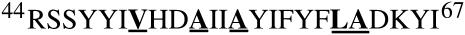Abstract
This study reports the solid-state NMR spectroscopic characterization of the amino-proximate transmembrane domain (TM-A) of a diverged microsomal Δ12-desaturase (CREP-1) in a phospholipid bilayer. A series of TM-A peptides were synthesized with 2H-labeled side chains (Ala-53, -56, and -63, Leu-62, Val-50), and their dynamic properties were studied in 1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine (DMPC) bilayers at various temperatures. At 6 mol % peptide to lipid, 31P NMR spectra indicated that the peptides did not significantly disrupt the phospholipid bilayer in the Lα phase. The 2H NMR spectra from Ala-53 and Ala-56 samples revealed broad Pake patterns with quadrupolar splittings of 16.9 kHz and 13.3 kHz, respectively, indicating restricted motion confined within the hydrocarbon core of the phospholipid bilayer. Conversely, the deuterated Ala-63 sample revealed a peak centered at 0 kHz with a linewidth of 1.9 kHz, indicating increased side-chain motion and solvent exposure relative to the spectra of the other Ala residues. Val-50 and Leu-62 showed Pake patterns, with quadrupolar splittings of 3.5 kHz and 3.7 kHz, respectively, intermediate to Ala-53/Ala-56 and Ala-63. This indicates partial motional averaging and supports a model with the Val and Leu residues embedded inside the lipid bilayer. Solid-state NMR spectroscopy performed on the 2H-labeled Ala-56 TM-A peptide incorporated into magnetically aligned phospholipid bilayers indicated that the peptide is tilted 8° with respect to the membrane normal of the lipid bilayer. Snorkeling and anchoring interactions of Arg-44 and Tyr-60, respectively, with the polar region or polar hydrophobic interface of the lipid bilayer are suggested as control elements for insertional depth and orientation of the helix in the lipid matrix. Thus, this study defines the location of key residues in TM-A with respect to the lipid bilayer, describes the conformation of TM-A in a biomembrane mimic, presents a peptide-bilayer model useful in the consideration of local protein folding in the microsomal desaturases, and presents a model of arginine and tyrosine control of transmembrane protein stability and insertion.
INTRODUCTION
Dehydrogenation performed by the fatty acid desaturases is critical for the modulation of membrane fluidity, which impacts membrane protein function, thermal adaptation, organelle integrity, the formation of signaling molecules, and insect defense in plants, animals, and fungi (1–4). Substantial diversity in oxidative function has been reported among the diverged desaturases homologs, particularly among members that regioselectively catalyze reactions at or near carbon-12. Δ12-desaturase homologs create a variety of functional groups among the higher plants seed oils, including both (E)- and (Z)-alkenes, acetylenes, epoxides, alcohols, and conjugated alkenes (5–8). Industrial oils, enriched in γ-linolenic acid, docosahexenoic acid, or unusual fatty acids may be produced in transgenic plants engineered to include these enzymes substituting for or even replacing natural sources (9–11). In particular, genes cloned from plant species that are not widely cultivated may allow access to lipids or lipid compositions that may provide dietary health benefits or be useful as nutritional supplements (9,12). In addition to the above uses, these fatty acids have potential for fine chemical feedstocks useful to the polymer and specialty chemical industries, providing an alternative to petrochemicals (7,13–15). To fully control the biosynthesis and accumulation of unsaturated seed oils, an improved structural and functional model of the desaturases and related enzymes is needed.
CREP-1 is a functionally diverged microsomal Δ12 desaturase that possesses Δ12 acetylenase (acetylenic bond-forming) activity. CREP-1 activity yields an acyl chain composition in Crepis alpina seed storage lipids dominated by crepenynate ((Z)-9-octadecen-12-ynoate) (7). This enzymatic activity corresponds to the first committed step in the metabolic pathway to most fatty acid-derived acetylenic secondary natural products ((16,17); B. J. Blacklock and R. E. Minto, unpublished results). Recently, further examples of Asteraceae acetylenases and a second set of phylogenetically distinct acetylenases have been isolated from the plant families Apiaceae and Araliaceae, which in certain cases have been shown to be upregulated in plants in response to fungal elicitation and are believed to be involved in the defensive phytoalexin response pathway (3,17).
Structural characterization of CREP-1 is valuable for the design of an experimental model of the microsomal Δ12 desaturase homologs. There is currently no detailed structure-function relationship at atomic resolution for any microsomal desaturase; these enzymes are membrane bound and have low apparent stabilities, therefore are difficult to solubilize and purify (16,18–20). The desaturases and their diverged relatives typically use acyl phosphatidylcholine or enzyme substrates and contain three functionally required iron-binding histidine-rich motifs with conserved spacing (21). The locations of the N-termini of two plant microsomal desaturases, FAD2 and FAD3, have been shown by immunofluorescence microscopy in transgenic tobacco (22).
All published studies are consistent with the diverged desaturases having similar protein folds. The collective TM domains are thought to organize the cytosolic domains. The location of the TM domains and the iron-coordinating sites are particularly relevant because the hydrophobic substrates and products will likely be transferred from the membrane. The TM domains have been proposed to play a direct role in chain length selectivity in other desaturases (23). The TM domains may also control interaction between the desaturases and the amphipathic auxiliary proteins: NADH:cytochrome b5 reductase and cytochrome b5. It is for these reasons that we have defined regions of CREP-1 that will serve as autonomous folding domains and outline the local fold of other similar enzymes (24). CREP-1 is predicted to encode a microsomal enzyme that contains four transmembrane helices—TM-A through TM-D—and three essential histidine boxes (25). A pictorial representation of CREP-1 and TM-A can be found in a previous study (25). By analyzing the structure and modes of peptide interaction, the purpose of TM-A (the autonomously folding domain of CREP-1 studied herein) may be further illuminated.
Obtaining crystals, needed for x-ray crystallography, of membrane-bound proteins like CREP-1 is extremely difficult due to the very hydrophobic nature of the proteins (26,27). One alternative for studying membrane-bound proteins is solid-state NMR spectroscopy. 2H NMR spectroscopic methods can be used to investigate the dynamic properties of synthetic hydrophobic peptides at specific residues and, therefore, determine their location with respect to the lipid bilayer (28–40). By selectively labeling amino acids with 2H, the locations and dynamic properties of side chains can be drawn from the results. NMR results for the amino-proximate transmembrane region (TM-A) are discussed here, which is the first of the CREP-1 folds to be studied.
2H solid-state NMR experiments will reveal the dynamic properties of many residues along the length of TM-A. This information will be analyzed to complete a model of CREP-1 TM-A inside a membrane including a discussion of insertion depth, tilt, and a hypothetical mode of stabilization inside a bilayer through arginine snorkeling and Phe-Tyr-Phe anchoring of TM-A to the bilayer. Most importantly, the mechanism discussed here that stabilizes the peptide in the bilayer can be used as a model for other proteins secured in similar manners.
MATERIALS AND METHODS
Materials
DMPC (1,2-dimyristoyl-sn-glycero-3-phosphatidylcholine), DHPC (1,2-dihexanoyl-sn-glycero-3-phophatidylcholine), and POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphatidylcholine) were purchased from Avanti Polar Lipids (Alabaster, AL). All phospholipids were dissolved in chloroform and stored at −20°C before use. HEPES, YbCl3 (ytterbium(III) hexahydrate), and TFE (2,2,2-trifluoroethanol) were purchased from Sigma-Aldrich (St. Louis, MO). NaCl and EDTA disodium salt were purchased from Fisher (Pittsburg, PA). Deuterium-depleted water, deuterated L-alanine ([d3]Ala), deuterated L-leucine ([d3]Leu), and perdeuterated L-valine ([d8]Val) were purchased from Isotec (Miamisburg, OH). Peptides were synthesized and purified as previously stated (25).
Sample preparation
The DMPC-rich bilayer samples, containing 6 mol % peptide to phospholipid, were made following the protocol in Rigby et al. (41) with the following changes. DMPC was dissolved in chloroform and added to a 12 × 75-mm test tube. The chloroform was removed by heating the sample in a 50°C aluminum block under nitrogen gas. The tube was placed under vacuum overnight to remove residual chloroform. The deuterated peptide (20 mg) was added to the tube, and an 80% TFE/water solution (4 mL) was used to dissolve both the peptide and the DMPC (76 mg). The TFE solution was evaporated using a heated aluminum block and stream of nitrogen gas and was then placed under vacuum overnight. The peptide/lipid mixture was resuspended in 380 μL HEPES buffer (5 mM EDTA, 20 mM NaCl, and 30 mM HEPES, pH 7.0) by heating in a water bath at ∼50°C, along with very slight sample agitation to avoid frothing the mixture. After all the phospholipids were fully dissolved, the sample was transferred to a flat-bottom 21-mm NMR tube (5 mm outer diameter) using a syringe and needle.
The 2H bicelle sample was prepared using DMPC and DHPC phospholipids at a q ratio of 3.5. The q ratio represents the mole ratio of the long chain phospholipid (DMPC) to the short chain phospholipid (DHPC). DMPC (72 mg) was placed into a pear-shaped flask and DHPC (15 mg) was added to a second pear-shaped flask. Deuterium-labeled Ala-56 TM-A (2 mg) was dissolved in TFE (200 μL) and added to the second flask. Both flasks were concentrated to dryness using a rotary evaporator and placed under vacuum overnight. HEPES buffer, pH 7.0 (270 μL, 100 mM), made from deuterium-depleted water was added to the second flask. The flask was vortexed, sonicated, frozen, and thawed as necessary to homogenize the sample. The solubilized sample was then transferred to the first flask, and the same steps were repeated until the sample was translucent. The sample was then transferred to a flat-bottom 21-mm NMR tube (5 mm outer diameter) using a Pasteur pipette. Ytterbium(III) chloride, prepared with deuterium-depleted water, was added to 5 mol % with respect to DMPC for experiments in which the bilayer normal of the phospholipids was aligned parallel to the static magnetic field.
Solid-state NMR spectroscopy
A modified Bruker AVANCE 7.05 T narrow bore 300/54 magnet equipped to conduct high-power solid-state NMR studies was used in all solid-state NMR experiments. The solid-state 2H NMR spectra were collected using a static double-tuned 5-mm round-coil solid-state NMR probe purchased from Doty Scientific (Columbia, SC). The solid-state 31P NMR spectra were collected using a standard 5-mm broadband solution NMR probe purchased from Bruker. The resonance frequencies used for 2H and 31P were 46.07 MHz and 121.5 MHz, respectively. The 2H NMR spectra were recorded using a standard quadrupole-echo pulse sequence (3.0-μs 90° pulses, 45-μs interpulse delay, 5.12-ms acquisition time, and a 0.4-s recycle delay). The 31P NMR spectra were recorded with 1H decoupling using a 7.0-μs 90° pulse for 31P and a 3.5-s recycle delay. For the 2H NMR data, 245,700 scans were taken and the free induction decay was processed using 200 Hz of exponential line broadening. For the 31P NMR data, 2048 scans were signal averaged. The aligned 2H bicelle data were obtained by signal averaging 491,400 scans utilizing a standard quadrupole-echo pulse sequence and 300 Hz of line broadening. For the 2 mol % peptide sample, 491,520 scans were acquired. The spectral width was set to 60.06 kHz for 2H and 150.20 ppm for 31P NMR. H3PO4 (85%) was used to reference all 31P chemical shifts. All NMR data were processed on a Power Macintosh computer using Igor Pro 3.12 and MacNUTS (Acorn NMR, Livermore, CA) software.
NMR data analysis
Deuterium NMR lineshapes were simulated using two different models. The first model employed the MXQET software program (42) to replicate tetrahedral three-site trideuteromethyl group hopping in which the deuterons were tilted 75° with respect to the rotation axis and with jumps of 120° at rates indicated in text. The second model used the MXQET program to create a second independent axis in which the former three-site axis makes two-site jumps of 109.5° and are tilted by 75°. The three-site jumping model simulates the lone trideuteromethyl group rotation, and the two-site jumping model simulates reorientation of the long backbone of leucine between its predominant rotamers. Although this does not simulate the predominant rotamers of valine, it simulates motions that a motionally unrestricted valine would be allowed due to the flexibility of its backbone. The asymmetry parameter (η) was 0.05 for all of the 2H NMR simulations (43).
In randomly oriented bilayer spectra, a characteristic 2H “Pake” powder pattern is expected to result from the rapid, axially symmetric rotation of deuterated side-chain molecules in a membrane perpendicular to the normal of the bilayer. Rotational motion around the C-CD3 bond yields a single Pake doublet for the deuterated methyl groups. The greater the motion of the probe, the greater the extent of motional averaging that occurs. Thus, a narrowing of the Pake powder pattern and lineshapes is observed for side chains when they do not have restricted motions. The magnitude of the quadrupolar splittings and lineshapes gives an indication of where the residue with the probe is located with respect to the lipid bilayer. Residues located outside the bilayer will be more motionally averaged and yield a smaller quadrupolar splitting which may resemble a broad isotropic peak. However, residues buried inside the phospholipid bilayer will experience less motional averaging and give rise to a broad Pake pattern, consisting of a larger quadrupolar splitting with the broadness determined by the degree of motional averaging experienced by the CD3 group. If the 2H3-methyl probe is immobile in a randomly dispersed sample, the resultant spectrum will consist of a Pake pattern with a 40-kHz quadrupolar splitting (41).
Although the lineshape of the methyl group must be simulated and its overall width is dependent upon its motion, the magnitude of the quadrupolar splitting at the most intense spots is also slightly influenced by the angle of the residue inside the bilayer (44). The corresponding 2H quadrupole splitting for aligned bilayers can be related to the motional characteristics of the deuterated methyl group using the following equation:
 |
(1) |
where e2Qq/h is the nuclear quadrupole coupling constant (165–170 kHz for a C-D bond) (28,30,31), Smol is the molecular order parameter (assuming axially symmetric order) describing orientational fluctuations of the C-D bond relative to the bilayer normal, and θi is the average orientation of each C-D bond relative to the bilayer normal.
The angle θi is a very important quantity and can be used to determine the peptide tilt within a bilayer. A derivation described thoroughly by Strandberg et al. (45) provides an easy way to relate α-helix geometries inside a lipid bilayer to the splitting observed for different residues experiencing environments that are dependant upon their tilt in the bilayer:
 |
(2) |
where Δvq is the quadrupolar splitting, S is the order parameter taking into account the molecular motion, e2qQ/h is the quadrupolar coupling constant, ɛ‖ is the angle between the helical axis and the alanine methyl group, ɛperp is the constant angle between the methyl group axis and a line coming from the helical axis when looking from the top of the helix down, ρ is an arbitrary value that quantifies the rotation of the peptide around its helical axis, φ is the angle of rotation between a reference amino acid and another amino acid (φ = (n − 1) × 100), and τ is the angle of the helical axis and the membrane normal (see Fig. 1 for reference). An alanine methyl group has a reported value for ɛperp of –43.3° and for e2qQ/h of 167 kHz for deuterons, but motional averaging caused by the three methyl deuteron rotation reduces this constant to 56 kHz. S is composed of the common order parameter of 0.875 multiplied by a parameter accounting for wobble the bicelles experience as compared to the stable glass plates used to determine the 0.875 equal to 0.84 (45,46).
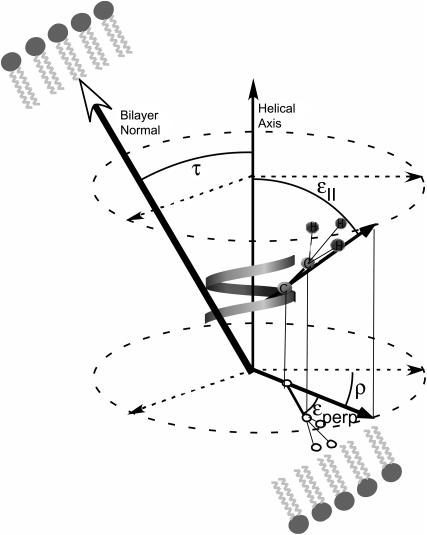
FIGURE 1.
Diagram explaining how the helical tilt was determined in a [d3]Ala peptide. ɛ‖ is the angle formed by the vector originating from the helical axis that intersects the Cα-Cβ alanine bond at the helix backbone, and the helical axis, ɛperp, is the angle between the Cα-Cβ alanine bond and the vector from the helical axis to the helix backbone as seen in the planar projection of the bond and vector. ρ is the angle of rotation around the helical axis as seen in the planar projection of the vector from the helical axis to the alanine residue to the arbitrary axis in the plane. τ is the tilt angle of the peptide which is shown between the helical axis and the bilayer normal vectors.
RESULTS AND DISCUSSION
In this study, there are three key pieces of information that are derived directly from the 2H solid-state NMR measurements: the depth of insertion, the possible creation of oligomers or peptide aggregates, and the tilt of TM-A CREP-1 peptide in a lipid bilayer. The lineshape comparisons between the randomly oriented bilayer sample spectra generated with different labeled amino acids bracket the insertion depth for the peptide. Temperature trends and the observed broad shoulders in spectra of the randomly oriented samples are used to posit the existence of oligomeric peptides. Finally, the use of the oriented bilayer sample augmented with the randomly oriented data allows for the estimation of the CREP-1 peptide tilt with respect to the lipid bilayer normal. Taking all of these discoveries under consideration as well as the experimental evidence that the TM-A peptide does not insert properly into a POPC bilayer, it is shown that CREP-1 is held in the bilayer specifically through “snorkeling” and “anchoring” interactions (47). We begin with a brief general discussion about the sequence of TM-A and its insertion into lipid bilayers.
General
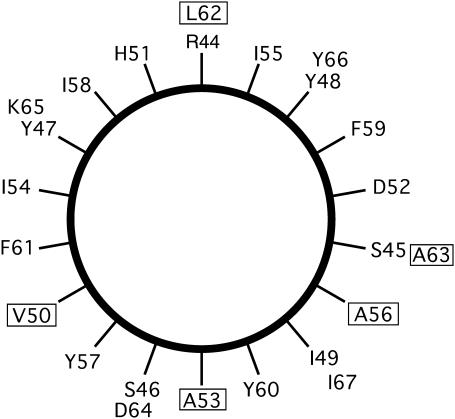
The amino acid sequence for TM-A consists of 24 amino acids and is shown in Fig. 2. This includes the first putative transmembrane region of CREP-1, consisting of residues I-49 to I-67, based upon secondary structure and transmembrane helix predictions (48–51). The TM-A peptide used in these studies included residues R-44 to I-67, which contains residues at the N-terminal end that are expected to be exposed to the cytosol (25). The boldfaced and underlined amino acids shown in Fig. 2 represent the residues selected for isotopic labeling with 2H. These residues include Ala-53, Ala-56, Ala-63, and Leu-62 with deuterated side-chain methyl groups (-CD3) groups and Val-50, which possessed a perdeuterated alkyl group (-CDCD(CD3)2). The locations of these residues in a helical wheel representation are shown in Fig. 3.
FIGURE 2.
Amino acid sequence of CREP-1 TM-A, where each amino acid is symbolized by its single-letter code. Peptides were synthesized that contained a single deuterated residue. Labeling locations are shown by the bold, underlined letters.
FIGURE 3.
Helical wheel projection of the TM-A CREP1 peptide, with the first amino acid synthesized at the top. The deuterated residues are marked with a box.
31P NMR spectra of DMPC-rich phospholipid bilayers containing the TM-A peptide and a control sample consisting of only DMPC-rich phospholipid bilayers were taken as a function of temperature (25–45°C) (data not shown). The spectra from samples prepared with 6 mol % TM-A peptide with respect to DMPC and DMPC without the peptide consisted of axially symmetric powder patterns, typical of unaligned DMPC phospholipid bilayers in the Lα phase. The 31P NMR spectra of the phospholipid bilayers incorporated with the TM-A peptides were not significantly different in shape when compared to DMPC-rich phospholipid bilayers prepared in the absence of TM-A. This indicated that the TM-A peptide does not significantly disrupt the phospholipid bilayers upon incorporation at these concentrations.
Depth of insertion
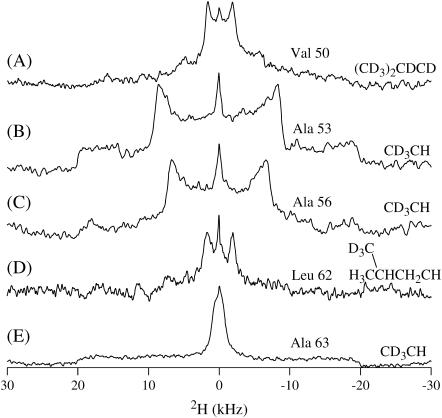
To gain a qualitative view of how deeply the first transmembrane domain of CREP-1 inserted into the lipid bilayers, the lineshapes of 2H-labeled CD3 Ala-53 and Ala-56 were compared to those of the unoriented Ala-63. Additionally, the lineshape of unoriented Leu-62 was compared to the lineshape of unoriented Val-50. Fig. 4 displays all of the spectra for specifically labeled TM-A inserted into randomly oriented DMPC bilayers at a single temperature (45°C) for comparison. Fig. 5, A–C, provides lineshape simulations to be able to discuss accurately the dynamics exhibited by all side chains.
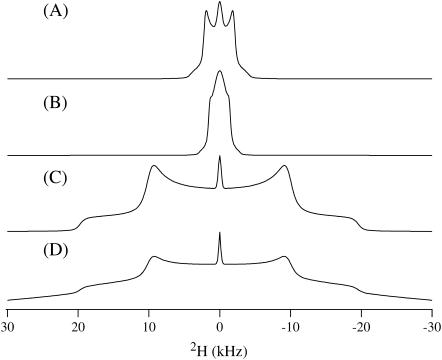
FIGURE 4.
2H solid-state NMR spectra of a DMPC bilayer incorporated with 6 mol % deuterated TM-A at 45°C. Labeling sites were (A) Val-50, (B) Ala-53, (C) Ala-56, (D) Leu-62, and (E) Ala-63.
FIGURE 5.
Simulated spectra of (A) Val-50 and Leu-62, (B) Ala-63, (C) Ala-56 and Ala-53, and (D) low temperature Ala-56 and Ala-53. Simulation parameters are described in the text.
First, Fig. 4 C shows the solid-state 2H NMR spectra from the [d3]Ala-56 TM-A peptide incorporated into DMPC-rich phospholipid bilayers. NMR spectra for this sample and all subsequent samples were collected over a temperature range from 25°C to 55°C. A quadrupolar splitting of 13.3 kHz was present in all of the spectra. At higher temperatures, this component diminished and the 13.3-kHz Pake pattern increased in intensity. The small isotropic peak centered at 0 kHz, seen in Fig. 4, was due to the rapid motional averaging of residual HOD in the deuterium-depleted water used to prepare the samples. The 2H solid-state NMR spectrum from the [d3]Ala-53 TM-A peptide incorporated into DMPC-rich phospholipid bilayers is displayed in Fig. 4 B. The resultant quadrupolar splitting of 16.9 kHz indicates that this residue is located in a slightly different environment then Ala-56.
Ala-53 and Ala-56 (Fig. 4, B and C) have nearly the same lineshape, which is accurately simulated by assuming that the deuterated methyl group of the alanine residue rotates along the Cα-Cβ bond and allows the deuterons to make jumps between three sites described by tetrahedral geometry (52). This simulation required a frequency of rotation of 1 × 107 Hz. The reduced quadrupolar splitting of ∼10 kHz in these spectra is evidence of that increased motion (Fig. 5 C). The typical Pake doublet pattern of an unoriented sample (a powder spectrum) for a methyl group would have a splitting of ∼40 kHz, but experimental splittings of this size only occur when all motion of the deuterons has nearly been frozen (typically at temperatures of −100°C) (52–54). There is a drastic change of the lineshape farther along the peptide helix as seen in the 2H solid-state NMR spectrum of the [d3]Ala-63 TM-A peptide incorporated into DMPC-rich phospholipid bilayers (Fig. 4 E). The 2H NMR linewidth was measured to be 1.9 kHz for [d3]Ala-63 TM-A. To simulate this motion, a very fast frequency of >1 × 108 Hz three-site jumping and an additional two-site reorientation frequency of the alanine residue of >1 × 107 Hz was used with an isotropic peak (52–55) (Fig. 5 B). In addition to the lineshape simulation needing high frequencies of two axes of rotation, the quadrupolar splitting is extremely decreased and is almost not discernible outside of the breadth of the small isotropic peak. This indicates greatly increased motion allowing the possible existence of more modes of rotation that affect the lineshape in ways not seen because of the isotropic peak. To get such a small quadrupolar splitting, there needs to be more than just the two rotations simulated although two axes are an adequate means to roughly describe the small, uncovered lineshape present outside of the isotropic component breadth. It should also be noted that alanine motions other than methyl group rotation have been observed to have a large energy barrier in the crystal structure of alanine; thus, there must be a significant reason Ala-63 rotates so freely (52).
Before the true significance of this unusual spectrum can be stated, the positioning of the residues around the helix must be mentioned (Fig. 3). It is very possible for alanine residues on opposite sides of a helix to be exposed to radically different chemical environments and thus exhibit very different lineshapes dependent upon whether they are facing the hydrophobic core of the bilayer (45). But if an ideal α-helix is assumed of 3.6 amino acids per turn, Ala-63 is aligned only 20° away from Ala-56 and, thus, should have nearly the same lineshape and splitting if both are in similar environments. Obviously then Ala-53 and Ala-56 are in a different environment than Ala-63. Because Ala-63 has much more motion it is most likely located outside of the lipid bilayer, whereas Ala-53 and Ala-56 are within the hydrophobic region of the bilayer experiencing restricted motions because of lipid acyl chain interactions and stabilization from hydrophobic interactions.
Further insertion information is gained by comparing the Val-50 and Leu-62 dynamics (Fig. 4, A and D, respectively). The 2H solid-state NMR spectrum from the [d3]Leu-62 TM-A peptide incorporated into DMPC-rich phospholipid bilayers is shown in Fig. 4 D. The [d3] probe was located on one of the two methyl side-chain groups of the leucine residue. The 3.7-kHz splitting was narrower than that for Ala-53 and Ala-56, yet wider than the splitting observed for Ala-63. The 2H solid-state NMR spectrum from the [d8]Val-50 TM-A peptide incorporated into DMPC-rich bilayers is displayed in Fig. 4 A. The deuterium labels are located on the two methyl groups at the end of the side chain together with the corresponding α and β carbons. Because the methyl groups are diastereotopic and not expected to be degenerate, one resolved doublet and one unresolved, smaller doublet was observed instead of a single doublet. As the temperature was increased, the definition of the peaks became clearer (data not shown).
Spectra examined over a range of temperatures are extremely similar for both of these residues and can both be simulated by the same model of motion including the three-site jumps of the methyl deuterons and the two-site reorientation of methyl groups at frequencies of 1 × 107 Hz and 1 × 106 Hz, respectively (Fig. 5 A) (41,56–58). There are two main differences in the spectra of Leu-62 and Val-50 which arise from the fact that Val-50 is completely deuterated and Leu-62 is only deuterated at one methyl group. The Val-50 spectrum is expected to have a peak at ∼±10 kHz on a 300 MHz NMR, which corresponds to the CβD that moves slower than the methyl deuterons. The CαD is not clearly visible at current signal/noise levels and it is not further addressed in this study. The second difference is the apparent “filling in” of the two peaks of Val-50 as compared to Leu-62. This arises from the presence of a second Pake doublet with very small quadrupolar splitting from one of the two inequivalent deuterated valine methyl groups superimposed on the isotropic peak observed as peak broadening (41,57,58). The 3.8-kHz quadrupolar splitting of the valine undeniably places Val-50 within the lipid bilayer because valine residues outside of the lipid bilayer exhibit almost isotropic peaks (57). Increased flexibility and freedom of motion are expected as the leucine residue has a longer side chain. However, the simulation of Leu-62 uses nearly the same frequencies of rotation and quadrupolar splitting as the valine residue, so the leucine residue must be in an environment with strongly restricted motion. Leu-62 is most likely not in the hydrophobic region but in the lipid-water interface. Tiburu and co-workers (59) have shown that leucine residues of transmembrane α-helices near or in the lipid headgroup experience restricted motions. In that study, a methyl-deuterated leucine residue was near the hydrophobic region and had a quadrupolar splitting of ∼6 kHz with a lineshape similar to TM-A Leu-62 that was attributed to motional hindrances from the phospholipid headgroups. The slightly smaller quadrupolar splitting of Leu-62 than the earlier-reported system perhaps indicates that Leu-62 is nearer the edge of the lipid-water interface, allowing marginally increased motion. This fits well with the conclusions gathered from the alanine analysis because if Leu-62 is close to the edge of the lipid-water interface, Ala-63 would be outside the bilayer as indicated by its increased motion.
It is useful to describe the length of the protein inside the hydrophobic region now that the insertion depth has been studied. If the TM-A peptide was 100% α-helical, it would be a total of 36 Å long as determined by the equation
 |
(3) |
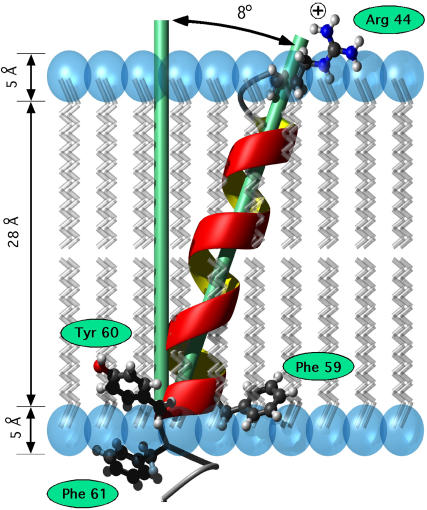
where p is the pitch (5.4 Å), AA is the number of residues of the peptide (24), and n is the number of residues per turn (60). However, we know that Leu-62 is the residue at the edge of the bilayer, thus maximally 19 residues, corresponding to a length of ∼28.5 Å, are inside the bilayer. Phospholipid headgroups occupy the outer ∼5 Å region of the bilayer, which means that there are ∼16 residues within the hydrophobic region if the N-terminal, Arg-44 residue stops before entering the other headgroup region (61). Additionally it should be noted that with this current model, at the interface between the hydrophobic region and the headgroup region, there is the pattern of F-Y-F amino acids.
Oligomerization of the peptide
There are two more spectral features that appear in the unoriented powder spectra that lead to very interesting conclusions. The first feature is the presence of very broad shoulders located at ±20 kHz independent of the shoulder components in Fig. 4, B and C, which are part of their deuterium lineshape simulation (41,58). This is most clearly seen in Fig. 4 E where the splitting Ala-63 is confined to the center region. As temperature increases, the shape of these broad shoulders does not change, but their intensity decreases (data not shown). As described repeatedly in the literature, this phenomenon can be attributed to small concentrations of peptide aggregates (41,62). It is common to see this spectral feature at high peptide concentrations (4–6%), and it is also common for it to decrease with increased temperature (41,62). The broad lineshape must be composed of many different rates of methyl group motion, implying that the peptides are interacting in many nonspecific and unstructured ways such as would occur in randomly assembled aggregates. The fact that the broad shoulder lineshapes stay constant as the temperature increases arises because no specific motional restriction is favored inside a randomly assembled aggregate. It is most likely that these aggregates are formed on the periphery of the lipid bilayer because of the excess amount of protein used to create the samples. Ala-53 and Ala-56 have nearly the same lineshape, but their shoulders at ±20 kHz have differing heights due to another source of signal that lies underneath the powder pattern. This other broad shoulder source of signal differs in intensity in Ala-53 and Ala-56 (Fig. 3, B and C, respectively). Because the aggregates may be outside the bilayer and are dependent upon peptide concentration, it is very conceivable that sample preparation could create slightly different amounts of aggregates and affect the height of the broad shoulders when comparing different samples.
The second important feature is the broadening of all spectra at 25°C (an example of which is found in the Supplementary Materials). For Ala-56, a simulation is provided in Fig. 5 D of the normal Ala-56 lineshape superimposed on a broad isotropic peak. A very similar broadening of 2H spectra has been described by Jones et al. (57) and is attributed to peptide oligomerization within the bilayer.
There arises a second explanation for these heterogeneous effects if phase separation occurs within the lipid bilayer (59,63). Phase separation results in the simultaneous presence of two or more phospholipid populations. If this is the case, TM-A could form several distinct populations, one in an environment that allows for more peptide-peptide interactions (44–47,55,59). Multiphasic samples are likely because the DMPC bilayer has extensive heterogeneity only very near to its lipid transition temperature of 23°C. It is entirely possible that not all of the lipids have transitioned into the Lα phase at that low temperature, especially if the peptide minimizes its energy by oligomerizing and forcing the lipid acyl chains to remain ordered in the gel phase. In summary, we postulate that two different types of peptide-peptide interactions exist within this DMPC bilayer system: aggregates localized outside the lipid bilayer and affected by sample preparation, and oligomers within the bilayer generated by phase separation at 25°C.
Tilt determination
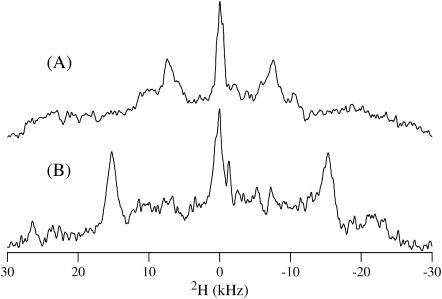
To determine the orientation of TM-A with respect to the bilayer, the peptide was aligned in the magnetic field. Fig. 6 A shows the 2H solid-state NMR spectrum for the [d3]Ala-56 TM-A peptide incorporated into DMPC/DHPC magnetically aligned phospholipid bilayers known as bicelles. Under these conditions, the phospholipid bilayers were aligned such that the membrane normal was perpendicular (no lanthanide) to the static magnetic field. Fig. 4 B represents a 2H solid-state NMR spectrum taken from the same sample except that Yb3+ had been added to flip the bicelles 90° such that the bilayer normal reoriented collinear with the direction of the static magnetic field. Upon addition of Yb3+ to the bicelles, the quadrupolar splitting doubled from 15 kHz to 30 kHz. The doubling of the quadrupolar splitting has been observed when the phospholipids' acyl chains are deuterated in magnetically aligned phospholipid bilayers utilizing DMPCd54 (54–67). This doubling of the quadrupolar splitting indicates that the TM-A peptide rotates rapidly about the bilayer normal (45). The 2H NMR spectra and lineshape arising from the aligned phospholipid bilayers (Fig. 6) differ dramatically from the unoriented DMPC-rich phospholipid bilayers observed in Fig. 4 C but have intense peaks at about the same point as the edges of the Pake-like patterns of the powder spectra.
FIGURE 6.
2H solid-state NMR spectra of a DMPC/DHPC bicelle samples containing 2 mg TM-A deuterated at Ala-56 acquired at 38°C without (A) and with (B) ytterbium ions. The quadrupolar splitting approximately doubles when comparing the sample with the bilayer normal perpendicular to the magnetic field normal (A) versus the sample with the bilayer normal parallel to the magnetic field normal (B).
We know that the protein does not rotate on its helical axis because then all residues in the bilayer would be averaged to the same value, which is not the case between Ala-53 and Ala-56 (44–46,55). However, in the aligned samples of Ala-56 in Fig. 6 there is exactly a doubling of the quadrupolar splitting, indicating that the protein rotates quickly around the bilayer normal. If the protein rotates around the bilayer normal that is distinct from its helical axis, then the peptide is not collinear with the bilayer and must be tilted (44). To quantify the tilt angle, we used the data from the aligned spectrum of Ala-56 (Fig. 6) that, within quadrupolar splitting measurement error, corresponded to the most intense peaks of the powder pattern. This is no coincidence because the most intense peaks are those that are perpendicularly aligned in the unoriented powder spectrum (44). So, with only minimal loss of accuracy, we can use the unaligned bilayer splitting of Ala-53 between its most intense peaks as a close estimate of the aligned Ala-53 splitting (Fig. 4 B). Using Insight II to quantify ɛ‖ to be between 55° and 60° and using a literature value of ɛperp for alanine of 43.3°, we were able to use a C++ program to acquire a τ-value of 8° ± 4° and a ρ-value of either 160° or 372° (45,46). It is of limited importance to know the value of ρ because it has no practical significance and is only useful to obtain the tilt angle (19). There is some error in this analysis because of the number of data points limited by the number of alanines on the transmembrane segment of the TM-A protein as well as the inaccuracy in predicting the quadrupolar splitting of an Ala-53 oriented sample. This 8° tilt has a minimal effect on the length of the hydrophobic region and decreases the 28.5 Å hydrophobic length to ∼28 Å.
Insertion dynamics
To combine all the data described above, two recently discovered protein interactions with lipid bilayers must be presented: snorkeling of lysine and arginine, and anchoring of tryptophan residues (47). Snorkeling occurs when a long positively charged side chain is near the lipid-water interface of a lipid bilayer. The polar positively charged amino acid such as lysine will stretch and put its polar charged segment into the polar lipid-water interface while keeping its hydrophobic backbone Cα carbon buried with the hydrophobic region (47,68–70). In this manner, the protein is able to stay at a specific location within the bilayer and is stabilized by this attachment. This snorkeling can even allow a lysine residue (slightly longer than an Arg residue) to stretch 3–4 Å to properly align itself (55). The second method of interaction is very similar except that it uses the aromatic properties of residues such as tryptophan to anchor its aromatic ring within the polar region and its backbone within the hydrophobic region, thereby securing it in the bilayer (44,47,69,71). This anchoring preferentially secures on the C-terminal region of a protein as determined by statistical analysis of transmembrane proteins (69). To date, tryptophan and lysine have been almost exclusively studied, but the similar properties of Tyr and perhaps Phe to Trp and Arg to Lys have resulted in suggestions of similar roles as anchors and snorkels, respectively (69,72).
It is hypothesized that in the transmembrane segment A of CREP-1, Arg-44 at the N-terminus snorkels into the polar headgroup and Tyr-60 at the C-terminal region, flanked on either side by phenylalanine anchors to the bilayer. This is very similar to the mattress model proposed by Mouritsen et al. (62). To show support for this hypothesis, the reanalysis of all of the former conclusions must be put into this new context beginning with the peptide insertion. The position of Leu-62 one amino acid into the lipid-water interface places Tyr-60 directly in the correct spot to be an anchor between the hydrophobic core and the polar headgroup region. Arg-44 is then 16 amino acids away and very near to the other polar region. With its ability to stretch, it can firmly insert into the polar region. To understand the tilt and oligomer formation, we must first make the following observation: there are 16–17 amino acids in the hydrophobic core, which only has a perpendicular capacity in DMPC of 15 amino acids (23 Å in the fluid phase) (61).
Hydrophobic mismatch occurs when the length of the hydrophobic region of a protein does not match that of the hydrophobic length of the bilayer (44,47). It is logical that to properly insert in the bilayer, a few extra residues could be “squeezed in” to accommodate the specific positioning of Arg-44 and Tyr-60, providing more evidence for the importance of these residues as the methods for protein stabilization in the bilayer. More importantly though, this provides a reason the protein is tilted nearly 10°. It is quite common for proteins to tilt to match their length with the length of the hydrophobic bilayer (47,68). But a second possibility exists because there is no evidence to restrict arginine or tyrosine from anchoring at slightly variable depths (1–2 Å), thereby compensating for the small hydrophobic mismatch directly without tilting. It has also been shown that an 8° tilt is required for the WALP protein to properly anchor the tryptophan residues of the alanine-tryptophan-alanine motif (44). Thus, the tilt of the TM-A peptide in DMPC could be due to the need for the tyrosine residue to anchor the peptide.
It is proposed that Arg-44 and Tyr-60 residues need to be in an exact position that creates hydrophobic mismatch that requires the peptide to tilt (Fig. 7). To explore this, a reanalysis of the conclusion made previously is needed: that there are oligomers formed because of phase separation. Studies performed on tryptophan- and phenylalanine-anchored peptides that had hydrophobic mismatch showed that the peptides oligomerized in a phase-separated state and gathered in striated arrays often between the boundaries of the lipid phases (47,72). Similar studies on lysine-anchored peptides demonstrated that they also oligomerized in phase-separated lipids but existed farther away from each other in disordered arrays due to the electrostatic repulsion (47,72). There is very qualitative evidence in the powder spectra at 25°C to show that these two oligomer formation patterns are followed, thus lending support to the hydrophobic mismatch conclusion. To form a striated array of proteins that do not interact at the charged ends, a pattern of α-helices might arise in which Arg-44 is pointing north then south repetitively to maximize the distance apart. The residues at the east and west positions in Fig. 3 would then interact most strongly. In other words, to create a line of peptide chains in an oligomeric-type formation, residues at nearly the same vertical position of Val-50 and Ala-56 will interact most heavily to minimize unfavorable charged side-chain interactions. It can be noticed by comparing powder spectra that the effects of broadening powder lineshapes are most pronounced in Ala-56 and Val-50 (data not shown). This supports the statement that east and west positions in Fig. 3 interact most heavily and that hydrophobic mismatch is a significant cause of the peptide tilt.
FIGURE 7.
Model for CREP-1 TM-A in a membrane bilayer based on 2H solid-state NMR spectra of DMPC bilayers infiltrated by TM-A peptides. Residues are labeled assuming a rigid α-helix with 3.6 residues per turn.
One powerful piece of support for Arg-44 and Tyr-60 being anchors is the fact that TM-A did not properly insert into POPC bilayers. A homogenous lipid/peptide mixture was never achieved when TM-A was added to POPC, indicating that TM-A did not properly insert into the POPC liposomes/vesicles while being vortexed. This inability to homogenize in POPC but ability to homogenize in DMPC with the same preparation conditions is quite different from many hydrophobic proteins such as phospholamban, which homogenizes/incorporates indiscriminately in the very similar DMPC and POPC lipids ((59) and E. S. Karp, E. K. Tiburn, and G. A. Lorigan, unpublished results). The main difference between DMPC and POPC is that POPC has a hydrophobic core of ∼3 Å larger than DMPC. It is hypothesized that because of the need for such a specific spacing of the Arg-44 snorkel and the Tyr-60 anchor, the POPC bilayer was too long. It has been shown that placement of tryptophan residues in the middle of transmembrane proteins inhibits proper insertion of the proteins possibly due to the abortive anchoring of the tryptophan residue arresting peptide insertion (73,74). Perhaps in TM-A, Tyr-60 anchors first and then the Arg-44 residue cannot snorkel properly and the conformation and dynamics of the peptide are disturbed. So, TM-A's inability to insert in POPC is a very convincing sign supporting the employment of both anchoring and snorkeling by TM-A.
The results of the insertion and tilt of TM-A can also be pertinent to the biological environment in which it natively exists. In vivo, CREP-1 and other FAD2-like desaturases are located in the endoplasmic reticulum (ER) membrane (22). Plant ER membranes have been estimated to be 50–60 Å in thickness; however, the morphology and thickness of the biological membranes can be substantially convoluted by lipid heterogeneity and the presence of other membrane proteins (75). The extension of the TM-A peptide to incorporate additional cytosolic residues could be attempted to minimize perturbations at the water-lipid interface of thicker bilayers. Whereas DMPC worked best in this study, POPC could be a better phospholipid in future studies of longer desaturase transmembrane segments or pairs of TM domains (e.g., TM-A + TM-B) due to the closer physicochemical match to a typical ER membrane. In the CREP-1 holoenzyme, interactions resulting from the association of TM-A with one or more of the remaining transmembrane domains from the desaturase are likely germane to the precise positioning of the TM domain with the lipid bilayer, altering the match between the hydrophobic domain length and the bilayer thickness of pure phospholipids and the organization of the His box domains. The next step in CREP-1 research is to study the properties of CREP-1 TM-A in the presence of other TM sections to see if TM-A alters its positioning and incorporates into POPC or if it differs its orientation in DMPC. By performing this future study, an important feature of how autonomously folding peptides act as a unit and interact with each other to perform their necessary function may be gained.
In conclusion, 2H solid-state NMR data on a variety of amino acids on a transmembrane segment of CREP-1 were investigated. A slight tilt angle resulting from hydrophobic mismatch, oligomers resulting from phase separation, and motions of amino acids that are greatly different were all uncovered and reconciled to build a fascinating model on a membrane protein. Most interesting is evidence for the participation of Arg-44 as a snorkel and Tyr-60 as an anchor in attaching the transmembrane helix in a lipid bilayer. This new discovery about a model transmembrane protein will be very useful for further studies of peptide-lipid interactions.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
Acknowledgments
Rafal R. Toczylowski is acknowledged for assisting with the calculations to estimate the angle that the average C-D bond makes with the TM-A backbone. Dr. Paul Scholten is acknowledged for help with the MXQET program to create the lineshape simulations.
The NMR spectrometer was obtained though National Science Foundation Grant CHE9012532. E.S.K. acknowledges the support of the Arnold Beckman Foundation. Startup funds for R.E.M. and G.A.L. were provided by Miami University. G.A.L. acknowledges funding by the Petroleum Research Fund (ACS PRF 35352-G4), a National Science Foundation Career Award (CHE-0133433), and the National Institutes of Health (GM60259-01). R.E.M. gratefully acknowledges funding by the National Institutes of Health (R15 GM 069493-01A1) and the Ohio Plant Biotechnology Consortium.
References
- 1.McConn, M., R. A. Creelman, E. Bell, J. E. Mullet, and J. Browse. 1997. Jasmonate is essential for insect defense in Arabidopsis. Proc. Natl. Acad. Sci. USA. 94:5473–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cook, H. W. 1991. Fatty acid desaturation and chain elongation in eukaryotes. In Biochemistry of Lipids, Lipoproteins and Membranes. D. E. Vance and J. Vance, editors. Elsevier, New York. 141–169.
- 3.Kirsch, C., K. Halbrock, and I. E. Somssich. 1997. Rapid and transient induction of a parsley microsomal Δ12 fatty acid desaturase mRNA by fungal elicitor. Plant Physiol. 115:282–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singer, S. J. 1974. Molecular organization of membranes. Annu. Rev. Biochem. 43:805–833. [DOI] [PubMed] [Google Scholar]
- 5.Broun, P., J. Shanklin, E. Whittle, and C. Sommerville. 1998. Catalytic plasticity of fatty acid modification enzymes underlying chemical diversity of plant lipids. Science. 282:1315–1317. [DOI] [PubMed] [Google Scholar]
- 6.Cahoon, E. B., K. G. Ripp, S. E. Hall, and J. A. Kinney. 2000. Formation of conjugated Δ8, Δ10-double bonds by Δ12-oleic-acid desaturase-related enzymes. J. Biol. Chem. 276:2637–2643. [DOI] [PubMed] [Google Scholar]
- 7.Lee, M., M. Lenman, A. Banas, M. Bafor, S. Singh, M. Schweizer, R. Nilsson, C. Liljenber, A. Dahlqvist, P.-O. Gummeson, S. Sjodahl, A. Green, and S. Stymne. 1998. Identification of non-heme diiron proteins that catalyze triple bond and epoxy group formation. Science. 280:915–918. [DOI] [PubMed] [Google Scholar]
- 8.Dyer, J. M., D. C. Chapital, J.-C. W. Kuan, R. T. Mullen, C. Turner, T. A. McKeon, and A. B. Pepperman. 2002. Molecular analysis of a bifunctional fatty acid conjugase/desaturase from tung. Implications for the evolution of plant fatty acid diversity. Plant Physiol. 130:2027–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Napier, J. A., and L. V. Michaelson. 2001. Towards the production of pharmaceutical fatty acids in transgenic plants. J. Sci. Food Agric. 81:883–888. [Google Scholar]
- 10.Napier, J. A., L. V. Michaelson, and A. K. Stobart. 1999. Plant desaturases: harvesting the fat of the land. Curr. Opin. Plant Biol. 2:123–127. [DOI] [PubMed] [Google Scholar]
- 11.Moon, H., J. Hazebroek, and D. F. Hildebrand. 2000. Changes in fatty acid composition in plant tissues expressing a mammalian delta 9 desaturase. Lipids. 35:471–479. [DOI] [PubMed] [Google Scholar]
- 12.Horrobin, D. F. 1990. Gamma linolenic acid: an intermediate in essential fatty acid metabolism with potential as an ethical pharmaceutical and as a food. Rev. Contemp. Pharmaco. 1:1–45. [Google Scholar]
- 13.Hitz, W. 1999. Economic aspects of transgenic crops which produce novel products. Curr. Opin. Plant Biol. 2:135–138. [DOI] [PubMed] [Google Scholar]
- 14.Murphy, D. J. 1999. Production of novel oils in plants. Curr. Opin. Biotechnol. 10:175–180. [DOI] [PubMed] [Google Scholar]
- 15.Singh, S., S. Thomaeus, M. Lee, S. Stymne, and A. Green. 2001. Transgenic expression of a Δ12-epoxygenase gene in Arabidopsis seeds inhibits accumulation of linoleic acid. Planta. 212:872–879. [DOI] [PubMed] [Google Scholar]
- 16.Bohlmann, F., T. Burkhardt, and C. Zdero. 1973. Naturally Occurring Acetylenes. Academic Press, London.
- 17.Cahoon, E. B., J. A. Schnurr, E. A. Huffman, and R. E. Minto. 2003. Fungal responsive fatty acid acetylenases occur widely in evolutionarily distant plant families. Plant J. 34:671–683. [DOI] [PubMed] [Google Scholar]
- 18.Enoch, H. G., A. Catala, and P. Strittmatter. 1976. Mechanism of rat liver microsomal stearyl-CoA desaturase. J. Biol. Chem. 251:5095–5103. [PubMed] [Google Scholar]
- 19.Schmidt, H., and E. Heinz. 1993. Direct desaturation of intact galactolipids by a desaturase solubilized from spinach (Spinacia oleracea) chloroplast envelopes. Biochem. J. 289:777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galle, A.-M., A. Oursel, M. Joseph, and J.-C. Kader. 1997. Solubilization of membrane bound Δ12- and Δ6-fatty acid desaturases from borage seeds. Phytochemistry. 45:1587–1590. [Google Scholar]
- 21.Shanklin, J., and E. B. Cahoon. 1998. Desaturation and related modifications of fatty acids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49:611–641. [DOI] [PubMed] [Google Scholar]
- 22.Dyer, J. M., and R. T. Mullen. 2001. Immunocytological localization of two plant fatty acid desaturases in the endoplasmic reticulum. FEBS Lett. 494:44–47. [DOI] [PubMed] [Google Scholar]
- 23.Libisch, B., L. V. Michaelson, M. J. Lewis, P. R. Shewry, and J. A. Napier. 2000. Chimeras of Δ6-fatty acid and Δ8-sphingolipid desaturases. Biochem. Biophys. Res. Commun. 279:779–785. [DOI] [PubMed] [Google Scholar]
- 24.Pattus, F. 1990. Membrane protein structure. Curr. Opin. Cell Biol. 2:681–685. [DOI] [PubMed] [Google Scholar]
- 25.Minto, R. E., W. J. Gibbons Jr., T. B. Cardon, and G. A. Lorigan. 2002. Synthesis and conformational studies of a transmembrane domain from a diverged microsomal Δ12-desaturase. Anal. Biochem. 308:134–140. [DOI] [PubMed] [Google Scholar]
- 26.Doyle, D. A., J. M. Cabral, R. A. Pfuentzner, A. L. Kuo, J. M. Gulbis, S. L. Cohen, B. T. Chait, and R. MacKinnon. 1998. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 280:69–77. [DOI] [PubMed] [Google Scholar]
- 27.Chang, G., R. H. Spencer, A. T. Lee, M. T. Barclay, and D. C. Rees. 1998. Structure of the Mscl homolog from Mycobacterium tuberculosis. Science. 282:2220–2226. [DOI] [PubMed] [Google Scholar]
- 28.Seelig, J. 1977. Deuterium magnetic resonance: theory and applications to lipid membranes. Q. Rev. Biophys. 10:353–418. [DOI] [PubMed] [Google Scholar]
- 29.Davis, J. H. 1991. Deuterium nuclear magnetic resonance spectroscopy in partially ordered systems. In Isotopes in the Physical and Biomedical Sciences, Vol. 2: Isotopic Applications in NMR Studies. E. Buncel and J. R. Jones, editors. Elsevier Science Publishers, Amsterdam, The Netherlands. 99–157.
- 30.Davis, J. H. 1983. The description of membrane lipid conformation, order, and dynamics by 2H-NMR. Biochim. Biophys. Acta. 737:117–171. [DOI] [PubMed] [Google Scholar]
- 31.Smith, I. C. P. 1984. Conformational and motional properties of lipids in biological membranes as determined by deuterium magnetic resonance. Biomembranes. 12:133–168. [Google Scholar]
- 32.Opella, S. J., Y. Kim, and P. McDonnell. 1994. Experimental nuclear magnetic resonance studies of membrane proteins. Methods Enzymol. 239:536–560. [DOI] [PubMed] [Google Scholar]
- 33.Opella, S. J. 1986. Protein dynamics by solid-state NMR. Methods Enzymol. 131:327–361. [DOI] [PubMed] [Google Scholar]
- 34.Siminovitch, D. J. 1998. Solid-state NMR studies of proteins: the view from static 2H NMR experiments. Biochem. Cell Biol. 76:411–422. [DOI] [PubMed] [Google Scholar]
- 35.Opella, S. J., and P. L. Stewart. 1989. Solid-state nuclear magnetic resonance structural studies of proteins. Methods Enzymol. 176:242–275. [DOI] [PubMed] [Google Scholar]
- 36.Smith, S. O., and O. B. Peersen. 1992. Solid-state NMR approaches for studying membrane protein structure. Annu. Rev. Biophys. Biomol. Struct. 21:25–47. [DOI] [PubMed] [Google Scholar]
- 37.Cross, T. A., and S. J. Opella. 1994. Solid-state NMR structural studies of peptides and proteins in membranes. Curr. Opin. Struct. Biol. 4:574–581. [Google Scholar]
- 38.Henry, G. D., and B. D. Sykes. 1994. Methods to study membrane protein structure in solution. Methods Enzymol. 239:515–535. [DOI] [PubMed] [Google Scholar]
- 39.Prosser, R. S., S. I. Daleman, and J. H. Davis. 1994. The structure of an integral membrane peptide: a 2H NMR study of gramicidin. Biophys. J. 66:1415–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, S. O., and B. J. Bormann. 1995. Determination of helix-helix interactions in membranes by rotational resonance NMR. Proc. Natl. Acad. Sci. USA. 92:488–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rigby, A. C., K. R. Barber, G. S. Shaw, and C. W. M. Grant. 1996. Transmembrane region of the epidermal growth factor receptor: behavior and interactions via 2H NMR. Biochemistry. 35:12591–12601. [DOI] [PubMed] [Google Scholar]
- 42.Greenfield, M., A. Ronemus, R. Vold, P. Ellis, and T. Raidy. 1987. Deuterium quadrupole-cho NMR spectroscopy. III. Practical aspects of linshape calculations for multiaxis rotational processes. J. Magn. Reson. 72:89–107. [Google Scholar]
- 43.Ying, W., S. E. Irvine, R. A. Beekman, D. J. Siminovitch, and S. O. Smith. 2000. Deuterium NMR reveals helix packing interactions in phospholamban. J. Am. Chem. Soc. 122:11125–11128. [Google Scholar]
- 44.van der Wel, P. C. A., E. Strandberg, J. A. Killian, and R. E. I. Koeppe. 2002. Geometry and intrinsic tilt of a tryptophan-anchored transmembrane α-helix determined by 2H NMR. Biophys. J. 83:1479–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strandberg, E., S. Ozdirekcan, D. T. S. Rijkers, P. C. A. van der Wel, R. E. I. Koeppe, R. M. J. Liskamp, and J. A. Killian. 2004. Tilt angles of transmembrane model peptides in oriented and non-oriented lipid bilayers as determined by 2H solid-state NMR. Biophys. J. 86:3709–3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Whiles, J. A., K. J. Glover, R. R. Vold, and E. A. Komives. 2002. Method for studying transmembrane peptides in bicelles: consequences of hydrophobic mismatch and peptide sequence. J. Magn. Reson. 158:149–156. [DOI] [PubMed] [Google Scholar]
- 47.de Planque, M. R. R., and J. A. Killian. 2003. Protein-lipid interactions studies with designed transmembrane peptides: role of hydrophobic matching and interfacial anchoring. Mol. Membr. Biol. 20:271–284. [DOI] [PubMed] [Google Scholar]
- 48.Juretic, D., and A. Lucin. 1998. The preference functions method for predicting protein helical turns with membrane propensity. J. Chem. Inf. Comput. Sci. 38:575–585. [Google Scholar]
- 49.Rost, B., R. Cassadio, P. Fariselli, and C. Sander. 1995. Transmembrane helices predicted at 95% accuracy. Protein Sci. 4:521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altschul, S. F., T. L. Madden, A. A. Schaffer, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1994. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry. 33:3038–3049. [DOI] [PubMed] [Google Scholar]
- 52.Batchelder, L. S., C. H. Niu, and D. A. Torchia. 1983. Methyl reorientation in polycrystalline amino acids in peptides: a 2H NMR spin-lattice relaxation study. J. Am. Chem. Soc. 105:2228–2231. [Google Scholar]
- 53.Gu, Z., K. Ebisawa, and A. McDermott. 1996. Hydrogen bonding effects on amine rotation rates in crystalline amino acids. Solid State Nucl. Magn. Reson. 7:161–172. [DOI] [PubMed] [Google Scholar]
- 54.Hing, A. W., S. P. Adams, D. F. Silbert, and R. E. Norberg. 1990. Deuterium NMR of 2HCO-Val1···gramicidin A and 2HCO-Val1-D-Leu2···gramicidin A in oriented DMPC bilayers. Biochemistry. 29:4156–4166. [DOI] [PubMed] [Google Scholar]
- 55.Sharpe, S., K. R. Barber, C. W. M. Grant, D. Goodyear, and M. R. Morrow. 2002. Organization of model helical peptides in lipid bilayers: insight into behavior of single-span protein transmembrane domains. Biophys. J. 83:345–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Batchelder, L. S., C. E. Sullivan, L. W. Jelinksi, and D. A. Torchia. 1982. Characterization of leucine side-chain reorientation in collagen fibrils by solid-state 2H NMR. Proc. Natl. Acad. Sci. USA. 79:386–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jones, D. H., A. C. Rigby, K. R. Barber, and C. W. M. Grant. 1997. Oligomerization of the EGF receptor transmembrane domain: a 2H NMR study in lipid bilayers. Biochemistry. 36:12616–12624. [DOI] [PubMed] [Google Scholar]
- 58.Lovell, S. C., J. M. Word, J. S. Richardson, and D. C. Richardson. 2000. The penultimate rotamer library. Proteins. 40:389–408. [PubMed] [Google Scholar]
- 59.Tiburu, E. K., E. S. Karp, P. C. Dave, K. Damodaran, and G. A. Lorigan. 2004. Investigating the structural and dynamics properties of the transmembrane segment of phospholamban incorporated into phospholipid bilayers utilizing 2H and 15N solid-state NMR spectroscopy. Biochemistry. 43:13899–13909. [DOI] [PubMed] [Google Scholar]
- 60.Voet, D., and J. G. Voet. 1995. Biochemistry. John Wiley & Sons, New York.
- 61.Lewis, B. A., and D. M. Engleman. 1983. Lipid bilayer thickness varies linearly with acyl chain length in fluid phosphatidylcholine vesicles. J. Mol. Biol. 166:211–217. [DOI] [PubMed] [Google Scholar]
- 62.Mouritzen, O. G., and M. Bloom. 1984. Mattress model of lipid protein interactions in membrane. Biophys. J. 46:141–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morrow, M. R., and C. W. M. Grant. 2000. The EGF receptor transmembrane domain: peptide-peptide interactions in fluid bilayer membranes. Biophys. J. 79:2024–2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hing, A. W., S. P. Adams, D. F. Silbert, and R. E. Norberg. 1990. Deuterium NMR of Val1···(2–2H) Ala3···gramicidin A in oriented DMPC bilayers. Biochemistry. 29:4144–4156. [DOI] [PubMed] [Google Scholar]
- 65.Hing, A. W., J. Schaefer, and G. S. Kobayashi. 2000. Deuterium NMR investigation of an amphotericin B derivative in mechanically aligned lipid bilayers. Biochim. Biophys. Acta. 1463:323–332. [DOI] [PubMed] [Google Scholar]
- 66.Prosser, R. S., S. A. Hunt, J. A. DiNatale, and R. R. Vold. 1996. Magnetically aligned membrane model systems with positive order parameter: switching the sign of Szz with paramagnetic ions. J. Am. Chem. Soc. 118:269–270. [Google Scholar]
- 67.Seelig, J. 1978. 31P nuclear magnetic resonance and the head group structure of phospholipids in membranes. Biochim. Biophys. Acta. 515:105–140. [DOI] [PubMed] [Google Scholar]
- 68.Harzer, U., and B. Bechinger. 2000. Alignment of lysine-anchored membrane peptides under conditions of hydrophobic mismatch: a CD, 15N and 31P solid-state NMR spectroscopy investigation. Biochemistry. 39:13106–13114. [DOI] [PubMed] [Google Scholar]
- 69.Killian, J. A., and G. von Heijne. 2000. How proteins adapt to a membrane-water interface. Trends Biochem. Sci. 25:429–434. [DOI] [PubMed] [Google Scholar]
- 70.Mishra, V. K., and M. N. Palgunachari. 1996. Interaction of model class A1, class A2, and class y amphipathic peptides with membranes. Biochemistry. 35:11210–11220. [DOI] [PubMed] [Google Scholar]
- 71.Yau, W.-W., W. C. Wimley, K. Gawrisch, and S. H. White. 1998. The preference of tryptophan for membrane proteins. Biochemistry. 37:14713–14718. [DOI] [PubMed] [Google Scholar]
- 72.Rinia, H. A., J.-W. P. Boots, D. T. S. Rijkers, R. A. Kik, M. M. E. Snel, R. A. Demel, J. A. Killian, J. P. J. M. van der Eerden, and B. de Kruijff. 2002. Domain formation in phosphatidylcholine bilayers containing transmembrane peptides: specific effects of flanking residues. Biochemistry. 41:2814–2824. [DOI] [PubMed] [Google Scholar]
- 73.de Planque, M. R. R., E. Goormaghtigh, D. V. Greathouse, R. E. Koeppe II, J. A. W. Kruijtzer, R. M. J. Liskamp, B. de Kruijff, and J. A. Killian. 2001. Sensitivity of single membrane-spanning alpha-helical peptides to hydrophobic mismatch with a lipid bilayer: effects on backbone structure, orientation, and extent of membrane incorporation. Biochemistry. 40:5000–5010. [DOI] [PubMed] [Google Scholar]
- 74.Braun, P., and G. von Heijne. 1999. The aromatic residues Trp and Phe have different effects on the positioning of a transmembrane helix in the microsomal membrane. Biochemistry. 38:9778–9782. [DOI] [PubMed] [Google Scholar]
- 75.Morré, D. J., A. O. Brightman, and A. S. Sandelius. 1987. Biological membranes. In Biological Membranes: A Practical Approach. J. B. C. Findlay and W. H. Evans, editors. IRL Press, Oxford, England. 37–72.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.