Abstract
Different surface organelles contribute to specific interactions of a pathogen with host tissues or infectious partners. Multiple pilus gene clusters potentially encoding different surface structures have been identified in several gram-positive bacterial genomes sequenced to date, including actinomycetales, clostridia, corynebacteria, and streptococci. Corynebacterium diphtheriae has been shown to assemble a pilus structure, with sortase SrtA essential for the assembly of a major subunit SpaA and two minor proteins, SpaB and SpaC. We report here the characterization of a second pilus consisting of SpaD, SpaE, and SpaF, of which SpaD and SpaE form the pilus shaft and SpaF may be located at the pilus tip. The structure of the SpaDEF pilus contains no SpaABC pilins as detected by immunoelectron microscopy. Neither deletion of spaA nor sortase srtA abolishes SpaDEF pilus formation. The assembly of the SpaDEF pilus requires specific sortases located within the SpaDEF pilus gene cluster. Although either sortase SrtB or SrtC is sufficient to polymerize SpaDF, the incorporation of SpaE into the SpaD pili requires sortase SrtB. In addition, an alanine in place of the lysine of the SpaD pilin motif abrogates pilus polymerization. Thus, SpaD, SpaE, and SpaF constitute a different pilus structure that is independently assembled and morphologically distinct from the SpaABC pili and possibly other pili of C. diphtheriae.
Protruding out of the bacterial surface are proteinaceous filaments, named pili or fimbriae, which mediate bacterial attachment and colonization of host tissues (24, 31, 32, 42). Pili are also known to interact with other bacteria in a microbiofilm (45), are involved in the translocation of DNA across biological membranes (4, 16), and can serve as phage receptors (6). The structure and function of pili in gram-negative bacteria have been studied in great detail (30, 34). In contrast, the mechanisms of pilus assembly in gram-positive bacteria are less well understood, although pili or fimbriae have been identified in several gram-positive organisms, including Actinomyces spp. (8), Arthrobacter photogonimos (11), Corynebacterium diphtheriae (43), Ruminococcus albus (28), Streptococcus parasanguis (42), and, more recently, Streptococcus agalactiae (15). It has been proposed that gram-positive bacterial pili are covalently linked to peptidoglycan and require sortase (18, 25, 37, 45), a transpeptidase which is found in all gram-positive organisms (21, 26, 35). Studies by Cisar et al. and Yeung et al. in Actinomyces revealed two fimbrial types, designated 1 and 2 (5, 46, 47). FimP and FimA precursor proteins, which harbor the C-terminal cell wall sorting signal, assemble type 1 and 2 fimbriae, respectively, in a manner that requires the expression of sortase-like genes (44, 48, 49). The assembly of S. parasanguis FW213 fimbriae involves the major structural subunit Fap1, which contains a C-terminal sorting signal (29, 41, 42), suggesting the role of sortase in fimbrial biogenesis.
In C. diphtheriae (12, 20), we found that corynebacterial pili are composed of three pilin subunits, SpaA, SpaB, and SpaC (designated the SpaABC pilus; Spa stands for “sortase-mediated pilus assembly”) (38). SpaA, the major pilus protein, seems to be distributed uniformly along the pilus shaft, while SpaB is observed at regular intervals and SpaC appears positioned at the tip (38). The assembly of SpaABC precursors into a high-molecular-weight complex requires sortase SrtA, which functions to cleave the sorting signal of the precursors to generate covalent linkages between pilin subunits at the cleaved polypeptides and the side chain amino groups of pilin motif sequences (36, 38). The incorporation of SpaB into polymerized SpaA fibers involves the E box element of SpaA (36). Homologs of SpaA, SpaB, and SpaC are present in the genome of several gram-positive pathogens (35). Recently, an operon containing three predicted surface proteins, GBS80, GBS52, and GBS104, with the C-terminal sorting signal has been identified in S. agalactiae (15). Pilus structures were detected by electron microscopy using a specific antibody against GBS80. Based on the protein sequence similarity, we now know that GBS80, GBS52, and GBS104 are SpaA, SpaB, and SpaC homologs, respectively. Deletion of a pilus-specific sortase abolished pilus formation of GBS80 (15), demonstrating that GBS pilus assembly is sortase dependent. As the fimbrial subunit proteins of actinomycetales, corynebacteria, and streptococci contain C-terminal sorting signals and require sortase enzymes for assembly, it is clear that a common pathway of pilus biogenesis exists in gram-positive bacteria.
The genes encoding corynebacterial pilin subunits and putative sortases are organized in three clusters (38). Based on sequence features and clustering, C. diphtheriae strain NCTC13129 has been suggested to assemble three pilus structures, designated according to the major pilin subunit SpaA-, SpaD-, and SpaH-type pili (38). Different types of pili presumably allow bacteria to interact with different host cell targets or with other infectious partners in their ecological niche, as in the case of the gram-positive pathogen Actinomyces naeslundii (45) (13, 14, 27). Unlike the two Actinomyces fimbrial operons (45), the second and third gene clusters of C. diphtheria each encode two putative sortases (38). The presence of multiple sortases has raised questions of redundancy, requirements for subsets of surface proteins, and/or perhaps a temporal expression of sortases for their functions during bacterial growth or infections (26, 37). Previous studies of S. aureus (23, 40), Listeria monocytogenes (2), and Streptococcus pyogenes sortases (1) support the hypothesis that each sortase is required for a specific set of substrates. This hypothesis remains to be tested for the assembly of corynebacterial pili.
Here we describe the characterization of a second pilus gene cluster of C. diphtheria containing srtB-spaD-srtC-spaE-spaF. We show that spaD encodes major pilin protein SpaD, while spaE and spaF encode two minor proteins, SpaE and SpaF. We further show the SpaDEF pilus is morphologically distinct from SpaABC pili, and sortase SrtB is specifically required for the incorporation of SpaE into SpaDF pili, whose assembly requires either SrtB or SrtC.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
C. diphtheriae NCTC13129 was obtained from the American Type Culture Collection. C. diphtheriae strains (Table 1) were grown on heart infusion broth (HIB), heart infusion agar (HIA), or trypticase soy agar supplemented with 5% sheep blood (TSASB). Escherichia coli strains were grown on Luria broth. Kanamycin was added at 50 μg/ml for C. diphtheriae and E. coli strains as needed. Reagents were purchased from Sigma unless otherwise indicated.
TABLE 1.
Corynebacterium diphtheriae strains used in this study
Generation of rabbit-raised polyclonal antibodies.
Appropriately synthesized oligonucleotide primers (Table 2) and C. diphtheriae chromosomal DNA were used for the PCR amplification of coding sequences for surface proteins and sortases, both lacking signal peptide and sorting signal sequences. DNA fragments were cloned between appropriate sites of the expression vector pQE30 (QIAGEN), thereby generating expression signals and coding sequences for six-histidine-tagged recombinant proteins (N-terminal tag). The recombinant plasmids were transformed into E. coli XL1-Blue. DNA sequencing was used to verify the recombinant plasmids. To purify the recombinant proteins, plasmid-bearing E. coli XL1-Blue cultures were induced with 1 mM isopropyl-β-d-thiogalactopyranoside for 3 h, and cells were harvested by centrifugation. Bacteria were broken in a French pressure cell, and membranes as well as insoluble proteins were sedimented by ultracentrifugation. The supernatant, containing cleared lysate, was subjected to affinity chromatography on nickel-nitrilotriacetic acid Sepharose, washed twice with wash buffer (150 mM NaCl, 10% glycerol, and 50 mM Tris-HCl, pH 7.5), and eluted with 0.5 M imidazole in wash buffer. Eluted protein was dialyzed against the same buffer without imidazole for 16 h. To generate antibodies, each rabbit was immunized with 1 mg of each protein mixed in Freund's complete adjuvant, followed by two booster injections after 3 weeks, each with 1 mg of protein mixed in Freund's incomplete adjuvant. Three weeks after the third injection, the animal were euthanized and exsanguinated by cardiac puncture. The specificity of each antibody was determined by immunoblotting against purified antigen as well as crude corynebacterial cell extracts prepared from wild-type and mutant strains.
TABLE 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| SpaD-Ab-5′ | AAGGATCCAAGGGGCACGAAACTAGTAC |
| SpaD-Ab-3′ | AAGGATCCTTACTGCTTGATGTTTTTA ATCTC |
| SpaE-Ab-5′ | AAGGATCCAAGGTAGCCGGGCTCAGT |
| SpaE-Ab-3′ | AAGGATCCCTATTCGCGTTCTTTCTCATC |
| SpaF-Ab-5′ | AAGGATCCTTTTATCTCGCTCCAAACGAT |
| SpaF-Ab-3′ | AAGGATCCTTATCCACCTACGGTCAGGT |
| SrtB-Ab-5′ | AAGGATCCCAGCAATACAACGCCAATCTC |
| SrtB-Ab-3′ | AAGGATCCTTACTCTCCCTGCGGATCC |
| SrtC-Ab-5′ | AAGGATCCCACGAGTACAACGAGAACC |
| SrtC-Ab-3′ | AAGGATCCCTAGTGATCTTCCGGGGC |
| SpaD-5′ | AAAAAGCTTAAGGAATTCACTCCAGTGAAG |
| SpaD-3′ | AAAAAGCTTATTCAGCGGGCTATTAGTTCT |
| SrtB-5′ | GAAGATCTCTTTACATCAGTAGTGACTGG |
| SrtB-3′ | GAAGATCTAGCCCGTTAACTCTCCGTG |
| SrtC-1 | CGCGGATCCTTGCAGGCGGAGAAATATGAA |
| SrtC-2 | CGGGGTACCATCGTCCATCTCCTTGCCG |
| SrtC-3 | CGGGGTACCGTGCTCGTGATGCTGTACC |
| SrtC-4 | AAACTGCAGGCGTGGCTAGTCATCTGC |
| SpaD-A | CGCGGATCCCTCCTCCCAGGCCTTATT |
| SpaD-B | CCCATCCACTAAACTTAAACAACCAGCAACA TCGTCCAT |
| SpaD-C | TGTTTAAGTTTAGTGGATGGGATCGTGGCA GCCGGTGCA |
| SpaD-D | CGCGGATCCTTCTTCAGTAGCCACCGC |
Generation of C. diphtheriae deletion mutants.
C. diphtheriae deletion mutants were generated by homologous recombination and verified by PCR and Western and Southern blotting techniques as previously described (38). Briefly, gene deletions were constructed by crossover PCR (19) and cloned between appropriate restriction sites of pK18mobsacB, and recombinant plasmids were transformed into E. coli S17-1 (7). For crossover PCR, two primers primer sets were generated. For example, to generate a deletion of spaD, we synthesized SpaD-A, SpaD-B, SpaD-C, and SpaD-D (Table 2). SpaD-A and SpaD-B as well as SpaD-C and SpaD-D were used to amplify two 1-kb fragments, which were then used as templates for another PCR amplification with primers A and D. As the tail ends of primers SpaD-B and SpaD-C anneal to one another, a 2-kb fused PCR product was obtained and digested overnight with BamHI, and the restricted DNA was purified after agarose gel electrophoresis. The fragment was cloned into the BamHI site of pK18mobsacB, and recombinant plasmids were purified, characterized, and transformed into E. coli DH5α (9). The transformants were again subjected to plasmid purification and finally transformed into E. coli S17-1 (7). Overnight cultures of E. coli S17-1 and C. diphtheriae were mixed in equal volumes and spread on agar plates at 30°C for 16 h. Corynebacterial cointegrates were isolated by plating conjugal bacteria on HIA plates supplemented with 35 μg ml−1 nalidixic acid and 50 μg ml−1 kanamycin. C. diphtheriae deletion mutants were selected by plating cointegrates on HIA plates supplemented with 10% sucrose and 35 μg ml−1 nalidixic acid. For complementation analysis, DNA fragments containing promoter regions and open reading frames of interest were cloned into pCGL0243, and the recombinant plasmids were electroporated into C. diphtheriae.
Plasmid construction. (i) Plasmid pSpaD.
Two primers were synthesized and used in a PCR amplification with chromosomal template DNA of C. diphtheriae strain NCTC13129 to amplify a DNA segment encompassing the spaD promoter, 5′ untranslated region (UTR), and spaD coding sequence with SpaD-5′ and SpaD-3′, each of which contained a HindIII site for cloning purposes (Table 2). The PCR-amplified DNA fragment was cut with HindIII and ligated with the cleaved HindIII sites of the E. coli/Corynebacterium shuttle vector pCGL0243 (38) to generate pSpaD.
(ii) Plasmid pSrtB. A DNA segment encompassing the srtB promoter, UTR, and srtB coding sequence was amplified using two primers (Table 2), SrtB-5′ and SrtB-3′, providing the amplified corynebacterial DNA segment with flanking BglII sites for cloning purposes. This fragment was cut with BglII and ligated with the cleaved BglII sites of the E. coli/Corynebacterium shuttle vector pCGL0243 to generate pSrtB.
(iii) Plasmid pSrtC. To construct pSrtC, the 5′ promoter sequence and UTR of spaD was fused to the coding sequence of srtC. For PCR amplification, four primers were synthesized (Table 2). SrtC-1 and SrtC-2 amplified the 5′ promoter sequence and UTR of spaD while providing the amplified corynebacterial DNA segment with flanking BamHI and KpnI sites for cloning purposes. SrtC-3 and SrtC-4 amplified from chromosomal DNA of C. diphtheriae NTCT13129 the coding region of srtC while appending the KpnI and PstI sites. The two PCR fragments were fused via the central KpnI site while inserting the recombinant gene into the BamHI and KpnI sites of the E. coli/Corynebacterium shuttle vector pCGL0243.
Site-directed mutagenesis of recombinant plasmids.
A PCR-based site-directed mutagenesis of double-stranded DNA was employed in this study (39). Plasmid DNA was used as a template for PCR amplification with Pfu DNA polymerase using primer sets (5′ and 3′) flanking six codons on both sides of the mutation K107. Following PCR, the amplified plasmids were digested overnight at 37°C with DpnI to select against parental DNA molecules. The digested plasmids were transformed into E. coli. Mutant plasmids were verified by DNA sequencing and transformed into C. diphtheriae by electroporation following a published protocol (10), with an exception that all corynebacterial strains were grown in HIB medium supplemented with 0.2% Tween-80.
Extraction of C. diphtheriae pili.
Extractions of C. diphtheriae pili were carried out as previously described (38). Briefly, C. diphtheriae strains were scraped from TSASB plates after overnight growth and washed in SMM buffer (0.5 M sucrose, 10 mM MgCl2, and 10 mM maleate, pH 6.8). Cell pellets were suspended in the same buffer, treated with mutanolysin (300 U ml−1) at 37°C for 6 h, or left untreated (mock). An equal amount of bacterial sediment was suspended in 70% formic acid and incubated at 65°C for 30 min. Solubilized pili were isolated from the supernatant after centrifugation at 16,000 × g, followed by trichloroacetic acid precipitation, acetone washing, and drying protein samples under vacuum. Pilus preparations were boiled in sodium dodecyl sulfate (SDS) sample buffer, separated by SDS-polyacrylamide gel electrophoresis (PAGE), subjected to immunoblotting with rabbit antisera (1:20,000 for α-SpaD, 1:5,000 for α-SpaE, and 1:1,000 for α-SpaF), and detected with chemiluminescence.
Electron microscopy and immunogold labeling.
Electron microscopic experiments were carried out as previously described (38). Briefly, bacterial strains were grown overnight on agar plates (TSBSB), washed in 0.1 M NaCl, and stained with 1% uranyl acetate. For immunogold labeling, a drop of bacterial suspension was placed on carbon grids, washed three times with phosphate-buffered saline (PBS) containing 2% bovine serum albumin (BSA), and blocked for 1 h in PBS with 0.1% gelatin. Pili were stained with primary antibody diluted 1:100 in PBS with 2% BSA for 1 h, followed by washing and blocking. Pili were stained with gold goat anti-rabbit immunoglobulin G (IgG) (Jackson ImmunoResearch) diluted 1:20 in PBS with 2% BSA for 1 h followed by washing in PBS with 2% BSA. The grids were washed five times with water before staining with 1% uranyl acetate. Samples were viewed in a Jeol 100CX electron microscope.
RESULTS
Characterization of a second pilus in Corynebacterium diphtheriae.
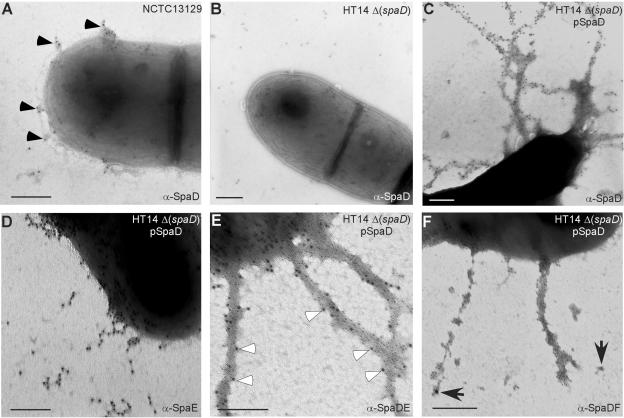
To investigate whether the spaD cluster encoded a pilus structure, we cloned each gene into an expression vector, pQE30 (QIAGEN). The proteins SpaD, SpaE, SpaF, SrtB, and SrtC were each expressed in E. coli XL1-Blue, purified, and injected into rabbits to raise specific antisera according to a published protocol (38). To determine whether spaD encoded a pilus protein, we examined corynebacteria by immunoelectron microscopy (IEM) using a specific antibody raised against purified SpaD as well as gold particles conjugated with IgG. When cells were reacted with α-SpaD and gold-labeled IgG, we observed immunogold labeling of stubby fibers external to the cell body (Fig. 1A, filled arrows). Control rabbit sera did not result in labeling of pilus fibers (data not shown). Moreover, no SpaD-labeled structures were observed in the isogenic strain of C. diphtheriae HT14 (ΔspaD) which lacks the spaD gene (Fig. 1B). Complementation of the ΔspaD mutant HT14 with wild-type spaD on a multicopy plasmid not only restored the SpaD-positive pilus but led to the formation of extended fibers (Fig. 1C), similar to the extended pilus phenotype previously observed with overproduced SpaA (38). Thus, overexpression of major subunits (SpaD and SpaA) allows us to easily observe longer pili than ones that are expressed from the chromosome. When cells with overproduced SpaD were stained with α-SpaE and gold-labeled IgG, we observed SpaE staining at intervals along the pilus shaft (Fig. 1D), a similar phenotype that has been reported with SpaB (38).
FIG. 1.
Identification of the second pilus of Corynebacterium diphtheriae. C. diphtheriae strain NCTC12139 (A) or its isogenic derivatives with a deletion of the spaD gene [ΔspaD (B)] transformed with plasmid pSpaD encoding wild-type spaD (C to F) were immobilized on carbon grids and stained with a specific antiserum. Single-labeling experiments were carried out by staining cells with α-SpaD (A to C, E, and F) or α-SpaE (D) and IgG-labeled 12 nm (A to D) or 6 nm (E and F) gold particles. In double-labeling experiments (E and F) α-SpaD/IgG-6-nm gold-stained cells were then stained with α-SpaE (E) or α-SpaF (F) and 12-nm gold-labeled IgG. Labeled SpaE pilins (open arrows) are spotted along the SpaD fibers (E). Few labeled SpaF pilins (arrows) are found at the pilus tip (F). Samples were viewed by transmission electron microscopy. Bars indicate a distance of 0.2 μm.
To determine if SpaD, SpaE, and SpaF are constituents of the same pilus structure, we carried out a double-labeling experiment (38). Cells were stained with α-SpaD followed by 6-nm gold-labeled IgG and then with α-SpaE followed by 12-nm gold-labeled IgG. SpaE staining occurred along the SpaD pilus structures (open arrows), suggestive of SpaD and SpaE forming the pilus shaft (Fig. 1E). A similar experiment was performed with α-SpaD and α-SpaF. No immunogold labeling of SpaF was observed along the SpaD fibers, but few labeled SpaF pilins were spotted at the end of the pili (Fig. 1F, arrows). In addition, we also present evidence below which shows that SpaF is indeed a pilus subunit (see Fig. 4D and 5D). Together the data indicate that SpaD and SpaE constitute the pilus shaft and SpaF may be positioned at the pilus tip, as in the case of SpaC (38).
FIG. 4.
Assembly of SpaD, SpaE, and SpaF requires a specific sortase. (A) Graphic representation of the second gene cluster in the chromosome of C. diphtheriae NCTC13129 that encode sortases (srtB and srtC) and sortase-mediated pilus assemblies (spaDEF). Location of designated genes in the genome is shown by numbers. Arrows indicate the position of predicted promoters as well as the direction of transcription. Corynebacterial strains were treated with muramidase (M) or formic acid (F) or were left untreated (−) prior to extraction with hot SDS sample buffer. Proteins were separated on SDS-PAGE and detected by immunoblotting with α-SpaD (B), α-SpaE (C), or α-SpaF (D). wt, wild type.
FIG. 5.
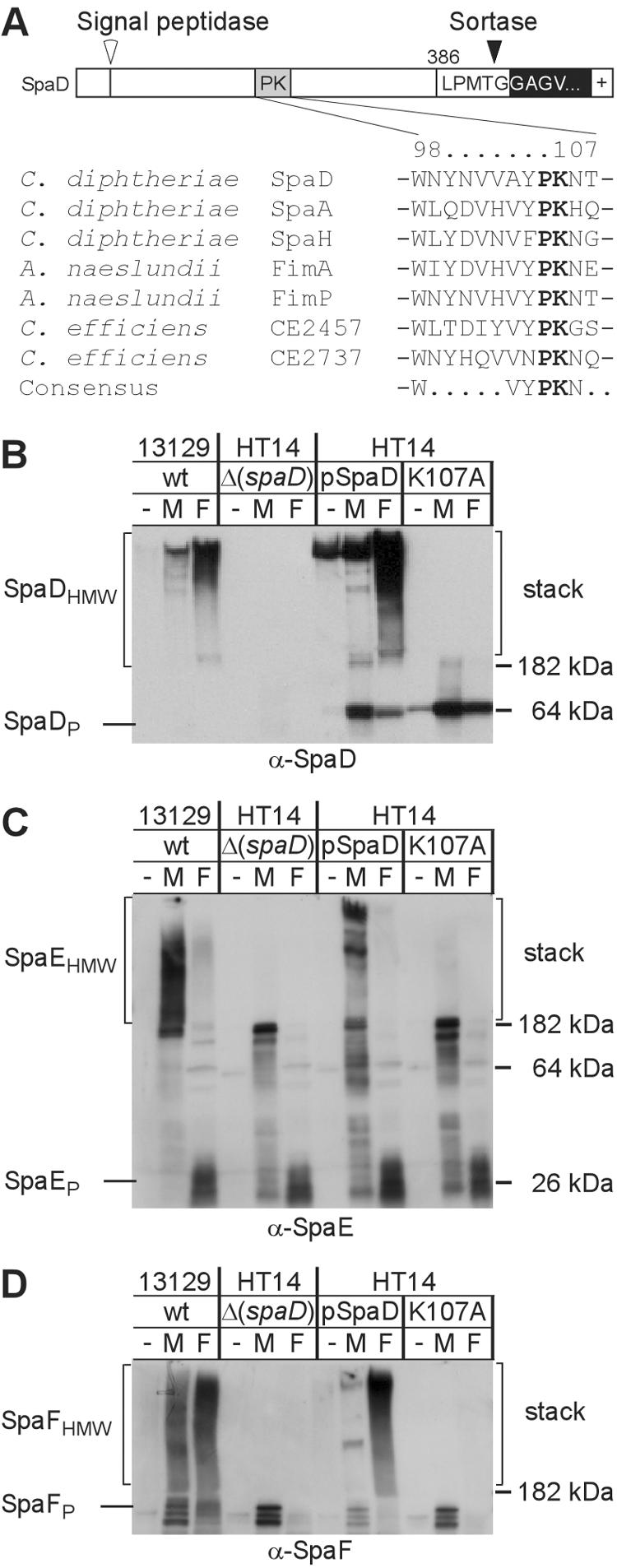
Conserved lysine (K) residue in the pilin motif of SpaD is required for pilus formation. The primary structure of SpaDP precursor with an N-terminal signal peptide, C-terminal sorting signal, and cleavage sites for signal peptidase and sortase is shown. The precursors of C. diphtheriae SpaA, SpaD, and SpaH harbor a pilin motif (WxxxVxVYPK) with a conserved lysine (K) residue. Similar pilin motifs are found in FimA and FimP of A. naeslundii and C. efficiens CE2457 and CE2737 (A). Corynebacterial strains were treated with muramidase (M) or formic acid (F) or left untreated (−) prior to extraction with hot SDS sample buffer. Proteins were separated on SDS-PAGE and detected by immunoblotting with specific antiserum [α-SpaD (B), α-SpaE (C), and α-SpaF (D)]. Replacement of K107 with alanine (A) abrogates SpaDEF pilin polymerization. wt, wild type.
SpaDEF pilus is morphologically distinct from other corynebacterial pili.
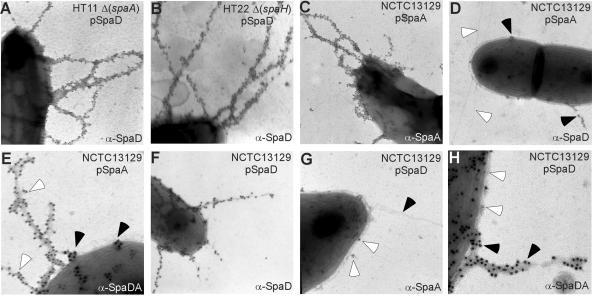
As previously mentioned, C. diphtheriae may encode three types of pili with SpaA, SpaD, and SpaH as major subunits. To examine if the formation of SpaD pili was independent of SpaA and SpaH pilus organelles (unpublished data), we introduced into the ΔspaA mutant HT11 or the ΔspaH mutant HT22 the wild-type spaD gene on a multicopy plasmid and observed the pilus formation using IEM. Deletion of spaA or spaH did not abrogate SpaD pilus structures (Fig. 2A and B). An identical phenotype was observed when srtA was deleted from the wild-type chromosome (data not shown). To differentiate two types of pili, the plasmid pSpaA was introduced into the wild-type corynebacterial strain NCTC13129. As a result, extended filaments of SpaA were observed by IEM using α-SpaA (Fig. 2C). Conversely, distinct, shorter SpaD pili were detected using α-SpaD, whereas unstained SpaA pili were visible (Fig. 2D, open arrows). We next confirmed that these SpaA and SpaD structures are different through double-labeling experiments. We observed two separate pilus types, extended SpaA pili labeled with 12-nm gold particles and shorter SpaD pili labeled with 18-nm gold particles (Fig. 2E). Likewise, when SpaD was overexpressed with the pSpaD plasmid, two different pili were detected using IEM with α-SpaA and α-SpaD in the wild-type strain (Fig. 2F to H), long pili labeled by α-SpaD (filled arrows) in this case. In addition, no labeled SpaA, SpaB, or SpaC was detected along the SpaD fibers using α-SpaA, α-SpaB, or α-SpaC, respectively (Fig. 2G and data not shown).
FIG. 2.
Distinct structure encoded by the second pilus gene cluster. C. diphtheriae strain NCTC12139 (C to H) or its isogenic derivatives carrying deletion of specific genes [Δ(spaA) (A) and Δ(spaH) (B)] transformed with plasmid pSpaD carrying wild-type spaD (A, B, and F to H) or plasmid pSpaA carrying wild-type spaA (C to E) were immobilized on carbon grids, stained with specific antiserum α-SpaD (A, B, D, E, and F) or α-SpaA (C, G and H), and IgG-labeled with 12-nm (A to D and G to H) or 18-nm (E and F) gold-labeled IgG. Double-labeling experiments (E and H) were followed by staining cells with α-SpaA and 12-nm gold particles (E) or with α-SpaD and 18-nm gold particles (H). Samples were viewed by transmission electron microscopy. Arrows indicate structures of SpaA (opened) or SpaD (filled).
To further test our proposal that SpaD pilus is independently expressed and assembled, we examined the assembly of pili with a previously developed assay (38), whereby corynebacterial proteins isolated by boiling cell extracts in SDS sample buffer with reducing agent are separated via electrophoresis in polyacrylamide gels and then subjected to immunoblotting with rabbit antisera and detected with chemiluminescence (38). Boiling corynebacterial extracts in SDS released very little SpaD (lanes “−” in Fig. 3A). However, muramidase and formic acid treatment solubilized pili of strain NCTC13129, and SpaD species were identified by immunoblotting with α-SpaD (Fig. 3A). Immunoreactive high-molecular-weight SpaD (SpaDHMW) was detected within the SDS-PAGE stack with a mass greater than 200 kDa. Muramidase-released SpaDHMW migrated as discrete species on SDS-PAGE, whereas formic acid extraction solubilized SpaDHMW as a smear of immunoreactive material (Fig. 3A). These observations are consistent with the hypothesis that pili are composed of covalently linked (SDS, formic acid resistant) pilin subunits, some of which may be attached to cell wall fragments (38). Deletion of spaD (strain HT14) abolished the polymerization of SpaD precursors as expected (Fig. 3A); the spaD deletion also abolished the polymerization of SpaE and SpaF (see Fig. 5C and D). Introduction of wild-type SpaD into the strain HT14 via an expression plasmid not only restored the synthesis and assembly of SpaDEF pili (Fig. 3A; see also Fig. 5C and D) but also led to the formation of extended fibers (Fig. 1C). Deletion of spaF, spaA, spaB, or spaC did not affect the polymerization of SpaD precursor proteins (Fig. 3A). Conversely, the formation of SpaABC pili was independent of SpaD pilus synthesis and assembly, as deletion of spaD did not abolish SpaABC pili (Fig. 3B to D). Together these data demonstrate that SpaDEF pilus is morphologically distinct from other corynebacterial pili.
FIG. 3.
SpaD pilins are independently synthesized and assembled. C. diphtheriae strain NCTC12139 or its isogenic derivatives carrying deletion of spa genes were treated with muramidase (M) or formic acid (F) or were left untreated (−) prior to extraction with hot SDS sample buffer. Proteins were separated on SDS-PAGE and detected by immunoblotting with the specific antisera α-SpaD (A), α-SpaA (B), α-SpaB (C), and α-SpaC (D). The precursor (e.g., SpaP) and high-molecular-weight mature products (e.g., SpaHMW) of pilus assembly, the position of molecular mass markers, and the stacking gel portion of SDS-PAGE (stack) are indicated. wt, wild type.
A specific sortase is required for the formation of SpaDEF pilus.
C. diphtheriae strain NCTC13129 has six sortase genes: srtA, srtB, srtC, srtD, srtE, and srtF (38). To investigate which of these sortases is involved in the assembly of SpaD pilus, we analyzed strain HT21 that contains a deletion of six sortase genes (srtA, srtB, srtC, srtD, srtE, and srtF) as well as strains lacking individual sortase genes (38). The polymerization of pili was examined by SDS-PAGE as described above. Surprisingly, individual deletion of neither srtA, srtB, srtC, srtD, srtE, nor srtF abolished SpaD pilus assembly and the production of SpaDHMW (Fig. 4B and data not shown). Nevertheless, deletion of six sortase genes in strain HT21 abolished the polymerization of SpaD precursor into high-molecular-weight species; only the SpaD precursor was observed migrating as a 64-kDa protein (data not shown). This phenotype for SpaD is similar to that observed for SpaA in strain HT21 (38), and it demonstrates that the polymerization of SpaD pilins requires a sortase.
Two sortases (SrtB and SrtC) are encoded by genes within the SpaD pilus gene cluster, and they are divergently transcribed (Fig. 4A). To examine whether these two sortases are required for SpaD pilus assembly, we generated strain HT17 that lacks sortases SrtB and SrtC using allelic replacement (38). In this strain, SpaD synthesis is not affected, but the polymerization of SpaD into high-molecular-weight species is abolished (Fig. 4B). Transformation of strain HT17 with a plasmid carrying srtB or srtC restored the polymerization of SpaD to a level similar to that of the strain that lacks either srtB or srtC, although less SpaD was detected in the muramidase treated sample (lane “M”). More SpaD polymerization in the muramidase-treated sample could be readily detected if the blot was exposed for longer times (Fig. 4B). Identical results were obtained for SpaF, demonstrating that the assembly of SpaF into high-molecular-weight SpaFHMW complexes required the presence of either the srtB or srtC gene but not srtA, srtD, srtE, and srtF (Fig. 4D).
Immunoblotting of corynebacterial extracts with α-SpaE revealed the incorporation of SpaE into high-molecular-weight complexes (SpaEHMW) that could be solubilized by muramidase (Fig. 4C). Surprisingly, treatment with formic acid released fewer SpaEHMW species than treatment with muramidase; its precursor SpaEP was mainly observed migrating around 26 kDa. Deletion of sortase srtB alone (strain HT6) or both srtB and srtC (strain HT17) abrogated SpaE incorporation into SpaD pili (Fig. 4C). A similar phenotype was observed in the strain that lacked spaD (see Fig. 5C). Strain HT6 (ΔsrtB) or HT17 (ΔsrtBC) containing the srtB plasmid restored SpaE inclusion (Fig. 4C). Together these results demonstrate that either SrtB or SrtC is sufficient for the polymerization of SpaD and SpaF, while SrtB is specifically required for the incorporation of SpaE into SpaD pili. The fact that sortases SrtA, SrtD, SrtE, and SrtF are dispensable for the formation of SpaDEF pilus indicates the involvement of a pilus-specific sortase in the assembly of specific pili.
Requirement of the pilin motif for SpaD pilus polymerization.
Previous work identified the pilin motif, a sequence element with a conserved lysine residue (K) that is conserved between the homologs of C. diphtheriae SpaA (38 and Fig. 5A). We suspected that the conserved lysine residue of SpaD might also be required for pilin polymerization, as this lysine's amino group could participate in sortase-catalyzed amide bond formation. Indeed, replacement of lysine K107 with alanine (A) abrogated polymerization of SpaDHMW (Fig. 5B). In addition, no high-molecular-weight species containing SpaE and SpaF was observed in this strain (Fig. 5C and D). Thus, the pilin motif sequence (with its conserved lysine) is required for the polymerization of SpaD precursor and in turn the incorporation of SpaE and SpaF into SpaD pili.
DISCUSSION
In this study we have characterized a second pilus in C. diphtheriae encoded by a gene cluster separate from the SpaABC pilus. Consistent with the molecular structure of corynebacterial SpaABC pilus (38), the SpaDEF pilus is composed of SpaD, SpaE, and SpaF, with SpaD and SpaE forming the pilus shaft and SpaF presumably located at the tip. Similar to factors required for the formation of SpaABC pili, the pilin motif and sortases are necessary for the SpaDEF pilus assembly; the incorporation of SpaE into SpaD pili specifically requires sortase SrtB, while either SrtB or SrtC is competent for the assembly of SpaDF pili. Because SrtB and SrtC are divergently transcribed and either can function to catalyze the SpaD pilus formation (Fig. 4), it is conceivable that their expression may be regulated, as is the case of S. aureus SrtB, a sortase responsible for the cell wall anchoring of receptive machinery for the acquisition of iron (22). If this were the case, there might be a condition whereby sortase SrtB would not be expressed; consequently, the resulting pili would not contain SpaE. Interestingly, unlike SpaB of the SpaABC pilus (36), SpaE pilins are released from the polymeric assembly by formic acid treatment (Fig. 4C and 5C), suggesting that the linkage between SpaD and SpaE may be different than that between SpaA and SpaB (36). In fact, the E box mutations of E334 of SpaD do not eliminate SpaE inclusion into SpaD pili (data not shown). Current experiments are examining this linkage.
BLAST homology searches revealed homologs of corynebacterial SpaA, SpaB, and SpaC or SpaD, SpaE, and SpaF (i.e., precursors harboring pilin motifs [Fig. 5A] and sorting signals) in A. naeslundii, Clostridium perfringens, Enterococcus faecalis, Streptococcus agalactiae, and Streptococcus pneumoniae (38). Importantly, the lysine within the pilin motif is a conserved feature of all analogs of SpaD; a plasmid containing SpaD with Lys107 replaced by alanine did not restore the pilus formation by SpaDEF in a strain that lacks SpaD (Fig. 5). A comparative analysis of these major subunits revealed a strong similarity between Actinomyces FimA/FimP and Corynebacterium SpaA/SpaD/SpaH, especially between FimA and SpaH (or FimP and SpaD; data not shown). Interestingly, FimA, when expressed under control of the spaA promoter, is polymerized in C. diphtheriae, a process that requires the pilin motif and sortase SrtD presumably catalyzing the formation of SpaH pili (36). Likewise, one would predict the SpaD precursors, when expressed in A. naeslundii, could be polymerized by Actinomyces sortase SrtB (or OrfB [18]). Genomic analysis of the C. diphtheriae strain NCTC13129 offers an explanation for the observed phenomenon, which is that the corynebacterial and actinomyces pilus gene clusters might have originated from the same ancestor (3).
Two or more pilus gene clusters are identified in each genome of several gram-positive pathogens sequenced to date, including A. naeslundii (45), C. diphtheriae (38), Corynebacterium efficiens, C. perfringens (data not shown), and S. agalactiae (15). Pilus gene clustering may be a result of horizontal gene transfer, whereby selective traits are beneficial to respective hosts (17). Like A. naeslundii, C. diphtheriae harbors multiple pilus gene clusters that may confer on the bacterium its ability to interact with multiple host cell receptors. Two pilus gene clusters are also found in its relative, C. efficiens (data not shown), a nonpathogenic bacterium isolated from soil, suggesting that the acquisition of gene clusters encoding adhesive organelles is not limited to bacterial pathogens. As genome sequencing continues, we expect to find more such clusters as recently reported for Corynebacterium jeikeium, a causative agent of many nosocomial infections (33). Common denominators for the assembly of proteinacious pili found in these organisms are sortase and the major pilus subunit, SpaD or SpaA homologs, whose conserved elements, the pilin motif and sorting signal, are structurally unique (37, 38). A specific sortase recognizes its own pilin substrate for the polymerization of each of the different pilus types (Fig. 6). However, what governs the specificity of each sortase to its substrate still remains to be answered.
FIG. 6.
Model representation of two different types of pili in Corynebacterium diphtheriae. Shown are two pilus gene clusters in the chromosome of C. diphtheriae (omitted is the third one). Precursor pilin proteins are synthesized in the cytoplasm, transported across the membrane by the Sec machinery, and recognized, polymerized, and anchored by a specific sortase. Sortase SrtA is required for the formation of SpaABC pilus, whereas sortases SrtB and SrtC are specific for the SpaDEF pilus. Pilin proteins are color coded to represent a morphological distinction.
Acknowledgments
We thank Arthur R. Hand (University of Connecticut Health Center) for help with electron microscopy and Asis Das, Mary J. Osborn, Peter Setlow, Sandra K. Weller (University of Connecticut Health Center), Sarkis K. Mazmanian (Harvard Medical School), Luciano A. Marraffini (University of Chicago), and members of our laboratory for critical review of the manuscript and discussion.
This work was supported by an institutional start-up fund to H.T.-T. from the University of Connecticut Health Center.
REFERENCES
- 1.Barnett, T. C., and J. R. Scott. 2002. Differential recognition of surface proteins in Streptococcus pyogenes by two sortase gene homologs. J. Bacteriol. 184:2181-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bierne, H., C. Garandeau, M. G. Pucciarelli, C. Sabet, S. Newton, F. Garcia-del Portillo, P. Cossart, and A. Charbit. 2004. Sortase B, a new class of sortase in Listeria monocytogenes. J. Bacteriol. 186:1972-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cerdeno-Tarraga, A. M., A. Efstratiou, L. G. Dover, M. T. Holden, M. Pallen, S. D. Bentley, G. S. Besra, C. Churcher, K. D. James, A. De Zoysa, T. Chillingworth, A. Cronin, L. Dowd, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Moule, M. A. Quail, E. Rabbinowitsch, K. M. Rutherford, N. R. Thomson, L. Unwin, S. Whitehead, B. G. Barrell, and J. Parkhill. 2003. The complete genome sequence and analysis of Corynebacterium diphtheriae NCTC13129. Nucleic Acids Res. 31:6516-6523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christie, P. J. 2004. Type IV secretion: the Agrobacterium VirB/D4 and related conjugation systems. Biochim. Biophys. Acta 1694:219-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cisar, J. O., and A. E. Vatter. 1979. Surface fibrils (fimbriae) of Actinomyces viscosus T14V. Infect. Immun. 24:523-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis, B. M., and M. K. Waldor. 2003. Filamentous phages linked to virulence of Vibrio cholerae. Curr. Opin. Microbiol. 6:35-42. [DOI] [PubMed] [Google Scholar]
- 7.de Lorenzo, V., E. Eltis, B. Kessler, and K. N. Timmis. 1993. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123:17-24. [DOI] [PubMed] [Google Scholar]
- 8.Girard, A. E., and B. H. Jacius. 1974. Ultrastructure of Actinomyces viscosus and Actinomyces naeslundii. Arch. Oral Biol. 19:71-79. [DOI] [PubMed] [Google Scholar]
- 9.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-572. [DOI] [PubMed] [Google Scholar]
- 10.Haynes, J. A., and M. L. Britz. 1989. Electrotransformation of Brevibacterium lactofermentum and Corynebacterium glutamicum: growth in tween 80 increases transformation frequencies. FEMS Microbiol. Lett. 61:329-333. [Google Scholar]
- 11.Hoober, J. K. 1978. Kinetics of accumulation of a photodynamically induced cell-surface polypeptide in a species of Arthrobacter. J. Bacteriol. 136:359-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klebs, E. 1883. Über Diphtherie. Verh. Cong. Inn. Med. 2:139-154. [Google Scholar]
- 13.Kolenbrander, P. E. 1988. Intergeneric coaggregation among human oral bacteria and ecology of dental plaque. Annu. Rev. Microbiol. 42:627-656. [DOI] [PubMed] [Google Scholar]
- 14.Kolenbrander, P. E., P. G. Egland, P. I. Diaz, and R. J. Palmer, Jr. 2005. Genome-genome interactions: bacterial communities in initial dental plaque. Trends Microbiol. 13:11-15. [DOI] [PubMed] [Google Scholar]
- 15.Lauer, P., C. D. Rinaudo, M. Soriani, I. Margarit, D. Maione, R. Rosini, A. R. Taddei, M. Mora, R. Rappuoli, G. Grandi, and J. L. Telford. 2005. Genome analysis reveals pili in group B streptococcus. Science 309:105. [DOI] [PubMed] [Google Scholar]
- 16.Lawley, T. D., W. A. Klimke, M. J. Gubbins, and L. S. Frost. 2003. F factor conjugation is a true type IV secretion system. FEMS Microbiol. Lett. 224:1-15. [DOI] [PubMed] [Google Scholar]
- 17.Lawrence, J. G., and J. R. Roth. 1996. Selfish operons: horizontal transfer may drive the evolution of gene clusters. Genetics 143:1843-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, T., M. K. Khah, S. Slavnic, I. Johansson, and N. Stromberg. 2001. Different type 1 fimbrial genes and tropisms of commensal and potentially pathogenic Actinomyces spp. with different salivary acidic proline-rich protein and statherin ligand specificities. Infect. Immun. 69:7224-7233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Link, A. J., D. Phillips, and G. M. Church. 1997. Methods for generating precise deletions and insertions in the genome of wild-type Escherichia coli: application to open reading frame characterization. J. Bacteriol. 179:6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loeffler, F. 1884. Untersuchungen über die Bedeutung der Mikroorganismen für die Enstehung der Diphtherie beim Menschen, bei der Taube und beim Kalbe. Mitt. Klin. Gesundheitsamte (Berlin) 2:421-499. [Google Scholar]
- 21.Mazmanian, S. K., G. Liu, H. Ton-That, and O. Schneewind. 1999. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285:760-763. [DOI] [PubMed] [Google Scholar]
- 22.Mazmanian, S. K., E. P. Skaar, A. H. Gasper, M. Humayun, P. Gornicki, J. Jelenska, A. Joachimiak, D. M. Missiakas, and O. Schneewind. 2003. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299:906-909. [DOI] [PubMed] [Google Scholar]
- 23.Mazmanian, S. K., H. Ton-That, and O. Schneewind. 2001. Sortase-catalyzed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40:1049-1057. [DOI] [PubMed] [Google Scholar]
- 24.Merz, A. J., and M. So. 2000. Interactions of pathogenic neisseriae with epithelial cell membranes. Annu. Rev. Cell Dev. Biol. 16:423-457. [DOI] [PubMed] [Google Scholar]
- 25.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and the mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pallen, M. J., A. C. Lam, M. Antonio, and K. Dunbar. 2001. An embarrassment of sortases - a richness of substrates? Trends Microbiol. 9:97-102. [DOI] [PubMed] [Google Scholar]
- 27.Palmer, R. J., Jr., S. M. Gordon, J. O. Cisar, and P. E. Kolenbrander. 2003. Coaggregation-mediated interactions of streptococci and actinomyces detected in initial human dental plaque. J. Bacteriol. 185:3400-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pegden, R. S., M. A. Larson, R. J. Grant, and M. Morrison. 1998. Adherence of the gram-positive bacterium Ruminococcus albus to cellulose and identification of a novel form of cellulose-binding protein which belongs to the Pil family of proteins. J. Bacteriol. 180:5921-5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sadler, J. E. 1998. Biochemistry and genetics of von Willebrand factor. Annu. Rev. Biochem. 67:395-424. [DOI] [PubMed] [Google Scholar]
- 30.Sauer, F. G., M. A. Mulvey, J. D. Schilling, J. J. Martinez, and S. J. Hultgren. 2000. Bacterial pili: molecular mechanisms of pathogenesis. Curr. Opin. Microbiol. 3:65-72. [DOI] [PubMed] [Google Scholar]
- 31.Schilling, J. D., M. A. Mulvey, and S. J. Hultgren. 2001. Structure and function of Escherichia coli type 1 pili: new insight into the pathogenesis of urinary tract infections. J. Infect. Dis. 183(Suppl. 1):S36-S40. [DOI] [PubMed] [Google Scholar]
- 32.Swanson, M. S., and B. K. Hammer. 2000. Legionella pneumophila pathogesesis: a fateful journey from amoebae to macrophages. Annu. Rev. Microbiol. 54:567-613. [DOI] [PubMed] [Google Scholar]
- 33.Tauch, A., O. Kaiser, T. Hain, A. Goesmann, B. Weisshaar, A. Albersmeier, T. Bekel, N. Bischoff, I. Brune, T. Chakraborty, J. Kalinowski, F. Meyer, O. Rupp, S. Schneiker, P. Viehoever, and A. Puhler. 2005. Complete genome sequence and analysis of the multiresistant nosocomial pathogen Corynebacterium jeikeium K411, a lipid-requiring bacterium of the human skin flora. J. Bacteriol. 187:4671-4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thanassi, D. G., and S. J. Hultgren. 2000. Assembly of complex organelles: pilus biogenesis in gram-negative bacteria as a model system. Methods 20:111-126. [DOI] [PubMed] [Google Scholar]
- 35.Ton-That, H., L. A. Marraffini, and O. Schneewind. 2004. Protein sorting to the cell wall envelope of gram-positive bacteria. Biochim. Biophys. Acta 1694:269-278. [DOI] [PubMed] [Google Scholar]
- 36.Ton-That, H., L. A. Marraffini, and O. Schneewind. 2004. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol. Microbiol. 53:251-261. [DOI] [PubMed] [Google Scholar]
- 37.Ton-That, H., and O. Schneewind. 2004. Assembly of pili in gram-positive bacteria. Trends Microbiol. 12:228-234. [DOI] [PubMed] [Google Scholar]
- 38.Ton-That, H., and O. Schneewind. 2003. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 50:1429-1438. [DOI] [PubMed] [Google Scholar]
- 39.Weiner, M. P., G. L. Costa, W. Schoettlin, J. Cline, E. Mathur, and J. C. Bauer. 1994. Site-directed mutagenesis of double-stranded DNA by the polymerase chain reaction. Gene 151:119-123. [DOI] [PubMed] [Google Scholar]
- 40.Weiss, W. J., E. Lenoy, T. Murphy, L. Tardio, P. Burgio, S. J. Projan, O. Schneewind, and L. Alksne. 2004. Effect of srtA and srtB gene expression on the virulence of Staphylococcus aureus in animal models of infection. J. Antimicrob. Chemother. 53:480-486. [DOI] [PubMed] [Google Scholar]
- 41.Wu, H., and P. M. Fives-Taylor. 1999. Identification of dipeptide repeats and a cell wall sorting signal in the fimbriae-associated adhesin, Fap1, of Streptococcus parasanguis. Mol. Microbiol. 34:1070-1081. [DOI] [PubMed] [Google Scholar]
- 42.Wu, H., and P. M. Fives-Taylor. 2001. Molecular strategies for fimbrial expression and assembly. Crit. Rev. Oral Biol. Med. 12:101-115. [DOI] [PubMed] [Google Scholar]
- 43.Yanagawa, R., and E. Honda. 1976. Presence of pili in species of human and animal parasites and pathogens of the genus corynebacterium. Infect. Immun. 13:1293-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yeung, M. K. 2000. Actinomyces: surface macromolecules and bacteria-host interactions, p. 583-593. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 45.Yeung, M. K. 1999. Molecular and genetic analyses of Actinomyces spp. Crit. Rev. Oral Biol. Med. 10:120-138. [DOI] [PubMed] [Google Scholar]
- 46.Yeung, M. K., and J. O. Cisar. 1988. Cloning and nucleotide sequence of a gene for Actinomyces naeslundii WVU45 type 2 fimbriae. J. Bacteriol. 170:3803-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yeung, M. K., and J. O. Cisar. 1990. Sequence homology between the subunits of two immunologically and functionally distinct types of fimbriae of Actinomyces spp. J. Bacteriol. 172:2462-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yeung, M. K., J. A. Donkersloot, J. O. Cisar, and P. A. Ragsdale. 1998. Identification of a gene involved in assembly of Actinomyces naeslundii T14V type 2 fimbriae. J. Bacteriol. 66:1482-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yeung, M. K., and P. A. Ragsdale. 1997. Synthesis and function of Actinomyces naeslundii T14V type 1 fimbriae require the expression of additional fimbria-associated genes. Infect. Immun. 65:2629-2639. [DOI] [PMC free article] [PubMed] [Google Scholar]