Abstract
Translation initiation in many eukaryotic mRNAs is modulated by an interaction between the cap binding protein complex, bound to the 5′ end of the mRNA, and the polyadenosine binding protein, bound to the 3′-terminal polyadenosine sequences. A few cellular and viral mRNAs, such as the hepatitis C virus (HCV) mRNA genome, lack 3′-terminal polyadenosine sequences. For such mRNAs, the question of whether their 3′-end sequences also regulate the initiation phase of protein synthesis via an interaction with their 5′ ends has received intense scrutiny. For HCV mRNA, various experimental designs have led to conflicting interpretations, that the 3′ end of the RNA can modulate translation initiation either in a positive or in a negative fashion. To examine the possibility of end-to-end communication in HCV in detail, mRNAs containing the HCV internal ribosome entry site linked to a luciferase coding region, followed by different 3′ noncoding regions, were expressed in the cytoplasm of cultured cells by T7 RNA polymerase. The intracellular translation efficiencies, steady-state levels, stabilities, and 3′-end sequences of these chimeric RNAs were examined. It was found that the HCV 3′ noncoding region modulates neither the translation nor the stability of the mRNAs. Thus, there is no detectable end-to-end communication in cytoplasmically expressed chimeric mRNAs containing the HCV noncoding regions. However, it remains an open question whether end-to-end communication occurs in full-length HCV mRNAs in the infected liver.
Hepatitis C virus (HCV) has been classified as the main agent causing non-A, non-B hepatitis (8). HCV is a member of the Flaviviridae, a family of viruses containing single-stranded RNA genomes that are translated into viral polyproteins, which are processed into the viral structural and nonstructural proteins (reviewed in references 2 and 9). Because there is no cell culture system in which HCV can be propagated, many functions of the HCV genome have been studied using either individually expressed genes or cell culture-adapted viral replicons (4, 18, 19, 26). These studies have shown that the viral 5′ and 3′ noncoding regions (NCR) are, as expected, important in translation and amplification of the viral RNA. The viral 5′ NCR contains an internal ribosome entry site (IRES) (31, 32) that can recruit 40S ribosomal subunits without additional translation initiation factors (25, 28). The viral 3′ NCR is conserved among several viral genotypes and does not contain terminal poly(A) sequences like other cellular mRNAs (29, 30). For other viral IRES elements, the poly(A) binding protein, associated with the 3′-terminal poly(A) sequences, can stimulate the efficiency of translation initiation (3, 21). Therefore, it was of interest to examine whether the 3′ NCR of HCV, lacking polyadenosine sequences, could stimulate the HCV IRES.
The 3′ NCR located in the six genotypes of HCV can be divided into three regions. Immediately following the stop codon of the viral polyprotein resides a 40-nucleotide variable region, followed by an approximately 80 nucleotide polypyrimidine-tract sequence element and a highly conserved 100-nucleotide sequence element, known as the X region, which contains three RNA hairpin structures (17, 29, 30). Full-length mRNAs that carry an alteration in the polypyrimidine tract or X region did not produce infectious virus after inoculation into susceptible chimpanzees; however, mRNAs with small mutations in the 3′ NCR variable region produced infectious virus in this assay (33). While this study clearly showed a role for the 3′ NCR in viral propagation, it did not address whether the deleterious mutations affected viral mRNA translation, RNA replication, RNA stability, or RNA packaging.
To test possible roles of the 3′-end sequences in viral mRNA translation, the translational efficiencies of chimeric mRNAs containing the HCV IRES linked to a reporter open reading frame followed by the viral 3′ NCR or other sequences were examined in cell-free translation systems, in cultured cells, and in transfected mouse livers (14, 15, 20-22). However, conflicting results have been obtained, pointing both to stimulatory and inhibitory effects of the HCV 3′ NCR on translation. Thus, a role of the viral end sequences in mRNA translation and stability awaits further proof. In this study, we have employed an expression system in which chimeric HCV NCR-containing mRNAs were synthesized in the cytoplasm of cultured cells instead of being delivered by transfection or synthesized in the nucleus. Analyses revealed that the 3′ NCR of HCV did not have a significant effect on the translatability or stability of the chimeric mRNAs either in cells of nonliver origin or in cells of liver origin.
MATERIALS AND METHODS
Cells.
The Huh7 cell line (obtained from H. Greenberg, Stanford University School of Medicine) was maintained in 50% Dulbecco's modified Eagle medium (DMEM) and 50% RPMI medium (Life Technologies, Inc.) supplemented with 10% fetal calf serum, 1% penicillin (10 U/ml)-streptomycin (10 μg/ml) (Life Technologies, Inc.), and 1% l-glutamine (200 μM; Life Technologies, Inc.). Huh7 cells were passaged fewer than 25 times prior to use. 293T cells were maintained in DMEM supplemented with 10% calf serum, 1× penicillin-streptomycin, and 1× l-glutamine. KJT7 cells, monkey kidney CV-1 cells that constitutively express T7 RNA polymerase (23), were maintained in DMEM supplemented with 10% calf serum and 1% penicillin-streptomycin.
Plasmid constructs.
To create pT7 5′HCVluc, the HindIII-to-BstEII fragment from plasmid pT7 CAT-HCV IRES luc (13) was subcloned into the SalI-to-BstEII site of pT7 lucAn (13). The HCV 3′ NCR was obtained from plasmid pTET/T7HCVcons/ΔApaI/BsmI (5) after PCR amplification using primers 5′-CTCCCCAACCGATGAAGGTTG-3′ (HCV11) and 5′CGGGATCCACATGATCTGCAGAGGCCAG-3′ (HCV8) and Advantage II polymerase (Clontech). The amplified DNA fragment was gel purified and subcloned into the BamHI site of pT7 5′HCVluc to create pT7 5′HCVluc3′NCR. The 3′ sequence was verified by sequencing.
Individual sequence elements of the viral 3′ end were obtained in a similar manner. For example, to create pT7 5′HCVluc3′X, primers HCV 11 and 5′-AATGGTGGCTCCATCTTAGCCC-3′ were used in a PCR to amplify the 3′ X region; to generate pT7 5′HCVluc3′UC, primers 5′-GCTCTAGATTTCCTGTTTTTTTTTTTTTTTTTTTT-3′ and 5′-CGGAATCCAAAGAAGGAAGGAAAAGAAAGG-3′ were utilized; to produce pT7 5′HCVluc 3′var, primer 5′-GCTCTAGAAGGTTGGGGTAAACACTCCGGCCTCTTAGGCCATTTCCTGGGATCCC-3′ and its complementary strand were annealed and subcloned into the BamHI site of pT7 5′HCVluc.
To create pT3 luc3′XRPA, 101 nucleotides of the HCV 3′ NCR were amplified using oligonucleotides HCV8 and 5′-GAGTCTAATGGTGGCTCCATCTTAGCCC-3′ and then subcloned into plasmid pCR4 (Invitrogen) by using the TopoTA cloning kit (Invitrogen). The resulting plasmid was confirmed by sequencing.
pRevT7RNApoly was generated by excising the T7 RNA polymerase gene from plasmid pAR3126 (11) with BamHI and HindIII and then inserting the fragment into the ClaI-to-HindIII sites of pRev Tet-ON (Promega Biotec). Thus, plasmid pRevT7RNApoly encodes a T7 RNA polymerase which lacks a nuclear localization signal.
In vitro RNA synthesis.
Firefly luciferase-encoding plasmids were linearized with BamHI, or with BsmI in the case of pT7 5′HCVluc165orf-3′HCV. Reaction mixtures were incubated at 37°C for 4 h in transcription buffer (80 mM HEPES-KOH [pH 7.5], 12 mM MgCl2, 2 mM spermidine, 10 mM nucleoside triphosphates, and T7 RNA polymerase [a gift from Jody Puglisi, Stanford University School of Medicine]). Production of full-length RNA was verified by electrophoresis using denaturing polyacrylamide gels containing Century Marker size standards (Ambion).
Transient transfections.
Huh7 and 293T cells were plated the day before transfection into 6-well plates and were transfected with 500 ng of the linearized reporter plasmid, 125 ng of the pRevT7 polymerase plasmid, and 10 ng of pRL by using the Fugene 6 (Roche, Mannheim, Germany) transfection reagent in a 6:1 (vol/vol) ratio. The medium was changed after 18 h of incubation. At the indicated time points, Renilla and firefly luciferase activities were assayed using the Dual Luciferase Assay system (Promega Biotec).
KJT7 cells were transfected with Effectene (Qiagen Inc.). Briefly, 1 μg of DNA and 250 ng of PRL were mixed in a 10:1 molar ratio with Effectene and an 8:1 (vol/vol) ratio with Enhancer. After 3 h, the medium was changed, followed by two washes with phosphate-buffered saline. Fresh medium was added and luciferase activity was assayed, as described above, after an additional 7 h of incubation.
RNA isolation and Northern blot analysis.
At the indicated time points, total RNA was harvested using the TRIzol reagent (Life Technologies, Inc.) according to the manufacturer's instructions. To obtain polysome-associated RNA, cells were lysed in 1% Triton X-100-300 mM NaCl-15 mM Tris-HCl (pH 7.5)-15 mM MgCl2-0.1 mg of cycloheximide/ml-1 mg of heparin/ml and were fractionated over sucrose gradients as described previously (16). Northern blot analysis was performed as described previously (7). Briefly, the RNA was separated in a 1% denaturing agarose gel in the presence of formaldehyde and morpholinepropanesulfonic acid (MOPS) buffer (1) and then transferred to a Zeta-probe membrane (Bio-Rad). Northern blots were hybridized in ExpressHyb (Clontech Inc.) to specific radiolabeled nucleic acid probes. Radiolabeled probes that can detect actin (nucleotides 685 to 1171) and firefly luciferase (nucleotides 99 to 1354) mRNAs were generated using the RadPrime Kit (Life Technologies, Inc.). ImageQuant (Molecular Dynamics) was used to quantify the images.
RNase protection assays.
Total RNA was harvested at the indicated time points and subjected to RNase protection assays using the HybSpeed RPA (Ambion Inc.) protocol at an optimized RNase A/T1 dilution of 1:100. The strand-specific RNA probe was generated by using NotI-linearized plasmid p3′XRPA as a template in the MAXIscript T3 (Ambion Inc.) labeling system, resulting in the synthesis of a 157-nucleotide radiolabeled RNA. Following hybridization and RNase digestion, RNA samples were precipitated and separated on a denaturing 5% polyacrylamide gel in the presence of 8 M urea and Tris-borate-EDTA buffer (1) and were then transferred to a Zeta-probe membrane by using the HEP-1 Panther semidry electroblotter system (Owl Inc.).
RNase H assays.
Total RNA was harvested at the indicated time points and hybridized to oligodeoxynucleotide 5′-TCTAGAAAGAAGGAAGGAAAAGAAAGG-3′. Briefly, RNA was denatured at 70°C for 3 min and incubated at 45°C for 30 min in Tris-EDTA buffer (1). Reaction mixtures were treated with 1 U of RNase H (Life Technologies, Inc.) at 37°C for 20 min. After extraction with phenol and precipitations with ethanol, the digested RNA was separated on denaturing 5% polyacrylamide gels. Northern blot analysis was performed with radiolabeled strand-specific RNA probes derived from p3′XRPA as described in the preceding section.
RESULTS
Effects of the 3′ NCR of HCV on IRES-mediated translation in a cell-based system.
Translation studies with HCV NCR-containing mRNAs performed in a variety of cell-free and cell-based translation systems have yielded different results with respect to the effect of the HCV 3′ NCR on translational efficiency. In some instances, a stimulatory role was observed for the viral 3′ NCR in HCV IRES-mediated translation (14, 15, 20). However, inhibitory roles for the 3′-end sequences have been reported as well (22). While the reasons for these inconsistencies are not known, it is likely that different concentrations of translation factors in the individual assay systems, different intracellular mRNA concentrations, and misfolding of in vitro-generated RNA templates could contribute to the discrepancies noted. To eliminate many of these variables, we established a cell-based assay in which both the translational efficiencies and the stabilities of newly synthesized cytoplasmic RNAs could be measured.
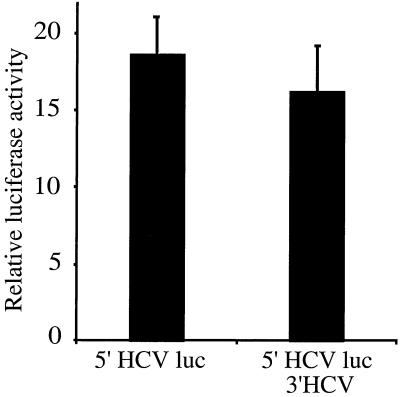
A series of plasmids was generated that contained the promoter for T7 RNA polymerase linked to the luciferase coding region surrounded by specific NCR (Fig. 1). Plasmids were linearized and transfected into KJT7 monkey kidney cells, which constitutively express the gene for T7 RNA polymerase (23). Because the expressed T7 RNA polymerase resides in the cytoplasm (23), the linearized plasmids can be used as templates by the RNA polymerase to direct the synthesis of uncapped luciferase mRNAs with predicted 5′ and 3′ NCR (Fig. 1). Cells transfected with DNAs encoding the HCV IRES were translated, and luciferase activities could be measured (Fig. 2). In contrast, transfection of a construct lacking an IRES produced only 1% normalized luciferase activity (data not shown). Surprisingly, RNAs containing different parts of the HCV 3′ NCR were all translated with similar efficiencies (Fig. 2). These findings suggest that the various 3′ NCR fail to regulate HCV IRES-mediated translation. Alternatively, the various viral 3′ NCR could have effects on the intracellular abundances of the expressed mRNAs; for example, 5′HCVluc RNAs might be more abundant than 5′HCVluc3′HCV RNAs but may be less efficiently translated. In addition, effects of the HCV 3′ NCR on translation may be observed only in liver cells.
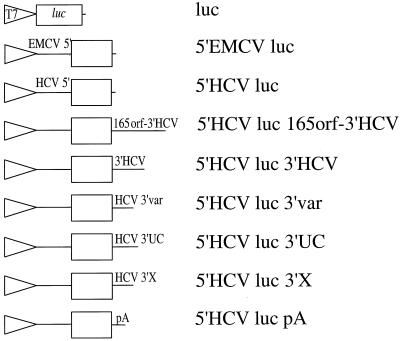
FIG. 1.
Schematic representation of the constructs used to synthesize chimeric mRNAs. DNA plasmids containing the T7 promoter (T7) and the luciferase (luc) open reading frame surrounded by various 5′ and 3′ NCR are shown. 5′EMCV and 5′HCV denote the IRES elements from EMCV and HCV, respectively. 3′ NCR consist of the following: the 165 terminal sequences of the HCV open reading frame followed by the HCV 3′ NCR (165orf-3′HCV), the 3′ NCR of HCV (3′HCV), the 3′ variable sequence (HCV 3′var), the 3′ polypyrimidine-rich element (HCV 3′UC), the 3′ X region (HCV 3′X), or a polyadenosine element (pA). See the text for details.
FIG. 2.
Translational activities of chimeric RNAs in DNA-transfected KJT7 cells constitutively expressing T7 RNA polymerase. Linearized reporter plasmids (see Fig. 1 for details) were transfected into KJT7 cells as described in Materials and Methods. Luciferase activities after a 10-h incubation are shown. The mean activity of the construct 5′HCV luc was set to a value of 1. Error bars, standard deviations from duplicate experiments.
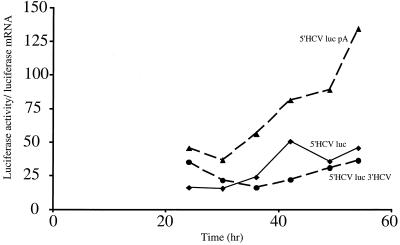
To distinguish between these possibilities, the accumulation of both intracellularly expressed mRNAs and translation products was examined. Chimeric RNAs could not be detected in DNA-transfected KJT7 cells. Therefore, a transient expression assay was developed in which human kidney 293T cells were cotransfected with linearized, reporter-encoding plasmids (see above) and a plasmid that expresses T7 RNA polymerase. Figure 3 displays a kinetic analysis in which both luciferase activity and luciferase RNA abundance were measured at different times after DNA transfection. Luciferase activities divided by the relative amounts of cytoplasmic luciferase mRNA, determined by Northern blot analyses, are displayed. In this analysis, HCV-luciferase mRNAs lacking or containing viral 3′ NCRs were translated with roughly equal efficiencies throughout the time course of the experiment. Importantly, the overall intracellular abundances of HCV-luciferase mRNAs lacking or containing the viral 3′ NCRs were the same, as can be seen in Fig. 5D and C, respectively. In contrast, HCV mRNAs that contained additional template-encoded 3′-terminal polyadenosine sequences were translated with higher efficiencies than the nonpolyadenylated mRNAs, especially at later times of infection. Thus, our assays were not performed under conditions where components of the translation apparatus were limiting. These results suggest that the HCV 3′ NCR has no significant enhancing or inhibitory effect on cytoplasmically synthesized mRNAs.
FIG. 3.
Translational efficiencies of chimeric RNAs expressed by T7 RNA polymerase in DNA-transfected 293T cells. The indicated linearized plasmids were cotransfected with a T7 RNA polymerase-expressing plasmid into 293T cells. At the indicated times after transfection, enzymatic luciferase activities were measured and luciferase mRNA levels were quantitated after Northern blot analysis. Levels of actin mRNA were determined as a control. The ratio of luciferase activity to luciferase mRNA is plotted versus time.
FIG. 5.
Association of luciferase RNA with polysomes in DNA-transfected 293T cells. Polysome-associated RNA from cells transfected with the indicated linearized plasmids was isolated at 39 h after transfection. Lysates were fractionated by sucrose gradient centrifugation. The fractions from the top to the bottom of the sucrose gradient are displayed from left to right. The absorbance profile at 254 nm is shown, and arrows indicate the migration of 80S monosomes. The distributions of chimeric luciferase and endogenous actin mRNAs in each fraction, obtained after Northern blot analysis, are shown. The fraction of the luciferase RNA signal associated with polysomal fractions 7 to 10, as quantified by ImageQuant, is indicated.
Cytoplasmically synthesized mRNAs containing or lacking the 3′ NCR of HCV are translated with similar efficiencies in transfected liver cells.
To examine the possibility that end-to-end communication in HCV mRNA molecules was restricted to liver cells, the translational activities of cytoplasmically synthesized mRNAs that contained or lacked viral 3′ NCRs were examined by transfection into Huh7 cells. The results show that chimeric mRNAs containing the HCV 3′ NCR were translated with similar efficiencies as mRNAs lacking a viral 3′ NCR (Fig. 4). Together with the results in Fig. 3, these findings indicate that the HCV 3′ NCR has no effect on translational efficiency or mRNA stability at any RNA concentration in cultured nonliver or liver cells.
FIG. 4.
Translational efficiencies of chimeric RNAs expressed by T7 RNA polymerase in DNA-transfected Huh7 cells. Linearized reporter plasmids (see Fig. 1 for details) were transfected into Huh7 cells as described in Materials and Methods. Relative luciferase activities after 48 h are expressed as ratios to Renilla luciferase activities expressed from cotransfected plasmids. Error bars, standard deviations of experiments performed in triplicate.
Polysomal association of IRES-containing mRNAs.
To more rigorously substantiate this notion, the intracellular translation-competent mRNA pool was examined. Cytoplasmic extracts were prepared from DNA-transfected cells and were fractionated on sucrose gradients. The positions of 80S monosomes and polysomes were determined from the absorbance profile at 254 nm (Fig. 5). Accordingly, RNA in fractions 7 to 10, derived from polysomes, was designated translation-competent RNA, while RNA in fractions 1 to 6, derived from monosomes or smaller complexes, was designated translationally inactive. Northern blot analysis showed that endogenous host cell actin mRNA was quantitatively associated with high-molecular-weight polysomes in fractions 9 and 10 (Fig. 5). Cytoplasmically expressed luciferase mRNA lacking an IRES element was found, as expected, predominately in the translationally inactive pool (Fig. 5A). Surprisingly, only a very small fraction of encephalomyocarditis virus (EMCV) IRES- or HCV IRES-containing mRNAs was associated with polysomes. We can only speculate that a nuclear experience might guarantee efficient association of an mRNA with the cytoplasmic translation apparatus, because HCV IRES-containing reporter mRNAs, which were expressed at a similar cytoplasmic abundance by nuclear RNA polymerase II, were associated with polysomes as efficiently as native actin mRNA (data not shown). In any case, the nature of the 3′ NCR element did not affect the representation of the HCV IRES-containing RNAs on polysomes.
Cytoplasmically expressed RNAs contain the predicted HCV 3′ NCR.
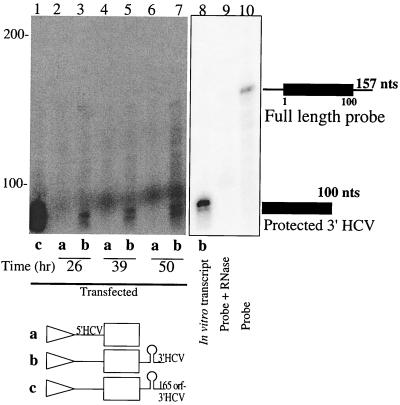
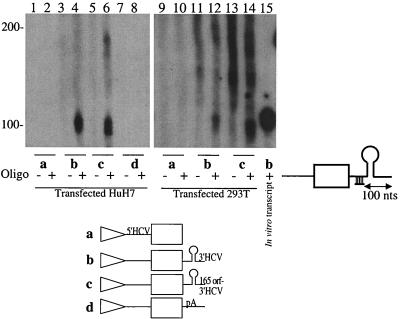
Of course, the apparent lack of effect of the HCV 3′ NCR could be explained if the 3′ ends of the RNAs were different from those predicted. To test this possibility, a RNase protection assay was used to determine whether T7 RNA polymerase-synthesized mRNAs contained the predicted viral 3′ NCR in DNA-transfected cells. To this end, a radiolabeled 157-nucleotide RNA, which contained the 100-nucleotide X region, was hybridized to cytoplasmic RNA and digested with single-strand-specific nucleases. Presence of a 3′-terminal X region should result in the appearance of a 100-nucleotide protected RNA species. Indeed, cells expressing the predicted viral 3′ NCR (Fig. 6, lanes b) displayed a doublet of approximately 95 to 100 nucleotides, while RNAs predicted to lack a viral 3′ NCR failed to display this doublet (Fig. 6, lanes a). These findings show that the viral 3′ NCR were expressed as predicted.
FIG. 6.
Determination of 3′-end sequences by RNase protection assays. Total RNA was harvested at the indicated times after linearized DNA transfection. After hybridization of the 157-nucleotide (nt) full-length radiolabeled RNA to the isolated RNA, an RNase protection assay was performed as described in Materials and Methods. Lane 8, positive control, obtained after an RNase protection assay using an in vitro-generated RNA as the template; lane 9, RNase protection assay performed in the absence of template RNA; lane 10, migration of the 157-nt radiolabeled probe. Migration of RNAs of known sizes is indicated on the left. An autoradiograph of the gel is shown.
Cytoplasmically expressed mRNAs are not polyadenylated.
Lastly, to test for the possibility that all expressed mRNAs received additional polyadenosine in the cytoplasmic sequences, which could override any effects of viral 3′ NCR sequences, we employed RNase H cleavage assays to search for additional polyadenosine sequences on the expressed RNAs. Oligodeoxynucleotides with sequences complementary to the 3′ end of the luciferase coding region were annealed to the expressed RNAs and treated with RNase H, which should endonucleolytically cleave the DNA-RNA sequence element. The length of the 3′-terminal RNA segment was then visualized by Northern blot analysis (Fig. 7). Addition of polyadenosine nucleotides to the 3′ ends of the RNAs should result in the appearance of RNA fragments larger than 100 nucleotides. Figure 7 shows that 3′ HCV-containing RNA preparations each released a predominantly 100-nucleotide fragment after RNase H digestion. RNA fragments of 150 to 200 nucleotides were sometimes seen in lanes containing reactions that were performed either in the presence or in the absence of olgideoxynucleotides, and therefore, they do not represent poly(A) sequences. These findings show that 3′ HCV-encoding RNAs were not extensively polyadenylated by cytoplasmic poly(A) polymerases in liver (Huh7) and nonliver (293T) cells.
FIG. 7.
Examination of 3′-end sequences by RNase H protection assays. Total RNA was harvested after linearized DNA transfection of Huh7 or 293T cells. RNase H digestion assays were performed as described in Materials and Methods. The last lane on the right shows migration of the predicted 100-nucleotide 3′-end fragment released after digestion with RNase H. Migration of RNAs of known sizes is indicated on the left. An autoradiograph of the gel is shown.
DISCUSSION
A variety of assay systems have been employed to study a role of the 3′ NCR of HCV in modulation of HCV IRES activity (14, 15, 20-22). However, depending on the assay, investigators have reported conflicting results. The system described here enabled us to measure translational efficiencies and intracellular stabilities of the chimeric mRNAs. Because sufficient amounts of mRNA accumulated in the transfected cells, sensitive RNase protection assays could be used to examine the integrity of 3′-terminal end sequences in the RNAs. Our results indicated that the 3′-terminal end sequences in HCV do not significantly enhance or diminish translational efficiencies or stabilities in the chimeric RNAs.
Why have studies on the HCV 3′ NCR produced such conflicting results (14, 15, 20-22)? First, studies performed in cell-free systems can lack regulatory functions. For example, it has long been known that the IRES elements in poliovirus and rhinovirus require additional host factors to mediate efficient translation in rabbit reticulocyte lysate (6, 10). Similarly, the IRES in hepatitis A virus requires liver-specific extracts for efficient function in cell-free lysates (12). Thus, there is evidence that IRES-containing RNAs can be regulated by cell-specific factors. Second, it is likely that not all RNA molecules synthesized in vitro acquired the proper folding to ensure optimal translation initiation. Third, most studies performed in cultured cells have relied on the expression of viral RNA either by cellular polymerase II or by T7 RNA polymerase expressed from recombinant vaccinia virus. Of course, polymerase II-derived transcripts encounter a nuclear experience before they are bound by cytoplasmic ribosomes, which could change the associated proteins, secondary structure, or both, of the normally cytoplasmic RNAs. More importantly, polymerase II-derived transcripts contain a 5′-terminal cap structure, and whether HCV mRNAs contain a cap structure at their 5′ ends remains unknown. To circumvent these variables, many investigators have delivered T7 RNA polymerase to the cytoplasm of cells by using recombinant vaccinia viruses. Vaccinia virus-derived T7 RNA polymerase could then transcribe IRES-containing mRNAs from plasmids containing the target gene under the control of a T7 RNA polymerase promoter. However, vaccinia virus infection is known to have profound effects on IRES-mediated translation (24). Therefore, we chose to deliver T7 RNA polymerase via transfection of DNA plasmids and not by recombinant vaccinia viruses in this study.
Clearly, we do not know whether effects of the HCV 3′ NCR on mRNA translation or stability can be observed in full-length viral mRNA molecules or in an infected liver. One can begin to answer these questions if a mutation in the HCV 3′ NCR can be obtained which has an effect on translation but not on RNA replication. The effects of such a mutation on translation initiation could then be studied in the cell-based replicon system (19). It is also conceivable that certain viral protein products are essential to mediate end-to-end communication in the viral mRNA. For example, the rotavirus-encoded protein NSP3 can enhance viral mRNA translation by promoting end-to end-communication in the viral RNA (27). So far, we have examined whether the viral core protein affects translational efficiencies in our chimeric mRNA, yet we have failed to note any effects of the core on mRNA translation or stability (data not shown). However, functional roles for virus-encoded proteins in the translation and stability of HCV RNA in the infected liver remain possible.
Acknowledgments
We are grateful to Karla Kirkegaard for critical reading of the manuscript.
This work was supported by NIH grant AI47365, the Hutchison Foundation, and Eli Lilly, Inc. L.K.K. was supported by a Walter V. and Idun Y. Berry Post-Doctoral Fellowship.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl. 1989. Current protocols in molecular biology. Greene Publishing Associates, Inc., and John Wiley & Sons, Inc., New York, N.Y.
- 2.Bartenschlager, R., and V. Lohmann. 2000. Replication of hepatitis C virus. J. Gen. Virol. 81:1631-1648. [DOI] [PubMed] [Google Scholar]
- 3.Bergamini, G., T. Preiss, and M. W. Hentze. 2000. Picornavirus IRESs and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 6:1781-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 5.Blight, K. J., and C. M. Rice. 1997. Secondary structure determination of the conserved 98-base sequence at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 71:7345-7352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borman, A. M., J. L. Bailly, M. Girard, and K. M. Kean. 1995. Picornavirus internal ribosome entry segments: comparison of translation efficiency and the requirements for optimal internal initiation of translation in vitro. Nucleic Acids Res. 23:3656-3663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter, M. S., and P. Sarnow. 2000. Distinct mRNAs that encode La autoantigen are differentially expressed and contain internal ribosome entry sites. J. Biol. Chem. 275:28301-28307. [DOI] [PubMed] [Google Scholar]
- 8.Choo, Q. L., G. Kuo, A. J. Weiner, L. R. Overby, D. W. Bradley, and M. Houghton. 1989. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science 244:359-362. [DOI] [PubMed] [Google Scholar]
- 9.Clarke, B. 1997. Molecular virology of hepatitis C virus. J. Gen. Virol. 78:2397-2410. [DOI] [PubMed] [Google Scholar]
- 10.Dorner, A. J., B. L. Semler, R. J. Jackson, R. Hanecak, E. Duprey, and E. Wimmer. 1984. In vitro translation of poliovirus RNA: utilization of internal initiation sites in reticulocyte lysate. J. Virol. 50:507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dunn, J. J., B. Krippl, K. E. Bernstein, H. Westphal, and F. W. Studier. 1988. Targeting bacteriophage T7 RNA polymerase to the mammalian cell nucleus. Gene 68:259-266. [DOI] [PubMed] [Google Scholar]
- 12.Glass, M. J., and D. F. Summers. 1993. Identification of a trans-acting activity from liver that stimulates hepatitis A virus translation in vitro. Virology 193:1047-1050. [DOI] [PubMed] [Google Scholar]
- 13.Iizuka, N., L. Najita, A. Franzusoff, and P. Sarnow. 1994. Cap-dependent and cap-independent translation by internal initiation of mRNAs in cell extracts prepared from Saccharomyces cerevisiae. Mol. Cell. Biol. 14:7322-7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ito, T., and M. M. Lai. 1999. An internal polypyrimidine-tract-binding protein-binding site in the hepatitis C virus RNA attenuates translation, which is relieved by the 3′-untranslated sequence. Virology 254:288-296. [DOI] [PubMed] [Google Scholar]
- 15.Ito, T., S. M. Tahara, and M. M. Lai. 1998. The 3′-untranslated region of hepatitis C virus RNA enhances translation from an internal ribosomal entry site. J. Virol. 72:8789-8796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johannes, G., and P. Sarnow. 1998. Cap-independent polysomal association of natural mRNAs encoding c-myc, BiP, and eIF4G conferred by internal ribosome entry sites. RNA 4:1500-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolykhalov, A. A., S. M. Feinstone, and C. M. Rice. 1996. Identification of a highly conserved sequence element at the 3′ terminus of hepatitis C virus genome RNA. J. Virol. 70:3363-3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohmann, V., F. Korner, A. Dobierzewska, and R. Bartenschlager. 2001. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J. Virol. 75:1437-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 20.McCaffrey, A. P., K. Ohashi, L. Meuse, S. Shen, A. M. Lancaster, P. J. Lukavsky, P. Sarnow, and M. A. Kay. 2002. Determinants of hepatitis C translational initiation in vitro, in cultured cells and mice. Mol. Ther. 5:676-684. [DOI] [PubMed] [Google Scholar]
- 21.Michel, Y. M., A. M. Borman, S. Paulous, and K. M. Kean. 2001. Eukaryotic initiation factor 4G-poly(A) binding protein interaction is required for poly(A) tail-mediated stimulation of picornavirus internal ribosome entry segment-driven translation but not for X-mediated stimulation of hepatitis C virus translation. Mol. Cell. Biol. 21:4097-4109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murakami, K., M. Abe, T. Kageyama, N. Kamoshita, and A. Nomoto. 2001. Down-regulation of translation driven by hepatitis C virus internal ribosomal entry site by the 3′ untranslated region of RNA. Arch. Virol. 146:729-741. [DOI] [PubMed] [Google Scholar]
- 23.Nugent, C. I., K. L. Johnson, P. Sarnow, and K. Kirkegaard. 1999. Functional coupling between replication and packaging of poliovirus replicon RNA. J. Virol. 73:427-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pal-Ghosh, R., and C. D. Morrow. 1993. A poliovirus minireplicon containing an inactive 2A proteinase is expressed in vaccinia virus-infected cells. J. Virol. 67:4621-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pestova, T. V., I. N. Shatsky, S. P. Fletcher, R. J. Jackson, and C. U. Hellen. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12:67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pietschmann, T., V. Lohmann, G. Rutter, K. Kurpanek, and R. Bartenschlager. 2001. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J. Virol. 75:1252-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Piron, M., P. Vende, J. Cohen, and D. Poncet. 1998. Rotavirus RNA-binding protein NSP3 interacts with eIF4GI and evicts the poly(A) binding protein from eIF4F. EMBO J. 17:5811-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spahn, C. M., J. S. Kieft, R. A. Grassucci, P. A. Penczek, K. Zhou, J. A. Doudna, and J. Frank. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science 291:1959-1962. [DOI] [PubMed] [Google Scholar]
- 29.Tanaka, T., N. Kato, M. J. Cho, and K. Shimotohno. 1995. A novel sequence found at the 3′ terminus of hepatitis C virus genome. Biochem. Biophys. Res. Commun. 215:744-749. [DOI] [PubMed] [Google Scholar]
- 30.Tanaka, T., N. Kato, M. J. Cho, K. Sugiyama, and K. Shimotohno. 1996. Structure of the 3′ terminus of the hepatitis C virus genome. J. Virol. 70:3307-3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsukiyama-Kohara, K., N. Iizuka, M. Kohara, and A. Nomoto. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang, C., P. Sarnow, and A. Siddiqui. 1993. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 67:3338-3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yanagi, M., M. St Claire, S. U. Emerson, R. H. Purcell, and J. Bukh. 1999. In vivo analysis of the 3′ untranslated region of the hepatitis C virus after in vitro mutagenesis of an infectious cDNA clone. Proc. Natl. Acad. Sci. USA 96:2291-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]