Abstract
Yersinia pestis is an important human pathogen that is maintained in flea-rodent enzootic cycles in many parts of the world. During its life cycle, Y. pestis senses host-specific environmental cues such as temperature and regulates gene expression appropriately to adapt to the insect or mammalian host. For example, Y. pestis synthesizes different forms of lipid A when grown at temperatures corresponding to the in vivo environments of the mammalian host and the flea vector. At 37°C, tetra-acylated lipid A is the major form; but at 26°C or below, hexa-acylated lipid A predominates. In this study, we show that the Y. pestis msbB (lpxM) and lpxP homologs encode the acyltransferases that add C12 and C16:1 groups, respectively, to lipid IVA to generate the hexa-acylated form, and that their expression is upregulated at 21°C in vitro and in the flea midgut. A Y. pestis ΔmsbB ΔlpxP double mutant that did not produce hexa-acylated lipid A was more sensitive to cecropin A, but not to polymyxin B. This mutant was able to infect and block fleas as well as the parental wild-type strain, indicating that the low-temperature-dependent change to hexa-acylated lipid A synthesis is not required for survival in the flea gut.
The cell envelope of gram-negative bacteria includes two lipid bilayers, an inner membrane composed primarily of phospholipids, and an outer membrane containing primarily phospholipids in the inner leaflet and lipopolysaccharide (LPS) in the outer leaflet. Tight stacking of the long-chain fatty acids of lipid A, the hydrophobic anchor of LPS, creates a permeability barrier against toxic compounds encountered in the environment and the host (25). In Escherichia coli and Salmonella enterica serovar Typhimurium, the final steps of lipid A synthesis occur in the inner membrane, where two acyl groups are added to the tetra-acylated Kdo-lipid IVA before the mature hexa-acylated lipid A is exported to the outer membrane. At normal growth temperatures, the late acyltransferases HtrB (LpxL) and MsbB (LpxM) consecutively add lauroyl (C12) and myristoyl (C14) groups to the tetra-acylated intermediate (7, 8, 31). At 12°C, however, the cold-temperature-specific late acyltransferase LpxP acts instead of LpxL to add palmitoleate (C16:1) (4). Mutation of htrB and msbB leads to growth defects and hypersensitivity to rifampin and vancomycin in E. coli, and to decreased virulence and resistance to macrophage killing in E. coli and S. enterica (37).
Yersinia pestis, the zoonotic agent of bubonic and pneumonic plague in humans, is primarily a pathogen of rodents that is transmitted by fleas (27). Upon transmission to a mammalian host and an increase in temperature to 37°C, Y. pestis upregulates the expression of virulence factors that aid colonization and invasion. Conversely, after Y. pestis is taken up in a blood meal by a flea vector, the decrease in temperature results in downregulation of virulence factors and activation of genes and gene products important for survival in the flea midgut and transmission to a new mammalian host (5, 11, 17, 19, 24a).
One example of a temperature-dependent phenotype in Y. pestis is lipid A structural variation (23, 24, 28). At the mammalian host temperature of 37°C, Y. pestis produces and exports primarily tetra-acylated lipid A to the outer membrane. However, at a temperature (21°C) typical of the flea vector, Y. pestis generates primarily hexa-acylated lipid A modified with C12 and C16:1, a form that resembles lipid A produced by E. coli at 12°C (4). The increased acylation of lipid A at 21°C correlates with resistance of Y. pestis to cationic antimicrobial peptides (CAMPs) (28). These observations led us to hypothesize that expression of the Y. pestis late acyltransferases is temperature controlled, and that the shift from tetra-acylated lipid A at 37°C to hexa-acylated lipid A at 21°C is required for CAMP resistance and for the ability to survive in the flea digestive tract.
To test these hypotheses, we identified Y. pestis homologs of the msbB and lpxP acyltransferase genes and characterized their role in the temperature-dependent change from tetra-acylated to hexa-acylated lipid A. We also examined the role of these late acyltransferases and lipid A variation on outer membrane permeability, resistance to CAMPs, and the ability to colonize and produce a transmissible infection in fleas.
MATERIALS AND METHODS
Protein sequences and phylogenetic analysis.
Predicted Y. pestis homologs of the E. coli and Salmonella MsbB, LpxP, and HtrB acyltransferases were identified in the Y. pestis genome sequence by means of a TBlastN (protein versus. translated DNA) search of the Y. pestis CO92 genome (http://www.sanger.ac.uk/Projects/Y_pestis/). The open reading frame numbers of the Y. pestis msbB and lpxP homologs are YPO2063 and YPO3632, respectively (26). Phylogenetic analysis was performed using the neighbor-joining and bootstrap programs of MacVector version 7.2.3 (Accelrys, San Diego, CA).
Bacterial strains and mutagenesis.
The bacterial strains and plasmids used are listed in Table 1. To create the acyltransferase mutants, the predicted Y. pestis msbB and lpxP genes were amplified by PCR, gel purified, and cloned into the pCR-2.1 TOPO vector (Invitrogen, Carlsbad, CA). The resulting recombinant plasmids were used to transform E. coli TOP-10 (Invitrogen) by electroporation. Internal 391-bp and 276-bp deletions in the msbB and lpxP homologs were generated by inverse PCR of the recombinant TOPO 2.1 plasmids (38), followed by blunt-end ligation to yield a TOPO 2.1-ΔmsbB or -ΔlpxP product. The mutated alleles were subcloned into the suicide vector pCVD442 and electroporated into E. coli S-17. The plasmids were then introduced into Y. pestis KIM6+ by conjugation, and clones in which allelic exchange had occurred were selected (10).
TABLE 1.
Bacteria and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| Y. pestis KIM6+ | pYV-negative derivative of strain KIM | 35 |
| Y. pestis ΔmsbB | KIM6+ with a 391-bp deletion in msbB | This study |
| Y. pestis ΔlpxP | KIM6+ with a 276-bp deletion in lpxP | This study |
| Y. pestis ΔmsbB ΔlpxP | KIM6+ double deletion mutant | This study |
| Y. pestis ΔmsbB(pLGmsbB) | Complemented msbB mutant | This study |
| Y. pestis ΔmsbB ΔlpxP(pLGlpxP) | Double mutant complemented with lpxP | This study |
| E. coli TOP-10 | Plasmid host strain | Invitrogen |
| E. coli S17-1 | recA λpir; host for suicide vector pCVD442 | 29 |
| Plasmids | ||
| pCR 2.1-TOPO | Cloning vector | Invitrogen |
| pLG338 | Low-copy-number vector used for complementation | 32 |
| pCVD442 | Suicide vector used for allelic exchange | 10 |
| TOPO-msbB | 1.6-kb msbB locus cloned into pCR 2.1-TOPO | This study |
| TOPO-lpxP | 1.2-kb lpxP locus cloned into pCR 2.1-TOPO | This study |
| TOPO-ΔmsbB | 1.2-kb msbB deletion allele cloned in pCR 2.1-TOPO | This study |
| TOPO-ΔlpxP | 1.0-kb lpxP deletion allele cloned into pCR 2.1-TOPO | This study |
| pCVD442-ΔmsbB | 1.2-kb msbB deletion allele cloned into pCVD442 | This study |
| pCVD442-ΔlpxP | 1.0-kb lpxP deletion allele cloned into pCVD442 | This study |
| pLGmsbB | 1.4-kb wild-type msbB locus cloned into pLG338 | This study |
| pLGlpxP | 1.7-kb wild-type lpxP locus cloned into pLG338 | This study |
A Y. pestis ΔmsbB ΔlpxP double mutant was generated by allelic exchange of the ΔlpxP allele into the Y. pestis ΔmsbB strain. DNA sequencing confirmed the deletions. Complementation plasmids were generated by PCR amplification of the msbB and lpxP open reading frames and promoter-operator regions using primers flanked with KpnI and EcoRI sites. The complementation vector pLG338 and the Y. pestis msbB and lpxP PCR fragments were digested with KpnI and EcoRI, and the products of the reactions were gel-purified and ligated to generate pLGmsbB and pLGlpxP, which were introduced by electroporation into the Y. pestis ΔmsbB and ΔmsbB ΔlpxP mutant strains, respectively. Primer sequences used for cloning, inverse PCR, complementation, and sequencing are listed in Table 2.
TABLE 2.
Sequences of primers and probes used in this study
| Target gene | Use | Sequence (5′ to 3′) |
|---|---|---|
| msbB | Cloning | TTTGGATGAACCAGCAAGCG |
| GATAGGCAGAGGAGTAAAGCGTCC | ||
| Deletion | GGATCACGGAATTTGGGTGGAATATAAGCGAGCGCCGC | |
| GCTTTACATTTCGGTGGTCGGATCCATGCCCGCG | ||
| Complementation | GCGCGGTACCCGCAGTTACAACTGGATTAGCAGTG | |
| GCGCGAATTCCTATCACTAGCGGGCCTTTTACC | ||
| Taq Man primers | ACCCAAATTCCGTGATCCTTTA | |
| ACGGGCGCTTTTAGCAAAT | ||
| TaqMan probea | CTGGCATAGGGCGTCTCGCTGG | |
| Sequencing | AGAAAACCTTCCTCCATCC | |
| lpxP | Cloning | TGCGTCAATTTTCGTCCTCAC |
| TGGCGGGTATCAGTAATGCTAAG | ||
| Deletion | CCGGAGACGGAAAACCAGCGCTGTATCCGGGTATCGGACC | |
| CCCGCGCGGCAGCGTTTTTGCACCACTGTTCGCCG | ||
| Complementation | GCGCGGTACCGTTTTCTTTCAGGTAACGGAACGG | |
| GCGCGAATTCTGTGATTCCTACGACCCCAACG | ||
| TaqMan primers | CACCACTGTTCGCCGTAGAA | |
| AACGGGCGAGCATAAAGGT | ||
| TaqMan probe | ATGCGGCAACCACCAGCGG | |
| Sequencing | CTTGAGCAGACTATCATCGG | |
| ymt | TaqMan primers | CACCAATCAACGATACAAGAATGAC |
| TGTCCACCAACAAGAGCTTCAG | ||
| TaqMan probe | TGGAATCACACAAAAATAATGGCCTCAGATG | |
| proS | TaqMan primers | ACGCGCACCGGCTACA |
| CTCGGCGATGGTTTTTGC | ||
| TaqMan probe | AGAGCTGCGAATCGTTGACACCCC |
TaqMan probes contain 5′-6-carboxyfluorescein reporter and 3′-6-carboxy-tetramethyl-rhodamine quencher.
Lipid A isolation and structural analysis.
Y. pestis strains were grown in Luria broth (LB), pH 7.4, at 21°C without aeration and harvested in late exponential phase (optical density at 600 nm [OD600] ∼1.0) and analyzed as described (28). LPS was purified by Mg2+-ethanol precipitation (9). Lipid A was isolated by mild acid hydrolysis of LPS in 1% sodium dodecyl sulfate at pH 4.5 (3). Negative-ion spectra were acquired from a delayed extraction matrix-assisted laser desorption ionization-time-of-flight (DE-MALDI-TOF) mass spectrometer (Biflex III, Bruker Daltonics; Billerica, MA) (12). Briefly, lipid A was dissolved in 10 μl of a mixture of 5-chloro-2-mercaptobenzothiazole (20 mg/ml) in chloroform/methanol 1:1 (vol/vol), and 1-μl samples were analyzed by MALDI-TOF MS. Acyl groups from LPS samples were derivatized to fatty methyl esters with 2 M methanolic HCl at 90°C for 18 h and were identified and quantified by gas chromatography (GC) using an HP 5890 series II with a 7673 autoinjector (Hewlett Packard; Palo Alto, CA) (31). MS and GC analyses were performed on a minimum of two independent samples for each bacterial strain.
RNA isolation.
For RNA extraction, bacteria were grown in LB, pH 7.4, at 21°C with aeration and harvested at an OD600 of 0.6. RNA for real-time PCR analysis was obtained using the RNeasy minikit (QIAGEN; Valencia, CA). Bacteria in 2 ml of culture were harvested by centrifugation for 1 min at 16,000 × g, resuspended in 1.0 ml ice-cold RLT buffer (QIAGEN), flash-frozen in liquid nitrogen, and stored at −80°C. The suspensions were transferred to chilled tubes containing lysing matrix B (Q-BIOgene; Carlsbad, CA) and the bacteria were disrupted by agitation for 30 seconds at speed 6.0 in a FastPrep device (Q-BIOgene). Lysates were then mixed with 0.4 ml of 100% ethanol, and the total RNA was isolated by means of RNeasy minicolumns (QIAGEN). Contaminating DNA in RNA samples was removed by using the DNA-free DNase kit (Ambion, Austin, TX).
Total RNA was also extracted from samples of 50 infected fleas that had been placed in lysing matrix B tubes (Q-BIOgene), flash frozen in liquid nitrogen, and stored at −80°C; 1 ml of ice-cold RLT buffer mix was added to each tube containing the frozen, infected fleas. Release of bacteria from the flea midguts and cell disruption was accomplished by two consecutive 30 sec FastPrep cycles at speed 6.0; tubes were cooled on ice for 1 min after each agitation cycle. Lysates were passed through a QIAshredder column (QIAGEN) to remove particulate matter and RNA was isolated and DNase-treated as described above.
Real-time quantitative reverse transcription-PCR (TaqMan).
Total RNA samples obtained from three biological replicates for each condition tested were diluted to 1 to 10 μg/ml after DNase treatment and A260 quantitation. Reverse transcription and TaqMan PCR analysis of Y. pestis transcript levels were performed in triplicate for sample as previously described (6) using an ABI 7700 thermocycler (Applied Biosystems; Foster City, CA) and primer-probe sets listed in Table 2. The relative quantities of msbB, lpxP, and ymt mRNA were normalized to the amount of proS mRNA present in the samples.
Antimicrobial susceptibility assays.
Susceptibility to polymyxin B (ICN Biochemicals, Aurora, OH), deoxycholate, rifampin, vancomycin, and cecropin A (Sigma-Aldrich, St. Louis, MO) was determined by bactericidal assays. Y. pestis KIM6+, ΔmsbB, ΔlpxP, and ΔmsbBΔlpxP strains were grown in LB supplemented with 1 mM MgCl2 at 21°C. Overnight cultures were diluted in Mueller Hinton broth to ∼5 × 105 CFU/ml. Antimicrobial agents were dissolved in 0.1% bovine serum albumin and 0.01% acetic acid, and serial dilutions were prepared. In a 96-well plate, 11 μl of each dilution was added to 100 μl of bacterial suspension and incubated at 21°C for 2 h. Samples were then serially diluted, plated on LB agar plates, and CFU were counted after 48 h incubation at 37°C. Percent survival was calculated relative to CFU counts from wells with no added antimicrobial agent. All assays were performed in triplicate.
Flea infections.
Xenopsylla cheopis fleas were infected with Y. pestis KIM6+ or Y. pestis KIM6+ ΔmsbB ΔlpxP by allowing them to feed on heparinized mouse blood containing approximately 5.0 × 108 bacteria per ml using a membrane feeder apparatus (16). Fleas that took the infected blood meal were maintained at 21°C and 75% relative humidity, and fed twice weekly on uninfected mice. At three different times after infection, samples of 50 fleas were collected for RNA extraction and reverse transcription (RT)-PCR analysis. Additional samples of 50 female and 50 male fleas were monitored over a 4-week period for proventricular blockage, indicated by the presence of fresh blood in the esophagus and absence of fresh blood in the midgut after feeding (16). At 1 h and 1, 7, and 24 days after infection, samples of 20 female fleas were collected and stored at −70°C. The infection rate and bacterial load in these fleas were determined by CFU count (16).
RESULTS
Identification of lipid A late acyltransferases in Y. pestis.
Two potential homologs of lipid A late acyltransferases were identified in the Y. pestis genome (26). The predicted product of one (YPO2063) has 65% amino acid identity and 79% similarity to E. coli MsbB (LpxM), and the second (YPO3632) has 67% identity and 80% similarity to E. coli LpxP (Fig. 1). No obvious Y. pestis HtrB (LpxL) homolog was detected, although the predicted product of YPO3632 also shared 59% identity and 78% similarity, to the E. coli HtrB (Fig. 1). No other Y. pestis open reading frames were similar to the three E. coli late acyltransferase genes. Y. pestis mutant strains deleted of the msbB and lpxP homologs were used to determine whether these genes were responsible for the temperature-dependent change from tetra-acylated to hexa-acylated lipid A.
FIG. 1.
Cladogram comparing the two Y. pestis late acyltransferases to HtrB, MsbB, and LpxP of E. coli and S. enterica. Salmonella PagP, an unrelated lipid A acyltransferase, was used to root the tree. The scale bar indicates the calculated evolutionary distances.
Role of the Y. pestis MsbB and LpxP homologs in temperature-dependent lipid A variation.
Lipid A produced by the Y. pestis KIM6+, ΔmsbB, and ΔmsbB ΔlpxP strains grown at 21°C was purified for structural analysis by MALDI-TOF MS and gas chromatography. As previously reported (28), lipid A from Y. pestis KIM6+ grown at 21°C was predominantly hexa-acylated (m/z 1824), containing four 3-OH-C14, one C12 and one C16:1 (Fig. 2A). Other identifiable minor lipid A species produced at 21°C included previously described tetra-acylated (m/z 1405), penta-acylated (m/z 1588), and C10-containing hepta-acylated molecules (m/z 1977) (28). The hexa-acylated and hepta-acylated species were further modified by aminoarabinose addition (m/z 1955 and 2108, respectively [Fig. 2A]) (28). Fatty acid quantitation by GC corroborated the interpretation of the MALDI-TOF spectra (Table 3).
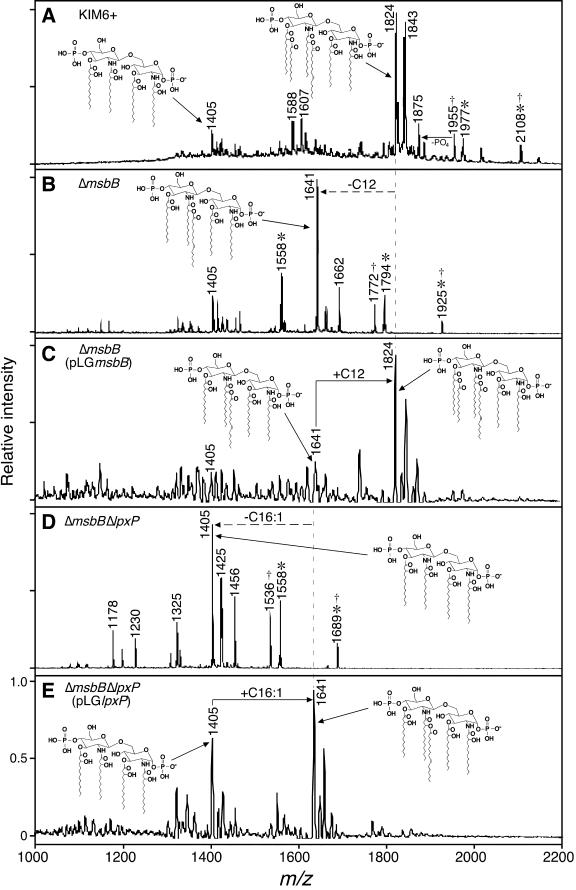
FIG.2.
Negative-ion MALDI-TOF MS spectra of lipid A isolated from Y. pestis KIM6+ (A), Y. pestis ΔmsbB (B), Y. pestis ΔmsbB (pLGmsbB) (C), Y. pestis ΔmsbB ΔlpxP (D), and Y. pestis ΔmsbB ΔlpxP(pLGlpxP) (E) grown at 21°C in LB. Peaks corresponding to lipid A species predicted to contain C10 (*), and aminoarabinose (†) are indicated (28). Acyl group positions are as proposed before (2, 23, 24).
TABLE 3.
Relative amount of different acyl groups (expressed as a percentage of the total) in lipid A of Y. pestis strains grown at 21°C
| Fatty acid | Relative amt (% of total)
|
||||
|---|---|---|---|---|---|
| Y. pestis KIM6+ | Y. pestis ΔmsbB | Y. pestis ΔmsbB (pLGmsbB) | Y. pestis ΔmsbB ΔlpxP | Y. pestis ΔmsbB ΔlpxP (pLGlpxP) | |
| C10 | 4.8 | 9.0 | 9.0 | 15.7 | 9.6 |
| C12 | 17.3 | 0.0 | 16.6 | 0.0 | 0.0 |
| C14 | 0.7 | 1.5 | 0.6 | 0.5 | 0.7 |
| 2-OH-C14 | 1.6 | 1.7 | 1.5 | 2.4 | 1.7 |
| 3-OH-C14 | 51.0 | 53.5 | 47.1 | 79.2 | 55.3 |
| C16 | 1.5 | 5.4 | 1.3 | 2.1 | 2.0 |
| C16:1 | 23.1 | 28.9 | 23.9 | 0.0 | 30.7 |
| Total | 100 | 100 | 100 | 99.9 | 100 |
The MS spectra of the two mutant strains were different from that of the parental KIM6+ strain and from each other. Notably, lipid A from Y. pestis ΔmsbB grown at 21°C contained no detectable C12 (0% compared to 17.3% for the parental strain; Fig. 2B, Table 3). Instead, a new predominant peak (m/z 1641) corresponding to a penta-acylated species that contained four 3-OH-C14 and one C16:1 was observed. Minor peaks corresponding to lipid A forms containing C10 (m/z 1558, 1794) and aminoarabinose (m/z 1772, 1925) modification were not affected by the msbB mutation (Fig. 2B). When the Y. pestis ΔmsbB mutant was complemented with a wild-type msbB gene controlled by its native promoter, the normal level of C12 addition to lipid A was restored and the wild-type hexa-acylated lipid A (m/z 1824) was again the predominant form (Fig. 2C, Table 3).
Lipid A produced by the Y. pestis ΔmsbBΔlpxP strain grown at 21°C lacked both C12 and C16:1 and was primarily the tetra-acylated species containing four 3-OH-C14 (Fig. 2D). This is also the predominant form produced by the wild-type Y. pestis KIM6+ parent strain at 37°C (28). GC analysis confirmed that lipid A produced by the Y. pestis ΔmsbB ΔlpxP strain contained <0.1% of either C12 or C16:1 (Table 3). MS peaks consistent with C10 (m/z 1558), aminoarabinose (m/z 1536), or both modifications (m/z 1689) were also detected (Fig. 2D). As predicted, Y. pestis ΔmsbB ΔlpxP complemented with a wild-type copy of lpxP synthesized primarily penta-acylated lipid A (m/z 1641) that contained 30.7% C16:1, comparable to the wild-type level (Fig. 2E, Table 3). These results showed that the Y. pestis msbB and lpxP homologs add C12 and C16:1, respectively, and account for the production of hexa-acylated lipid A from tetra-acylated lipid A when Y. pestis is shifted from 37 to 21°C (28).
In vitro and in vivo transcriptional regulation of msbB and lpxP.
The previous observation that Y. pestis produces primarily tetra-acylated lipid A at 37°C and hexa-acylated lipid A at 21°C suggested that expression of late acyltransferase genes of Y. pestis is regulated by temperature (28). To test this hypothesis, transcription of msbB and lpxP relative to the housekeeping gene proS in bacteria grown at 37 and 21°C was analyzed by real-time quantitative RT-PCR. As a positive control, relative expression of ymt, a gene known to be regulated by temperature at the transcriptional level (11), was also assayed. Expression of the msbB, lpxP, and ymt genes was more than twofold higher in bacteria grown at 21°C (Fig. 3).
FIG. 3.
Relative expression of msbB (A), lpxP (B), and ymt (C) genes in Y. pestis. Samples were obtained from Y. pestis KIM6+ grown in LB at 37°C (black bars), LB at 21°C (gray bars), and infected fleas maintained at 21°C (white bars). The mean and standard deviation of the relative transcript level compared to the transcript level of the rpoS housekeeping gene in three independent experiments are indicated.
To determine if the Y. pestis late acyltransferases were expressed in the flea, transcript levels of msbB, lpxP, and ymt were determined from total RNA extracted from infected fleas. Transcription of all three genes was threefold higher in infected fleas than in cultures grown at 37°C (P < 0.05, Fig. 3).
Role of Y. pestis msbB and lpxP homologs in resistance to antimicrobial agents.
Loss of C14 and C16:1 modification of E. coli lipid A results in increased sensitivity to rifampin and vancomycin (37). We compared the susceptibility of the Y. pestis KIM6+, ΔmsbB, and ΔmsbB ΔlpxP strains to these antibiotics and to the CAMPs polymyxin B and cecropin A in bactericidal assays. Lack of C12 and/or C16:1 modification of lipid A modestly increased the susceptibility of Y. pestis to cecropin A and deoxycholate (Fig. 4), but not to polymyxin B, vancomycin, or rifampin (data not shown).
FIG. 4.
Effect of lipid A acylation on Y. pestis viability after 2 h of exposure to increasing concentrations of (A) cecropin A or (B) deoxycholate.
In vivo phenotype of a Y. pestis ΔmsbB ΔlpxP mutant.
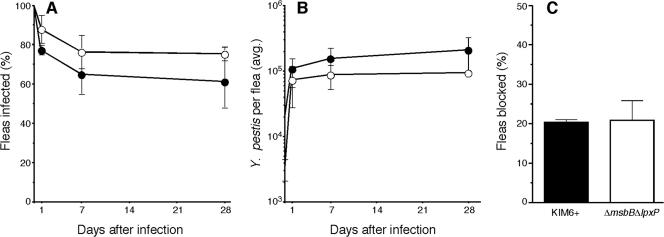
Fleas were infected with Y. pestis KIM6+ or with the ΔmsbB ΔlpxP strain to determine if the ability to make hexa-acylated lipid A conferred protection in the flea gut environment. The Y. pestis ΔmsbB ΔlpxP mutant was able to infect fleas as well as the Y. pestis KIM6+ parental strain (Fig. 5). The bacterial load in fleas infected with the two strains did not differ significantly during a 28-day period after the infectious blood meal, reaching a maximum of ∼105 CFU/flea for the mutant and ∼2.2 ×105 CFU/flea for the parental strain. Furthermore, the percentage of fleas that developed proventricular blockage after infection with Y. pestis KIM6+ or Y. pestis ΔmsbB ΔlpxP did not differ significantly (Fig. 5C).
FIG. 5.
Mutational loss of the Y. pestis acyltransferases MsbB and LpxP does not affect flea infection. The percentage of fleas infected (A), average Y. pestis CFU per infected flea (B), and percentage of fleas that developed proventricular blockage (C) were equivalent after infection with either Y. pestis KIM6+ (•) or Y. pestis ΔmsbB ΔlpxP (○). The mean and standard deviation of two independent experiments are indicated.
DISCUSSION
The pathogenic yersiniae differ from E. coli, Salmonella, and other Enterobacteriaceae that have been examined in temperature-dependent lipid A acylation. At 37°C, Y. pestis synthesizes tetra-acylated lipid A and smaller amounts of penta-acylated forms modified with C10 or C12 acyl groups, and LPS from cells grown at 37°C has limited ability to induce tumor necrosis factor alpha (TNF-α) secretion from human and murine monocytes. At temperatures below 26°C, however, Y. pestis lipid A is hexa-acylated and more endotoxic, resembling the lipid A of other Enterobacteriaceae produced at temperatures above 30°C (23, 24, 28). The first aim of this study was to identify the genes in Y. pestis responsible for the temperature-dependent generation of hexa-acylated lipid A at 21°C, and to determine if their expression is upregulated by a temperature shift from 37 to 21°C.
Homologs of the lpxP and msbB (lpxM) late acyltransferase genes of E. coli were identified in the Y. pestis chromosome, and loss-of-function deletion mutations were introduced into the two genes. Comparative MALDI-TOF and GC structural analyses showed clearly that the Y. pestis msbB and lpxP homologs encode acyltransferases responsible for the addition of C12 and C16:1 acyl groups, respectively, to lipid A at 21°C (Fig. 2; Table 3). Notably, the lipid A spectrum of the Y. pestis ΔmsbB ΔlpxP mutant grown at 21°C was identical to that of wild-type Y. pestis grown at 37°C (Fig. 2D) (28). Thus, the msbB and lpxP homologs are responsible for the temperature-dependent phenotypic variation from tetra- to hexa-acylated lipid A in Y. pestis.
RT-PCR analyses indicated that expression of these two genes was upregulated more than twofold in Y. pestis at 21°C both in vitro and in infected fleas (Fig. 3), suggesting that they are regulated by temperature. Transcript levels were low in all conditions, however. Thus, although decreased transcription is consistent with the lack of acylation at 37°C, temperature-sensitive catalytic activity or other posttranscriptional effects could also be responsible.
Y. pestis MsbB and LpxP likely transfer acyl groups to the 3′-3-OH-C14 and 2′-3-OH-C14 positions of the lipid A disaccharide, respectively, as in E. coli (2, 24). Some differences were apparent between the Y. pestis late acyltransferases and their E. coli counterparts, however. Y. pestis MsbB adds C12 only at growth temperatures below 26°C (23, 28), whereas E. coli MsbB adds a C14 group at all growth temperatures (8). Thus, although the sequence of Y. pestis MsbB is most similar to that of MsbB of E. coli and S. enterica, it more closely resembles that of E. coli HtrB in having C12 substrate specificity.
Bacterial acyltransferases can discriminate among acyl group substrates of different chain lengths via molecular measuring devices termed hydrocarbon rulers (1, 39), but the preferred chain length of homologous acyltransferases from different bacteria can differ as a result of even a single amino acid change (39). In addition, although Y. pestis LpxP adds a C16:1 acyl group to lipid A, as does E. coli LpxP, this modification occurs only under cold shock conditions in E. coli (4). At growth temperatures above 12°C, a different E. coli late acyltransferase, HtrB (LpxL), adds a saturated C12 group (7, 8).
An htrB (lpxL) homolog could not be identified in Y. pestis. The lack of an htrB homolog and the temperature regulation of msbB may explain why Y. pestis produces primarily tetra-acylated lipid IVA at 37°C; in E. coli, addition of C14 by MsbB follows C12 addition by HtrB, and in the absence of HtrB activity, MsbB catalysis is much less efficient (8). Consistent with this interpretation, complementation of the Y. pestis ΔmsbB ΔlpxP strain with msbB did not result in C12-modified, penta-acylated lipid A (R. Rebeil, unpublished data).
A second aim of this study was to test the hypothesis that the hexa-acylated lipid A generated by Y. pestis at 21°C is required for outer membrane integrity and survival in the environment where Y. pestis would normally experience low temperature, the digestive tract of the flea vector. Of the agents used to assess increased outer membrane permeability, the detergent deoxycholate but not the hydrophobic antibiotics rifampin and vancomycin had increased bactericidal activity when fewer fatty acids were present in Y. pestis lipid A. In contrast, E. coli msbB and lpxP mutants are much more sensitive to rifampin and vancomycin (36, 37). Likewise the absence of C16:1 and C12 modification of Y. pestis lipid A did not affect susceptibility to polymyxin B; slightly increased susceptibility to cecropin A was detected in the bactericidal assay (Fig. 4), but not in a MIC assay (data not shown). This was surprising, because wild-type Y. pestis is much more sensitive to both of these CAMPs in MIC assays at 37°C, when predominantly tetra-acylated lipid A lacking C16:1 and C12 is produced, than at 21°C, when C16:1- and C12-containing hexa-acylated lipid A predominates (28).
Other temperature-dependent LPS modifications besides C12 and C16:1 addition may account for the increased resistance of wild-type Y. pestis to CAMPs at 21°C. For example, aminoarabinose and C10 modification of Y. pestis LPS did not appear to be affected by msbB or lpxP mutation (Fig. 2). Positive surface charge associated with aminoarabinose addition to lipid A is critical for resistance to polymyxin B (13, 14), and aminoarabinose modification increases with lower growth temperature in Y. pestis (24). Alternatively, temperature-dependent differences in LPS core structure (24), or an as yet unknown temperature-mediated factor might be responsible for increased resistance at 21°C.
Ultimately, we wanted to test the hypothesis that the change from primarily tetra-acylated lipid A at 37°C to hexa-acylated lipid A at <26°C is important to the arthropod-borne life cycle of Y. pestis. To produce a transmissible infection in the flea vector, Y. pestis grows to large numbers in the flea midgut and eventually forms a biofilm that obstructs the proventriculus, a valve between the midgut and esophagus (20). Little is known about the flea gut environment. However, digestion and solubilization of the blood meal occurs in the midgut, which presumably contains proteases, lipases, and surfactants (34), and it seemed likely that full acylation of lipid A might enhance bacterial survival in that environment. Although it was not feasible to isolate sufficient quantities of LPS for structural analysis from infected fleas, the increased expression of the Y. pestis late acyltransferase genes indicates that hexa-acylated lipid A is produced in fleas (Fig. 3). Nevertheless, we observed no difference in the ability of Y. pestis KIM6+ and Y. pestis ΔmsbB ΔlpxP to infect or block the proventriculus (Fig. 4), indicating that hexa-acylated lipid A is not necessary for survival in the X. cheopis digestive tract or to produce a transmissible infection. In contrast, the late acyltransferases are essential for normal in vitro and in vivo growth of E. coli and Salmonella spp. (21, 22, 30, 37).
The change from tetra-acylated lipid A at 37° to hexa-acylated lipid A at 21°C is characteristic of the enteropathogenic yersiniae as well as Y. pestis (18, 28). Thus, the hexa-acylated lipid A produced by Y. pestis at temperatures below 26°C may simply be an evolutionary inheritance from Y. pseudotuberculosis with a neutral effect in the flea. The fully acylated lipid A form made at ambient temperatures may be important for survival in the environment outside of a eukaryotic host, and downregulation of the late acyltransferases at 37°C to produce the less immunostimulatory tetra-acylated lipid A may be important for pathogenesis. This hypothesis remains to be tested.
The hexa-acylated lipid A by Y. pestis might also play a protective role immediately after flea-borne transmission. Upon entering a mammalian host, Y. pestis must survive the innate immune response and resist killing by dermal phagocytes until the expression of known antiphagocytic factors such as the F1 capsule and the Yop proteins is induced (5, 19, 33). In other gram-negative bacteria, increased acylation of lipid A confers protection against mammalian defensins (12, 15). Thus, the fully acylated lipid A produced by Y. pestis in the flea might help complete the transmission cycle by increasing resistance to phagocytes or defensins encountered in the skin upon transmission.
Acknowledgments
This work was supported by the Division of Intramural Research, NIAID, NIH; the Ellison Medical Foundation (New Scholars Award in Global Infectious Diseases to B.J.H.); and by the WWAMI Regional Center of Excellence for Biodefense and Emerging Infectious Disease, PHS grant U54 AI057141 (R.K.E. and S.I.M.).
REFERENCES
- 1.Ahn, V. E., E. I. Lo, C. K. Engel, L. Chen, P. M. Hwang, L. E. Kay, R. E. Bishop, and G. G. Privé. 2004. A hydrocarbon ruler measures palmitate in the enzymatic acylation of endotoxin. EMBO J. 23:2931-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aussel, L., H. Therisod, D. Karibian, M. B. Perry, M. Bruneteau, and M. Caroff. 2000. Novel variation of lipid A structures in strains of different Yersinia species. FEBS Lett. 465:87-92. [DOI] [PubMed] [Google Scholar]
- 3.Caroff, M., A. Tacken, and L. Szabo. 1988. Detergent-accelerated hydrolysis of bacterial endotoxins and determination of the anomeric configuration of the glycosyl phosphate present in the “isolated lipid A” fragment of the Bordetella pertussis endotoxin. Carbohydr. Res. 175:273-282. [DOI] [PubMed] [Google Scholar]
- 4.Carty, S. M., K. R. Sreekumar, and C. R. H. Raetz. 1999. Effect of cold shock on lipid A biosynthesis in Escherichia coli: Induction at 12°C of an acyltransferase specific for palmitoleoyl-acyl carrier protein. J. Biol. Chem. 274:9677-9685. [DOI] [PubMed] [Google Scholar]
- 5.Cavanaugh, D. C., and R. Randall. 1959. The role of multiplication of Pasteurella pestis in mononuclear phagocytes in the pathogenesis of flea-borne plague. J. Immunol. 83:348-363. [PubMed] [Google Scholar]
- 6.Chaussee, M. S., R. O. Watson, J. C. Smoot, and J. M. Musser. 2001. Identification of Rgg-regulated exoproteins of Streptococcus pyogenes. Infect. Immun. 69:822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clementz, T., J. J. Bednarski, and C. R. H. Raetz. 1996. Function of the htrB high temperature requirement gene of Escherichia coli in the acylation of lipid A: HtrB catalyzed incorporation of laurate. J. Biol. Chem. 271:12095-12102. [DOI] [PubMed] [Google Scholar]
- 8.Clementz, T., Z. Zhou, and C. R. H. Raetz. 1997. Function of the Escherichia coli msbB gene, a multicopy suppressor of htrB knockouts, in the acylation of lipid A: acylation by MsbB follows laurate incorporation by HtrB. J. Biol. Chem. 272:10353-10360. [DOI] [PubMed] [Google Scholar]
- 9.Darveau, R. P., and Hancock. 1983. Procedure for isolation of bacterial lipopolysaccharides from both smooth and rough Pseudomonas aeruginosa and Salmonella typhimurium strains. J. Bacteriol. 155:831-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Du, Y., E. Galyov, and Å. Forsberg. 1995. Genetic analysis of virulence determinants unique to Yersinia pestis. Contrib. Microbiol. Immunol. 13:321-324. [PubMed] [Google Scholar]
- 12.Ernst, R. K., E. C. Yi, L. Guo, K. B. Lim, J. L. Burns, M. Hackett, and S. I. Miller. 1999. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 286:1561-1565. [DOI] [PubMed] [Google Scholar]
- 13.Groisman, E. A., J. Kayser, and F. C. Soncini. 1997. Regulation of polymyxin resistance and adaptation to low-Mg2+ environments. J. Bacteriol. 179:7040-7045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunn, J. S., K. B. Lim, J. Krueger, K. Kim, L. Guo, M. Hackett, and S. I. Miller. 1998. PmrA-PmrB-regulated genes necessary for 4-aminoarabinose lipid A modification and polymyxin resistance. Mol. Microbiol. 27:1171-1182. [DOI] [PubMed] [Google Scholar]
- 15.Guo, L., K. B. Lim, C. M. Poduje, M. Daniel, J. S. Gunn, M. Hackett, and S. I. Miller. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95:189-198. [DOI] [PubMed] [Google Scholar]
- 16.Hinnebusch, B. J., R. D. Perry, and T. G. Schwan. 1996. Role of the Yersinia pestis hemin storage (hms) locus in the transmission of plague by fleas. Science 273:367-370. [DOI] [PubMed] [Google Scholar]
- 17.Hinnebusch, B. J., A. E. Rudolph, P. Cherepanov, J. E. Dixon, T. G. Schwan, and Å. Forsberg. 2002. Role of the Yersinia murine toxin in the survival of Yersinia pestis in the midgut of the flea vector. Science 296:733-735. [DOI] [PubMed] [Google Scholar]
- 18.Holst, O. 2003. Lipopolysaccharides of Yersinia. Adv. Exp. Med. Biol. 529:219-228. [DOI] [PubMed] [Google Scholar]
- 19.Hudson, B. W., L. Kartman, and F. M. Prince. 1966. Pasteurella pestis detection in fleas by fluorescent antibody staining. Bull. W.H.O. 34:709-714. [PMC free article] [PubMed] [Google Scholar]
- 20.Jarrett, C. O., E. Deak, K. E. Isherwood, P. C. Oyston, E. R. Fischer, A. R. Whitney, S. D. Kobayashi, F. R. DeLeo, and B. J. Hinnebusch. 2004. Transmission of Yersinia pestis from an infectious biofilm in the flea vector. J. Infect. Dis. 190:783-792. [DOI] [PubMed] [Google Scholar]
- 21.Jones, B. D., W. A. Nichols, B. W. Gibson, M. G. Sunshine, and M. A. Apicella. 1997. Study of the role of the htrB gene in Salmonella typhimurium virulence. Infect. Immun. 65:4778-4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karow, M., O. Fayet, A. Cegielska, T. Ziegelhoffer, and C. Georgopoulos. 1991. Isolation and characterization of the Escherichia coli htrB gene, whose product is essential for bacterial viability above 33°C in rich media. J. Bacteriol. 173:741-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawahara, K., H. Tsukano, H. Watanabe, B. Lindner, and M. Matsuura. 2002. Modification of the structure and activity of lipid A in Yersinia pestis lipopolysaccharide by growth temperature. Infect. Immun. 70:4092-4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knirel, Y. A., B. Lindner, E. V. Vinogradov, N. A. Kocharova, S. N. Senchenkova, R. Z. Shaikhutdinova, S. V. Dentovskaya, N. K. Fursova, I. V. Bakhteeva, G. M. Titareva, S. V. Balakhonov, O. Holst, T. A. Gremyakova, G. B. Pier, and A. P. Anisimov. 2005. Temperature-dependent variations and intraspecies diversity of the structure of the lipopolysaccharide of Yersinia pestis. Biochemistry 44:1731-1743. [DOI] [PubMed] [Google Scholar]
- 24a.Motin, V. L., A. M. Georgescu, J. P. Fitch, P. P. Gu, D. O. Nelson, S. L. Mabery, J. B. Garnham, B. A. Sokhansanj, L. L. Oh, M. A. Coleman, J. M. Elliott, L. M. Kegelmeyer, A. J. Wyrobek, T. R. Slezak, R. R. Brubaker, and E. Garcia. 2004. Temporal global changes in gene expression during temperature transition in Yersinia pestis. J. Bacteriol. 186:6298-6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nikaido, H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67:593-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 27.Perry, R. D., and J. D. Fetherston. 1997. Yersinia pestis—etiologic agent of plague. Clin. Microbiol. Rev. 10:35-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rebeil, R., R. K. Ernst, B. B. Gowen, S. I. Miller, and B. J. Hinnebusch. 2004. Variation in lipid A structure in the pathogenic yersiniae. Mol. Microbiol. 52:1363-1373. [DOI] [PubMed] [Google Scholar]
- 29.Simon, R., U. Priefer, and A. Pülher. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 30.Somerville, J. E., L. Cassiano, and R. P. Darveau. 1999. Escherichia coli msbB gene as a virulence factor and a therapeutic target. Infect. Immun. 67:6583-6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Somerville, J. E. J., L. Cassiano, B. Bainbridge, M. D. Cunningham, and R. P. Darveau. 1996. A novel Escherichia coli lipid A mutant that produces an antiinflammatory lipopolysaccharide. J. Clin. Investig. 97:359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stoker, N. G., N. F. Fairweather, and B. G. Spratt. 1982. Versatile low-copy-number plasmid vectors for cloning Escherichia coli. Gene 18:335-341. [DOI] [PubMed] [Google Scholar]
- 33.Straley, S. C., and R. D. Perry. 1995. Environmental modulation of gene expression and pathogenesis in Yersinia. Trends Microbiol. 3:310-317. [DOI] [PubMed] [Google Scholar]
- 34.Terra, W. R., B. P. Ferreira, and R. J. Dillon. 1996. Digestive enzymes, p. 153-194. In M. J. Lehane and P. F. Billingsley (ed.), Biology of the insect midgut. Chapman & Hall, London, England.
- 35.Une, T., and R. R. Brubaker. 1984. In vivo comparison of avirulent Vwa− and Pgm− or Pstr phenotypes of yersiniae. Infect. Immun. 43:895-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaara, M., and M. Nurminen. 1999. Outer membrane permeability barrier in Escherichia coli mutants that are defective in the late acyltransferases of lipid A biosynthesis. Antimicrob. Agents Chemother. 43:1459-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vorachek-Warren, M. K., S. Ramirez, R. J. Cotter, and C. R. H. Raetz. 2002. A triple mutant of Escherichia coli lacking secondary acyl chains on lipid A. J. Biol. Chem. 277:14194-14205. [DOI] [PubMed] [Google Scholar]
- 38.Wang, J., and M. F. Wilkinson. 2001. Deletion mutagenesis of large (12-kb) plasmids by a one step PCR protocol. BioTechniques 31:722-724. [DOI] [PubMed] [Google Scholar]
- 39.Wyckoff, T. J. O., S. Lin, R. J. Cotter, G. D. Dotson, and C. R. H. Raetz. 1998. Hydrocarbon rulers in UDP-N-acetylglucosamine acyltransferases. J. Biol. Chem. 273:32369-32372. [DOI] [PubMed] [Google Scholar]