Abstract
Anew Escherichia coli phage, named Rtp, was isolated and shown to be closely related to phage T1. Electron microscopy revealed that phage Rtp has a morphologically unique tail tip consisting of four leaf-like structures arranged in a rosette, whereas phage T1 has thinner, flexible leaves that thicken toward the ends. In contrast to T1, Rtp did not require FhuA and TonB for infection. The 46.2-kb genome of phage Rtp encodes 75 open reading frames, 47 of which are homologous to phage T1 genes. Like phage T1, phage Rtp encodes a large number of small genes at the genome termini that exhibit no sequence similarity to known genes. Six predicted genes larger than 300 nucleotides in the highly homologous region of Rtp are not found in T1. Two predicted HNH endonucleases are encoded at positions different from those in phage T1. The sequence similarity of rtp37, -38, -39, -41, -42, and -43 to equally arranged genes of lambdoid phages suggests a common tail assembly initiation complex. Protein Rtp43 is homologous to the λ J protein, which determines λ host specificity. Since the two proteins differ most in the C-proximal area, where the binding site to the LamB receptor resides in the J protein, we propose that Rtp43 contributes to Rtp host specificity. Lipoproteins similar to the predicted lipoprotein Rtp45 are found in a number of phages (encoded by cor genes) in which they prevent superinfection by inactivating the receptors. We propose that, similar to the proposed function of the phage T5 lipoprotein, Rtp45 prevents inactivation of Rtp by adsorption to its receptor during cells lysis. Rtp52 is a putative transcriptional regulator, for which 10 conserved inverted repeats were identified upstream of genes in the Rtp genome. In contrast, the much larger E. coli genome has only one such repeat sequence.
Phages form the largest group of “organisms” on earth and display a huge genetic diversity. To date, 228 phage genomes have been sequenced and the 252 sequenced bacterial genomes contain many prophages and phage-like elements. Horizontal gene transfer between phages and their hosts largely contributes to the genetic variety of the phages and their hosts. During evolution, phages endowed bacteria with new functions enabling them to occupy new environmental niches. Phage genomics provides data for tracing phage and bacterial evolution. It is therefore desirable to determine the sequences of as many phages as possible.
Escherichia coli and a selected group of E. coli phages paved the way into molecular biology. Among the phages was T1 (1, 29), whose genome sequence was, however, not determined until 2004, over 60 years after its discovery (75). The phage T1 genome encodes 77 open reading frames (ORFs), 37 of which are homologous to described phage ORFs. Functions have been assigned to 27 ORFs. The largest functional group consists of tail proteins. Twenty-two ORFs encoding putative proteins of fewer than 100 amino acid (aa) residues lie predominantly close to the termini. These putative proteins display no sequence similarity to known phage and prophage proteins.
We report on a novel E. coli phage with a genome similar in gene order and sequence to phage T1. This phage, named Rtp, lysed an E. coli production culture in an industrial fermentation plant. The bacterial resistance spectrum of phage Rtp was different from that of known E. coli phages that use outer membrane proteins as receptors or coreceptors. This finding prompted an electron microscopy study of the phage which revealed a rosette-like morphology at the tip of the tail. To our knowledge, such a morphology has not been hitherto described.
Here we report our analysis of the morphology, receptor specificity, and genome sequence of phage Rtp and its similarities to and differences from phage T1. Unlike phage T1, which uses the FhuA outer membrane protein as a receptor and depends on TonB function for infection, we show here that phage Rtp infection is independent of FhuA and TonB and requires a rough lipopolysaccharide (LPS).
MATERIALS AND METHODS
E. coli strains and phage Rtp.
The E. coli strains used are listed in Table 1. Phage Rtp (rosette-type phage) was isolated from a lysed E. coli industrial fermentation culture. The phage was propagated on E. coli K-12 W3110 on tryptone-yeast extract (Difco Laboratories) agar plates or in liquid culture. For phage propagation, 200 ml of an E. coli liquid culture was inoculated with 0.4 ml of a phage suspension with a titer of 10 when the culture reached an optical density at 578 nm of 0.1. The cells continued to grow and started to lyse after 4 h. After the optical density had decreased from 1.66 to 0.26, carbenicillin was added to prevent growth of phage-resistant E. coli mutants, 0.1% Triton X-100 was added to suspend the phage particles, and DNase (12 μg/ml) was added to degrade residual cell DNA. Cell debris was pelleted by centrifugation twice for 10 min at 8,000 × g. Phages were precipitated at 0°C with one-fifth of the lysate volume of 30% polyethylene glycol 6000-3 M NaCl for 1 day. Phages were suspended in 5 ml of phage buffer (10 mM Tris-HCl, 0.15 M NaCl, 1 mM CaCl2, 1 mM MgSO4, pH 7.4). Precipitation with polyethylene glycol was repeated, and the phages were suspended in phage buffer and centrifuged at 12,000 × g to remove residual cell debris. The phage titer of the supernatant was 1013; this preparation was used for sequencing of the phage DNA. The sensitivity of E. coli strains was determined with freshly prepared cell lysates with titers of 105 to 106 on the sensitive strain E. coli KO483.
TABLE 1.
E. coli strains used for determination of phage Rtp receptor specificity
| Strain | Relevant genotypea | Titerb | Source |
|---|---|---|---|
| E. coli B | |||
| B | R | This institute | |
| BL21(DE3) | ompC ompT hsdS | 8 | This institute |
| E. coli K-12 | |||
| W3110 | Wild type | 8 | This institute |
| MC 4100 | 8 | 16 | |
| MH225 | MC4100 ompC | 8 | 43 |
| KB5 | W3110 ompC phoA phoE | 8 | K. Brass |
| KB429 | W3110 ompC | 8 | H. Krieger-Brauer |
| JWC30 | AB2847 ompC ompF phoE fepA cir | 8 | J. W. Coulton |
| H345 | AB2847 ompC fhuA fepA btuB | 8 | K. Hantke |
| MH513 | MC4100 ompF | 8 | 43 |
| KB426C | ompA | 8 | H. Krieger-Brauer |
| KB419 | W3110 lamB | 8 | H. Krieger-Brauer |
| AB2847 | tsx | 8 | This institute |
| KO483 | H1443 fhuA | 8 | This institute |
| BR158 | AB2847 tonB | 8 | This institute |
| WA1013 | AB2847 fecA tsx | 8 | This institute |
| CO1031 | WA1031 tonB | 8 | This institute |
| AA93 | H1443 ΔfecABCDE | 8 | This institute |
| CO93 | AA93 tonB | 8 | This institute |
| HK97 | MC4100 fhuA | 8 | This institute |
| H1880 | MC4100 fhuA fepA | 8 | K. Hantke |
| GM1 | 6 | 82 | |
| HE11 | GM1 tonB | 6 | 14 |
| TPS13 | GM1 tolQ (tolR) | 6 | 82 |
| HE1 | GM1 exbB (exbD) | 6 | 11 |
| HE2 | GM1 exbB (exbD) tolQ | 7 | 11 |
| A592 | tolA | 8 | B. J. Bachmannc |
| A593 | tolB | 8 | B. J. Bachmann |
| 2602 | C600 tolC::Tn10 | 8 | C. Wandersman |
| 2603 | C600 tolC::Tn5 | 8 | C. Wandersman |
| H1388 | exbB | 8 | 12 |
| D21 | 8 | 9 | |
| D21e7 | D21 rfa-1 | 5 | 9 |
| D21f1 | D21e7 rfa-1 rfa-21 | 4 | 9 |
| F464 | O8:K27− | 2 | 79, 87 |
| F470 | Same as F464 but O8−, R1 core type | R | 79, 87 |
Relevant genotype indicates the mutation that was tested for conferring phage resistance.
R, resistant.
Barbara J. Bachmann, E. coli Genetic Stock Center, Department of Biology, Yale University.
Adsorption studies.
Exponentially growing cells or stationary-phase cells cultivated in nutrient broth or tryptone-yeast extract were incubated in the growth medium or in λ SM buffer (8 mM MgSO4, 100 mM NaCl, 25 mM Tris, pH 7.5) with 106 PFU of phage Rtp for 10 min at 37°C and then for 10 min at 0°C. Cells were pelleted at 12,000 × g for 1 min, and the PFU counts in the supernatant were determined.
Outer membranes were prepared as described previously (60). Exponentially growing cells in 100 ml of tryptone-yeast extract were harvested at a density of 5 × 108 cells/ml and suspended in a solution containing 2.15 ml of 0.2 M Tris-HCl (pH 8), 0.2 ml of 10 mM EDTA, 0.845 ml of 80% sucrose, 0.2 ml of lysozyme (4 mg/ml), 6 μl of phenylmethylsulfonyl fluoride (37 mg/ml), and 6 μl of aminobenzamidine (43 mg/ml). The cells were lysed after a 10-min incubation; 0.2 ml each of DNase (1 mg/ml) and RNase (1 mg/ml) and 6.4 ml of double-distilled water were then added. Ten milliliters of buffer containing Triton X-100 was added, and the suspension was centrifuged for 1 h at 30,000 × g at 4°C. The outer membrane fraction in the sediment was washed once with 0.2 ml of water and then with 0.4 ml of 0.2 M Tris-HCl (pH 8) and finally suspended in 0.4 ml of buffer. One sample was supplemented with 10 μl of proteinase K (10 mg/ml), and the control sample received 10 μl of 0.2 M Tris-HCl (pH 7)-0.1% sodium dodecyl sulfate. Both samples were shaken for 1 h at 40°C. The outer membranes were pelleted, washed four times with 0.1 ml of water, and suspended in 0.1 ml of 0.2 M Tris-HCl (pH 7). The membranes were incubated with 106 PFU of phage Rtp for 10 min at 37°C with gentle shaking. The control contained the same concentration of phage Rtp in the same buffer without outer membranes. The sample was centrifuged to pellet the outer membranes, and the phage titer in the supernatant was determined by plating on E. coli W3110.
Electron microscopy.
Phage morphology was examined by electron microscopy of high-titer phage samples prepared as described above and adsorbed to freshly glow-discharged grids coated with a carbon support film. The mounted phages were washed with water and negatively stained with 1% aqueous uranyl acetate. Micrographs were taken at a primary magnification of ×52,000 with a Philips CM10 transmission electron microscope.
Isolation of phage Rtp DNA.
Phage Rtp DNA was prepared by the protocol of QIAGEN (Hilden, Germany) for λ DNA. A purified phage preparation (0.2 ml) was diluted with 100 ml of buffer containing 100 mM Tris-HCl (pH 7.5), 10 mM EDTA, 300 mM NaCl, 20 mg/ml RNase A, 6 mg/ml DNase, and 0.2 mg/ml bovine serum albumin. After incubation for 30 min at 37°C, the mixture was cooled to 0°C and mixed with 20 ml of ice-cold 30% polyethylene glycol 6000-3 M NaCl. After incubation overnight, the suspension was centrifuged for 30 min at 38,000 × g. The sediment was suspended in 6 ml of 100 mM Tris-HCl (pH 7.5)-100 mM NaCl-25 mM EDTA. Sodium dodecyl sulfate (6 ml, 4%) was added, and the mixture was incubated for 10 min at 70°C. Then, 2.4 mg of proteinase K was added and the incubation continued for 10 min at 60°C. The mixture was cooled to 0°C and mixed with 6 ml of ice-cold 3 M sodium acetate (pH 5.5). After 15 min of incubation on ice, the mixture was centrifuged for 30 min at 38,000 × g. During all incubation steps, the mixtures were gently shaken. The supernatant was filtered through a QIA filter and then placed on a QIAGEN-tip 100 equilibrated with 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS; pH 7.0)-750 mM NaCl-15% ethanol-0.15% Triton X-100. The tip was washed twice with 10 ml of 50 mM MOPS (pH 7.0)-1 M NaCl-15% ethanol; the DNA was then eluted with 10 ml of 50 mM Tris-HCl (pH 8.5)-1.25 M NaCl-15% ethanol. The DNA solution was collected in 7 ml of isopropanol, and the precipitated DNA was centrifuged for 30 min at 38,000 × g. The DNA pellet was washed with 70% ethanol, dried in vacuo for 10 min, dissolved in 80 μl of 10 mM Tris-HCl (pH 8.0)-1 mM EDTA, and used for DNA sequencing.
DNA sequencing.
The genome of phage Rtp was sequenced by MWG (Ebersberg, Germany) by the dideoxy chain termination method. Sequencing involved generation of a random genomic library in the plasmid sequencing vector pBluescript SK+, bulk sequencing of 384 shotgun clones, and closure of the remaining gaps. The average size of the hydrodynamically sheared DNA inserts was 1.4 kb. A single gap of approximately 5.0 kb remained. This region was amplified by using PCR with the 24-mer oligonucleotides 5′-GAGGTTCAGATTCAGTCAGGTGAG and 5′-TGCACTGAATGTAACCTCTAGGGC and the Roche Expand High Fidelity PCR kit according to the manufacturer's instructions. The sequence of the PCR product on both strands was determined by GATC (Constance, Germany) by primer walking with the purified PCR fragment as a sequencing template. PhredPhrap and Consed (39) were used to assemble the sequencing reads into a single circular contig.
DNA manipulations and restriction mapping.
Standard methods were used for DNA manipulations and restriction mapping (78). To test for cohesive end sites in the chromosome of phage Rtp, one sample of a complete restriction enzyme DNA digest was heated to 80°C for 10 min to disrupt the annealing of potential complementary overhangs and then rapidly cooled on ice prior to gel electrophoresis. A second sample was heated to 55°C and allowed to cool slowly to room temperature to promote annealing of potential complementary ends before being loaded onto a gel next to the heated sample. HindIII-digested λ DNA was used as a positive control to detect annealing of cohesive ends. The 23.1-kb and 4.4-kb fragments annealed at the 12-nucleotide (nt) cohesive ends and together formed a 27.5-kb fragment.
Sequence analysis software
Protein coding genes were predicted with GeneMarkS (8) and Zcurve (42). The predicted translational start sites were corroborated by inspection of E. coli ribosome-binding sites (consensus sequence, AAGGAGGT; 62, 99). tRNAscan-SE was used to search for tRNA genes (61). BLAST, PSI-BLAST, and Pfam (2, 6) were used for similarity searches. Multiple-sequence alignments were generated with ClustalW (83; http://www.ebi.ac.uk/clustalw). Protein motifs were scanned at the Prosite server (50; http://www.expasy.org/prosite), and DNA motifs were examined with a local installation of PatScan (34). DNA was restriction mapped in silico at http://www.restrictionmapper.org/. BetaWrap (10; http://betawrap.lcs.mit.edu) was used for the prediction of β-helix folds in proteins, and 3D-PSSM (58; www.sbg.bio.ic.ac.uk/∼3dpssm) was used for protein fold recognition and remote homolog detection. DNA-binding helix-turn-helix motifs were predicted by using the service for the Dodd-Egan method at the Pôle Bioinformatique Lyonnais (32; http://pbil.univ-lyon1.fr). Relevant genomic data of completely sequenced phages mentioned in this report were inspected with the Protein View facility of the Genome database section at the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/).
Nucleotide sequence accession number.
The complete nucleotide sequence of the phage Rtp genome has been deposited in the EMBL nucleotide sequence database under accession number AM156909.
RESULTS AND DISCUSSION
The tail morphologies of phages Rtp and T1 differ.
After growth in E. coli K-12 W3110, parallel cultures of Rtp and T1 yielded similar numbers of PFU (approximately 100 per cell). Electron micrographs of phage Rtp revealed a rosette-like morphology of the tail tip (Fig. 1, A). Three or four rosette leaves were observed. In the former case, the fourth leaf was probably shielded by the rest of the tail tip. The rosettes are robust and were seen in all phages in the micrographs. To our knowledge, such phage tail tip structures have not been observed previously.
FIG. 1.
Electron micrographs of purified phages after negative staining with uranyl acetate. A, phage Rtp; B, phage T1. Note the thinner and longer tail fibers of phage T1 in comparison to those of phage Rtp. Bar, 100 nm.
Since analysis revealed a high sequence similarity between the genomes of phage Rtp and phage T1, we determined the morphology of phage T1 under identical conditions. In both phages, the tail fibers extend from a knob-like structure at the tip of the phage tail. However, the tail fibers of phage T1 (Fig. 1B) are very thin at the base and became thicker toward the end but do not reach the diameter of the Rtp fibers. The T1 fibers assumed various orientations on the micrographs, presumably because of the flexibility of the thin segments. This made it difficult to obtain a representative high-resolution picture of the T1 tail tip.
The receptor specificities of phage Rtp and phage T1 differ.
The unusual morphology of the phage Rtp tail tip prompted us to identify the E. coli receptor. We tested whether Rtp, like phage T1, uses the FhuA protein as a receptor and requires TonB for infection and ExbB and ExbD for energization of FhuA (Table 1) since the sequence of the Rtp genome had high similarity to that of phage T1 (see below). Three fhuA mutants of E. coli K-12—KO483, HK97, and H1880—were examined and found to be fully sensitive. Three tonB mutants derived from three different parental strains and exbB and exbD mutants were also fully sensitive. The functions of ExbB and ExbD can be partially replaced by TolQ and TolR (14). However, tolQ and tolR mutants were as sensitive as parental strain GM1 (Table 1), which for unknown reasons poorly supported Rtp propagation. These results showed that the receptor specificity of phage Rtp differs from that of phage T1.
A series of other E. coli K-12 mutants with defects in defined, well-studied outer membrane proteins and LPS were tested (Table 1). All of the protein mutant strains were sensitive to phage Rtp. In contrast, E. coli B was completely resistant. Since E. coli B lacks the OmpC porin, which is present in the K-12 strains tested, OmpC was a candidate for the phage Rtp receptor. However, the E. coli B derivative BL21(DE3) and the E. coli K-12 ompC mutants KB429, KB5, and JWC30 were sensitive, showing that OmpC did not serve as the phage Rtp receptor. As will be shown below, resistance of E. coli B is probably caused by restriction of the phage genome.
To test whether LPS serves as a receptor, the sensitivity of rfa and rfb mutants of E. coli D21 was determined. The rfa-1 mutant D21e7 was 3 orders of magnitude less sensitive, and the rfa-1 rfa-21 mutant D21f1 was 4 orders of magnitude less sensitive than parental strain D21 (Table 1), which suggests that LPS contributes to binding of phage Rtp to E. coli. The major core sequence of LPS in strain D21 has been proposed to be GlcNAc-(Glc)2-Glc(Gal)(Hep)4-KDO-lipid A (9, 46), whereKDO is 2-keto-3-deoxyoctulosonic acid. Strain D21e7 lacks GlcNAc-(Glc)2, and strain D21f1 lacks, in addition, Glc(Gal). Additional evidence for involvement of LPS in phage Rtp infection came from the partial sensitivity of E. coli F464 and the complete resistance of its R1 core LPS mutant F470 (Table 1). Because of the lack of chemically well-defined E. coli LPS mutants and the variations in LPS structure, e.g., heterogeneous glycoforms in the core sequence and side chains with various amounts of phosphate and ethanolamine, we did not further test which LPS substructure contributes to phage Rtp binding.
Attempts to determine phage Rtp binding to viable cells failed under various experimental conditions. Exponential-phase cells (108 cells/ml) and stationary-phase cells in nutrient broth and in phage adsorption buffer were incubated at 37°C for 15 min with freshly prepared phage suspensions (106 PFU). No reduction in the number of PFU was observed. The experiments were repeated with outer membrane preparations of sensitive strains W3110 and MH225 and their spontaneous Rtp-resistant derivatives. Tenfold fewer PFU formed with the outer membranes of both the sensitive and resistant strains. Degradation of outer membrane proteins by proteinase K did not decrease, but rather increased, phage inactivation. These results are consistent with LPS playing a role in phage Rtp binding; i.e., presumably, proteinase K increased access of the phage to LPS. Since proteins embedded in the outer membrane are largely resistant to proteases, the Rtp receptor can be a protein. We propose that LPS is not the only receptor but facilitates adsorption, as has been shown for other phages, e.g., phage T5, where the L-shaped tail fibers bind to the LPS O9 antigen (47) and protein pb2 binds to the FhuA protein (48). Reversible binding to O9 accelerates T5 phage infection 15-fold, but infection occurs only through FhuA, which determines host specificity. Similarly, phage Rtp binding may be facilitated through LPS, to which the phage binds in wild-type and mutant outer membrane preparations. The receptor required for infection would then be another outer membrane component, presumably a less-well-characterized protein of which no mutant form was in our collection. Apparently, phage Rtp adsorbs poorly to cells, like phage T5 lacking the L-shaped tail fibers or when incubated with cells lacking the O antigen.
The Rtp genome is linear but circularly permutated.
Restriction mapping of phage Rtp DNA revealed a double-stranded molecule of 46 to 48 kb. Despite the high coverage of the shotgun library (11.8-fold), commercial shotgun sequencing of the Rtp phage DNA and additional genome walking yielded a contig of only 41,234 bp. The remaining gap was covered through PCR amplification with primers placed close to the ends of the known sequence. The amplified DNA fragment of approximately 5.2 kb was sequenced directly on both strands by primer walking. The sequence obtained overlapped the ends of the 41,235-bp sequence and contained an additional 4,985 bp. The final assembly of the Rtp genome produced a 46,219-bp circular sequence. Restriction sites in the genome predicted in silico agreed with the experimentally determined restriction sites (data not shown), confirming correct genome assembly and supporting an apparently circular physical map. In addition to the expected fragments of the circular map, a single DNA fragment was observed in each digest that was present in submolar concentrations. The size of the fragment was dependent on the restriction enzyme used. These observations were consistent with a linear but circularly permutated genome of phage Rtp generated by a headful packaging mechanism (17, 52; see the later section on terminase and DNA packaging).
Overall features of the genome
Phage Rtp multiplied on E. coli K-12 host strains but not on the restriction-proficient, hsdS+ E. coli B strain. Restriction-deficient E. coli B strain BL21(DE3) hsdS was sensitive. A computer search for strain-specific restriction sites in the Rtp genome sequence (76; http://rebase.neb.com) provided a rationale for this observation. We found 10 target sites for the EcoBI restriction system [TGA(N8)TGCT] but none for the EcoKIrestriction system [AAC(N6)GTGC]. Apparently, the genome was degraded by the EcoB1 endonuclease in E. coli B hsdS+.
The G+C content of the genome is 44.3 mol%, which is lower than the 51 mol% of the E. coli host. Phage Rtp does not carry any tRNA genes, as determined by tRNAscan analysis (61).
Seventy-five ORFs were identified with the complementary gene prediction programs GeneMarkS and Zcurve1.0 (8, 42). Agreement between the two programs was high; 70 of 75 genes found by GeneMarkS were also predicted by Zcurve 1.0, albeit sometimes with an alternative start codon. A large proportion of the predicted genes, i.e., 34 of 75 (45%), mostly located near the ends of the genome map as defined in Fig. 2, code for proteins of fewer than 100 aa. Only eight of these short gene products are significantly similar to proteins in the NCBI nonredundant protein database or could be predicted as translation products by TBlastN searches of the GenBank nucleotide database.
FIG. 2.
Comparative maps of the genomes of E. coli phages Rtp (top, 46,219 bp) and T1 (bottom, 48,836 bp). Both phages contain circularly permutated genomes, which are shown with the predicted locations of the pac sites close to the left end. The scale for the T1 genome starts 2 kb to the left of the scale for the genome of Rtp. The T1 map is redrawn to scale from the data presented by Roberts et al. (75). The relative locations of orthologous genes, shown by the dashed lines between the Rtp and T1 genes, reveal a high degree of synteny (see Table 2 for details). Gene numbering is shown above the ORFs. The locations of the HNH endonuclease genes (two genes in Rtp, three genes in T1) are not conserved between the two phages. Gene products: Pnk, polynucleotide kinase; TerS, terminase small subunit; TerL, terminase large subunit; tail tip proteins M, L, K, and I and fiber J, homologs of λ tail assembly proteins; Erf, essential recombination function; Ssb, single-stranded-DNA-binding protein. Regul., regulator.
Sequence similarity searches showed that the genome of phage Rtp, belonging to the family Siphoviridae, shared short runs of tail genes with other Siphoviridae family members, such as Enterobacteriaceae phages N15, HK022, and HK97 and Xanthomonas oryzae phage Xp10 (53, 73, 100), and frequently with uncharacterized prophages in sequenced genomes of gram-negative bacteria. For genes with other functions such as packaging, replication, or lysis, the distribution of related genes was not confined to the family Siphoviridae. Unfortunately, the genes were often not sufficiently annotated, and therefore the proteins were described as conserved or hypothetical. A prophage of Yersinia pestis strain CO92, located at genes YPO2084 to YPO2140, is well annotated and encodes the highest number of prophage gene products with similarity to putative Rtp proteins (12 genes in the regions from rtp30 to rtp44 and YPO2113 to YPO2132; http://www.sanger.ac.uk/Projects/Y_pestis) (67). In summary, phage Rtp was not closely related to any known phage before the genome sequence of phage T1 was published.
Rtp is closely related to coliphage T1
Genome comparison of phage Rtp with T1 showed that 47 (63%) of the Rtp genes were homologous to genes of phage T1, covering almost entirely the functionally annotated regions of the phage T1 genome, extending from genes encoding the terminase, head, tail, recombination, replication, and lysis proteins (Fig. 2 and Table 2). There is extensive synteny between the two genomes, and with the exception of three genes (rtp6, rtp44, and rtp59; discussed below), the closest homologs of Rtp genes are all found in phage T1.
TABLE 2.
List and annotation of Rtp ORFs
| ORF | Nucleotides | Strand | Protein length (aa) | Related protein | BlastPa
|
Description of related protein | |||
|---|---|---|---|---|---|---|---|---|---|
| Score | E value | ID% | Overlap (aa) | ||||||
| rtp1 | 546-1064 | + | 172 | None | |||||
| rtp2 | 1064-1123 | + | 19 | None | |||||
| rtp3 | 1120-1383 | + | 87 | None | |||||
| rtp4 | 1444-1983 | + | 179 | YP_003880 | 184 | 7e-46 | 59 | 156 | Putative polynucleotide kinase/phosphatase gp65 (phage T1) |
| AAQ15454 | 106 | 3e-22 | 35 | 155 | Hypothetical protein ORF229c (enterobacterial phage RB49) | ||||
| YP_006560 | 65 | 6e-10 | 31 | 151 | Putative acid phosphatase Pap (phage P1) | ||||
| rtp5 | 2102-2362 | + | 86 | None | |||||
| rtp6 | 2432-2524 | + | 30 | AAD42660 | 47 | 1e-04 | 79 | 24 | Stp activator of host PrrC lysyl-tRNA endonuclease (phage T4) |
| rtp7 | 2521-2745 | + | 74 | None | |||||
| rtp8 | 2742-2930 | + | 62 | None | |||||
| rtp9 | 2934-3080 | + | 48 | YP_003941 | 54 | 1e-06 | 57 | 40 | Hypothetical protein gp04 (phage T1)b |
| rtp10 | 3152-3442 | + | 96 | None | |||||
| rtp11 | 3534-3713 | + | 59 | None | |||||
| rtp12 | 3765-3887 | + | 40 | None | |||||
| rtp13 | 3887-4030 | + | 47 | None | |||||
| rtp14 | 4101-4283 | + | 60 | None | |||||
| rtp15 | 4299-4535 | + | 78 | YP_003886 | 58 | 7e-08 | 48 | 74 | Hypothetical protein gp59 (phage T1)b |
| rtp16 | 4997-5182 | + | 61 | None | |||||
| rtp17 | 5214-5366 | + | 50 | YP_003889 | 45 | 5e-04 | 40 | 44 | Hypothetical protein gp56 (phage T1)b |
| rtp18 | 5457-5738 | + | 93 | None | |||||
| rtp19 | 5813-6319 | + | 168 | YP_003891 | 171 | 5e-42 | 52 | 164 | Putative terminase small subunit gp54 (phage T1) |
| CAA09708 | 74 | 1e-12 | 30 | 110 | Packaging protein gp3 (phage PS34) | ||||
| rtp20 | 6388-6864 | + | 158 | YP_003930 | 97 | 2e-19 | 38 | 162 | Putative endonuclease gp15 (phage T1) |
| YP_003937 | 90 | 2e-17 | 40 | 132 | Putative endonuclease gp08 (phage T1) | ||||
| AAP58725 | 79 | 4e-14 | 40 | 132 | 57R (Xanthomonas oryzae phage Xp10) | ||||
| rtp21 | 6857-8428 | + | 523 | YP_003892 | 577 | e-163 | 54 | 528 | Putative terminase large subunit gp53 (phage T1) |
| ZP_00168230 | 327 | 7e-88 | 37 | 496 | COG5410; uncharacterized protein conserved in bacteria (Ralstonia eutropha JMP134) | ||||
| NP_852753 | 182 | 3e-44 | 30 | 468 | Putative terminase large subunit TerL (phage Aaphi23) | ||||
| rtp22 | 8500-8856 | + | 118 | None | |||||
| rtp23 | 8913-10181 | + | 422 | YP_003893 | 458 | e-127 | 56 | 401 | Putative portal protein gp52 (phage T1) |
| ZP_00321768 | 137 | 8e-31 | 26 | 372 | COG3567; uncharacterized protein conserved in bacteria (Haemophilus influenzae 86-028NP) | ||||
| rtp24 | 10234-10866 | + | 210 | YP_003894 | 150 | 3e-35 | 36 | 217 | Hypothetical protein gp51 (phage T1)b |
| NP_439559 | 61 | 2e-08 | 36 | 87 | Plasmid RP4 TraN-related protein (Haemophilus influenzae Rd KW20) | ||||
| rtp25 | 10880-11968 | + | 362 | YP_003895 | 353 | 3e-96 | 53 | 370 | Putative major head subunit precursor gp50 (phage T1) |
| ZP_00321766 | 182 | 1e-44 | 36 | 337 | COG3566; uncharacterized protein conserved in bacteria (Haemophilus influenzae 86-028NP) | ||||
| NP_108196 | 123 | 9e-27 | 34 | 347 | Hypothetical protein mlr8006 (Mesorhizobium loti MAFF303099) | ||||
| rtp26 | 11981-12460 | + | 159 | YP_003896 | 107 | 1e-22 | 41 | 160 | Hypothetical protein gp49 (phage T1) |
| YP_003897 | 92 | 6e-18 | 38 | 152 | Hypothetical protein gp48 (phage T1) | ||||
| rtp27 | 12574-13518 | + | 314 | YP_003898 | 296 | 5e-79 | 49 | 315 | Hypothetical protein gp47 (phage T1) |
| ZP_00321764 | 120 | 5e-26 | 28 | 307 | COG4834; uncharacterized protein conserved in bacteria (Haemophilus influenzae 86-028NP) | ||||
| rtp28 | 13610-13855 | + | 81 | YP_003899 | 31 | 25 | 82 | Hypothetical protein gp46 (phage T1) | |
| rtp29 | 14007-14405 | + | 132 | YP_003900 | 116 | 1e-25 | 47 | 127 | Hypothetical protein gp45 (phage T1)b |
| rtp30 | 14405-14776 | + | 123 | YP_003901 | 89 | 3e-17 | 39 | 123 | Hypothetical protein gp44 (phage T1) |
| NP_405661 | 79 | 2e-14 | 40 | 101 | Hypothetical protein YPO2113 (Yersinia pestis CO92) | ||||
| rtp31 | 14769-15206 | + | 145 | YP_003902 | 100 | 7e-21 | 41 | 139 | Hypothetical protein gp43 (phage T1) |
| NP_890030 | 72 | 3e-12 | 31 | 135 | Phage-related conserved hypothetical protein (Bordetella bronchiseptica RB50) | ||||
| rtp32 | 15208-15597 | + | 129 | YP_003903 | 118 | 3e-26 | 47 | 109 | Hypothetical protein gp42 (phage T1) |
| NP_890029 | 42 | 0.002 | 30 | 79 | Phage-related conserved hypothetical protein (Bordetella bronchiseptica RB50) | ||||
| rtp33 | 15613-16269 | + | 218 | YP_003904 | 233 | 2e-60 | 50 | 214 | Putative major tail protein gp41 (phage T1) |
| NP_405664 | 163 | 3e-39 | 41 | 208 | Hypothetical protein YPO2116 (Yersinia pestis CO92) | ||||
| rtp34 | 16380-16766 | + | 128 | None | |||||
| rtp35 | 16785-17099 | + | 104 | YP_003905 | 105 | 3e-22 | 47 | 105 | Hypothetical protein gp40 (phage T1) |
| NP_405665 | 49 | 4e-05 | 29 | 100 | Hypothetical protein YPO2117 (Yersinia pestis CO92) | ||||
| NP_881900 | 42 | 0.003 | 25 | 109 | Phage-related conserved hypothetical protein (Bordetella pertussis) | ||||
| rtp36 | 17108-17419 | + | 103 | YP_003906 | 66 | 2e-10 | 60 | 50 | Hypothetical protein gp39 (phage T1) |
| NP_890025 | 53 | 1e-06 | 34 | 96 | Phage-related conserved hypothetical protein (Bordetella bronchiseptica RB50) | ||||
| rtp37 | 17455-20220 | + | 921 | YP_003907 | 441 | e-122 | 31 | 984 | Putative tail tape-measure protein gp38 (phage T1) |
| AAC19052 | 335 | 4e-90 | 32 | 841 | gp16 (phage N15) | ||||
| BAB35066 | 284 | 8e-75 | 29 | 805 | Tail length tape measure protein precursor (Escherichia coli O157:H7) | ||||
| rtp38 | 20251-20601 | + | 116 | YP_003908 | 115 | 2e-25 | 42 | 113 | Putative minor tail protein gp37 (phage T1) |
| NP_405668 | 57 | 1e-07 | 30 | 115 | Hypothetical protein YPO2120 (Yersinia pestis CO92) | ||||
| AAC19053 | 54 | 1e-06 | 26 | 115 | gp17 (phage N15) | ||||
| rtp39 | 20641-21396 | + | 251 | YP_003909 | 345 | 5e-94 | 68 | 243 | Putative minor tail protein gp 36 (phage T1) |
| AAC19054 | 200 | 3e-50 | 46 | 241 | gp18 (phage N15) | ||||
| rtp40 | 21464-21979 | + | 171 | YP_003937 | 127 | 9e-29 | 40 | 168 | Putative HNH endonuclease gp08 (phage T1) |
| YP_003881 | 117 | 1e-25 | 43 | 161 | Putative HNH endonuclease gp63 (phage T1) | ||||
| AAP58684 | 100 | 2e-20 | 40 | 147 | 17R (Xanthomonas oryzae phage Xp10) | ||||
| rtp41 | 21960-22718 | + | 252 | YP_003910 | 284 | 1e-75 | 53 | 244 | Putative minor tail protein gp35 (phage T1) |
| AAC19055 | 201 | 1e-50 | 44 | 236 | gp19 (phage N15) | ||||
| YP_006598 | 199 | 4e-50 | 42 | 231 | gp18 (phage phiKO2) | ||||
| rtp42 | 22699-23271 | + | 190 | YP_003911 | 191 | 7e-48 | 50 | 193 | Putative tail assembly protein gp34 (phage T1) |
| NP_718510 | 156 | 3e-37 | 47 | 185 | Prophage LambdaSo, tail assembly protein (Shewanella oneidensis MR-1) | ||||
| AAC19056 | 142 | 5e-33 | 43 | 190 | gp20 (phage N15) | ||||
| rtp43 | 23313-26723 | + | 1136 | YP_003912 | 1358 | 0.0 | 57 | 1,163 | Putative tail fiber protein gp33 (phage T1) |
| AAF31093 | 870 | 0.0 | 40 | 1,180 | Tail fiber (phage HK97) | ||||
| AAF30375 | 867 | 0.0 | 40 | 1,186 | gp24 (phage HK022) | ||||
| rtp44 | 26752-27687 | − | 311 | NP_405680 | 103 | 6e-21 | 26 | 310 | Hypothetical protein YPO2132 (Yersinia pestis CO92) |
| NP_892069 | 70 | 6e-11 | 25 | 346 | Hypothetical protein (phage PY54) | ||||
| rtp45 | 27687-27917 | − | 76 | None | |||||
| rtp46 | 28260-28658 | + | 132 | None | |||||
| rtp47 | 28662-29627 | + | 321 | YP_003916 | 280 | 4e-74 | 44 | 348 | Exodeoxyribonuclease VIII gp29 (phage T1) |
| CAC33454 | 67 | 7e-10 | 24 | 313 | Unnamed protein product (Legionella pneumophila) | ||||
| rtp48 | 29701-30351 | + | 216 | YP_003917 | 183 | 2e-45 | 43 | 216 | Putative recombination protein gp28 (phage T1) |
| YP_011247 | 92 | 7e-18 | 37 | 130 | ERF family protein (Desulfovibrio vulgaris) | ||||
| YP_112530 | 82 | 7e-15 | 41 | 124 | Essential recombination protein (phage 11b) | ||||
| rtp49 | 30395-30826 | + | 143 | YP_003918 | 122 | 1e-27 | 45 | 146 | Single-stranded binding protein gp27 (phage T1) |
| AAO53202 | 60 | 1e-08 | 46 | 69 | Hypothetical protein (Dictyostelium discoideum) | ||||
| rtp50 | 30856-33738 | − | 960 | YP_003919 | 137 | 2e-30 | 42 | 166 | Putative tail fiber gp26 (phage T1) |
| CAD44528 | 68 | 1e-09 | 29 | 165 | Endo-alpha-sialidase (E. coli phage K1F) | ||||
| rtp51 | 33826-34749 | − | 307 | YP_003921 | 251 | 1e-65 | 42 | 312 | Putative DNA primase gp24 (phage T1) |
| NP_458910 | 70 | 5e-11 | 27 | 252 | Phage P4 DNA primase (Salmonella enterica subsp. enterica serovar lyphi strain CT18) | ||||
| AAG54598 | 70 | 9e-11 | 25 | 258 | Alpha replication protein of prophage CP-933 (E. coli O157:H7 EDL933) | ||||
| rtp52 | 34808-35281 | − | 157 | YP_003922 | 135 | 3e-31 | 51 | 133 | Hypothetical protein gp23 (phage T1)b |
| rtp53 | 35383-37377 | + | 664 | YP_003923 | 679 | 0.0 | 50 | 670 | Putative ATP-dependent helicase gp22 (phage T1) |
| NP_934217 | 155 | 4e-36 | 23 | 631 | DNA or RNA helicase (Vibrio vulnificus YJ016) | ||||
| rtp54 | 37379-37798 | + | 139 | YP_003924 | 142 | 2e-33 | 53 | 138 | Hypothetical protein gp21 (phage T1) |
| AAW67538 | 59 | 2e-08 | 31 | 121 | Hypothetical protein (Listonella pelagia phage phiHSIC) | ||||
| YP_006552 | 44 | 6e-04 | 30 | 118 | PmgM (phage P1) | ||||
| rtp55 | 37875-38075 | + | 66 | None | |||||
| rtp56 | 38075-38296 | + | 73 | YP_003926 | 37 | 0.12 | 48 | 39 | Hypothetical protein gp19 (phage T1)b |
| rtp57 | 38293-38481 | + | 62 | None | |||||
| rtp58 | 38552-38677 | + | 41 | YP_003927 | 37 | 43 | Hypothetical protein gp18 (phage T1)b | ||
| rtp59 | 38674-38922 | + | 82 | YP_070306 | 65 | 3e-10 | 44 | 58 | Hypothetical protein YPTB1780 (Yersinia pseudotuberculosis IP 32953) |
| rtp60 | 38925-39176 | + | 83 | YP_003928 | 42 | 0.003 | 34 | 83 | Hypothetical protein gp17 (phage T1)b |
| rtp61 | 39259-40395 | + | 378 | YP_003929 | 684 | 0.0 | 87 | 377 | Hypothetical protein gp16 (phage T1) |
| ZP_00303432 | 138 | 2e-31 | 26 | 376 | Hypothetical protein (Novosphingobium aromaticivorans) | ||||
| YP_025077 | 47 | 9e-04 | 22 | 203 | Hypothetical protein ORF50 (phage phi AT3) | ||||
| rtp62 | 40469-40642 | + | 57 | YP_003931 | 42 | 0.003 | 44 | 50 | Hypothetical protein gp14 (phage T1)b |
| rtp63 | 40773-40988 | + | 71 | YP_003932 | 93 | 1e-18 | 59 | 71 | Putative holin.gp13 (phage T1)b |
| rtp64 | 40989-41474 | + | 161 | YP_003933 | 130 | 1e-29 | 43 | 158 | Endolysin gp12 (phage T1) |
| NP_877484 | 120 | 1e-26 | 46 | 149 | Putative phage lysozyme (phage phiKMV) | ||||
| rtp65 | 41477-41839 | + | 120 | YP_003934 | 83 | 2e-15 | 39 | 116 | Hypothetical protein gp11 (phage T1)b |
| rtp66 | 41854-42129 | − | 91 | YP_003935 | 101 | 4e-21 | 50 | 90 | Hypothetical protein gp10 (phage T1)b |
| rtp67 | 42193-43776 | − | 527 | YP_003936 | 579 | e-164 | 56 | 521 | Hypothetical protein gp09 (phage T1) |
| NP_840326 | 92 | 4e-17 | 22 | 426 | Hypothetical protein NE0232 (Nitrosomonas europaea ATCC 19718) | ||||
| rtp68 | 43963-44202 | − | 79 | None | |||||
| rtp69 | 44199-44552 | − | 117 | YP_003938 | 75 | 4e-13 | 40 | 120 | Hypothetical protein gp07 (phage T1)b |
| rtp70 | 44552-44713 | − | 53 | None | |||||
| rtp71 | 44713-44886 | − | 57 | None | |||||
| rtp72 | 44950-45114 | − | 54 | None | |||||
| rtp73 | 45111-45350 | − | 79 | None | |||||
| rtp74 | 45361-45537 | − | 58 | None | |||||
| rtp75 | 45546-45749 | − | 67 | None | |||||
ID%, percent amino acid identity in region of BlastP alignment; overlap, length of BlastP alignment between query and target sequences.
The only homolog for the Rtp query sequence is found in phage T1.
We numbered the Rtp genes from left to right in ascending order, in agreement with the main direction of transcription and the standard lambdoid gene order of head, tail, and early genes. For consistency with early T1 gene numbering, Roberts et al. (75) chose to number the genes of T1 from left to right in descending order. The distributions of small genes, i.e., encoding putative proteins shorter than 100 aa, are similar in phages Rtp and T1; these genes are clustered at the ends of the chromosome maps and between the replication and lysis regions. The genome ends of phages Rtp and T1, according to the map in Fig. 2, have diverged, as shown by the lack of sequence similarity in the predicted small proteins encoded in the terminal regions. However, the high level of congruence, which includes very similar gene lengths for both small and longer genes in the “functional” part of the genomes, lends high credibility to the in silico gene identification.
In their discussion of the phage T1 genome, Roberts et al. (75) mentioned that the genome of a very close relative of phage T1, phage TLS (38), has been completely sequenced by Gregory German and Rajeev Misra at the University of Arizona, but this genome sequence has not been deposited in a public database. There are clear differences that distinguish phages Rtp and TLS from each other. (i) The genome of TLS contains a cytosine methylase (dcm), which is lacking in Rtp. (ii) TLS lacks ORFs 30, 31, and 32 (cor) found in T1, whereas Rtp carries three genes in that position (rtp44, rtp45, and rtp46), although these are not homologous to the T1 genes. (iii) TLS infection occurs through the TolC protein (5, 38, 66), in contrast to Rtp, which is independent of TolC. Therefore, the Rtp genome represents the second published sequence of three very closely related T1-like phages with three distinct receptor specificities.
Terminase and DNA packaging
The rtp19 and rtp21 genes code for the typical small and large subunits, respectively, of a phage terminase protein complex that initiates, drives, and terminates translocation of phage DNA into proheads (17, 21, 37). The small subunit has DNA-binding activity, and the large subunit provides ATP-binding, prohead-binding, and DNA cleavage activities (18, 20, 40, 41). Usually, the two subunits are encoded by adjacent genes, as in phage T1. The two predicted Rtp terminase genes show high end-to-end similarity to their T1 homologs (Table 2), but they are separated by rtp20, which encodes an HNH-type endonuclease (24, 27, 54). Casjens et al. (20) found a strong correlation between the phylogenetic relationships of phage terminase large subunits and the structure of the corresponding genome ends. In their comparison of 114 large terminase subunits, they distinguished the following types of phage chromosome packaging: (i) 5′-extended cos ends generated by terminase subunits related to the λ gene A product; (ii) 5′ cos ends generated by P2-like terminase subunits; (iii) 3′ cos ends (e.g., E. coli phages HK022 and HK97); (iv) T7-like packaging, which generates terminal direct repeats but no circular permutation; (v) P22-like headful terminases; (vi) T4-like headful terminases; (vii) Mu-like headful terminases, and (viii) GTA-like terminases of transduction-deficient prophages. The comparative analysis also shows that headful packaging terminases constitute a diverse group of sequences for which there is no evidence of a monophyletic origin. The closest homolog of the large terminase subunits of Rtp and T1 in a free-living phage is found in phage Aaphi23 of the periodontal pathogen Actinobacillus actinomycetemcomitans (Table 2). Phage T1 and Aaphi23 DNAs are packaged by the headful mechanism, resulting in terminal redundancy and circular permutation (72, 95). Restriction analysis of Rtp DNA argues against the presence of cohesive ends and for packaging of circularly permutated genome units. The agarose gel electrophoresis pattern of single digests of Rtp DNA with PstI (two recognition sites), BamHI (three sites), XbaI (seven sites), EcoRI (eight sites), or XhoI (nine sites) was not affected by heating to 80°C or by promoting annealing of potential complementary overlaps of terminal fragments prior to electrophoresis. Under the same conditions, annealing of the 12-nt-long complementary 5′ extensions in HindIII-digested λ DNA was readily detected (see Materials and Methods). The following Rtp restriction fragments that are not predicted by the in silico restriction map were found at submolar concentrations as sharp bands on agarose gels: XbaI (3.6 kb), XhoI (5.6 kb), EcoRI (6.0 kb), and PstI (10.2 kb). Undigested Rtp DNA produced only a single high-molecular-weight band. An additional restriction fragment of reduced concentration, the pac fragment, is a hallmark of the headful packaging mechanism. Therefore, packaging of Rtp appears to be initiated at a precise location on a concatemeric substrate (17, 52). This pac site is only used once in a packaging series, so that only the first packaged DNA of the series has a distinctive left end. The positions of the XbaI, XhoI, EcoRI, and PstI sites permitted unambiguous determination of the location of pac. The Rtp pac site is located approximately 3.6 kb (± 0.2 kb) to the left of the XbaI restriction site found at positions 3689 to 3694 of the genome sequence (Fig. 2). By pairwise alignment searches, we failed to identify any obvious DNA sequence conservation in the regions of the Rtp and T1 genomes that are predicted to contain the pac sites. The pac site of T1 was mapped 1 kb to the left of an EcoRI restriction site (EcoRI site at nt 1323 to 1328 in the T1 genome sequence; GenBank accession no. NC_005833) (71). The five repeats of the ATATA sequence mentioned by Roberts et al. (75) as a possible motif in the pac region of phage T1 are not present in phage Rtp. The finding that the virion DNA of phage Rtp is generated by the headful mechanism supports the hypothesis of Casjens et al. (20) that the packaging type can be predicted from the phylogeny of the large terminase subunit. In their analysis, the terminase of phage Aaphi23 formed a weakly supported group with only one other sequence from lactococcal phage TP901-1. The present data expand the cluster to four sequences (from T1, Rtp, Aaphi23, and TP901-1) that appear to form a subfamily of headful terminases distinct from the currently more numerous P22-like large subunits.
Head genes
The region from rtp23 to rtp43 contains genes for head and tail morphogenesis; this region is highly syntenic between phages Rtp and T1. The precise location of the border between the head and tail genes is difficult to predict from the available database information. Rtp23 is a putative portal protein. Close homologs in free phages are only found in phages T1 and Aaphi23. Rtp24 is related to the Pfam family phage_Mu_F (6). Another member of this family, gp7 of Bacillus subtilis phage SPP1, is a minor head protein that interacts with portal protein gp6 (51, 81). Rtp25 is a possible prohead protease. Rtp25 is similar to products of conserved gene cluster COG3566, which was shown by computational analysis to belong to a superfamily of prohead proteases that includes proteins of double-stranded DNA bacteriophages and herpesviruses (23). The downstream rtp26 gene is duplicated in phage T1 as orf49 and orf48, and these are the only clear homologs. Proteome analysis of phage T1 has shown that both gp49 and gp48 are abundant proteins (75), but their mutual similarity has not been noted previously. Rtp27 displays high end-to-end similarity to the putative major T1 head protein gp47.
Tail genes
Differences in an otherwise highly conserved region of tail genes may localize the distinct specificities of phages Rtp and T1 for cellular surface receptors used for infection. The gene order from rtp30 to rtp50 is the same as in phage T1, with two gene insertions in Rtp (rtp34, rtp40) and a potential module replacement where Rtp genes rtp44, rtp45, and rtp46 replace T1 genes 32, 31, and 30. rtp33 codes for a homolog of the putative major tail protein gp41 of T1. For the genes rtp30, rtp31, and rtp32, there is no sequence-based evidence that they are involved in tail formation. The 3′ end of the rtp35 coding sequence contains a slippery sequence motif (AAAAAAC-stop codon) that might permit a translational −1 shift, thus leading to the translation of an Rtp35-Rtp36 fusion protein across the 8-nt intergenic gap in addition to the expression of proteins Rtp35 and Rtp36. In tailed phages, such programmed translational frameshifting seems to be typical for genes corresponding to the λ G/GT region upstream of the gene encoding the tape measuring protein (97).
Rtp37 is a tail tape-measuring protein with many homologs in phages and prophages of gram-negative bacteria, including the λ gene H product. The rtp38 homolog orf37 in phage T1 and the three downstream T1 genes are conserved tail genes also found in λ-like phages and prophages of various members of the γ subgroup of proteobacteria, including T1, N15, HK022, HK97, λSo, PhiE125, Gifsy-1 and -2, Fels-1, CP933-O, CP933-K, and λ (genes M, L, K, and I; see reference 75 for a phylogenetic analysis). Notably, in phage Rtp, this run of four genes is separated by the insertion of a second HNH-type endonuclease gene (rtp40). This endonuclease gene overlaps downstream tail gene rtp41 by 20 bp without changing the amino acid conservation that exists between the N termini of Rtp41 and T1-gp35.
Sequence similarity searches for Rtp41 revealed not only homology to many putative tail assembly proteins of phages and prophages, including λ tail protein K, but also similarity to more distantly related bacterial endopeptidases and cell wall-associated hydrolases. In silico domain analysis suggests that the 252-aa Rtp41 protein is composed of two domains, both of which are related to proteases. The Conserved Domain search service integrated into NCBI BlastP searches (63) indicated the presence of an N-terminal COG1310 domain (predicted metal-dependent protease of the PAD1/JAB1 superfamily) and a C-terminal NlpC/C60 domain. The latter was also clearly identified in a Pfam search (6). NlpC/C60 is part of a superfamily of cysteine/histidine-dependent amidohydrolases/peptidases (4, 7, 74). Since the protease-defining residues of the CHAP family are well conserved in Rtp41 and its homologs in λ and T1, one can envisage an enzymatic role for this widespread and highly conserved tail protein of lambdoid phages, for example, in the assembly of the tail. In fact, tape-measuring protein H of λ is cleaved toward the end of tail assembly by an unidentified protease (55, 85). Alternatively, Rtp41 and related proteins may be involved in cell wall degradation during infection of the host.
Rtp42 is a homolog of conserved λ tail assembly protein I, and Rtp43 is homologous to the tail fiber J, which determines λ host specificity (36, 94). The presence of several λ-related genes in the tail region of Rtp prompted us to determine how many conserved genes are shared between Rtp and model phage λ. A BlastP search (E-value limit, 10E-5) with all of the predicted Rtp gene products against a λ protein database revealed that sequence similarity between the two genomes was limited to adjacent tail genes rtp37, -38, -39, -41, -42, and -43 and λ H, M, L, K, I, and J. These λ gene products and λ G form the tail assembly initiation complex, which precedes the polymerization of major tail protein V up to a shaft length that is determined by the sequence length of protein H (reviewed in references 55 and 56). It appears that phages Rtp and T1 have a conserved λ-like initiation mechanism for the assembly of their lambdoid tails and that the amino acid sequences of the major subunit proteins can be completely unrelated.
The putative tail fiber proteins Rtp43 and T1-gp33 are probably involved in phage adsorption to their E. coli outer membrane receptors. Differences between these highly similar proteins may indicate the differing receptor specificities. Sequence comparisons of the C-terminal part of Rtp43 with closely related tail fibers are of particular interest since the last 249 residues of λ J bind specifically to the cognate receptor LamB (90). A comparison of Rtp43 with its closest homologs is summarized in Table 3. The lengths of the related tail fibers from phages Rtp, T1, HK97, HK022, N15, phi1026b, phiE125, and ES18 vary from 1,061 to 1,296 residues. The very long J fiber homolog of phiKO2 (3,433 aa) is an exception. This protein can be viewed as an approximately 1,300-aa sequence with a long insertion made up of eight evenly spaced imprecise repeats of 130 to 180 aa (19). Multiple-sequence alignment of these tail fibers revealed that the N-proximal 800 to 900 aa residues are more strongly conserved than the C-proximal part, whose length also varies more than that of the N-proximal portion. Four of the nine closest fiber homologs in free phages belong to FhuA-dependent phages T1, HK022, N15, and ES18 (Table 3). The amino acid sequence conservation in the C-proximal part of the various λ J-like proteins is reduced to different degrees. The T1 fiber is very similar to Rtp43, whereas λ J has the lowest score. The fiber protein of ES18 has an overall score slightly above that of λ J, but it aligns much better with Rtp43 in the C-proximal region. We propose that Rtp43 has not undergone a major domain exchange in the C-proximal region to yield a new receptor specificity. If Rtp43 determines the host range of Rtp, which differs from that of T1, then the difference is likely caused by minor changes in the amino acid sequence, probably in the variable C-terminal region. This conclusion is supported by the finding that small sequence changes lead to new receptor specificities. For example, mutations in the hypervariable region of the long tail fibers of phage Ox2 change the receptor specificity from the wild-type OmpA protein to the OmpC and OmpX proteins, and even to LPS (33). Similar results were obtained with host range mutations in T-even-type phage M1, which changed specificity from OmpA in the wild type to OmpC and subsequently to OmpT (45).
TABLE 3.
Similarity of Rtp43 to λ J homologs
| Rank of overall similarity | Phage | Receptor | Length of λ J homolog (aa) | Overalla | BlastP score
|
|
|---|---|---|---|---|---|---|
| N-terminal 840 aab | C-terminal 296 aab | |||||
| 1 | T1 | FhuA | 1,172 | 1,358 | 1,103 | 224 |
| 2 | HK97 | LamB | 1,296 | 870 | 747 | 132 |
| 3 | HK022 | FhuA | 1,183 | 867 | 742 | 143 |
| 4 | N15 | FhuA | 1,061 | 852 | 729 | 129 |
| 5 | phiKO2 | Uc | 3,433 | 742 | 714 | 88 |
| 6 | phi1026b | U | 1,101 | 656 | 579 | 101 |
| 7 | phiE125 | U | 1,101 | 648 | 573 | 100 |
| 8 | ES18 | FhuAd | 1,051 | 386 | 303 | 97 |
| 9 | λ | LamB | 1,132 | 328 | 327 | 34 |
Full-length Rtp43 (1,136 aa) was used to search the virus section of the NCBI nonredundant protein database.
Only the 840 N-terminal residues or the 296 C-terminal residues of Rtp43 were used for the BlastP search.
U, unknown.
FhuA of Salmonella enterica serovar Typhimurium, formerly designated SidA (13, 59), but not FhuA of E. coli. phiE125 (96) and phi1026b (30) are temperate phages that infect Burkholderia mallei, both of which require LPS O antigen for infection. Information on additional protein receptors is not available.
rtp50 encodes a second putative tail fiber protein. The 960-aa protein displays similarity to predicted additional tail fiber proteins from phages T1 (728 aa), HK022 (371 aa), and HK97 (321 aa). However, the sequence conservation is limited to the N-proximal 170 to 190 residues. The large remainder of Rtp50 is rich in serine and glycine residues and is similar, although to a very low degree, to cell surface proteins of various bacteria. Three lines of evidence suggest that the unique C-proximal part of Rtp50 contains a β-helix domain, a structure that takes the shape of a three-sided prism (80, 98). (i) Sequence analysis with the BetaWrap algorithm gave a highly significant P value of 0.00026. (ii) An analysis based on hidden Markov models yielded an E value of 0.00037 (26). (iii) Fold recognition based on threading of predicted secondary-structure elements of Rtp50 detected the crystal structure of a polygalacturonase with a score (3D-PSSM E value, 0.0599) that represents more than 90% confidence for correct fold recognition (58). This polygalacturonase of Erwinia carotovora subsp. carotovora (PDB identifier, 1bhe) has a single-stranded right-handed β-helix fold (70). Additional lower-quality hits in the same fold library also supported the conclusion that Rtp50 contains a domain with a β-helix fold. The interesting aspect of the predicted fold of Rtp50 is that β-helix domains are typical constituents of viral adhesins, some phage tail proteins, and many carbohydrate-binding proteins (http://betawrap.lcs.mit.edu/list.txt). For a review of examples, e.g., P22 tailspike protein gp12, T4 short tail fiber gp12, and adenovirus penton fiber, see references 70 and 93. We found no evidence for a β-helix in the distinct C-proximal parts of the fiber proteins of phages T1, HK022, and HK97, pointing to tail functions which are distinct from Rtp50. The conserved N-terminal domain could serve as an anchor during tail assembly, and the nonconserved C-terminal domains might provide tail functions specific for each phage. The relative locations of rtp50 and the corresponding orf25 tail fiber gene in phage T1 are conserved in the two genomes. Both genes are separated by six genes from the last gene in the tail gene cluster, the λ J homolog. In phage HK97, the additional tail fiber gene, stf or orf23, is located immediately downstream of the λ J homolog. In HK022, the tail fiber genes are separated by three genes that are conserved among phages N15, ES18, and HK022. One of these conserved genes is the lysogenic conversion gene cor (88).
Are rtp45, cor, and the T5 gene llp functional analogues?
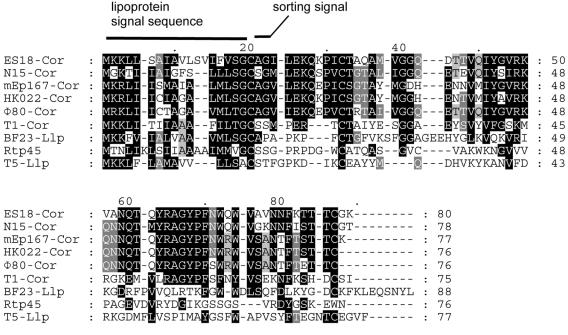
The most notable difference between the central regions of the Rtp and T1 genomes is the lack of sequence similarity of the rtp44, -45, and -46 genes in Rtp and orf32, -32, and -30 (cor) in T1 downstream of the tail gene operons. Homologs of orf32, -33, and -30 (cor) are found at the same genome location and in the same order in five phages—T1, HK022, N15, ES18, and Φ80—all of which use FhuA as a receptor. This set of three genes is strictly limited to these phages, as shown by comprehensive BlastP and TBlastN searches of all of the phage and bacterial sequences in GenBank. Lambdoid FhuA- and TonB-dependent phage mEp167 also contains a cor homolog, but sequence information is limited to this gene only (86). Related phages with different receptor specificities—HK97 (receptor, LamB; 31), TLS (receptor, TolC; 38), and Rtp (receptor unknown)—lack sequence similarities. The small cor gene was originally identified in Φ80 as a lysogenic conversion gene that prevents adsorption of superinfecting Φ80 to FhuA in E. coli lysogens carrying a Φ80 prophage (65, 88; note: the correct ORF for cor of Φ80 and N15 was identified by Vostrov and colleagues [88], but Cor database entry BAA00267 still contains the incorrect protein sequence). We noticed a number of similarities between the Cor family and the llp gene product of phage T5, which inactivates the phage T5 FhuA receptor (15, 28), a receptor-blocking protein from T5-related phage BF23 (GenBank accession number AAZ03643), and the predicted Rtp45 gene product. The proteins are putative outer membrane proteins (shown for Llp of phage T5). Rtp45 and the other lipoproteins contain an N-terminal sorting signal (positions 2 and 3 of the mature protein; Fig. 3) that favors translocation into the outer membrane (84). Their N termini conform to the Prosite consensus for prokaryotic lipoprotein lipid attachment sites (50; http://www.expasy.org/prosite). The genes are located next to tail fiber genes (Fig. 4). The lengths of the putative lipoproteins (a lipid has been shown for Llp of T5) are very similar, between 76 and 80 residues. Only Llp of phage BF23 is slightly longer, with 88 residues (Fig. 3). Although the amino acid sequences of the mature Cor proteins differ from the lipoproteins of Rtp, T5, and BF23, the hydrophobicity profile of Rtp45 is similar to those of the Cor proteins (60). In the cases studied, genetic and biochemical evidence demonstrates inactivation of the host phage receptor. Receptor inactivation is not only advantageous to prevent superinfection of lysogenic bacteria carrying prophages but may also serve to prevent phage inactivation by receptors when phages are released from lysing bacteria (28). This explains why nonlysogenic phages like Rtp, T1, and T5 synthesize receptor-inactivating lipoproteins. We propose that all known Cor homologs are outer membrane lipoproteins that inactivate receptors and that Rtp45 and Llp of BF23 share this property.
FIG. 3.
Alignment of Cor proteins with other predicted small phage lipoproteins. This multiple-sequence alignment of full-length protein sequences was generated with ClustalW. The fully conserved cysteine residue at position 20 of the alignment constitutes the N-terminal amino acid of the mature lipoproteins. It is the site for cleavage of the signal sequence and lipid modification. Llp of phage T5 is the only one of these proteins for which lipid modification and its functional importance have been experimentally proven (28, 68, 77).The Cor protein of phage T1 is the most distant member of the Cor family, which consists of a group of six homologous sequences. Llp of T5 and BF23 and Rtp45 show little sequence conservation with respect to the Cor proteins beyond the signal and sorting sequences. The correct amino acid sequence for Cor of Φ80 has been taken from Vostrov et al. (88). Accession numbers of the other Cor and Llp proteins: ES18, AAW70503; N15, NP_046919; mEp167, AAT11800; HK022, NP_037685; T1, AAP49969; T5, Q38162; BF23, AAZ03643.
FIG. 4.
Genetic map of the cor region in FhuA-dependent phages and the corresponding region of T1-related phage Rtp. Homologous genes of FhuA-dependent phages HK022, N15, ES18, Φ80, and T1 and T1-related phage Rtp are indicated by white arrows. J, homolog of λ tail fiber gene J; fibB, putative second tail fiber gene (the module of three recombination genes in T1 and Rtp is followed by fibB homologs in these phages); cor, lysogenic/lytic conversion gene (see text). The lengths of the gene products are indicated inside the arrows; values between the gene arrows indicate intergenic distances (negative values for overlapping genes). The genome of Φ80 has not been sequenced. Two partial ORFs encoding FibB and a homolog of the conserved but uncharacterized 212-, 225-, 217-, and 228-aa proteins of the other FhuA-dependent phages were identified by TBlastN searches of the nucleotide database entry for Φ80 cor (D00360). The genes of Rtp shown by gray shaded arrows, including that for the putative lipoprotein Rtp45, are not homologous to the genes of the FhuA-dependent phages. The latter display a conserved arrangement of the genes between fibJ and cor.
The two genes of unknown function to the left of cor are closely linked to the preceding tail fiber genes. They share end-to-end sequence similarity and were only found in FhuA-dependent phages. Thus, the cor region, including the two left genes, might constitute a phage module involved in specificity and inactivation of FhuA. Since the llp/cor homologs and the putative outer membrane lipoprotein Rtp45 are located close to the tail fiber genes (Fig. 4), horizontal cotransfer of the tail fiber and the receptor inactivator genes is favored. The only homologs of the neighboring gene in Rtp, rtp44, are genes of unknown function located next to tail fiber genes in Yersinia enterocolitica phage PY54 and in prophages of two Y. pestis strains, CO92 and KIM.
Recombination, replication, and lysis modules.
The genes rtp47, rtp48, and rtp49 show 44, 43, and 45% sequence identities, respectively, to the genes encoding the putative T1 recombination module (Table 2). Rtp47 is a homolog of RecE, a 5′ exonuclease (25). Rtp48 is a member of the Erf (essential recombination function) family. rtp49 and orf27 of T1 both encode putative single-stranded DNA-binding (Ssb) proteins. Roberts et al. (75) identified the biological function of the Ssb protein as replication. However, as Ssb of phage T7 is required for both replication and recombination (49), this might also be the case for T1 and Rtp. Rtp51 (DNA primase) and Rtp53 (ATP-dependent helicase) share 42 and 51% sequence identity, respectively, over their entire length with the predicted replication proteins gp24 and gp22 of T1. Similarity to other bacterial and phage primase and helicase homologs further supports these functional assignments. gp23 of T1, whose function is unknown, is also conserved in Rtp and T1. rtp52 and its homolog orf23 code for putative DNA-binding transcriptional regulators (see below).
Like T1, Rtp contains two closely linked lysis genes, coding for a holin and an endolysin. The first genes of the lysis cassette (encoding those for Rtp63 and T1-gp13) code for holins that form pores in the cytoplasmic membrane. The products of the downstream genes are phage endolysins (Rtp64 and T1-gp12) that cleave the β-1,4 linkages between N-acetyl-d-glucosamine and N-acetylmuramic acid in peptidoglycan heteropolymers of prokaryotic cell walls (Pfam PF00959, phage lysozyme) (75). The function of the surrounding genes that are conserved between Rtp and T1 is unknown (Table 2).
Identification of a putative transcriptional regulator in Rtp and T1.
The predicted proteins Rtp52 (157 aa) and T1-gp23 (150 aa) are 51% identical over 133 residues, and no other closely related sequences were detected by BlastP. In both phages, the transcription polarity of these genes is the opposite of that of the adjacent DNA helicase genes. The length of the intergenic regions, putative divergent promoter regions, is approximately 100 bp in both phages. Analysis of Rtp52 and T1-gp23 for the presence of a DNA-binding domain yielded helix-turn-helix motifs with confidence levels of close to 100% for Rtp52 and approximately 50% for T1-gp23 (32) (Fig. 5). The helix-turn-helix motifs were in the same location in the two homologs. Other sequences that are distantly related to Rtp52 and T1-gp23 were detected with significant E values by a PSI-BlastP search. The result of the fifth iteration is shown in Fig. 5. The best hits of the search are all transcriptional regulators, followed by a mixture of transposases and more transcriptional regulators. All of the homologs tested contain a predicted helix-turn-helix motif, with various degrees of confidence, in a position equivalent to that of the Rtp and T1 proteins. Conservation of certain amino acid residues was limited to the helix-turn-helix region. The data suggest that both Rtp52 and T1-gp23 are transcriptional regulators with DNA-binding activity.
FIG. 5.
Identification of a DNA-binding helix-turn-helix motif in Rtp52 and T1-gp23. (A) Alignment of the helix-turn-helix regions of Rtp52 and T1-gp23 with distantly related DNA-binding domains. The numbering of homologs refers to the PSI-BLAST results in panel B. Numbering of amino acids is shown at the right side of the alignment. A helix-turn-helix motif search by the Dodd-Egan method (32) identified the sequence GTKANIAKQLKVTPQAVEEWFK, starting at position 53 in Rtp52, with a score of 5.19, representing approximately 100% probability for a DNA-binding helix-turn-helix fold. The position of the predicted helix-turn-helix motif is shown by an arrow below the alignment. All of the other proteins in the alignment also contained a predicted helix-turn-helix motif in an equivalent position. (B) Result of PSI-BLAST iteration 5 with Rtp52 as the query sequence. The 10 best-scoring hits were tested for helix-turn-helix motifs. The probabilities for a helix-turn-helix motif based on the Dodd-Egan method are shown next to the PSI-BLAST expectation values. The annotation of each remote homolog was reexamined by BlastP database searches. In the PSI BLAST analysis of Rtp52, we excluded hits to the multidomain proteins of the nonribosomal peptide synthetase family from subsequent iterations of the search. Manual filtering was necessary since low-scoring PSI-BLAST search results tend to be drawn to large protein families.
Conserved inverted repeats upstream of predicted promoter regions.
After identification of a putative transcriptional regulator, we searched for potential cis-acting transcription signals that might be recognized by Rtp52. Transcriptional regulators are often autoregulated, and divergent promoter regions frequently function in the coordinated control of two transcription units. The gene arrangement in Rtp suggests (auto)regulation of Rtp52 and two replication proteins, the DNA helicase Rtp53 and the primase Rtp51. Additional target sites might exist elsewhere in the genome. Therefore, the 101-nt rtp52-rtp53 intergenic region was tested for similarity against the whole genomes of Rtp and T1 by local BlastN searches. The alignments with the best E values (<10−3) involved the section from nt 12 to nt 31 of the 101-nt intergenic region, corresponding to the 20-nt sequence AATAGCATTTTTTGTTAAAA. Since 20 continuous base pairs would be an unusually large target for typical transcriptional regulators, we attempted to divide the sequence into motif elements. The pattern TAGCA-NNNNN-TGCTAwas highly overrepresented in the genomes of Rtp and T1. Both phage genomes contain 10 perfect matches (for comparison, there are only 2 perfect matches in the 4.6-Mb E. coli genome). The expected random frequency would be 1 in every 410 bp = 1,048,576 bp. Allowing one mismatch in the pattern increases the number in Rtp to 19, 15 of which are located in noncoding regions. Figure 6 shows an alignment of these 15 repeat sequences, together with flanking DNA sequences and distances from start codons. Transcription of T1 genes is dependent on host RNA polymerase (89). One likely possibility for gene regulation by phage T1 or Rtp is therefore the activation or repression of σ70-dependent promoters. The multiple-sequence alignment shows that hexamers 22 bases downstream from the inverted repeat motif conform well to the −10 region of σ70 promoters (−12 to −7 region), especially at functionally important and highly conserved positions −12, −11, and −7 (35, 44, 64). The distance from predicted −10 boxes to the predicted start codons is in most cases between 20 and 40 nt, often close to 30 nt (Fig. 6B). In contrast, there are no obvious −35 boxes when promoter spacer regions of 16 to 18 nt are allowed. The right unit of the inverted repeats is close to the −35 box recognized by region 4.2 of σ70. It seems possible that the TAGCA inverted repeats with a 5-nt intervening sequence might be binding sites for a transcriptional activator. There are additional highly conserved bases outside the TAGCA repeats (marked with an asterisk in Fig. 6) which were not included in our interpretation.
FIG. 6.
Repeat sequences in putative promoter regions of the Rtp genome. (A) Alignment of upstream regions of Rtp genes containing the repeat sequence TAGCA-(N)5-TGCTA with one mismatch allowed. This 15-nt repeat sequence presents an inverted repeat with a 5-nt spacer. The inverted repeat (matching bases in uppercase letters) is followed downstream by a 22-nt spacer region and a hexamer that conforms to the −10 consensus sequence of E. coli σ70-dependent promoters (consensus-matching bases in uppercase letters). Additional sequences flanking these elements are also shown. The bases that occupy the positions of a canonical −35 hexamer 17 nt upstream of the aligned −10 regions are underlined. These bases do not resemble −35 sequences of σ70 promoters, even if a shift of 1 nt to the right or left is considered (16- or 18-nt promoter spacer). The consensus sequence of E. coli σ70-dependent promoters is shown below the alignment, highly conserved bases are in uppercase, and less conserved bases are in lowercase (35a, 44, 50a); asterisks below the sequence alignment mark further conserved nucleotide positions that are not discussed. (B) Positions of the putative transcriptional regulatory regions. The listed coordinates on the Rtp genome start at the left end of the hyphenated TAGCA inverted repeats and end with the last nucleotide of the predicted −10 elements shown in panel A. The upstream region of gene rtp75 contains two copies of the motif; both are located much farther away from the start codon than in the other genes.
rtp4 and rtp6 code for T4-like functions.
Rtp6 is a short protein of 30 aa that is highly similar to the 27-residue Stp protein of T4 (3, 22, 57) and related phages (79% identity over 24 residues). Rtp6 is the first Stp homolog outside the T4-like phages. The only other phages that are known to contain stp homologs are the T4-related phages TuIa, TuIb, LZ3, LZ5, SCI, Ox2, and Baker (all classified in the T4-like group of the family Myoviridae). Stp acts as an activator of a tRNA anticodon nuclease (PrrC) of certain E. coli strains which specifically cleaves the anticodon loop of tRNALys. Usually, the prrC-encoded anticodon nuclease is maintained in an inactive state by the PrrI restriction endonuclease. Stp acts as a negative effector of PrrI, thereby activating the PrrC anticodon nuclease. This results in depletion of the translational capacity in the infected host cell. T4 counteracts this by repairing cleaved tRNALys with the help of its polynucleotide kinase and RNA ligase (69). Rtp4 is homologous to PseT of T4 (T4 polynucleotide kinase; 91, 92). However, there is no counterpart in Rtp for a gene encoding RNA ligase. T1 also contains a polynucleotide kinase gene (orf64) but no stp or any obvious gene for a RNA ligase. The function of the stp homolog encoding Rtp6 in Rtp is therefore unclear.
Concluding remarks.
Comparison of the genomes of phages Rtp and T1 shows (i) a remarkable overall similarity of gene content and synteny; (ii) diverged genome ends—as defined in the chromosome maps in Fig. 2—with limited gene conservation to the left of the terminase genes (5.5 kb at the left end) and no sequence conservation across the rightmost 1.7-kb segment of the Rtp genome; (iii) distinct numbers of HNH-type endonuclease genes (two in Rtp, three in T1); (iv) modular exchanges of rtp44, -45, and -46 and orf32, -31, and -30 of phage T1; (v) putative secondary tail fibers Rtp50 and T1-gp26; and (vi) a single conserved putative transcriptional regulator in both phages.
Rtp shows a novel tail tip morphology which displays distant similarity to phage T1. The greater thickness and rigidity of the four leaf-like structures protruding at the tail tip of phage Rtp in comparison to the tail tip of phage T1 might be caused by amino acid substitutions in the FibJ homolog Rtp43. We favor the idea that the nonhomologous replacements involving rtp44 and rtp45, as well as the approximately 800-residue unique C-terminal domain of Rtp50, are responsible for the morphogenetic changes in the tail tip which probably cause the different receptor specificity of Rtp. The conservation of a putative three-gene cor module in FhuA-dependent phages (orf32, -31, and -30 of phage T1) warrants further studies.
Both Roberts et al. (75) and we found signs of multiple promoter regions. Studies of gene expression in T1 date back to 1977 (89) and were only directed at the level of translation. These analyses revealed three stages of T1 gene expression: early, early-late, and late. It will be interesting to find out where transcription start points are located, whether Rtp52 and T1-gp23 are indeed novel DNA-binding transcriptional regulators, whether they function as activators or repressors, and where their target sites might be. So far, T1-like phages could only be classified on the relatively weak basis of morphology and virulence. Now there are molecular criteria available for the classification of new members of the T1 group.
Acknowledgments
We thank Silke Patzer and Klaus Hantke for many helpful discussions and Karen A. Brune for critical reading of the manuscript.
This work was supported by the Fonds der Chemischen Industrie.
REFERENCES
- 1.Abedon, S. T. 2000. The murky origin of Snow White and her T-even dwarfs. Genetics 155:481-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amitsur, M., I. Morad, and G. Kaufmann. 1989. In vitro reconstitution of anticodon nuclease from components encoded by phage T4 and Escherichia coli CTr5X. EMBO J. 8:2411-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anantharaman, V., and L. Aravind. 2003. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 4:R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Austin, E. A., J. F. Graves, L. A. Hite, C. T. Parker, and C. A. Schnaitman. 1990. Genetic analysis of lipopolysaccharide core biosynthesis by Escherichia coli K-12: insertion mutagenesis of the rfa locus. J. Bacteriol. 172:5312-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bateman, A., and N. D. Rawlings. 2003. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem. Sci. 28:234-237. [DOI] [PubMed] [Google Scholar]
- 8.Besemer, J., A. Lomsadze, and M. Borodovsky. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boman, H. G., and D. A. Monner. 1975. Characterization of lipopolysaccharides from Escherichia coli K-12 mutants. J. Bacteriol. 121:455-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bradley, P., L. Cowen, M. Menke, J. King, and B. Berger. 2001. BETAWRAP: successful prediction of parallel beta-helices from primary sequence reveals an association with many microbial pathogens. Proc. Natl. Acad. Sci. USA 98:14819-14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Braun, V. 1989. The structurally related exbB and tolQ genes are interchangeable in conferring tonB-dependent colicin, bacteriophage, and albomycin sensitivity. J. Bacteriol. 171:6387-6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Braun, V., S. Gaisser, C. Herrmann, K. Kampfenkel, H. Killmann, and I. Traub. 1996. Energy-coupled transport across the outer membrane of Escherichia coli: ExbB binds ExbD and TonB in vitro, and leucine 132 in the periplasmic region and aspartate 25 in the transmembrane region are important for ExbD activity. J. Bacteriol. 178:2836-2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braun, V., K. Hantke, and W. Stauder. 1977. Identification of the sid outer membrane receptor protein in Salmonella typhimurium SL1027. Mol. Gen. Genet. 155:227-229. [DOI] [PubMed] [Google Scholar]
- 14.Braun, V., and C. Herrmann. 1993. Evolutionary relationship of uptake systems for biopolymers in Escherichia coli: cross-complementation between the TonB-ExbB-ExbD and the TolA-TolQ-TolR proteins. Mol. Microbiol. 8:261-268. [DOI] [PubMed] [Google Scholar]
- 15.Braun, V., H. Killmann, and C. Herrmann. 1994. Inactivation of FhuA at the cell surface of Escherichia coli K-12 by a phage T5 lipoprotein at the periplasmic face of the outer membrane. J. Bacteriol. 176:4710-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casadaban, M. J., and S. N. Cohen. 1979. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc. Natl. Acad. Sci. USA 76:4530-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Casjens, S., and M. Hayden. 1988. Analysis in vivo of the bacteriophage P22 headful nuclease. J. Mol. Biol. 199:467-474. [DOI] [PubMed] [Google Scholar]
- 18.Casjens, S., L. Sampson, S. Randall, K. Eppler, H. Wu, J. B. Petri, and H. Schmieger. 1992. Molecular genetic analysis of bacteriophage P22 gene 3 product, a protein involved in the initiation of headful DNA packaging. J. Mol. Biol. 227:1086-1099. [DOI] [PubMed] [Google Scholar]
- 19.Casjens, S. R., E. B. Gilcrease, W. M. Huang, K. L. Bunny, M. L. Pedulla, M. E. Ford, J. M. Houtz, G. F. Hatfull, and R. W. Hendrix. 2004. The pKO2 linear plasmid prophage of Klebsiella oxytoca. J. Bacteriol. 186:1818-1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casjens, S. R., E. B. Gilcrease, D. A. Winn-Stapley, P. Schicklmaier, H. Schmieger, M. L. Pedulla, M. E. Ford, J. M. Houtz, G. F. Hatfull, and R. W. Hendrix. 2005. The generalized transducing Salmonella bacteriophage ES18: complete genome sequence and DNA packaging strategy. J. Bacteriol. 187:1091-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Catalano, C. E. 2000. The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell. Mol. Life Sci. 57:128-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chapman, D., I. Morad, G. Kaufmann, M. J. Gait, L. Jorissen, and L. Snyder. 1988. Nucleotide and deduced amino acid sequence of stp: the bacteriophage T4 anticodon nuclease gene. J. Mol. Biol. 199:373-377. [DOI] [PubMed] [Google Scholar]
- 23.Cheng, H., N. Shen, J. Pei, and N. V. Grishin. 2004. Double-stranded DNA bacteriophage prohead protease is homologous to herpesvirus protease. Protein Sci. 13:2260-2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chevalier, B. S., and B. L. Stoddard. 2001. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 29:3757-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clark, A. J., V. Sharma, S. Brenowitz, C. C. Chu, S. Sandler, L. Satin, A. Templin, I. Berger, and A. Cohen. 1993. Genetic and molecular analyses of the C-terminal region of the recE gene from the Rac prophage of Escherichia coli K-12 reveal the recT gene. J. Bacteriol. 175:7673-7682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cowen, L., P. Bradley, M. Menke, J. King, and B. Berger. 2002. Predicting the beta-helix fold from protein sequence data. J. Comput. Biol. 9:261-276. [DOI] [PubMed] [Google Scholar]
- 27.Crutz-Le Coq, A. M., B. Cesselin, J. Commissaire, and J. Anba. 2002. Sequence analysis of the lactococcal bacteriophage bIL170: insights into structural proteins and HNH endonucleases in dairy phages. Microbiology 148:985-1001. [DOI] [PubMed] [Google Scholar]
- 28.Decker, K., V. Krauel, A. Meesmann, and K. J. Heller. 1994. Lytic conversion of Escherichia coli by bacteriophage T5: blocking of the FhuA receptor protein by a lipoprotein expressed early during infection. Mol. Microbiol. 12:321-332. [DOI] [PubMed] [Google Scholar]
- 29.Delbrueck, M., and S. E. Luria. 1942. Interference between bacterial viruses. I. Interference between two bacterial viruses acting upon the same host, and the mechanism of virus growth. Arch. Biochem. 1:111-141. [Google Scholar]
- 30.DeShazer, D. 2004. Genomic diversity of Burkholderia pseudomallei clinical isolates: subtractive hybridization reveals a Burkholderia mallei-specific prophage in B. pseudomallei 1026b. J. Bacteriol. 186:3938-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dhillon, E. K., T. S. Dhillon, A. N. Lai, and S. Linn. 1980. Host range, immunity and antigenic properties of lambdoid coliphage HK97. J. Gen. Virol. 50:217-220. [DOI] [PubMed] [Google Scholar]
- 32.Dodd, I. B., and J. B. Egan. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 18:5019-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Drexler, K., J. Dannull, I. Hindennach, B. Mutschler, and U. Henning. 1991. Single mutations in a gene for a tail fiber component of an Escherichia coli phage can cause an extension from a protein to a carbohydrate as a receptor. J. Mol. Biol. 219:655-663. [DOI] [PubMed] [Google Scholar]
- 34.Dsouza, M., N. Larsen, and R. Overbeek. 1997. Searching for patterns in genomic data. Trends Genet. 13:497-498. [DOI] [PubMed] [Google Scholar]
- 35.Fenton, M. S., and J. D. Gralla. 2001. Function of the bacterial TATAAT −10 element as single-stranded DNA during RNA polymerase isomerization. Proc. Natl. Acad. Sci. USA 98:9020-9025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Fenton, M. S., and J. D. Gralla. 2003. Roles for inhibitory interactions in the use of the −10 promoter element by sigma 70 holoenzyme. J. Biol. Chem. 278:39669-39674. [DOI] [PubMed] [Google Scholar]
- 36.Fuerst, C. R., and H. Bingham. 1978. Genetic and physiological characterization of the J gene of bacteriophage lambda. Virology 87:437-458. [DOI] [PubMed] [Google Scholar]
- 37.Fujisawa, H., and M. Morita. 1997. Phage DNA packaging. Genes Cells 2:537-545. [DOI] [PubMed] [Google Scholar]
- 38.German, G. J., and R. Misra. 2001. The TolC protein of Escherichia coli serves as a cell-surface receptor for the newly characterized TLS bacteriophage. J. Mol. Biol. 308:579-585. [DOI] [PubMed] [Google Scholar]
- 39.Gordon, D., C. Abajian, and P. Green. 1998. Consed: a graphical tool for sequence finishing. Genome Res. 8:195-202. [DOI] [PubMed] [Google Scholar]
- 40.Gual, A., and J. C. Alonso. 1998. Characterization of the small subunit of the terminase enzyme of the Bacillus subtilis bacteriophage SPP1. Virology 242:279-287. [DOI] [PubMed] [Google Scholar]
- 41.Gual, A., A. G. Camacho, and J. C. Alonso. 2000. Functional analysis of the terminase large subunit, G2P, of Bacillus subtilis bacteriophage SPP1. J. Biol. Chem. 275:35311-35319. [DOI] [PubMed] [Google Scholar]
- 42.Guo, F. B., H. Y. Ou, and C. T. Zhang. 2003. ZCURVE: a new system for recognizing protein-coding genes in bacterial and archaeal genomes. Nucleic Acids Res. 31:1780-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall, M. N., and T. J. Silhavy. 1981. The ompB locus and the regulation of the major outer membrane porin proteins of Escherichia coli K12. J. Mol. Biol. 146:23-43. [DOI] [PubMed] [Google Scholar]
- 44.Harley, C. B., and R. P. Reynolds. 1987. Analysis of E. coli promoter sequences. Nucleic Acids Res. 15:2343-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hashemolhosseini, S., Z. Holmes, B. Mutschler, and U. Henning. 1994. Alterations of receptor specificities of coliphages of the T2 family. J. Mol. Biol. 240:105-110. [DOI] [PubMed] [Google Scholar]
- 46.Havekes, L., J. Tommassen, W. Hoekstra, and B. Lugtenberg. 1977. Isolation and characterization of Escherichia coli K-12 mutants defective in conjugation with an I-type donor. J. Bacteriol. 129:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heller, K., and V. Braun. 1979. Accelerated adsorption of bacteriophage T5 to Escherichia coli F, resulting from reversible tail fiber-lipopolysaccharide binding. J. Bacteriol. 139:32-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Heller, K. J., and H. Schwarz. 1985. Irreversible binding to the receptor of bacteriophages T5 and BF23 does not occur with the tip of the tail. J. Bacteriol. 162:621-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hollis, T., J. M. Stattel, D. S. Walther, C. C. Richardson, and T. Ellenberger. 2001. Structure of the gene 2.5 protein, a single-stranded DNA binding protein encoded by bacteriophage T7. Proc. Natl. Acad. Sci. USA 98:9557-9562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hulo, N., C. J. Sigrist, S. Le, V., P. S. Langendijk-Genevaux, L. Bordoli, A. Gattiker, E. De Castro, P. Bucher, and A. Bairoch. 2004. Recent improvements to the PROSITE database. Nucleic Acids Res. 32:D134-D137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50a.Huerta, A. M., and J. Collado-Vides. 2003. Sigma70 promoters in Escherichia coli: specific transcription in dense regions of overlapping promoter-like signals. J. Mol. Biol. 333:261-278. [DOI] [PubMed] [Google Scholar]
- 51.Isidro, A., M. A. Santos, A. O. Henriques, and P. Tavares. 2004. The high-resolution functional map of bacteriophage SPP1 portal protein. Mol. Microbiol. 51:949-962. [DOI] [PubMed] [Google Scholar]
- 52.Jackson, E. N., D. A. Jackson, and R. J. Deans. 1978. EcoRI analysis of bacteriophage P22 DNA packaging. J. Mol. Biol. 118:365-388. [DOI] [PubMed] [Google Scholar]
- 53.Juhala, R. J., M. E. Ford, R. L. Duda, A. Youlton, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequences of bacteriophages HK97 and HK022: pervasive genetic mosaicism in the lambdoid bacteriophages. J. Mol. Biol. 299:27-51. [DOI] [PubMed] [Google Scholar]
- 54.Jurica, M. S., and B. L. Stoddard. 1999. Homing endonucleases: structure, function and evolution. Cell. Mol. Life Sci. 55:1304-1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katsura, I. 1983. Tail assembly and injection, p. 331-346. In R. W. Hendrix, J. W. Roberts, F. W. Stahl, and R. A. Weisberg (ed.), Lambda II. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 56.Katsura, I. 1990. Mechanism of length determination in bacteriophage lambda tails. Adv. Biophys. 26:1-18. [DOI] [PubMed] [Google Scholar]
- 57.Kaufmann, G., M. David, G. D. Borasio, A. Teichmann, A. Paz, and M. Amitsur. 1986. Phage and host genetic determinants of the specific anticodon loop cleavages in bacteriophage T4-infected Escherichia coli CTr5X. J. Mol. Biol. 188:15-22. [DOI] [PubMed] [Google Scholar]
- 58.Kelley, L. A., R. M. MacCallum, and M. J. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299:499-520. [DOI] [PubMed] [Google Scholar]
- 59.Killmann, H., C. Herrmann, H. Wolff, and V. Braun. 1998. Identification of a new site for ferrichrome transport by comparison of the FhuA proteins of Escherichia coli, Salmonella paratyphi B, Salmonella typhimurium, and Pantoea agglomerans. J. Bacteriol. 180:3845-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 61.Lowe, T. M., and S. R. Eddy. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25:955-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ma, J., A. Campbell, and S. Karlin. 2002. Correlations between Shine-Dalgarno sequences and gene features such as predicted expression levels and operon structures. J. Bacteriol. 184:5733-5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. DeWeese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matlock, D. L., and T. Heyduk. 2000. Sequence determinants for the recognition of the fork junction DNA containing the −10 region of promoter DNA by E. coli RNA polymerase. Biochemistry 39:12274-12283. [DOI] [PubMed] [Google Scholar]
- 65.Matsumoto, M., N. Ichikawa, S. Tanaka, T. Morita, and A. Matsushiro. 1985. Molecular cloning of φ80 adsorption-inhibiting cor gene. Jpn. J. Genet. 60:475-483. [Google Scholar]
- 66.Parker, C. T., A. W. Kloser, C. A. Schnaitman, M. A. Stein, S. Gottesman, and B. W. Gibson. 1992. Role of the rfaG and rfaP genes in determining the lipopolysaccharide core structure and cell surface properties of Escherichia coli K-12. J. Bacteriol. 174:2525-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 68.Pedruzzi, I., J. P. Rosenbusch, and K. P. Locher. 1998. Inactivation in vitro of the Escherichia coli outer membrane protein FhuA by a phage T5-encoded lipoprotein. FEMS Microbiol. Lett. 168:119-125. [DOI] [PubMed] [Google Scholar]
- 69.Penner, M., I. Morad, L. Snyder, and G. Kaufmann. 1995. Phage T4-coded Stp: double-edged effector of coupled DNA and tRNA-restriction systems. J. Mol. Biol. 249:857-868. [DOI] [PubMed] [Google Scholar]
- 70.Pickersgill, R., D. Smith, K. Worboys, and J. Jenkins. 1998. Crystal structure of polygalacturonase from Erwinia carotovora ssp. carotovora. J. Biol. Chem. 273:24660-24664. [DOI] [PubMed] [Google Scholar]
- 71.Ramsay, N., and D. A. Ritchie. 1980. A physical map of the permuted genome of bacteriophage T1. Mol. Gen. Genet. 179:669-675. [DOI] [PubMed] [Google Scholar]
- 72.Ramsey, N., and D. A. Ritchie. 1983. Uncoupling of initiation site cleavage from subsequent headful cleavages in bacteriophage T1 DNA packaging. Nature 301:264-266. [DOI] [PubMed] [Google Scholar]
- 73.Ravin, V., N. Ravin, S. Casjens, M. E. Ford, G. F. Hatfull, and R. W. Hendrix. 2000. Genomic sequence and analysis of the atypical temperate bacteriophage N15. J. Mol. Biol. 299:53-73. [DOI] [PubMed] [Google Scholar]
- 74.Rigden, D. J., M. J. Jedrzejas, and M. Y. Galperin. 2003. Amidase domains from bacterial and phage autolysins define a family of γ-d,l-glutamate-specific amidohydrolases. Trends Biochem. Sci. 28:230-234. [DOI] [PubMed] [Google Scholar]
- 75.Roberts, M. D., N. L. Martin, and A. M. Kropinski. 2004. The genome and proteome of coliphage T1. Virology 318:245-266. [DOI] [PubMed] [Google Scholar]
- 76.Roberts, R. J., T. Vincze, J. Posfai, and D. Macelis. 2005. REBASE—restriction enzymes and DNA methyltransferases. Nucleic Acids Res. 33:D230-D232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robichon, C., M. Bonhivers, and A. P. Pugsley. 2003. An intramolecular disulphide bond reduces the efficacy of a lipoprotein plasma membrane sorting signal. Mol. Microbiol. 49:1145-1154. [DOI] [PubMed] [Google Scholar]
- 78.Sambrook, S., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 79.Schmidt, G., B. Jann, and K. Jann. 1970. Immunochemistry of R lipopolysaccharides of Escherichia coli. Studies on R mutants with an incomplete core, derived from E. coli O8:K27. Eur. J. Biochem. 16:382-392. [DOI] [PubMed] [Google Scholar]
- 80.Steinbacher, S., S. Miller, U. Baxa, N. Budisa, A. Weintraub, R. Seckler, and R. Huber. 1997. Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage. J. Mol. Biol. 267:865-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stiege, A. C., A. Isidro, A. Droge, and P. Tavares. 2003. Specific targeting of a DNA-binding protein to the SPP1 procapsid by interaction with the portal oligomer. Mol. Microbiol. 49:1201-1212. [DOI] [PubMed] [Google Scholar]
- 82.Sun, T. P., and R. E. Webster. 1986. fii, a bacterial locus required for filamentous phage infection and its relation to colicin-tolerant tolA and tolB. J. Bacteriol. 165:107-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tokuda, H., and S. Matsuyama. 2004. Sorting of lipoproteins to the outer membrane in E. coli. Biochim. Biophys. Acta 1693:5-13. [DOI] [PubMed] [Google Scholar]
- 85.Tsui, L. C., and R. W. Hendrix. 1983. Proteolytic processing of phage lambda tail protein gpH: timing of the cleavage. Virology 125:257-264. [DOI] [PubMed] [Google Scholar]
- 86.Uc-Mass, A., E. J. Loeza, M. de la Garza, G. Guarneros, J. Hernandez-Sanchez, and L. Kameyama. 2004. An orthologue of the cor gene is involved in the exclusion of temperate lambdoid phages. Evidence that Cor inactivates FhuA receptor functions. Virology 329:425-433. [DOI] [PubMed] [Google Scholar]
- 87.Vinogradov, E. V., K. Van Der Drift, J. E. Thomas-Oates, S. Meshkov, H. Brade, and O. Holst. 1999. The structures of the carbohydrate backbones of the lipopolysaccharides from Escherichia coli rough mutants F470 (R1 core type) and F576 (R2 core type). Eur. J. Biochem. 261:629-639. [DOI] [PubMed] [Google Scholar]
- 88.Vostrov, A. A., O. A. Vostrukhina, A. N. Svarchevsky, and V. N. Rybchin. 1996. Proteins responsible for lysogenic conversion caused by coliphages N15 and φ80 are highly homologous. J. Bacteriol. 178:1484-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wagner, E. F., H. Ponta, and M. Schweiger. 1977. Development of E. coli virus T1: the pattern of gene expression. Mol. Gen. Genet. 150:21-28. [DOI] [PubMed] [Google Scholar]
- 90.Wang, J., M. Hofnung, and A. Charbit. 2000. The C-terminal portion of the tail fiber protein of bacteriophage lambda is responsible for binding to LamB, its receptor at the surface of Escherichia coli K-12. J. Bacteriol. 182:508-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang, L. K., C. D. Lima, and S. Shuman. 2002. Structure and mechanism of T4 polynucleotide kinase: an RNA repair enzyme. EMBO J. 21:3873-3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang, L. K., and S. Shuman. 2002. Mutational analysis defines the 5′-kinase and 3′-phosphatase active sites of T4 polynucleotide kinase. Nucleic Acids Res. 30:1073-1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Weigele, P. R., E. Scanlon, and J. King. 2003. Homotrimeric, β-stranded viral adhesins and tail proteins. J. Bacteriol. 185:4022-4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Werts, C., V. Michel, M. Hofnung, and A. Charbit. 1994. Adsorption of bacteriophage lambda on the LamB protein of Escherichia coli K-12: point mutations in gene J of lambda responsible for extended host range. J. Bacteriol. 176:941-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Willi, K., and J. Meyer. 1998. DNA analysis of temperate bacteriophage Aaphi23 isolated from Actinobacillus actinomycetemcomitans. Mol. Gen. Genet. 258:323-325. [DOI] [PubMed] [Google Scholar]
- 96.Woods, D. E., J. A. Jeddeloh, D. L. Fritz, and D. DeShazer. 2002. Burkholderia thailandensis E125 harbors a temperate bacteriophage specific for Burkholderia mallei. J. Bacteriol. 184:4003-4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Xu, J., R. W. Hendrix, and R. L. Duda. 2004. Conserved translational frameshift in dsDNA bacteriophage tail assembly genes. Mol. Cell 16:11-21. [DOI] [PubMed] [Google Scholar]
- 98.Yoder, M. D., and F. Jurnak. 1995. Protein motifs. 3. The parallel beta helix and other coiled folds. FASEB J. 9:335-342. [DOI] [PubMed] [Google Scholar]
- 99.Yusupova, G. Z., M. M. Yusupov, J. H. Cate, and H. F. Noller. 2001. The path of messenger RNA through the ribosome. Cell 106:233-241. [DOI] [PubMed] [Google Scholar]
- 100.Yuzenkova, J., S. Nechaev, J. Berlin, D. Rogulja, K. Kuznedelov, R. Inman, A. Mushegian, and K. Severinov. 2003. Genome of Xanthomonas oryzae bacteriophage Xp10: an odd T-odd phage. J. Mol. Biol. 330:735-748. [DOI] [PubMed] [Google Scholar]