Abstract
Archaeal preflagellin peptidases and bacterial type IV prepilin peptidases belong to a family of aspartic acid proteases that cleave the leader peptides of precursor proteins with type IV prepilin signal sequences. The substrate repertoire of PibD from the crenarchaeon Sulfolobus solfataricus is unusually diverse. In addition to flagellin, PibD cleaves three sugar-binding proteins unique to this species and a number of proteins with unknown function. Here we demonstrate that PibD contains two aspartic acid residues that are essential for cleavage activity. An additional pair of aspartic acids in a large cytoplasmic loop is also important for function and is possibly involved in leader peptide recognition. Combining the results of transmembrane segment predictions and cysteine-labeling experiments, we suggest a membrane topology model for PibD with the active-site aspartic acid residues exposed to the cytosol.
Type IV prepilin peptidases (TFPPs) are prokaryotic signal peptidases that are required for the correct secretion of affiliated substrates, which are termed type IV prepilins and type IV prepilin-like proteins (42). Several TFPPs from gram-negative and gram-positive bacteria have been previously characterized, and recently, archaeal homologues have also been described (6, 8). All TFPPs have in common that they recognize an unusual membrane-targeting sequence termed the type IV pilin signal peptide. It is located at the amino terminus of a precursor protein and consists of a short positively charged leader followed by a hydrophobic stretch of about 20 amino acids, with a conserved cleavage site located between these two domains. The extreme N terminus of membrane-inserted prepilins has been shown to face the cytoplasm (11, 41). Accordingly, the active site of the processing enzyme is expected to be on the cytoplasmic side of the membrane. After cleavage by the TFPP, the hydrophobic domain remains part of the mature protein. Additionally, most bacterial TFPPs transfer a methyl group to the new amino terminus, whereas the archaeal enzymes seem to possess only cleavage activity (4; unpublished results). Indeed, the peptidase and methyl transferase activities of PilD from Pseudomonas aeruginosa are separable, which suggests two independent active sites (33, 40, 42). Processed (i.e., cleaved and methylated) type IV pilins are assembled into the type IV pilus, a long, retractable surface organelle involved in diverse processes, such as surface attachment, twitching motility, and DNA uptake (16, 25, 30). Additionally, type IV pilin-like proteins play an important role in the general (type II) secretion pathway of gram-negative bacteria by forming subunits of the type II pseudopilus (12, 13, 49). In archaea, type IV pilin-like signal peptides were first identified in flagellins, the structural subunits of flagellar filament (15, 46). The hyperthermophilic acidophile Sulfolobus solfataricus, however, was shown to secrete a more diverse set of proteins with type IV pilin-like signal sequences (3, 6), including flagellin. In this organism, a cleaved type IV prepilin-like signal peptide was identified in three membrane-associated sugar-binding proteins which mediate solute uptake by ABC transporters (4, 14). Further analysis of the genome revealed that 12 additional proteins could be equipped with this signal peptide, including 4 putative solute-binding proteins and small proteins of unknown function (3, 6).
Recently, we identified PibD, the S. solfataricus enzyme responsible for cleavage of flagellin (FlaB) and glucose-binding protein (GlcS) precursors (pre-FlaB and pre-GlcS, respectively) (6). PibD was discovered on the basis of a phylogenetic relationship with bacterial TFPPs described in the COG database (44). This relationship could not be detected by BLAST searches. Indeed, a multiple linear alignment of whole sequences shows no significant homology between bacterial TFPPs and their archaeal counterparts. Therefore, it is important to determine whether the enzymes from these two prokaryotic domains have similar catalytic mechanisms, which would support the suggested evolutionary relationship. The proteolytic mechanism of TFPPs from Pseudomonas aeruginosa and Vibrio cholerae has been studied by use of site-directed mutagenesis of conserved residues in the corresponding enzymes. For PilD from P. aeruginosa, cysteine residues were shown to be important for the processing of prepilin (40). However, the related enzyme from Xanthomonas campestris, which does not contain cysteine residues, could complement a P. aeruginosa pilD mutant (19), suggesting that amino acids other than cysteine could be required for the cleavage reaction. Indeed, when either of two universally conserved aspartic acid residues was altered in the TFPP homologue TcpJ from V. cholerae, the enzyme showed no activity in vivo or in vitro (28). The same report shows that the domain containing the conserved cysteines is not required for cleavage activity. It is thus evident that TFPPs are aspartic acid proteases and, because the catalytic residues are conserved among bacteria, they should also be present in archaeal enzymes, provided that the evolutionary connection is true. Recently, the preflagellin peptidase FlaK from Methanococcus voltae was shown to contain two aspartic acid residues required for in vitro activity, and these residues could correspond to those identified in TcpJ (7). As these residues are found in the amino-terminal half of the protein, no function could be assigned to the carboxyl-terminal portion of the protein, which includes a large cytoplasmic loop.
Here we present further evidence for the relatedness of archaeal TFPPs, exemplified by PibD from S. solfataricus, and bacterial enzymes. By mutational analysis, two conserved aspartic acid residues are identified as essential for activity. Additionally, a pair of aspartic acid residues in the large cytoplasmic loop of the enzyme is shown to be important for enzyme function. Finally, we suggest a membrane topology model for PibD based on cysteine accessibility studies.
MATERIALS AND METHODS
Strains, growth conditions, and plasmids.
Escherichia coli strain DH5α was used for all cloning steps (17), and strains BL21(DE3) (43) and C43(DE3) (32) were used for the overproduction of protein. The two latter strains additionally carry the pACYC-RIL plasmid, which encodes additional tRNAs to compensate for rare E. coli codons (Stratagene, La Jolla, Calif.). Plasmid pZA1 encodes the flaB precursor protein, and pZA5 contains pibD; both are under the control of the T7 promoter and contain a carboxyl-terminal six-histidine tag. pBAD/HisA is a low-copy-number arabinose-inducible expression vector (Invitrogen, Breda, The Netherlands). A FlaB precursor with a carboxyl-terminal hemagglutinin (HA) tag was constructed by PCR with primers 5′-CCCCCCATGGTAAAGGAATACAAC-3′ and 5′-AAGCTTTTACGCGTAGTCCGGAACGTCATACGGGTAGGATCCCCCTATTACTGATAC-3′ (NcoI, BamHI, and HindIII restriction sites are underlined, and the coding region for the HA tag is in bold) and with pZA1 as the template. The 944-bp PCR product was digested with NcoI and HindIII and ligated into the compatible sites of pBAD/HisA, yielding pZA8. Point mutations in the pibD gene on pZA5 were introduced using QuikChange (Stratagene, La Jolla, CA), and primers were designed as recommended by the manufacturer. To produce single-cysteine mutants of PibD, the native cysteine at position 41 was first altered to a serine. Then, single cysteines were introduced at positions Glu28, Asn50, Thr63, Ser74, and Ile236. All mutants were verified by sequencing of the relevant DNA fragments.
Growth conditions for overproduction of six-histidine-tagged PibD and FlaB proteins were as described previously (6). To produce FlaB carrying a carboxyl-terminal HA tag, pZA8 was transformed into E. coli strain C43(DE3)-pACYC-RIL, and cells were grown for 16 h in LB medium supplemented with 50 μg/ml ampicillin at 30°C. This was used to inoculate a culture into dYT medium (1% yeast extract, 1.6% Bacto tryptone, 0.5% NaCl) supplemented with ampicillin and 30 μg/ml chloramphenicol. Cells were grown at 37°C to an optical density at 600 nm of 0.6. Protein production was induced with 0.2% l-arabinose, and cells were grown for a further 4 h.
Preparation of membrane vesicles and in vitro cleavage assay.
Preparation of E. coli inner membrane vesicles was done as described previously (6). Everted membrane vesicles were prepared by use of a French press. Cells from a 1-liter culture were harvested (8 min, 10,500 × g, 4°C) and resuspended in 10 ml buffer A (50 mM Tris-HCl [pH 7.5], 10 mM MgSO4, 20% [wt/vol] sucrose). After the addition of 0.1 mM phenylmethylsulfonyl fluoride and 0.1-mg/ml DNase I, the cells were passed once through a continuous cell disruptor (Constant Systems, Daventry, United Kingdom) at 20,000 lb/in2. Subsequently, the cells were flushed with 35 ml buffer A, which was then pooled with the disrupted cell material. Unbroken cells and cell debris were removed by low-speed centrifugation (10 min, 10,000 × g, 4°C), and the supernatant was spun in a Ti45 rotor for 90 min at 125,000 × g and 4°C. The crude membrane pellet was resuspended at a protein concentration of about 20 mg/ml in 50 mM Tris-HCl, pH 7.5, containing 20% (wt/vol) glycerol. The protein concentrations of the membrane preparations were determined using a DC protein assay kit (Bio-Rad, Hercules, CA) with bovine serum albumin as a standard. Activity of wild-type and mutant variants of PibD was tested using an in vitro cleavage assay as described previously (6). Because expression levels of PibD were too low to be directly visualized by Western immunoblotting, an enrichment step was introduced. Membranes in amounts corresponding to two times the amount used in each cleavage reaction were solubilized by the addition of 1% (vol/vol) Triton X-100 in a total volume of 0.5 ml. The mixture was incubated for 1 h at 37°C and centrifuged in a TLA100.2 rotor for 20 min at 220,000 × g and 4°C. Under these conditions, PibD was found exclusively in the pellet fraction (not shown). The pellet was resuspended in 40 μl sodium dodecyl sulfate (SDS) sample buffer, and proteins were separated on a 12% SDS-polyacrylamide gel electrophoresis (PAGE) gel and blotted onto a polyvinylidene difluoride membrane (Roche, Mannheim, Germany). The C-terminal six-histidine tag on PibD was detected using an anti-His tag antibody (Dianova, Hamburg, Germany).
Sulfhydryl labeling with maleimides and protein purification.
4-Acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS; Molecular Probes, Eugene, OR) was freshly prepared as a 100 mM stock solution and added to a final concentration of 0.2 mM to 3 mg of everted membranes containing the cysteineless or single-cysteine mutants of PibD in a total volume of 1 ml buffer (50 mM Tris-HCl, pH 7.5). Membranes were incubated for 5 min at 30°C, and the reaction was stopped by the addition of 2 mM dithiothreitol (DTT) diluted to 3 ml with 50 mM Tris-HCl, pH 7.5, and followed by ultracentrifugation (TLA100.4 rotor, 20 min, 288,000 × g, 4°C). The pellet was resuspended at a protein concentration of 1.5 mg/ml in 50 mM sodium phosphate, pH 8.0, 150 mM NaCl, 10% (wt/vol) glycerol, and the nonionic detergent dodecylmaltoside (DDM) (Anatrace, Maumee, OH) at 1% (wt/vol). Membranes were solubilized for 15 min at ambient temperature, whereupon the solubilized material was cleared by ultracentrifugation (MLA 80 rotor; 15 min, 20,500 × g, 4°C). The supernatant was mixed with 10 mM imidazole, pH 8.0, and 40 μl HIS-select nickel affinity agarose resin (Sigma) equilibrated with wash buffer (50 mM sodium phosphate, pH 8.0, 150 mM NaCl, 10% glycerol, 20 mM imidazole, 0.1% DDM). The mixture was incubated 16 h at 4°C with gentle shaking. Next, the resin was collected by centrifugation and washed twice with 1 ml wash buffer. Washed resin was incubated 5 min with 35 μl elution buffer (50 mM sodium phosphate, pH 8.0, 150 mM NaCl, 10% glycerol, 100 mM Na2-EDTA, 0.1% DDM) under gentle shaking and collected by centrifugation. Eluted protein was labeled with fluorescein maleimide (Molecular Probes, Leiden, The Netherlands) as follows. A fresh stock solution was prepared by diluting 50 mM fluorescein maleimide dissolved in dimethyl formamide 10-fold with deionized water. Fluorescein maleimide was added to elution fractions at a final concentration of 0.1 mM. Samples were incubated 5 min at 30°C in the dark, and the reaction was quenched by the addition of 1 mM DTT. For analysis, purified and labeled samples were mixed with SDS sample buffer and separated on a 12% SDS-PAGE gel according to the method of Laemmli (27). Fluorescent bands were detected using an F1 Lumi-Imager (Roche Diagnostics, Almere, The Netherlands) with a 520-nm fluorescence filter. Subsequently, the same gel was fixed and stained with Coomassie brilliant blue R-250 (CBB), destained, and recorded with an F1 Lumi-Imager in the transillumination mode. Densitometric analysis was carried out using the public-domain software ImageJ (developed by W.S. Rasband, U.S. National Institutes of Health, Bethesda, Maryland). Pixel densities of each fluorescent band were measured and corrected for background signal by use of an area of identical size in the same lane that did not contain protein. Bands in the CBB-stained gel were quantified in the same way. Then, the labeling efficiency ratio (expressed as a ratio of fluorescent band signal and CBB band signal) of AMS-treated versus mock-treated samples was calculated.
Predictions of TMS.
The PibD primary sequence was submitted to the World Wide Web versions of the PsiPred/MEMSAT2 (21, 22, 31), HMMTOP (47, 48), TMHMM2.0 (38) (26), and Phobius (23) programs; default settings were used. In the case of Phobius, predictions were also run in the constrained mode with the amino-terminal noncytoplasmic loop setting. However, this did not change the number of transmembrane segments (TMS) predicted by the program.
RESULTS
Chemical inactivation of PibD.
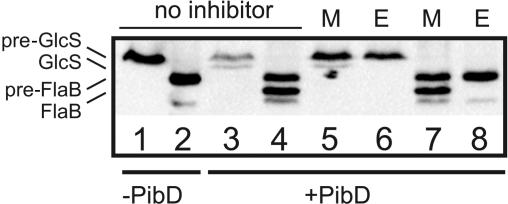
To establish whether aspartic acids are involved in the reaction catalyzed by the type IV pilin-like signal peptidase PibD, the inhibitor 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide hydrochloride (EDAC) in the presence of glycinamide was tested. EDAC is known to modify acidic amino acid residues (i.e., aspartic acid and glutamic acid) and has been used previously to inhibit aspartic acid proteases (18, 51), including TcpJ. E. coli inner membranes containing expressed PibD were treated with EDAC-glycinamide and subsequently tested for in vitro cleavage of precursors of flagellin (FlaB) and glucose-binding protein (GlcS) (pre-FlaB and pre-GlcS, respectively) (Fig. 1). Treatment with the inhibitor completely abolished the cleavage of both precursors, whereas mock-treated membranes retained activity. FlaB was cleaved more efficiently than GlcS, consistent with our previous observations (6). Treatment of the membranes with phenylmethylsulfonyl fluoride, an inhibitor of serine proteinases, or with EDTA, an inhibitor of metal proteases, had no effect on the PibD activity (data not shown). These data therefore show that acidic residues, presumably aspartate residues, are essential for its activity. Because cleavage of both tested substrates was affected, in the studies described below, only the cleavage of the flagellin precursor is shown.
FIG. 1.
Chemical inhibition of PibD activity. E. coli membranes with expressed PibD were incubated with EDAC and glycinamide (E) or mock treated (M) and subsequently tested for cleavage of the glucose-binding protein (lanes 1, 3, 5, and 6) or flagellin (lanes 2, 4, 7, and 8) precursors as indicated. In lanes 1 to 4, controls for uncleaved and cleaved substrates are shown.
Identification of catalytic residues in PibD by site-directed mutagenesis.
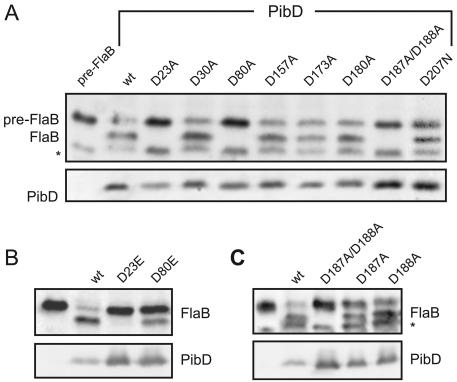
To determine which residues are essential for catalysis, each of the nine aspartate residues was changed to alanine by site-directed mutagenesis. The adjacent residues D187 and D188 were replaced simultaneously. Mutant proteins were expressed in E. coli, and membranes containing the enzymes were tested for cleavage of pre-FlaB in vitro (Fig. 2) (6). Expression of the D207A mutant could not be detected by Western immunoblot analysis with antibodies directed against the carboxyl-terminal epitope tag of PibD (data not shown). However, a mutant with an asparagine (D207N) at this position was stable upon expression in E. coli. The amounts of PibD protein present in the membrane preparations were estimated by Western immunoblot analysis. Based on this estimation, comparable amounts of enzyme source were used in all experiments. The activity levels of five single mutants (D30A, D157A, D173A, D180A, D207N) were comparable to that of the wild-type enzyme (Fig. 2A). However, no cleavage of preflagellin was observed in D23A, D80A, and D187A/D188A mutants. Therefore, these amino acids represent potential candidates for catalytic residues of the enzyme. The aspartate residues at positions 23 and 80 are part of putative signature motifs that are also found around catalytic residues of other TFPPs and intramembrane aspartic acid proteases (28, 39, 52). Asp23 is embedded between hydrophobic residues, and Asp80 is preceded by an alanine and two glycines. This resembles the characteristic GxGD motif found in TFPPs and in presenilin aspartic proteases (39). An aspartate-to-glutamate mutation in catalytic sites of aspartic peptidases might restore enzyme activity. However, a D23E mutant of PibD was also inactive, whereas a D80E mutant retained partial activity (Fig. 2B). In order to dissect the defect of the D187A/D188A mutant, the respective single mutants were generated. PibD containing the D187A or D188A mutation was active, although there was an apparent reduction in activity in the D187A mutant (Fig. 2C). In conclusion, the data suggest that Asp23 and Asp80 are the catalytic residues in PibD.
FIG. 2.
In vitro activity of PibD wild-type (wt) and mutant proteins. Western immunoblot analysis of pre-FlaB cleavage by PibD wild-type and mutant proteins (upper panels). The amounts of PibD in membranes corresponding to those used in the assays were detected with six-His antibodies (lower panels). The asterisks mark a degradation product of FlaB observed in some membrane preparations (6). (A) Aspartate-to-alanine mutants of PibD were tested for cleavage of pre-FlaB. (B) Activity of aspartate-to-glutamate mutants of catalytic residues. (C) Activity of the single-alanine mutants of the pair of aspartates at positions Asp187 and Asp188.
Membrane topology analysis of PibD.
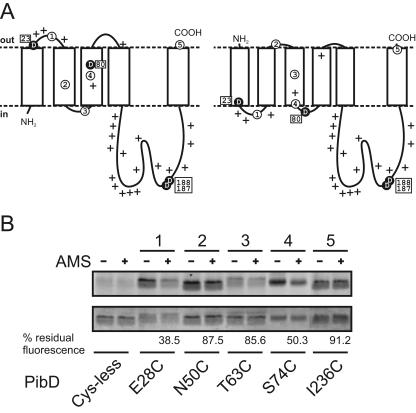
Previously, a topology model of PibD was suggested based on the prediction by TMHMM 2.0 (6, 38). In this model, the amino terminus is cytoplasmic and followed by five transmembrane segments and an extracellular carboxyl terminus (Fig. 3A). However, in this case the two catalytic residues are predicted to be located in an extracellular loop and a transmembrane segment, respectively. Since cleavage by TFPPs is expected to occur at the cytoplasmic face of the membrane (10), the membrane topology of PibD was investigated. We first compared transmembrane topology predictions for PibD obtained from World Wide Web-based versions of commonly used algorithms (see Materials and Methods). All programs predict the carboxyl terminus to be extracellular and also predict a large cytoplasmic loop between the last two transmembrane helices. However, the amino terminus is predicted by different programs to be either intra- or extracellular, leading to a model with either five or six transmembrane segments, respectively. The two representative models are shown in Fig. 3A. To be able to assign a correct model, we investigated the topology of PibD expressed in E. coli experimentally by cysteine accessibility studies. A cysteineless mutant was generated and used to construct single-cysteine mutants in the predicted loops. We focused on regions close to the catalytic aspartates. Both the cysteineless and single-cysteine mutants were active in vitro (not shown). To determine whether a particular cysteine residue is cytoplasmic, everted membrane vesicles were treated with 4-acetamido-4′-maleimidylstilbene-2,2′-disulfonic acid (AMS), a nonfluorescent, membrane-impermeable, thiol-reactive probe. Residues exposed to the cytoplasmic side of the membrane are modified under these conditions. Control membranes received deionized water instead of AMS. Subsequently, the reactions were quenched by addition of DTT, membranes were solubilized with detergent, and the His-tagged PibD proteins were enriched by immobilized metal affinity chromatography. Eluted proteins were labeled with fluorescein maleimide and separated by SDS-PAGE, and the fluorescence intensities of prelabeled samples and mock-treated samples were compared. A strong decrease in fluorescence signal was observed for the E28C mutant by AMS treatment (Fig. 3B), suggesting that this position is cytoplasmic, as predicted by the six-transmembrane model. According to a model with five TMS, this residue should be located on the extracellular side of the membrane and therefore be protected. Similarly, the S74C mutant showed some reduction of fluorescence in AMS-treated membranes. For the N50C and I236C mutants, little decrease of fluorescence intensity was seen, consistent with these residues being inaccessible from the cytoplasmic side of the membrane. In the case of the T63C mutant, the labeling of the cysteine by fluorescein maleimide was very poor, even without pretreatment of the sample with AMS. This indicates that the residue is buried in the protein structure, possibly within a helical transmembrane segment. Moreover, AMS treatment of membranes containing this mutant had a minor effect on subsequent fluorescein maleimide labeling. Quantification of fluorescent band intensities further supports the observation that residues Glu28 and, most likely, Ser74 are exposed to the cytoplasmic side of the membrane (Fig. 3B). In conclusion, our data support a six-transmembrane model for PibD, with extracellular amino and carboxyl termini and the two catalytic residues located in or close to intracellular loops between TMS 1 and 2 and TMS 3 and 4, similar to the second model presented in Fig. 3A. This model also fits better with the distribution of positively charged residues in the protein (Fig. 3A). Taken together, these results indicate that PibD is an aspartyl protease with an unusual topology consisting of extracellular amino and carboxyl termini and six transmembrane segments.
FIG. 3.
Membrane topology models of PibD and accessibility of single-cysteine mutants to AMS and fluorescein maleimide in everted membrane vesicles. (A) Representative membrane topology models predicted with TMHMM2.0 (left) and Phobius (right). Critical aspartic acid residues are highlighted as black circles, and positively charged residues (lysine and arginine) are marked with plus signs. Amino acid positions that were altered to cysteine are Glu28 (circled 1), Asn50 (circled 2), Thr63 (circled 3), Ser74 (circled 4), and Ile236 (circled 5). (B) Fluorescence labeling of PibD single-cysteine mutants. Everted E. coli vesicles without or with preincubation with AMS were solubilized, and protein was purified by immobilized metal affinity chromatography. Eluted protein was labeled with fluorescein maleimide and separated by SDS-PAGE. Fluorescent bands were visualized using an F1 Lumi-Imager (upper panel). Subsequently, the same gel was stained with Coomassie brilliant blue (lower panel). Numbers above lanes correspond to those used in panel A. Residual fluorescence of bands is expressed for each mutant as a ratio of AMS prelabeled samples to mock-treated samples after correction for protein loading and the background signal of the cysteineless mutant.
DISCUSSION
The family of type IV prepilin peptidases is divided into two subfamilies of bacterial and archaeal enzymes (MEROPS database identifiers A24A and A24B, respectively) (34). The archaeal subfamily has been named preflagellin peptidases because the flagellin subunits were presumed to be the only substrate of the archaeal enzymes. However, PibD from S. solfataricus is involved not only in preflagellin processing but also in the processing of a set of binding proteins with prepilin-like signal sequences and of small pilin-like proteins (2, 3). By screening archaeal genomes for putative type IV pilin-like signal sequences, we found that this type of secretory sequence is in fact more widely used by archaea than previously assumed (Z. Szabo, O. de Andrade Oliveira, J. C. Kissinger, S.-V. Albers, A. J. M. Driessen, and M. Pohlschroder, unpublished results). In the present work, we demonstrate that PibD is an aspartic acid protease, as suggested by the inactivation of the enzyme by EDAC and glycinamide, a procedure previously reported to inhibit bacterial TFPPs (28). Site-directed mutagenesis of the nine aspartic acid residues showed that Asp23 and Asp80 are essential for cleavage. The aspartate-to-alanine mutants were inactive, whereas a glutamate substitution at position 80 resulted in an enzyme with partial activity. The D23E mutant, however, did not cleave preflagellin. In aspartate-to-glutamate mutations, amino acid side chain length increases but the negative charge is conserved. It can be concluded that structural constraints are less tolerated at position Asp23 than at position Asp80. Interestingly, similar mutations in TcpJ led to opposite results: a glutamate mutation at the position corresponding to Asp23 in PibD was tolerated better than a mutation corresponding to D80E (28). For TcpJ, it was concluded that the first catalytic amino acid might coordinate a water molecule necessary for hydrolysis and therefore be structurally more flexible. The second would then directly participate in chemical reduction of the peptide bond. However, taking our results into consideration, no common theme concerning the local structural environments of the two catalytic residues can be deduced, and therefore it is difficult to access their specific function during catalysis.
Site-directed mutagenesis of aspartate residues in PibD also identified a pair of amino acids, Asp187/Asp188, that appeared essential for enzyme function. A double-alanine mutant was inactive, whereas single mutations did not critically affect activity. These residues localize within the large cytoplasmic domain that is typically found in the archaeal PibD homologs. Together with the distal transmembrane segment, this domain defines a unique protein family, “Archaeal Peptidase A24 C terminus Type II” (Pfam accession number, PF06847) (9), which was based on an alignment of 10 archaeal sequences, including those of PibD and M. voltae FlaK. Although no absolutely conserved residues can be found in the cytoplasmic domain, it is conspicuous that they all contain small clusters of from two to four successive and negatively charged residues (aspartate and/or glutamate). M. voltae FlaK also contains a double aspartate at position 185 and 186 which could correspond to Asp187 and 188 in PibD. A D186A mutant was reported to be active in vitro (7), but the role of Asp185 was not investigated. It is therefore not clear whether these aspartate residues are functionally equivalent to those in PibD. Nevertheless, one may speculate that clusters of acidic residues in the cytoplasmic loop play an important role in archaeal TFPP homologs. They might fulfill a structural function by participating in intramolecular salt bridges. Alternatively, these residues could be essential for recognition of the positively charged leader sequence of the substrate protein. An involvement of negatively charged residues of a protease in recognition of positively charged amino acids in the substrate was reported for the yeast (Saccharomyces cerevisiae) mitochondrial processing peptidase (MPP). MPP is a heterodimeric metalloprotease that shows a functional similarity to TFPPs, as it cleaves a positively charged leader peptide of precursor proteins imported into mitochondria (for a review, see reference 20). Biochemical and structural studies of MPP revealed that negatively charged residues in the substrate-binding pocket of the enzyme might form stabilizing electrostatic interactions with the substrate (24, 37, 45). If such electrostatic interactions are also required for the interaction of PibD with its substrates, then Asp187 and 188 are likely to play a role.
Cleavage of substrate proteins by prepilin/preflagellin peptidases is expected to occur at the cytoplasmic side of the membrane (10). To define the topology of PibD, we employed cysteine accessibility studies with everted membrane vesicles containing single-cysteine mutants. Two positions (Glu28 and Ser74) were found to be exposed to the cytoplasmic side of the membrane. Two mutants (N50C and I236C) were not accessible to AMS, and therefore it can be concluded that these residues are extracellular or within a transmembrane segment. One mutant (T63C) was inefficiently labeled by fluorescein maleimide in the solubilized state of the protein, even without pretreatment with AMS. This residue is likely to be buried within the protein structure, which prevents access of the bulky sulfhydryl-modifying reagents. In none of the mutants could fluorescein maleimide labeling be completely blocked by pretreatment with AMS. This can be explained by the fact that pibD is expressed very poorly in E. coli, a fact which is also reflected by the low protein yield (Fig. 3). Therefore, PibD represents only a minor fraction of the total membrane protein, which in turn leads to very high background labeling of endogenous cysteine containing proteins. This might be compensated for by increasing the concentration of AMS or extending labeling times beyond those used in previously published work. Attempts to modify the labeling protocol in this way, however, led to the modification of all cysteine mutations, suggesting that the probe leaked through the membrane (not shown). In conjunction with the positive-inside rule for membrane proteins (50), our results are consistent with a model of six transmembrane segments and extracellular amino and carboxyl termini. Membrane topology data based on reporter gene fusions are available for three bacterial TFPPs, PilD from P. aeruginosa, OutO from Erwinia carotovora, and PilU encoded by plasmid R64 (1, 29, 36, 40). OutO contains eight TMS, and PilU, which belongs to a distinct group of smaller prepilin peptidases, contains six TMS. Interestingly, in all cases the N- and C-terminal ends of the proteins were shown to be exposed to the periplasmic side of the membrane. This feature therefore seems to be conserved among the TFPP homologues from eubacteria and archaea.
It is now evident that bacterial type IV prepilin peptidases and archaeal preflagellin/prepilin-like peptidases are functionally homologous and that they work by the same catalytic mechanism (6, 7, 28). An important question that remains is how these enzymes interact with, and recognize, the substrate proteins. To answer this question, further biochemical studies are necessary; more importantly, it would be desirable to obtain structural information. Hyperthermophilic proteins are generally more robust and are therefore attractive targets for crystallization studies (35). The major challenge in this respect is to produce sufficient amounts of protein, which is not a trivial task for membrane proteins. Because expression of PibD in E. coli is relatively low, we have recently developed new tools for homologous protein expression in S. solfataricus (5). This method results in the overexpression of various S. solfataricus proteins in their native state, and it will be of interest to use homologous expression to isolate PibD from the native host.
Acknowledgments
This work was supported by a VENI grant to S.-V.A. from the Dutch Organization for Scientific Research (NWO) and by the Van der Leeuw Programme of the Earth and Life Sciences Foundation (ALW), which is subsidized by NWO.
REFERENCES
- 1.Akahane, K., D. Sakai, N. Furuya, and T. Komano. 2005. Analysis of the pilU gene for the prepilin peptidase involved in the biogenesis of type IV pili encoded by plasmid R64. Mol. Genet. Genomics 273:350-359. [DOI] [PubMed] [Google Scholar]
- 2.Albers, S. V., and A. J. Driessen. 2005. Analysis of ATPases of putative secretion operons in the thermoacidophilic archaeon Sulfolobus solfataricus. Microbiology 151:763-773. [DOI] [PubMed] [Google Scholar]
- 3.Albers, S. V., and A. J. M. Driessen. 2002. Signal peptides of secreted proteins of the archaeon Sulfolobus solfataricus: a genomic survey. Arch. Microbiol. 177:209-216. [DOI] [PubMed] [Google Scholar]
- 4.Albers, S. V., M. G. Elferink, R. L. Charlebois, C. W. Sensen, A. J. M. Driessen, and W. N. Konings. 1999. Glucose transport in the extremely thermoacidophilic Sulfolobus solfataricus involves a high-affinity membrane-integrated binding protein. J. Bacteriol. 181:4285-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albers, S. V., M. Jonuscheit, S. Dinkelaker, T. Urich, A. Kletzin, R. Tampé, A. J. M. Driessen, and C. Schleper. 2006. Production of recombinant and tagged proteins in the hyperthermophilic archaeon Sulfolobus solfataricus. Appl. Environ. Microbiol. 72:102-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Albers, S. V., Z. Szabo, and A. J. Driessen. 2003. Archaeal homolog of bacterial type IV prepilin signal peptidases with broad substrate specificity. J. Bacteriol. 185:3918-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bardy, S. L., and K. F. Jarrell. 2003. Cleavage of preflagellins by an aspartic acid signal peptidase is essential for flagellation in the archaeon Methanococcus voltae. Mol. Microbiol. 50:1339-1347. [DOI] [PubMed] [Google Scholar]
- 8.Bardy, S. L., and K. F. Jarrell. 2002. FlaK of the archaeon Methanococcus maripaludis possesses preflagellin peptidase activity. FEMS Microbiol. Lett. 208:53-59. [DOI] [PubMed] [Google Scholar]
- 9.Bateman, A., E. Birney, L. Cerruti, R. Durbin, L. Etwiller, S. R. Eddy, S. Griffiths-Jones, K. L. Howe, M. Marshall, and E. L. Sonnhammer. 2002. The Pfam protein families database. Nucleic Acids Res. 30:276-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dupuy, B., A. E. Deghmane, and M. K. Taha. 2004. Type IV prepilin peptidase, p. 204-208. In A. J. Barrett, N. D. Rawlings, and J. F. Woessner (ed.), Handbook of proteolytic enzymes, vol. 2. Elsevier, London, England. [Google Scholar]
- 11.Dupuy, B., M. K. Taha, A. P. Pugsley, and C. Marchal. 1991. Neisseria gonorrhoeae prepilin export studied in Escherichia coli. J. Bacteriol. 173:7589-7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durand, E., A. Bernadac, G. Ball, A. Lazdunski, J. N. Sturgis, and A. Filloux. 2003. Type II protein secretion in Pseudomonas aeruginosa: the pseudopilus is a multifibrillar and adhesive structure. J. Bacteriol. 185:2749-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durand, E., G. Michel, R. Voulhoux, J. Kurner, A. Bernadac, and A. Filloux. 2005. XcpX controls biogenesis of the Pseudomonas aeruginosa XcpT-containing pseudopilus. J. Biol. Chem. 280:31378-31389. [DOI] [PubMed] [Google Scholar]
- 14.Elferink, M. G., S. V. Albers, W. N. Konings, and A. J. Driessen. 2001. Sugar transport in Sulfolobus solfataricus is mediated by two families of binding protein-dependent ABC transporters. Mol. Microbiol. 39:1494-1503. [DOI] [PubMed] [Google Scholar]
- 15.Faguy, D. M., K. F. Jarrell, J. Kuzio, and M. L. Kalmokoff. 1994. Molecular analysis of archaeal flagellins: similarity to the type IV pilin-transport superfamily widespread in bacteria. Can. J. Microbiol. 40:67-71. [DOI] [PubMed] [Google Scholar]
- 16.Fernandez, L. A., and J. Berenguer. 2000. Secretion and assembly of regular surface structures in Gram-negative bacteria. FEMS Microbiol. Rev. 24:21-44. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan, D. 1985. Techniques for transformation in E. coli, p. 109-135. In D. Rickwood and B. D. Hames (ed.), DNA cloning, a practical approach, vol. 1. IRL Press, Oxford, England. [Google Scholar]
- 18.Hoare, D. G. and D. E. Koshland, Jr. 1967. A method for the quantitative modification and estimation of carboxylic acid groups in proteins. J. Biol. Chem. 242:2447-2453. [PubMed] [Google Scholar]
- 19.Hu, N. T., P. F. Lee, and C. Chen. 1995. The type IV pre-pilin leader peptidase of Xanthomonas campestris pv. campestris is functional without conserved cysteine residues. Mol. Microbiol. 18:769-777. [DOI] [PubMed] [Google Scholar]
- 20.Ito, A. 1999. Mitochondrial processing peptidase: multiple-site recognition of precursor proteins. Biochem. Biophys. Res. Commun. 265:611-616. [DOI] [PubMed] [Google Scholar]
- 21.Jones, D. T. 1998. Do transmembrane protein superfolds exist? FEBS Lett. 423:281-285. [DOI] [PubMed] [Google Scholar]
- 22.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1994. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry 33:3038-3049. [DOI] [PubMed] [Google Scholar]
- 23.Kall, L., A. Krogh, and E. L. Sonnhammer. 2004. A combined transmembrane topology and signal peptide prediction method. J. Mol. Biol. 338:1027-1036. [DOI] [PubMed] [Google Scholar]
- 24.Kitada, S., K. Kojima, K. Shimokata, T. Ogishima, and A. Ito. 1998. Glutamate residues required for substrate binding and cleavage activity in mitochondrial processing peptidase. J. Biol. Chem. 273:32547-32553. [DOI] [PubMed] [Google Scholar]
- 25.Koebnik, R. 2001. The role of bacterial pili in protein and DNA translocation. Trends Microbiol. 9:586-590. [DOI] [PubMed] [Google Scholar]
- 26.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 28.LaPointe, C. F., and R. K. Taylor. 2000. The type 4 prepilin peptidases comprise a novel family of aspartic acid proteases. J. Biol. Chem. 275:1502-1510. [DOI] [PubMed] [Google Scholar]
- 29.Lory, S., and M. S. Strom. 1997. Structure-function relationship of type-IV prepilin peptidase of Pseudomonas aeruginosa—a review. Gene 192:117-121. [DOI] [PubMed] [Google Scholar]
- 30.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 31.McGuffin, L. J., K. Bryson, and D. T. Jones. 2000. The PSIPRED protein structure prediction server. Bioinformatics 16:404-405. [DOI] [PubMed] [Google Scholar]
- 32.Miroux, B., and J. E. Walker. 1996. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260:289-298. [DOI] [PubMed] [Google Scholar]
- 33.Pepe, J. C., and S. Lory. 1998. Amino acid substitutions in PilD, a bifunctional enzyme of Pseudomonas aeruginosa. Effect on leader peptidase and N-methyltransferase activities in vitro and in vivo. J. Biol. Chem. 273:19120-19129. [DOI] [PubMed] [Google Scholar]
- 34.Rawlings, N. D., D. P. Tolle, and A. J. Barrett. 2004. MEROPS: the peptidase database. Nucleic Acids Res. 32:D160-D164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rees, D. C. 2001. Crystallographic analyses of hyperthermophilic proteins. Methods Enzymol. 334:423-437. [DOI] [PubMed] [Google Scholar]
- 36.Reeves, P. J., P. Douglas, and G. P. Salmond. 1994. Beta-lactamase topology probe analysis of the OutO NMePhe peptidase, and six other Out protein components of the Erwinia carotovora general secretion pathway apparatus. Mol. Microbiol. 12:445-457. [DOI] [PubMed] [Google Scholar]
- 37.Shimokata, K., S. Kitada, T. Ogishima, and A. Ito. 1998. Role of alpha-subunit of mitochondrial processing peptidase in substrate recognition. J. Biol. Chem. 273:25158-25163. [DOI] [PubMed] [Google Scholar]
- 38.Sonnhammer, E. L., G. von Heijne, and A. Krogh. 1998. A hidden Markov model for predicting transmembrane helices in protein sequences, p. 175-182. In J. Glasgow, T. Littlejohn, F. Major, R. Lathrop, D. Sankoff, and C. Sensen (ed.), Proceedings of the Sixth International Conference on Intelligent Systems for Molecular Biology. AAAI Press, Menlo Park, Calif. [PubMed]
- 39.Steiner, H., M. Kostka, H. Romig, G. Basset, B. Pesold, J. Hardy, A. Capell, L. Meyn, M. L. Grim, R. Baumeister, K. Fechteler, and C. Haass. 2000. Glycine 384 is required for presenilin-1 function and is conserved in bacterial polytopic aspartyl proteases. Nat. Cell Biol. 2:848-851. [DOI] [PubMed] [Google Scholar]
- 40.Strom, M. S., P. Bergman, and S. Lory. 1993. Identification of active-site cysteines in the conserved domain of PilD, the bifunctional type IV pilin leader peptidase/N-methyltransferase of Pseudomonas aeruginosa. J. Biol. Chem. 268:15788-15794. [PubMed] [Google Scholar]
- 41.Strom, M. S., and S. Lory. 1987. Mapping of export signals of Pseudomonas aeruginosa pilin with alkaline phosphatase fusions. J. Bacteriol. 169:3181-3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Strom, M. S., D. N. Nunn, and S. Lory. 1993. A single bifunctional enzyme, PilD, catalyzes cleavage and N-methylation of proteins belonging to the type IV pilin family. Proc. Natl. Acad. Sci. USA 90:2404-2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 44.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278:631-637. [DOI] [PubMed] [Google Scholar]
- 45.Taylor, A. B., B. S. Smith, S. Kitada, K. Kojima, H. Miyaura, Z. Otwinowski, A. Ito, and J. Deisenhofer. 2001. Crystal structures of mitochondrial processing peptidase reveal the mode for specific cleavage of import signal sequences. Structure (Cambridge) 9:615-625. [DOI] [PubMed] [Google Scholar]
- 46.Thomas, N. A., S. L. Bardy, and K. F. Jarrell. 2001. The archaeal flagellum: a different kind of prokaryotic motility structure. FEMS Microbiol. Rev. 25:147-174. [DOI] [PubMed] [Google Scholar]
- 47.Tusnady, G. E., and I. Simon. 1998. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J. Mol. Biol. 283:489-506. [DOI] [PubMed] [Google Scholar]
- 48.Tusnady, G. E., and I. Simon. 2001. Topology of membrane proteins. J. Chem. Infect. Comput. Sci. 41:364-368. [DOI] [PubMed] [Google Scholar]
- 49.Vignon, G., R. Kohler, E. Larquet, S. Giroux, M. C. Prevost, P. Roux, and A. P. Pugsley. 2003. Type IV-like pili formed by the type II secreton: specificity, composition, bundling, polar localization, and surface presentation of peptides. J. Bacteriol. 185:3416-3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Heijne, G. 1986. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 5:3021-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiner, H., W. N. White, D. G. Hoare, and D. E. Koshland, Jr. 1966. The formation of anhydrochymotrypsin by removing the elements of water from the serine at the active site. J. Am. Chem. Soc. 88:3851-3859. [DOI] [PubMed] [Google Scholar]
- 52.Wolfe, M. S., and D. J. Selkoe. 2002. Biochemistry. Intramembrane proteases—mixing oil and water. Science 296:2156-2157. [DOI] [PubMed] [Google Scholar]