Abstract
The twin-arginine translocation (TAT) system secretes fully folded proteins that contain a twin-arginine motif within their signal sequence across the cytoplasmic membrane in bacteria. Using a green fluorescent protein fused with a TAT signal sequence, we demonstrated that Mycobacterium smegmatis contains a TAT system. By inactivating individual genes, we showed that three genes (tatA, tatB, and tatC) are required for a functional TAT system in M. smegmatis. The tat mutants exhibited a decreased growth rate and altered colony morphology compared to the parent strain. Comparison of the secreted proteins of the ΔtatC and parent strain by two-dimensional polyacrylamide gel electrophoresis revealed an alteration in the secretion of at least five proteins, and one of the major TAT-dependent secreted proteins was identified as β-lactamase (BlaS). The genome of M. smegmatis was analyzed with the TATFIND program, and 49 putative TAT substrates were identified, including the succinate transporter DctP. Because disruption of the TAT secretion system has a direct effect on the physiology of M. smegmatis and homologs of the TAT proteins are also present in the genome of Mycobacterium tuberculosis, the TAT secretion system or its substrates may be good candidates for drug or vaccine development.
Bacteria possess several means of secreting proteins across the cytoplasmic membrane into the periplasm or extracellular environment. General secretory pathways include Sec-dependent and Sec-independent secretion systems. Some pathogenic bacteria also possess specialized secretion systems that allow the delivery of bacterial proteins directly into eukaryotic host cells. In mycobacteria, three distinct secretion pathways have been identified including the Sec, SecA2, and Snm (for secretion in mycobacteria) systems. The preproteins secreted by the Sec pathway do not share any highly conserved amino acid residues, but they do contain a characteristic signal sequence at the N terminal that is required for secretion. The signal sequence, on average, contains 24 amino acids, which form three characteristic regions including (i) a positively charged amino terminus, (ii) a hydrophobic core region, and (iii) a carboxy-terminal domain that contains the cleavage site for signal peptidase. Proteins secreted via the Sec system are threaded in an unfolded state through a channel in the cytoplasmic membrane, and SecA derives energy for secretion from the hydrolysis of ATP. Once across the membrane, the proteins fold into their correct conformation. The Sec pathway is considered the general housekeeping secretion system and is essential in all bacteria for which it has been studied. (For recent reviews, see references 10 and 13.)
The Sec secretory machinery is composed of several proteins, with SecA being a major component of the system. SecA drives the secretion of proteins through the hydrolysis of ATP and interacts with the other components of the Sec pathway. Most bacteria only contain one copy of the secA gene, but the genomes of all mycobacteria that have been sequenced contain two genes, secA1 and secA2, that encode SecA homologs (4, 7, 8, 16, 22). SecA1 resembles the general housekeeping SecA of other bacteria, while SecA2 is only 50% similar to SecA1 at the amino acid sequence level. SecA2 is involved in secretion but does not perform the same function as SecA1; unlike SecA1, it is dispensable for growth in axenic medium in mycobacteria (4, 5). However, deletion of secA2 results in the attenuation of the ability of Mycobacterium tuberculosis to grow in vivo (5). Overall, the SecA2 pathway is considered to be an alternate secretion system to the general housekeeping pathway.
Some pathogenic bacteria contain specialized secretion pathways that are Sec independent and are required for the export of virulence factors. Recently, the first pathway of this type in M. tuberculosis was identified and is referred to as the Snm pathway. The Snm system is composed of at least four genes (snm1 to snm4) that are located in the region of difference locus designated RD1 and encode two AAA ATPases, a putative chromosome partitioning ATPase, and an integral membrane protein, respectively. Each of the Snm proteins is required for the secretion of the immunodominant mycobacterial antigens, 6-kDa early secretory antigenic target (ESAT-6) and 10-kDa culture filtrate protein (CFP-10). Deletion of any of the snm genes results in the attenuation of M. tuberculosis, and the mutants exhibit the same lack of pathogenesis as the RD1 deletion strain (17, 36). Although the Snm secretion system is required for virulence in M. tuberculosis, the pathway is also conserved in nonpathogenic Mycobacterium smegmatis (9). Homologs for the snm genes have been identified and proven to be essential for the secretion of ESAT-6 and CFP-10. Four other genes (snm5, snm6, snm7, and mycp1) were also shown to be required for the secretion of these proteins in M. smegmatis (9). Homologs of these genes do exist in M. tuberculosis and are thought to be essential for secretion of ESAT-6 and CFP-10.
In this study, we identified a fourth secretion system in M. smegmatis that is homologous to the twin-arginine translocase (TAT) secretion pathway of other bacteria. The TAT system is a Sec-independent secretion system and is able to secrete completely folded proteins across the cytoplasmic membrane, unlike the Sec pathway. The preproteins secreted by the TAT pathway contain a signal sequence with a tripartite organization similar to the Sec signal sequence, as well as distinct features. The main difference is the conserved twin-arginine motif located between the N region and the hydrophobic region of the signal sequence. The energy required for secretion of proteins via the TAT pathway is derived solely from the proton motive force (3). We identified three genes that constitute the TAT pathway in M. smegmatis and have shown that each gene is essential for the secretion of TAT-dependent substrates. We have also demonstrated that the disruption of the TAT secretion system has an effect on the physiology of M. smegmatis.
MATERIALS AND METHODS
Bacterial strains, media, and DNA manipulations.
The strains and plasmids used in this study are listed in Table 1. Mycobacterium smegmatis LR222 was cultured on Trypticase soy agar (TSA) or Middlebrook 7H9 medium (Difco) containing albumin-dextrose-catalase (ADC) supplement (BBL) and 0.025% Tween 80 (7H9-ADC) or M63 medium [0.022 M KH2PO4, 0.04 M K2HPO4, 15 mM (NH4)2SO4, 1.6 μM FeSO4, 1.0 mM MgSO4, and 0.02% Tween 80]. Media were supplemented with kanamycin (25 μg/ml), gentamicin (5 μg/ml), ampicillin (25 μg/ml), X-galactosidase (X-Gal; 50 μg/ml), sucrose (10%), glycerol (2.0%), succinate (2.0%), or acetamide (2.0%) when required. All chemicals were purchased from Sigma. For growth curves, each strain was cultured in 7H9-ADC medium until mid-log phase, diluted to a starting optical density at 600 nm (OD600) of 0.03 in the same medium, and incubated at 37°C in a roller drum. To determine if the strains could utilize succinate as a carbon source, the strains were cultured in 7H9-ADC medium until mid-log phase, harvested, and washed two times with M63 medium. The strains were then diluted to a starting OD600 of 0.04 in M63-succinate and incubated at 37°C in a roller drum. Cell growth was monitored by measuring the change in OD600 values over time. The growth curves were repeated twice with samples in triplicate for each experiment. Standard protocols or the manufacturer's instructions were used for all DNA manipulations. Oligonucleotide primers were synthesized at the Biotechnology Core Facility, National Center for Infectious Diseases, Centers for Disease Control and Prevention (CDC), and sequences are available (see Table S1 in the supplemental material).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| M. smegmatis LR222 | Wild type | |
| M. smegmatis ΔtatA | ΔtatA | This study |
| M. smegmatis ΔtatB | ΔtatB::Genr | This study |
| M. smegmatis ΔtatC | ΔtatC | This study |
| M. smegmatis tatA+ | ΔtatA + pMVtatAC | This study |
| M. smegmatis tatB+ | ΔtatB + pMVtatB | This study |
| M. smegmatis tatC+ | ΔtatC + pMVtatAC | This study |
| E. coli TAM1 | mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Active motif |
| Plasmids | ||
| pCR-Blunt-II TOPO | Cloning vector; Kanr | Invitrogen |
| pCR2.1 TOPO | Cloning vector; Ampr Kanr | Invitrogen |
| pMV306 | Integrating vector; Kanr | 21 |
| pGOAL17 | lacZ sacB cassette | 27 |
| pDtatA | ΔtatA suicide vector | This study |
| pDtatB | ΔtatB suicide vector | This study |
| pDtatC | ΔtatC suicide vector | This study |
| pUGA61B | pMV306-Paceti::torA-gfp; Kanr | This study |
| pMVtatAC | pUGA61B + tatAC | This study |
| pMVtatB | pUGA61B + Phsp60::tatB | This study |
Construction of mutants and complementation strains.
Single unmarked deletions of the tatA and tatC genes were constructed using previously described methods (28). Briefly, approximately 1-kb regions upstream and downstream of each gene were amplified by PCR using Pfu (Stratagene) polymerase and ligated into pCR-Blunt II-TOPO (Invitrogen). The two fragments were ligated together, creating an internal deletion, and the counterselectable markers (lacZ and sacB) from pGOAL17 (28) were ligated into the EcoRV site of the plasmid to generate the suicide vectors pDtatA and pDtatC. The upstream region of tatA was amplified using the TatAUS5 and TatAUS3 primers and contained 52 bp of tatA and 953 bp of the region upstream of the start codon. The downstream region was amplified using the TatADS5 and TatADS3 primers and contained 58 bp of tatA and 848 bp of the region downstream of the stop codon. The cloning of the two fragments created a 211-bp internal deletion of tatA. For the ΔtatC construct, the upstream region was amplified using the TatCUS5 and TatCUS3 primers and contained 29 bp of tatC and 890 bp of the region upstream of tatC. The downstream region was amplified with the TatCDS5 and TatCDS3 primers and contained 90 bp of tatC and 836 bp of the region downstream of the stop codon. The cloning of these two fragments created a 763-bp internal deletion of tatC. M. smegmatis was electroporated (18) with 1 μg of each suicide vector, and initial transformants were selected on TSA plates containing kanamycin and X-Gal as previously described. A single-crossover event resulted in kanamycin-resistant and blue (β-galactosidase-positive) colonies. To select for double-crossover events, the initial transformants were grown in 7H9 broth (with no drugs) for 48 h and plated onto TSA plates containing sucrose and X-Gal. White colonies were patched onto TSA plates with and without kanamycin, and kanamycin-susceptible colonies were screened by PCR to determine if they contained the wild-type or mutant allele. Mutants were further confirmed by Southern blot analysis.
For complementation of ΔtatA and ΔtatC, the tatAC region including 500 bp upstream of tatA was amplified by PCR using primers TatAC5 and TatAC3 and ligated into pCR2.1 TOPO (Invitrogen) to generate pUGA86C. The tatAC region was removed from pUGA86C by digestion of the plasmid with HindIII, treatment with T4 DNA polymerase, and digestion with EcoRV. The fragment of interest was ligated into pUGA61B that had been digested with XbaI and treated with T4 DNA polymerase to generate pMVtatAC.
A marked deletion of tatB was constructed in a manner similar to that described above. The upstream fragment was amplified using the TatBUS5 and TatBUS3 primers and contained 10 bp of tatB and 1,158 bp of the region upstream of the start codon. The downstream fragment was amplified using the TatBDS5 and TatBDS3 primers and contained 51 bp of tatB and 1,016 bp of the region downstream of the stop codon. The ligation of the upstream and downstream fragment created a 430-bp internal deletion and a unique PacI site at the junction region. A gentamicin cassette was ligated into this site to create the suicide vector pDtatB. After transformation, the cells were plated onto TSA plates containing gentamicin. Transformants were patched onto TSA plates containing either gentamicin or kanamycin. The gentamicin-resistant, kanamycin-susceptible transformants were screened by PCR, and the mutants were confirmed by Southern blotting. Because tatB is the last gene of a four-gene operon,we used the hsp60 promoter to express this gene to complement the ΔtatB strain. A 500-bp fragment upstream and including the start codon of hsp60 was amplified by PCR from pMV261 (37) using primers Hsp60F and Hsp60R, and this amplicon was ligated into pCR2.1 TOPO to generate pHsp60. The coding sequence of tatB was also amplified by PCR using primers TatB5 and TatB3 and ligated into pCR2.1 TOPO to generate ptatB. The hsp60 promoter was cloned upstream of tatB by digestion of pHsp60 with XbaI and NdeI and ligation of this fragment into the SpeI and NdeI sites of ptatB to generate pHsp60tatB. The hsp60-tatB fusion was removed from pHsp60tatB by digestion of the plasmid with KpnI, and this fragment was ligated into the KpnI site of pUGA61B to generate pMVtatB. This construct was used to complement the ΔtatB strain.
GFP constructs and microscopy.
The gfp-mut2 gene was amplified by PCR from pFJS17 (23) using primers GFP-F and GFP-R and ligated into pCR2.1 TOPO to create pGFP. The first 150 bp of torA, which includes the TAT-dependent signal sequence and the first four amino acids of the mature protein, were amplified by PCR from Escherichia coli MC4100 using primers TorA5 and TorA3 and ligated into pCR2.1 TOPO to generate pTorA. To remove the downstream BamHI site of pGFP, the plasmid was digested with HindIII and religated to create pGFP-1. The truncated torA gene was removed from pTorA by digestion of the plasmid with BamHI and SpeI, and this fragment was ligated into the BamHI and XbaI sites of pGFP-1 to create pTorAGFP, which contains the trimethylamine N-oxide reductase (TorA)-green fluorescent protein (GFP) fusion. The TorA-GFP fusion was ligated downstream of the acetamide-inducible promoter of the acetamidase gene of M. smegmatis LR222 and inserted into the integrating mycobacterial plasmid pMV306 (21) to generate pUGA61B by the following procedure. The TorA-GFP fusion was amplified by PCR with Pfu and the TorA5P and M13R primers, and the amplicon was digested with HindIII. A 2.8-kb fragment containing the four open reading frames upstream of the acetamidase gene and the start codon was amplified by PCR using Paceti5 and Paceti3 primers, and the amplicon was digested with XbaI. These two fragments were ligated together and into the HindIII and XbaI sites of pMV306 via a three-way ligation to generate pUGA61B. All strains were electroporated with 100 ng of pUGA61B, and transformants were selected on TSA plates containing kanamycin.
The complemented and wild-type strains were cultured in M63 medium containing succinate and acetamide, while the mutant strains were cultured in 7H9-ADC supplemented with acetamide to induce the expression of the TorA-GFP fusion. To turn off expression, the cells were washed and suspended in the same medium lacking acetamide and incubated for 1 h. Cells were mounted on lysine-coated slides, and GFP was visualized by fluorescence microscopy.
Isolation and analysis of CFPs.
The cell filtrate proteins (CFPs) were isolated as previously described (12). Briefly, M. smegmatis strains were cultured in 500 ml of glycerol alanine salt medium (38) until the cells reached mid-log phase. Cells were removed by centrifugation, and the supernatant containing the CFPs was filtered through a 0.2-μm low-protein binding filter and concentrated to approximately 10 ml using a stirred cell concentrator (Amicon) equipped with a YM membrane with a 10,000-molecular-weight cutoff (Millipore). CFPs were dialyzed against 1 liter of dialysis buffer (10 mM ammonium bicarbonate, 1 mM dithiothreitol, pH 7.0) at 4°C for at least 16 h. At this time, the buffer was exchanged with another 1 liter of dialysis buffer without dithiothreitol and dialyzed at 4°C for 4 h. After dialysis, the CFP solutions were further concentrated to 1 ml with a spin column (Millipore). The CFPs were quantitated and stored at −80°C until needed. For two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) analysis, CFPs (125 μg) from each strain were lyophilized and suspended in 185 μl of solubilization buffer (5 M urea, 2 M thiourea, 2% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 2% SB 3-10, 40 mM Tris, 0.2% Bio-Lyte 3/10 ampholyte) and used to hydrate 11-cm IPG strips (Bio-Rad) with a pH 4 to 7 gradient. The strips were prepared and electophoresed according to the manufacturer's protocols. For the second dimension, the samples were analyzed on Criterion 12% Bis-Tris gels (Bio-Rad), and the proteins were visualized using Sypro Ruby (Molecular Probes) stain. Unique proteins were cut from the gels and analyzed by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) as previously described (6).
RESULTS
Identification and inactivation of the tat genes.
The genes encoding the TAT secretion system of M. tuberculosis were identified, based on amino acid homology to TAT proteins of other bacteria (3, 40). tatA and tatC are located in an operon and are designated Rv2094c and Rv2093c, respectively. tatB is the last gene of the sigE operon and is designated Rv1224. In database searches, we identified a homolog for each of these genes in the genome of M. smegmatis mc2155. The genes displayed the same genetic organization as that found in M. tuberculosis, and all tat genes displayed a high level of homology, approximately 70% amino acid identity in both strains. The gene designations for the tat genes are tatA (Msmeg3890), tatC (Msmeg3889), and tatB (Msmeg5057).
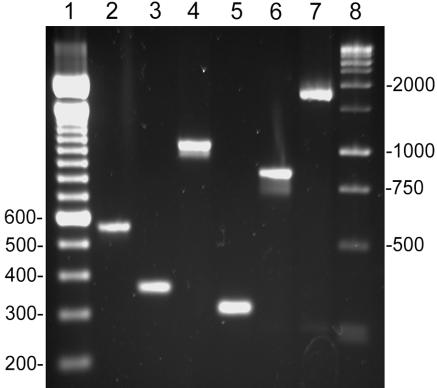
To determine if these genes encode proteins involved in a functional TAT secretion system, each gene in M. smegmatis was inactivated individually. Because the tatA and tatC genes are located in an operon, unmarked deletions of each gene were created individually, designated ΔtatA and ΔtatC. The ΔtatA and ΔtatC strains contain a 211-bp and 763-bp deletion of each gene, respectively, as analyzed by PCR (Fig. 1). The ΔtatB strain contained a 430-bp deletion, but the gentamicin cassette was 1,300 bp in length, resulting in the addition of approximately 870 bp to the PCR fragment (Fig. 1). For complementation studies, the tatAC region, including its native promoter, was used to complement the ΔtatA and ΔtatC strains, while the ΔtatB strain was complemented by cloning tatB downstream of the hsp60 promoter.
FIG. 1.
PCR analysis of the tat deletion strains. Primers specific for each tat gene were used to amplify the flanking deleted region from wild-type and the tat deletion strains. Lanes: 1, 100-bp marker; 2, 4, and 6, wild type; 3, ΔtatA; 5, ΔtatC; 7, ΔtatB; 8, 1-kb marker.
In vitro growth of tat mutants.
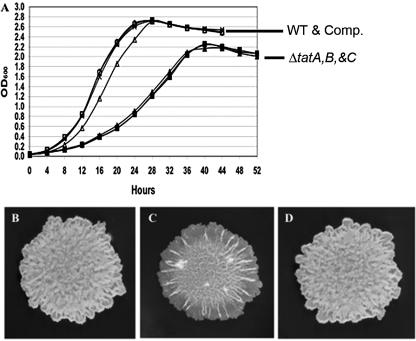
The colonies of the parent strain were raised, slightly pigmented, and exhibited a cord-like structure, a well-defined edge, and an overall rough appearance (Fig. 2B). The colonies of the ΔtatA strain displayed fewer cord-like structures, which only occurred in the center of the colony, and the edges of the colonies were more transparent and smooth compared to the wild type. The ΔtatA colonies were also less raised and not pigmented (Fig. 2C). The colony morphology of the ΔtatB and ΔtatC strains was the same as that of the ΔtatA strain (data not shown). Each of the complemented strains exhibited the same colony morphology as the wild-type strain (Fig. 2D). The growth of the tat deletion strains was also slower than the wild type and the complemented strains on solid medium and in liquid medium, as discussed below. The wild-type and complemented strains were photographed at 4 days, while the tat deletion strains required 7 days to reach the same size under these conditions.
FIG. 2.
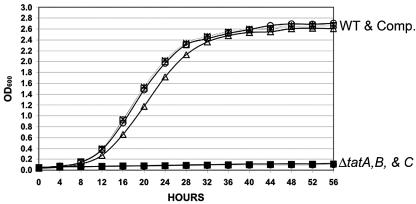
In vitro growth of wild-type, tat deletion, and complemented strains in broth (A) and on solid medium (B to D). Growth was monitored by measuring OD600 changes over time. The symbols for each strain are as follows: x, wild type; open symbols, tat deletion strains; solid symbols, complemented strains.
The growth of wild-type and the tat deletion strains in 7H9-ADC medium was compared by monitoring the change in optical densities over time (Fig. 2A). All mutants grew at the same rate (doubling time of approximately 6 h), while the wild-type and the complemented strains grew at a faster rate (doubling time of approximately 3.5 h). The wild-type and complemented strains reached a maximum cell density of 2.6 × 109 CFU/ml (OD600 = 2.7), while the tat deletion strains grew to a cell density of 1.2 × 109 CFU/ml (OD600 = 2.2). In multiple experiments, the tat deletion strains were only able to grow to a cell density of 50% of the wild type. We repaired the growth defect and colony morphology of the tat mutant strains by complementation and proved the differences were due to the specific gene deletions.
The tat mutants are unable to secrete TorA-GFP.
GFP can be used to localize proteins in bacteria by creating an in-frame fusion of GFP with a protein of interest or with a signal sequence. For example, the trimethylamine N-oxide reductase of E. coli is a molybdenum-containing protein that is involved in anaerobic respiration, contains a twin-arginine motif (RRRFL) within its N-terminal region, and is secreted across the cytoplasmic membrane via the TAT secretion system (32, 33). When the signal sequence of TorA is fused to GFP, the TorA-GFP fusion is secreted across the cytoplasmic membrane into the periplasm and maintains its activity (39).
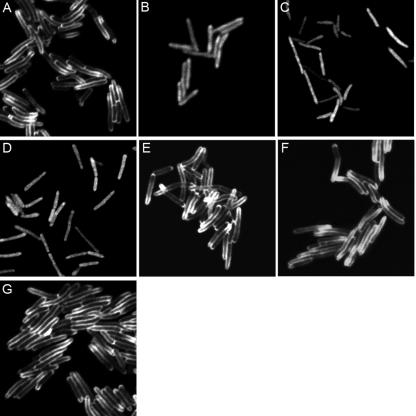
We constructed a TorA-GFP fusion using the first 43 amino acids of TorA fused in frame to GFP and cloned this construct downstream of an acetamide-inducible promoter (27) in a vector that integrates into the mycobacterial chromosome. The wild-type and complemented strains containing the inducible torA-gfp fusion were grown in M63 medium supplemented with succinate and acetamide to induce expression of the torA-gfp fusion. After induction for 12 h, the cells were washed and suspended in M63 medium lacking acetamide and incubated for 2 h to repress expression and allow TorA-GFP to localize. The TorA-GFP fusion was visualized by confocal microscopy. Green fluorescence was only observed at the periphery of the cells in these strains, and no detectable green fluorescence was seen in the cytoplasm (Fig. 3A, E, F, and G). If the repression step was omitted, green fluorescence could also be observed in the cytoplasm, but the majority of the fusion protein still localized to the periphery of the cells (data not shown). The TorA-GFP fusion was secreted and partially remained associated with either the cytoplasmic membrane or the cell wall in the wild-type and complemented strains.
FIG. 3.
Localization of TorA-GFP in the wild type (A), ΔtatA (B), ΔtatB (C), ΔtatC (D), tatA+ (E), tatB+ (F), and tatC+ (G) by fluorescent microscopy. Cells harboring the torA-gfp fusion were grown in the presence of acetamide, washed, and suspended in medium lacking acetamide for 1 h.
Initial attempts to culture the tat mutants in the M63 medium for the induction of torA-gfp were unsuccessful. However, we were able to culture the tat mutants in 7H9-ADC medium and induce expression with the addition of acetamide. Green fluorescence was observed in the cytoplasm of the tat mutants, and there was no green fluorescence along the periphery of the cell (Fig. 3B to D), suggesting that the tat mutants are not able to secrete the TorA-GFP fusion across the cytoplasmic membrane. We also tried to induce the expression of torA-gfp in the wild-type and complemented strains when they were grown in 7H9-ADC medium, but we did not detect any fluorescence after induction with acetamide. The acetamidase promoter in the wild-type strain was not induced when the cells were grown in medium containing glucose as a carbon source, even when acetamide was present (24), but this was not the case for the tat deletion strains. This suggests that the metabolism of the tat mutants is altered, compared to the wild type, since induction of the acetamidase promoter did occur in the presence of glucose.
Analysis of cell filtrate proteins.
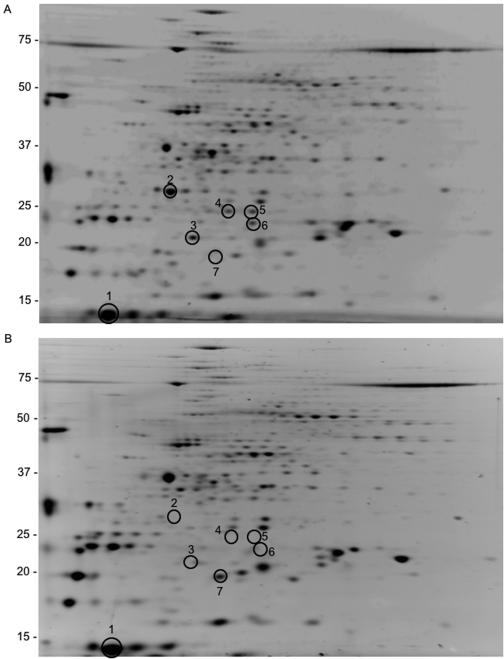
To identify proteins that are secreted by the TAT translocase system in M. smegmatis, the CFPs of the wild-type and ΔtatC strains were compared BY 2D-PAGE. Secreted proteins that are TAT dependent should be absent in the CFPs isolated from the ΔtatC strain.
At least five proteins were either absent or secreted in smaller quantities from the ΔtatC strain compared to the wild type (Fig. 4, spots 2 to 6 in both gels), and at least one protein was secreted at a higher level in the ΔtatC strain (Fig. 4, spot 7 in both gels). The expression level of these proteins was consistent in all experiments. These spots were further analyzed by MALDI-TOF to identify the proteins, and spot 1 from each gel was used as an internal control for MALDI-TOF analysis. This spot was identified as a flavin-containing monamine oxidase protein (Msmeg1187). We positively identified two of the remaining proteins. Spot 2 was identified as β-lactamase (Msmeg2657), and spot 3 was identified as a CaiB-BaiF homolog (Msmeg4864). The β-lactamase contains the twin-arginine signal sequence (SRRSVL) at its N-terminal region, followed by a hydrophobic region and a possible signal peptidase cleavage site (ASA). We were not able to locate a TAT signal sequence within the first 20 to 30 amino acids of the CaiB-BaiF homolog, but a possible TAT signal sequence is located at amino acids 101 to 105. The correct start site of the CaiB-BaiF homolog is not known. The CaiB-BaiF homolog is a coenzyme A transferase, and it would seem that this protein should not be secreted. However, an acetyl-coenzyme A-C acetyltransferase is also secreted from M. tuberculosis (25); therefore, the CaiB-BaiF homolog may also be a truly secreted protein.
FIG. 4.
Analysis of CFPs of the wild type (A) and ΔtatC (B) by 2D-PAGE. The proteins that consistently displayed an altered expression level are circled and were analyzed by MALDI-TOF.
The tat mutants are susceptible to β-lactam antibiotics.
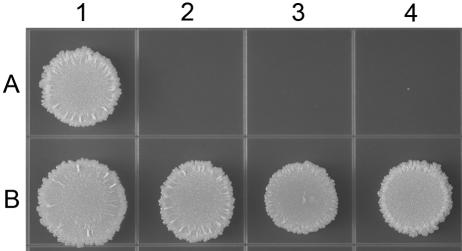
Because secretion of β-lactamase in M. smegmatis appears to be dependent on the TAT translocase system, the tat deletion strains were tested for their susceptibility to ampicillin. The cells were grown to mid-log phase, and 103 to 104 cells were spotted onto TSA plates containing ampicillin. After 4 days, the wild-type and complemented strains displayed growth in the presence of ampicillin (Fig. 5), while the tat deletion strains were not able to grow during this time frame (Fig. 5). Even after the plates were incubated for 14 days, the tat deletion strains did not exhibit any growth (data not shown).
FIG. 5.
The tat deletion strains are sensitive to ampicillin. Approximately 104 cells of each strain were spotted onto TSA plates containing ampicillin (25 μg/ml). (A) Lanes: 1, wild type; 2, ΔtatA; 3, ΔtatB; 4, ΔtatC. (B) Lanes: 1, wild type; 2, tatA+; 3, tatB+; 4, tatC+.
In silico analysis of M. smegmatis genome.
Dilks et al. developed the TATFIND program, which is able to identify putative TAT substrates from a bacterial genome (11). The TAT substrates have to meet specific criteria, including the presence of a TAT signal sequence within the first 35 amino acids of the protein and a stretch of at least 13 uncharged amino acids downstream of the signal sequence. The authors analyzed 84 bacterial genomes with TATFIND, and the number of putative TAT substrates ranged from 145 (Streptomyces coelicolor) to 1 (Listeria monocytogenes) in the different bacterial species (11). The annotated genome of M. smegmatis was analyzed with TATFIND, and 49 putative TAT substrates were identified which belong to five classes of proteins (Table 2). The protein classes are as follows: a transport and binding class (17/49), a cell envelope-associated class (10/49), a metabolism class (8/49), a class of hypothetical proteins (9/49), or a class of proteins involved in detoxification processes (5/49). The TATFIND program also falsely identified six proteins that are known to be located in the cytoplasm. These six proteins are not included in Table 2.
TABLE 2.
Putative TAT substrates
| Protein class and name | Function of substrate |
|---|---|
| Transport-solute binding | |
| Msmeg0430 | FecB2; iron transport |
| Msmeg0477 | ABC transporter |
| Msmeg0651 | Solute binding protein |
| Msmeg0839 | Na+-H+ antiporter |
| Msmeg1209 | ABC transporter |
| Msmeg2633 | Sulfonate transporter |
| Msmeg2994 | Short-chain amide transporter |
| Msmeg3293 | Solute binding protein |
| Msmeg3412 | Major facilitator transporter |
| Msmeg3547 | Amino acid transporter |
| Msmeg3998 | Dicarboxylate transporter |
| Msmeg4463 | Sugar ABC transporter |
| Msmeg4515 | Sulfonate ABC transporter |
| Msmeg5131 | Solute binding protein |
| Msmeg5348 | Amino acid transporter |
| Msmeg6629 | Spermidine ABC transporter |
| Msmeg6807 | DctP; succinate transporter |
| Cell envelope | |
| Msmeg0018 | ExiT protein |
| Msmeg0104 | Transcriptional attenuator |
| Msmeg0132 | LrpL lipoprotein |
| Msmeg0921 | LppS lipoprotein |
| Msmeg1135 | Mce family |
| Msmeg2252 | Lipoprotein |
| Msmeg2857 | Mce3 |
| Msmeg3424 | Membrane protein |
| Msmeg4298 | Proteinase |
| Msmeg4737 | LppS lipoprotein |
| Metabolism | |
| Msmeg0187 | Steroid monooxygenase |
| Msmeg0637 | Glucanase |
| Msmeg3180 | Asparaginase |
| Msmeg3558 | Dienelactone hydrogenase |
| Msmeg5008 | Polysaccharide deacetylase |
| Msmeg5257 | Lipase |
| Msmeg5827 | Oxidoreductase |
| Msmeg6510 | Cytochrome b6f |
| Detoxification | |
| Msmeg2657 | β-Lactamase |
| Msmeg3715 | Catalase |
| Msmeg5400 | Peroxidase |
| Msmeg6530 | Peroxidase |
| Msmeg6836 | β-Lactamase |
| Hypothetical-unknown | |
| Msmeg0402 | |
| Msmeg0866 | |
| Msmeg1517 | |
| Msmeg2624 | |
| Msmeg3234 | |
| Msmeg3624 | |
| Msmeg3930 | |
| Msmeg5842 | |
| Msmeg6321 |
The tat mutants cannot use succinate as a sole carbon source.
The succinate transporter, DctP, was identified as a putative TAT-dependent protein by in silico analysis. DctP is a periplasmic transport protein in gram-negative bacteria (15) and is anchored to the cytoplasmic membrane in gram-positive bacteria (1). DctP transports succinate into the cytoplasm of bacteria and allows bacteria to use succinate as a carbon source. The tat deletion strains were assayed for the ability to use succinate as the sole carbon source by growing the strains in M63 medium containing succinate (M63-succinate). The wild-type and complemented strains were able to grow in M63-succinate and exhibited a doubling time of approximately 5 h (Fig. 6). Unlike the wild type, the tat deletion strains did not exhibit any significant growth during the time course of the experiment (Fig. 6). The wild-type, complemented, and tat deletion strains were able to grow with a similar growth rate in M63 medium containing glycerol as the sole carbon source (data not shown), indicating that it was the inability of the tat deletion strains to use succinate as a sole carbon source that was responsible for the growth defect.
FIG. 6.
Succinate does not support the growth of the tat deletion strains. The strains were initially cultured in 7H9-ADC medium, and the cells were harvested, washed, and diluted into M63-succinate medium. Growth was monitored by measuring the change in OD600 values over time. The symbols for each strain are as follows: x, wild type; open symbols, tat deletion strains; closed symbols, complemented strains.
DISCUSSION
Initially, database searches revealed that mycobacteria contain homologs of each the three genes (tatA, tatB, and tatC) known to be essential for the secretion of proteins using the TAT system. To determine if mycobacteria actually had a functional TAT system, a TorA-GFP reporter system similar to that previously used with E. coli and Synechocystis was constructed. When the TorA-GFP fusion protein was expressed in wild-type M. smegmatis bacilli, green fluorescence was localized at the periphery of the cells. Native GFP, which does not contain a secretion signal sequence, remains in the cytoplasm. Therefore, the addition of the TorA TAT signal sequence to the GFP is sufficient to direct secretion of the fusion protein across the cytoplasmic membrane, which shows that M. smegmatis has a functional TAT system. In M. smegmatis as in other bacteria, each of the three tat gene products is necessary for TAT-dependent secretion. That is, strains carrying a mutation in any one of the tat genes are unable to secrete a TorA-GFP fusion protein across the cytoplasmic membrane.
The disruption of the TAT secretion system in E. coli and P. aeruginosa has a dramatic effect on physiology, including the metabolism of specific substrates under aerobic or anaerobic growth conditions and growth under iron-limiting conditions (19, 26). In E. coli, tat deletion strains are not able to divide correctly and form long chains, and the integrity of the outer membrane is altered (35). These phenotypes have been linked to the incorrect localization of two TAT substrates, AmiA and AmiC, which are cell wall amidases (20). The disruption of the TAT system in M. smegmatis also affects the physiology of the bacteria including growth rate and colony morphology. For example, tat mutant strains exhibited altered colony morphology (less cording; more transparency) when cultured on TSA plates, somewhat similar to that of an M. smegmatis secA2 mutant cultured on solid rich medium (4). Also, the tat mutant strains are not capable of using succinate or acetamide as a sole carbon source when cultured in minimal medium. The inability to use these substrates might be due to the lack of transport of succinate or acetamide into the cell given that the succinate transporter (DctP) and urea short-chain amide transporter (FmdD) contain putative TAT signal sequences.
Two approaches were used to identify proteins secreted by the TAT system. By comparing the culture filtrate proteins (i.e., primarily secreted proteins) of the ΔtatC strain and wild-type bacteria, we identified at least five proteins that were present in the filtrates of wild-type bacteria but not in filtrates of the ΔtatC strain. These proteins are likely secreted by the TAT system. A similar approach was used to show that SecA2 was required to secrete SodA in M. tuberculosis (5). One of the putative TAT-secreted proteins was subsequently identified as a β-lactamase (Msmeg2657-BlaS). BlaS contains the TAT signal sequence RRSVLI, followed by a hydrophobic domain and a possible cleavage site. BlaS belongs to the class A β-lactamases, which have a conserved S-X-X-K motif and are not metal cofactor dependent, as is the case for the class B lactamases. The lack of metal dependence of BlaS is interesting because the TAT system is often used for the secretion of cofactor-dependent enzymes, due to its ability to transport proteins in a folded state. Interestingly, failure to secrete BlaS confers ampicillin susceptibility upon strains with tatA, tatB, or tatC mutations. This is consistent with the observation that deletion of blaS confers ampicillin susceptibility (14), although M. smegmatis contains several lactamases (www.tigr.org.tdb/ofmg/).
We analyzed the genome of M. smegmatis with TATFIND (11) and identified 49 putative TAT substrates, which represent approximately 0.7% of the encoded proteins. The genome of M. tuberculosis has also been analyzed with TATFIND, and the authors identified 31 putative TAT substrates (11), which also represent 0.7% of the encoded proteins. The majority of the TAT substrates of M. smegmatis seem to be non-redox proteins, in contrast to E. coli in which the majority of TAT substrates are involved in redox reactions and contain some type of cofactor (2). The TAT substrates of other bacteria such as Bacillus subtilis (11), Caulobacter crescentus (11), Halobacterium (30), and Streptomyces coelicolor (34) are also mainly non-redox proteins. Of the 49 putative TAT substrates in M. smegmatis, only 12 appear to have a cleavable signal sequence that would allow secretion of the substrate into the extracellular environment, while the majority of proteins seem to be anchored to the cytoplasmic membrane, due to an uncleaved signal sequence or lipidation of the protein. This might explain why we only observed minor differences in the CFPs of the ΔtatC strain compared to the wild type, since most of the putative TAT substrates are not exported from the cell.
The TAT system has also been shown to be required for virulence in numerous bacteria including E. coli (29), P. aeruginosa (26), and Legionella pneumophila (31). Because M. smegmatis is a nonpathogenic soil-dwelling bacterium, we were not able to study this aspect. However, the TAT system is conserved in pathogenic M. tuberculosis. To begin to determine the role the TAT system plays in pathogenesis, we created a tatB deletion in M. tuberculosis, and studies are under way to evaluate the importance of the TAT system for pathogenesis.
Supplementary Material
Acknowledgments
We thank James Carroll for performing the MALDI-TOF analysis of the secreted proteins. We also thank Kieran Dilks and Mechthild Pohlschroder for analyzing the genome of M. smegmatis with TATFIND.
This work was supported by CDC funds and NIH grant AI49659 awarded to J.P.
Use of trade names is for identification only and does not constitute endorsement by the U.S. Department of Health and Human Services, the U.S. Public Health Service, or the CDC.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Asai, K., S. H. Baik, Y. Kasahara, S. Moriya, and N. Ogasawara. 2000. Regulation of the transport system for C4-dicarboxylic acids in Bacillus subtilis. Microbiology 146:263-271. [DOI] [PubMed] [Google Scholar]
- 2.Berks, B. C. 1996. A common export pathway for proteins binding complex redox cofactors? Mol. Microbiol. 22:393-404. [DOI] [PubMed] [Google Scholar]
- 3.Berks, B. C., F. Sargent, and T. Palmer. 2000. The Tat protein export pathway. Mol. Microbiol. 35:260-274. [DOI] [PubMed] [Google Scholar]
- 4.Braunstein, M., A. M. Brown, S. Kurtz, and W. R. Jacobs, Jr. 2001. Two nonredundant SecA homologues function in mycobacteria. J. Bacteriol. 183:6979-6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braunstein, M., B. J. Espinosa, J. Chan, J. T. Belisle, and W. R. Jacobs, Jr. 2003. SecA2 functions in the secretion of superoxide dismutase A and in the virulence of Mycobacterium tuberculosis. Mol. Microbiol. 48:453-464. [DOI] [PubMed] [Google Scholar]
- 6.Carroll, J. A., R. M. Cordova, and C. F. Garon. 2000. Identification of 11 pH-regulated genes in Borrelia burgdorferi localizing to linear plasmids. Infect. Immun. 68:6677-6684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 8.Cole, S. T., K. Eiglmeier, J. Parkhill, K. D. James, N. R. Thomson, P. R. Wheeler, N. Honore, T. Garnier, C. Churcher, D. Harris, K. Mungall, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. M. Davies, K. Devlin, S. Duthoy, T. Feltwell, A. Fraser, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, C. Lacroix, J. Maclean, S. Moule, L. Murphy, K. Oliver, M. A. Quail, M. A. Rajandream, K. M. Rutherford, S. Rutter, K. Seeger, S. Simon, M. Simmonds, J. Skelton, R. Squares, S. Squares, K. Stevens, K. Taylor, S. Whitehead, J. R. Woodward, and B. G. Barrell. 2001. Massive gene decay in the leprosy bacillus. Nature 409:1007-1011. [DOI] [PubMed] [Google Scholar]
- 9.Converse, S. E., and J. S. Cox. 2005. A protein secretion pathway critical for Mycobacterium tuberculosis virulence is conserved and functional in Mycobacterium smegmatis. J. Bacteriol. 187:1238-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Keyzer, J., C. van der Does, and A. J. Driessen. 2003. The bacterial translocase: a dynamic protein channel complex. Cell Mol. Life Sci. 60:2034-2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dilks, K., R. W. Rose, E. Hartmann, and M. Pohlschroder. 2003. Prokaryotic utilization of the twin-arginine translocation pathway: a genomic survey. J. Bacteriol. 185:1478-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobos, K. M., K. Swiderek, K. H. Khoo, P. J. Brennan, and J. T. Belisle. 1995. Evidence for glycosylation sites on the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. Infect. Immun. 63:2846-2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Economou, A. 2002. Bacterial secretome: the assembly manual and operating instructions. Mol. Membr. Biol. 19:159-169. [DOI] [PubMed] [Google Scholar]
- 14.Flores, A. R., L. M. Parsons, and M. S. Pavelka, Jr. 2005. Genetic analysis of the beta-lactamases of Mycobacterium tuberculosis and Mycobacterium smegmatis and susceptibility to beta-lactam antibiotics. Microbiology 151:521-532. [DOI] [PubMed] [Google Scholar]
- 15.Forward, J. A., M. C. Behrendt, N. R. Wyborn, R. Cross, and D. J. Kelly. 1997. TRAP transporters: a new family of periplasmic solute transport systems encoded by the dctPQM genes of Rhodobacter capsulatus and by homologs in diverse gram-negative bacteria. J. Bacteriol. 179:5482-5493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garnier, T., K. Eiglmeier, J. C. Camus, N. Medina, H. Mansoor, M. Pryor, S. Duthoy, S. Grondin, C. Lacroix, C. Monsempe, S. Simon, B. Harris, R. Atkin, J. Doggett, R. Mayes, L. Keating, P. R. Wheeler, J. Parkhill, B. G. Barrell, S. T. Cole, S. V. Gordon, and R. G. Hewinson. 2003. The complete genome sequence of Mycobacterium bovis. Proc. Natl. Acad. Sci. USA 100:7877-7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guinn, K. M., M. J. Hickey, S. K. Mathur, K. L. Zakel, J. E. Grotzke, D. M. Lewinsohn, S. Smith, and D. R. Sherman. 2004. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol. Microbiol. 51:359-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hinds, J., E. Mahenthiralingam, K. E. Kempsell, K. Duncan, R. W. Stokes, T. Parish, and N. G. Stoker. 1999. Enhanced gene replacement in mycobacteria. Microbiology 145:519-527. [DOI] [PubMed] [Google Scholar]
- 19.Ize, B., I. Porcelli, S. Lucchini, J. C. Hinton, B. C. Berks, and T. Palmer. 2004. Novel phenotypes of Escherichia coli tat mutants revealed by global gene expression and phenotypic analysis. J. Biol. Chem. 279:47543-47554. [DOI] [PubMed] [Google Scholar]
- 20.Ize, B., N. R. Stanley, G. Buchanan, and T. Palmer. 2003. Role of the Escherichia coli Tat pathway in outer membrane integrity. Mol. Microbiol. 48:1183-1193. [DOI] [PubMed] [Google Scholar]
- 21.Kong, D., and D. Y. Kunimoto. 1995. Secretion of human interleukin 2 by recombinant Mycobacterium bovis BCG. Infect. Immun. 63:799-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, L., J. P. Bannantine, Q. Zhang, A. Amonsin, B. J. May, D. Alt, N. Banerji, S. Kanjilal, and V. Kapur. 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis. Proc. Natl. Acad. Sci. USA 102:12344-12349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Limia, A., F. J. Sangari, D. Wagner, and L. E. Bermudez. 2001. Characterization and expression of secA in Mycobacterium avium. FEMS Microbiol. Lett. 197:151-157. [DOI] [PubMed] [Google Scholar]
- 24.Mahenthiralingam, E., P. Draper, E. O. Davis, and M. J. Colston. 1993. Cloning and sequencing of the gene which encodes the highly inducible acetamidase of Mycobacterium smegmatis. J. Gen. Microbiol. 139:575-583. [DOI] [PubMed] [Google Scholar]
- 25.Mattow, J., U. E. Schaible, F. Schmidt, K. Hagens, F. Siejak, G. Brestrich, G. Haeselbarth, E. C. Muller, P. R. Jungblut, and S. H. Kaufmann. 2003. Comparative proteome analysis of culture supernatant proteins from virulent Mycobacterium tuberculosis H37Rv and attenuated M. bovis BCG Copenhagen. Electrophoresis 24:3405-3420. [DOI] [PubMed] [Google Scholar]
- 26.Ochsner, U. A., A. Snyder, A. I. Vasil, and M. L. Vasil. 2002. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc. Natl. Acad. Sci. USA 99:8312-8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parish, T., E. Mahenthiralingam, P. Draper, E. O. Davis, and M. J. Colston. 1997. Regulation of the inducible acetamidase gene of Mycobacterium smegmatis. Microbiology 143:2267-2276. [DOI] [PubMed] [Google Scholar]
- 28.Parish, T., and N. G. Stoker. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969-1975. [DOI] [PubMed] [Google Scholar]
- 29.Pradel, N., C. Ye, V. Livrelli, J. Xu, B. Joly, and L. F. Wu. 2003. Contribution of the twin arginine translocation system to the virulence of enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 71:4908-4916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose, R. W., T. Bruser, J. C. Kissinger, and M. Pohlschroder. 2002. Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol. Microbiol. 45:943-950. [DOI] [PubMed] [Google Scholar]
- 31.Rossier, O., and N. P. Cianciotto. 2005. The Legionella pneumophila tatB gene facilitates secretion of phospholipase C, growth under iron-limiting conditions, and intracellular infection. Infect. Immun. 73:2020-2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santini, C. L., B. Ize, A. Chanal, M. Muller, G. Giordano, and L. F. Wu. 1998. A novel Sec-independent periplasmic protein translocation pathway in Escherichia coli. EMBO J. 17:101-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sargent, F., E. G. Bogsch, N. R. Stanley, M. Wexler, C. Robinson, B. C. Berks, and T. Palmer. 1998. Overlapping functions of components of a bacterial Sec-independent protein export pathway. EMBO J. 17:3640-3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schaerlaekens, K., L. Van Mellaert, E. Lammertyn, N. Geukens, and J. Anne. 2004. The importance of the Tat-dependent protein secretion pathway in Streptomyces as revealed by phenotypic changes in tat deletion mutants and genome analysis. Microbiology 150:21-31. [DOI] [PubMed] [Google Scholar]
- 35.Stanley, N. R., K. Findlay, B. C. Berks, and T. Palmer. 2001. Escherichia coli strains blocked in Tat-dependent protein export exhibit pleiotropic defects in the cell envelope. J. Bacteriol. 183:139-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanley, S. A., S. Raghavan, W. W. Hwang, and J. S. Cox. 2003. Acute infection and macrophage subversion by Mycobacterium tuberculosis require a specialized secretion system. Proc. Natl. Acad. Sci. USA 100:13001-13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stover, C. K., V. F. de la Cruz, G. P. Bansal, M. S. Hanson, T. R. Fuerst, W. R. Jacobs, Jr., and B. R. Bloom. 1992. Use of recombinant BCG as a vaccine delivery vehicle. Adv. Exp. Med. Biol. 327:175-182. [DOI] [PubMed] [Google Scholar]
- 38.Takayama, K., H. K. Schnoes, E. L. Armstrong, and R. W. Boyle. 1975. Site of inhibitory action of isoniazid in the synthesis of mycolic acids in Mycobacterium tuberculosis. J. Lipid Res. 16:308-317. [PubMed] [Google Scholar]
- 39.Thomas, J. D., R. A. Daniel, J. Errington, and C. Robinson. 2001. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol. Microbiol. 39:47-53. [DOI] [PubMed] [Google Scholar]
- 40.Yen, M. R., Y. H. Tseng, E. H. Nguyen, L. F. Wu, and M. H. Saier, Jr. 2002. Sequence and phylogenetic analyses of the twin-arginine targeting (Tat) protein export system. Arch. Microbiol. 177:441-450. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.