflux pump required for catecholate siderophore export. FEMS Microbiol. Lett. 228:211-216. [DOI] [PubMed] [Google Scholar]
- 51.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Parent, M. A., B. H. Bellaire, E. A. Murphy, R. M. Roop 2nd, P. H. Elzer, and C. L. Baldwin. 2002. Brucella abortus siderophore 2,3-dihydroxybenzoic acid (DHBA) facilitates intracellular survival of the bacteria. Microb. Pathog. 32:239-248. [DOI] [PubMed] [Google Scholar]
- 53.Park, M. Y., K. S. Ko, H. K. Lee, M. S. Park, and Y. H. Kook. 2003. Legionella busanensis sp. nov., isolated from cooling tower water in Korea. Int. J. Syst. Evol. Microbiol. 53:77-80. [DOI] [PubMed] [Google Scholar]
- 54.Paulsen, I. T., M. H. Brown, and R. A. Skurray. 1996. Proton-dependent multidrug efflux systems. Microbiol. Rev. 60:575-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paulsen, I. T. a. R. A. S. 1993. Topology, structure, and evolution of two families of proteins involved in antibiotic and antiseptic resistance in eukaryotes and prokaryotes-an analysis. Gene 124:1-11. [DOI] [PubMed] [Google Scholar]
- 56.Payne, S. M. 1994. Detection, isolation, and characterization of siderophores. Methods Enzymol. 235:329-344. [DOI] [PubMed] [Google Scholar]
- 57.Poch, M. T., and W. Johnson. 1993. Ferric reductases of Legionella pneumophila. Biometals 6:107-114. [DOI] [PubMed] [Google Scholar]
- 58.Poole, K., K. Krebes, C. McNally, and S. Neshat. 1993. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J. Bacteriol. 175:7363-7372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pope, C. D., W. O'Connell, and N. P. Cianciotto. 1996. Legionella pneumophila mutants that are defective for iron acquisition and assimilation and intracellular infection. Infect. Immun. 64:629-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 61.Reeves, M. W., L. Pine, S. H. Hutner, J. R. George, and W. K. Harrell. 1981. Metal requirements of Legionella pneumophila. J. Clin. Microbiol. 13:688-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reeves, M. W., L. Pine, J. B. Neilands, and A. Balows. 1983. Absence of siderophore activity in Legionella species grown in iron-deficient media. J. Bacteriol. 154:324-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robey, M., and N. P. Cianciotto. 2002. Legionella pneumophila feoAB promotes ferrous iron uptake and intracellular infection. Infect. Immun. 70:5659-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Robey, M., W. O'Connell, and N. P. Cianciotto. 2001. Identification of Legionella pneumophila rcp, a pagP-like gene that confers resistance to cationic antimicrobial peptides and promotes intracellular infection. Infect. Immun. 69:4276-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rossier, O., S. Starkenburg, and N. P. Cianciotto. 2004. Legionella pneumophila type II protein secretion promotes virulence in the A/J. mouse model of Legionnaires' disease pneumonia. Infect. Immun. 72:310-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roy, C. R. 2002. Exploitation of the endoplasmic reticulum by bacterial pathogens. Trends Microbiol. 10:418-424. [DOI] [PubMed] [Google Scholar]
- 67.Runyen-Janecky, L. J., S. A. Reeves, E. G. Gonzales, and S. M. Payne. 2003. Contribution of the Shigella flexneri Sit, Iuc, and Feo iron acquisition systems to iron acquisition in vitro and in cultured cells. Infect. Immun. 71:1919-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sambrook, J., E. F. Fritsch, and T. Maniatis (ed.). 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 69.Schweizer, H. P., and T. T. Hoang. 1995. An improved system for gene replacement and xylE fusion analysis in Pseudomonas aeruginosa. Gene 158:15-22. [DOI] [PubMed] [Google Scholar]
- 70.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 71.Shuman, H. A., M. Purcell, G. Segal, L. Hales, and L. A. Wiater. 1998. Intracellular multiplication of Legionella pneumophila: human pathogen or accidental tourist? Curr. Top. Microbiol. Immunol. 225:99-112. [DOI] [PubMed] [Google Scholar]
- 72.Sillen, L. G., and A. E. Martell. 1971. Stability constants of metal-ion complexes. Supplement no. 1. Part I. Inorganic ligands. Part II. Organic including macromolecule ligands. The Chemical Society, London, England.
- 73.Soderberg, M. A., O. Rossier, and N. P. Cianciotto. 2004. The type II protein secretion system of Legionella pneumophila promotes growth at low temperatures. J. Bacteriol. 186:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Solomon, J. M., and R. R. Isberg. 2000. Growth of Legionella pneumophila in Dictyostelium discoideum: a novel system for genetic analysis of host-pathogen interactions. Trends Microbiol. 8:478-480. [DOI] [PubMed] [Google Scholar]
- 75.Starkenburg, S. R., J. M. Casey, and N. P. Cianciotto. 2004. Siderophore activity among members of the Legionella genus. Curr. Microbiol. 49:203-207. [DOI] [PubMed] [Google Scholar]
- 76.Stintzi, A., Z. Johnson, M. Stonehouse, U. Ochsner, J. M. Meyer, M. L. Vasil, and K. Poole. 1999. The pvc gene cluster of Pseudomonas aeruginosa: role in synthesis of the pyoverdine chromophore and regulation by PtxR and PvdS. J. Bacteriol. 181:4118-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stone, B. J., and Y. Abu Kwaik. 1999. Natural competence for DNA transformation by Legionella pneumophila and its association with expression of type IV pili. J. Bacteriol. 181:1395-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stookey, L. L. 1970. Ferrozine—a new spectrophotometric reagent for iron. Anal. Chem. 42:779-781. [Google Scholar]
- 79.Swanson, M. S., and B. K. Hammer. 2000. Legionella pneumophila pathogenesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54:567-613. [DOI] [PubMed] [Google Scholar]
- 80.Tanabe, T., T. Funahashi, H. Nakao, S. Miyoshi, S. Shinoda, and S. Yamamoto. 2003. Identification and characterization of genes required for biosynthesis and transport of the siderophore vibrioferrin in Vibrio parahaemolyticus. J. Bacteriol. 185:6938-6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Viswanathan, V. K., P. H. Edelstein, C. D. Pope, and N. P. Cianciotto. 2000. The Legionella pneumophila iraAB locus is required for iron assimilation, intracellular infection, and virulence. Infect. Immun. 68:1069-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Viswanathan, V. K., S. Kurtz, L. L. Pedersen, Y. Abu-Kwaik, K. Krcmarik, S. Mody, and N. P. Cianciotto. 2002. The cytochrome c maturation locus of Legionella pneumophila promotes iron assimilation and intracellular infection and contains a strain-specific insertion sequence element. Infect. Immun. 70:1842-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vogel, J. P., and R. R. Isberg. 1999. Cell biology of Legionella pneumophila. Curr. Opin. Microbiol. 2:30-34. [DOI] [PubMed] [Google Scholar]
- 84.Zgurskaya, H. I., and H. Nikaido. 2000. Multidrug resistance mechanisms: drug efflux across two membranes. Mol. Microbiol. 37:219-225. [DOI] [PubMed] [Google Scholar]
- 85.Zhu, W., J. E. Arceneaux, M. L. Beggs, B. R. Byers, K. D. Eisenach, and M. D. Lundrigan. 1998. Exochelin genes in Mycobacterium smegmatis: identification of an ABC transporter and two nonribosomal peptide synthetase genes. Mol. Microbiol. 29:629-639. [DOI] [PubMed] [Google Scholar]