Abstract
Cadherins are multidomain adhesion proteins whose interactions direct cell sorting during histogenesis. They determine cell adhesion specificity, but prior studies failed to identify physical differences that could underlie cell sorting. These single molecule studies identify kinetic and strength differences between different cadherins. They further demonstrate that the modular extracellular architecture of cleavage stage C-cadherin supports a multistate binding mechanism. These multiple bonds exhibit a kinetic hierarchy of strengths that map to the different cadherin domains. The outer two N-terminal domains of C-cadherin form two bound states with dissociation rates of 3.9 and 0.02 s−1. The latter is 25-fold slower than between the corresponding epithelial cadherin segments. In addition to the two fast bonds, the five-domain fragment (CEC1-5) forms two additional stronger, longer-lived bonds with dissociation rates of 0.00039 and 0.00001 s−1. We further quantified the lifetimes of bonds subject to a constant force, and thus identified multiple dissociation events with rates that agree quantitatively with the force spectroscopy data. The qualitative features are similar to those reported for epithelial cadherin. However, the significant differences in the dissociation rates of the outer domains, which include the specificity-determining region, suggest that kinetic differences may determine cadherin discrimination, rather than adhesion energies.
INTRODUCTION
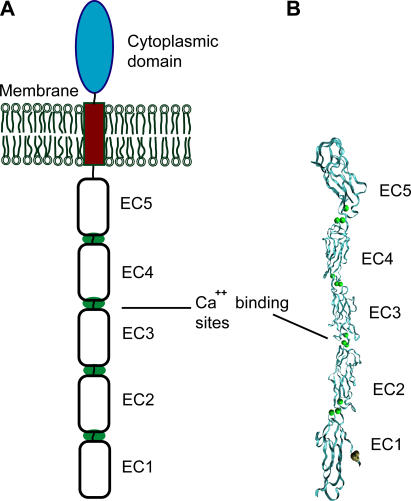
Intercellular adhesion in vertebrates is determined by the specificity and adhesive function of cell surface receptors. Many of these adhesion proteins are members of the immunoglobulin and cadherin families, and a distinct feature of their extracellular, adhesive regions is that they comprise multiple tandemly repeated domains. The general structural organization of these proteins is known, but establishing the mechanisms by which they form and stabilize adhesive junctions remains a challenge. Cadherins, in particular, play a pivotal role in tissue morphogenesis, neural development, and signal transduction (1,2). In particular, these proteins drive cell segregation in tissue formation by an undetermined mechanism. The overall architecture of classical cadherins comprises a highly conserved cytoplasmic domain, a single transmembrane segment, and an extracellular region consisting of five, tandemly arranged cadherin-like domains (3). The extracellular domains are numbered EC1 through EC5, starting from the outermost domain (see Fig. 1). In the presence of Ca2+ these domains form a relatively rigid structure.
FIGURE 1.
(A) Structure of the extracellular domain of C-cadherin. (B) Proposed trans adhesive interface observed in the crystal structure of the full C-cadherin ectodomain (8). The intermolecular bond involves EC1 domains from both molecules. The molecular representation was generated with VMD (36).
Because of its central importance in development, the mechanism of homophilic cadherin adhesion is a topic of intense study. Of primary interest are the mechanism of adhesion and the molecular basis of the tissue specificity of cadherin-mediated cell adhesion. Different experimental approaches generated different models for cadherin adhesion, and the molecular basis of cadherin selectivity is unknown. Several models of cadherin binding are based on NMR spectroscopy and on x-ray crystallography of the N-terminal cadherin fragments, as well as on electron microscopy and electron tomography studies of the intact protein (4–11). These models proposed the exclusive involvement of EC1 domains in both trans and cis interactions. Specifically, the trans interaction between the EC1 domains involves the insertion of the conserved Trp2 side chain of one protein into the hydrophobic pocket of the N-terminal domain on the opposed molecule (Fig. 1 B). This strand exchange was observed in the structures of the full-length C-cadherin extracellular domains (8) and of E-cadherin N-terminal domains (11). A recent cross-linking study also confirmed the formation of this strand dimer (12). Electron tomography images of fixed skin identified structures similar to the C-cadherin crystal structure (10), as well as many other bound and unbound configurations.
Intimately related to the structural characteristics of cadherin association, the molecular origin of the cadherin selectivity in cell adhesion is undetermined. Homophilic interactions among classical cadherins are widely believed to be the main determinant of cell sorting during development (1,2,13). Key studies mapped the specificity-determining region to the N-terminal domain (14). However, structures of the Trp2 in the hydrophobic pocket of an opposed EC1 domain of both C- and E-cadherin are very similar and do not display any obvious differences that could account for cadherin selectivity (8,11). Studies demonstrated that differences in neither the specificity nor the adhesion strength account for cell sorting (15). The expression of multiple cadherin subtypes by a single cell type also suggests that adhesive specificity in vivo results from the combinatorial action of multiple cadherins (1). However, because of the low affinity of the EC1-EC1 bond (11), it has been difficult to quantify physical differences between the cadherins that may contribute to their binding selectivity.
The EC1 domain is critical for adhesion, but several lines of evidence indicate that the modular cadherin structure can engage in multiple, trans-bonded associations that involve more than the EC1 domain. Molecular adhesion measurements with the surface force apparatus (SFA) demonstrated that cleavage stage cadherin (C-cadherin) forms either of three different trans-bonds (16–18). Besides interacting through the EC1 domains, the C-cadherin ectodomain formed two additional bound states that involve two different antiparallel alignments of the extracellular regions. The strongest adhesive bond corresponded to the full overlap of antiparallel ectodomains, and required EC3 (18). The third, weakest bond corresponded to contact between the N-terminal domains, and required EC1. Cell adhesion and bead aggregation assays with C-cadherin domain deletion mutants corroborated the requirement for multiple domains in homophilic cadherin binding (19). More recent single molecule force spectroscopy measurements further demonstrated that epithelial cadherin (E-cadherin) forms multiple bound states that involve multiple EC domains (20). Whereas the EC1-2 fragment exhibited two, weak bonds with rapid kinetics, the full ectodomain formed two additional, stronger long-lived states, in addition to the weaker bonds measured with EC1-2.
This work describes biomembrane force probe (BFP) measurements of the strengths and lifetimes of homophilic C-cadherin bonds. The approach is similar to atomic force microscopy, but the dynamic range of achievable loading rates is three orders-of-magnitude greater than the atomic force microscope and the force sensitivity is an order-of-magnitude greater. These investigations focused largely on the C-cadherin dissociation rates, which are linked to the bond strengths. Three different BFP measurement approaches were used to resolve the multiple bonds formed between C-cadherin ectodomains and to quantify their respective dissociation rates (or lifetimes). Measurements with the cadherin fragment CEC1-5 (full ectodomain) and CEC1-2 (domains EC1 and EC2) mapped the binding interactions to different regions of the C-cadherin structure. The trans-bonded CEC1-2 fragments form two weak, short-lived bound states. On the other hand, the full ectodomain exhibited two long-lived states, in addition to the fast bonds observed with the CEC12 fragment. Significantly, the N-terminal EC12 segments of CEC12 and EEC12 exhibit the greatest differences in kinetic rates. Additionally, new, independent information, obtained with the constant force mode of the BFP, directly gives the lifetimes of homophilic CEC1-5 bonds subject to a constant force of ∼40 pN. The resulting decay curves confirm the multistate nature of the cadherin interaction, and directly quantify the corresponding dissociation rates under force.
Importantly, while C-cadherin bond strengths are generally stronger than those of E-cadherin, the most striking finding is the significant difference in their dissociation rates. The differences in the lifetimes of the four C-cadherin bonds as well as differences relative to E-cadherin bonds suggest that kinetics may play an important role in cell sorting. These results, which simultaneously confirm the multistate nature of cadherin adhesion and reveal significant differences between EC12 binding kinetics, appear to reconcile different models of cadherin binding.
MATERIALS AND METHODS
Protein production and purification
The design of C-cadherin fragments with Fc domains fused at the C-termini of CEC1-5 (full ectodomain) and CEC1-2 were described previously (19). C-cadherin is expressed in the early embryo of Xenopus laevis, and plays an important role in the formation of a well-organized embryo (21,22). The soluble proteins were purified from cultures of stably transfected Chinese hamster ovary cells according to published procedures (18,19).
Preparation of the glass microspheres
Borosilicate spheres (Duke Scientific, Palo Alto, CA) with ∼2-μm diameters were chosen for probe and target beads. The spheres (500 μL, 10 g %) were first washed in a mixture of ammonium hydroxide, hydrogen peroxide, and water (pH ∼11) at boiling temperature. The beads were then washed by centrifugation and resuspension in 1 mL of methanol. After that, the beads were derivatized with thiol groups by mixing 1 mL bead solution with 45.6 mL methanol, 0.4 mL acetic acid (∼17.5 M), 1.85 mL H20, and 1.15 mL of 3-mercaptopropyltrimethoxysilane (United Chemical Technologies, Bristol, PA). The mixture was incubated for 3 h at room temperature in a rotary mixer. The beads were then washed by centrifugation and resuspension in 500 μL of methanol. This solution was divided into 20 dry, clean glass vials. Finally, the contents of each vial was dried and stored at room temperature. Before using the beads, they were washed with calcium-containing phosphate-buffered saline buffer (pH 7.5).
The proteins were immobilized to the beads by first reacting the soluble protein with the heterobifunctional cross-linker NHS-PEG-MAL (Nektar Therapeutics, Huntsville, AL). The activated protein was then linked to the beads by reacting the maleimide (MAL) groups with the thiol-terminated groups on the activated microspheres. Before this, the proteins were first desalted and placed in calcium-containing carbonate/bicarbonate buffer (pH 7, with 50 mM Ca2+). Solutions (∼70 μM) of the heterobifunctional polymers NHS-PEG-MAL and Biotin-PEG-MAL (Nektar Therapeutics, Huntsville, AL) were prepared with carbonate/bicarbonate buffer (pH 8.5). To prepare the probe beads, a mixture (5 μL) of NHS-PEG-MAL (10 mol%) and Biotin-PEG-MAL (90 mol%) was added to 5 μL of protein solution (∼0.5 mg/mL for CEC1-5 and ∼0.2 mg/mL for CEC1-2) and incubated for 30 min at room temperature. In this way, 10% (at the most) of the MAL-PEG linkers are attached to proteins whereas 90% of the PEG linkers are end-functionalized with biotin. To prepare the target beads, the proteins (5 μL, same concentrations as before) were derivatized with 5 μL of NHS-PEG-MAL solution for 30 min at room temperature. Then 5 μL of hydrolyzed NHS-PEG-MAL solution was added to obtain a final solution in which the ratio of PEG linkers to protein was ∼2:1. Once the proteins functionalized with the PEG-MAL linkers, the derivatized protein was immobilized onto glass beads by reacting the free maleimide (MAL) moiety with the thiol groups on the silanized beads. To do this, 10 μL of a suspension of silanized beads and 200 μL of phosphate buffer (pH 7, with 50 mM Ca2+) were mixed with the previously prepared solution of the protein-polymer conjugate, and the mixture was incubated overnight. With the thus-prepared beads, for every 100 bead-bead contacts, we measured 30 binding events.
Red blood cell preparation: the BFP force transducer
Fresh red blood cells (∼10 μL) (RBCs) were first washed with 0.1 M carbonate/bicarbonate buffer (pH 8.5). The washed RBCs (5 μL) were then coated with the heterobifunctional cross-linker NHS-LC-Biotin (Pierce, Rockford, IL) (95 μL, ∼0.8 mg/mL, carbonate/bicarbonate buffer, pH 8.5) by amide reaction (30 min at RT). After washing the RBCs with PBS buffer, they were reacted for 45 min at room temperature with streptavidin (10 μL, 2mg/mL) (Sigma, St. Louis, MO). Finally, the cells were washed with a solution containing 10 mM HEPES, 75 mM NaCl, 0.1% BSA, pH 7.5, and then stored at 4°C (∼150 μL). After this procedure, the cell capsule is ready to be used as a force transducer in the BFP measurements.
BFP measurements
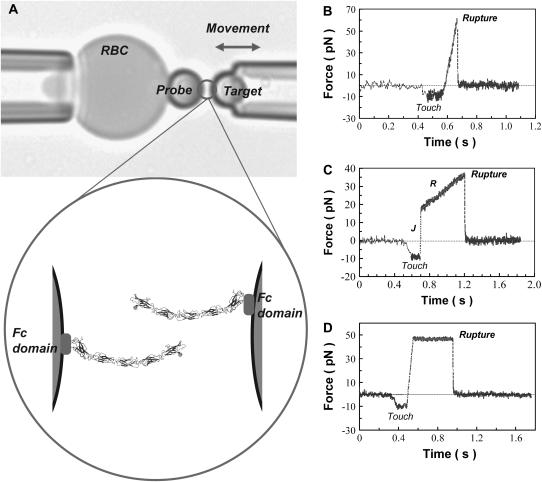
All the experiments were performed in a ∼1 × 5 × 25 mm3 chamber mounted on the stage of an inverted microscope. The chamber contained 75 mM NaCl, 10 mM HEPES, 2 mM CaCl2, and 0.1 wt % BSA (pH 7.5). For the measurements, the streptavidin-coated RBC was pressurized by micropipette suction into a spherical shape (see Fig. 2 A). The BFP spring constant kf (force/RBC extension) was preset in the range of 0.2 to 2 pN/nm by controlling the pipette suction (23). A biotinylated, protein-functionalized glass microsphere was then selected by micromanipulation, and maneuvered to form strong adhesive contact with the RBC. This formed the BFP (see Fig. 2 A). The latter sphere was kept fixed, whereas a target sphere, which was also held by a micropipette, was translated into and out of contact with the probe by precision-piezo displacements. The difference between the piezo-driven translation and the tip displacement was multiplied by the spring constant, to obtain the elastic force (fe) applied to the BFP. Under ultrafast detachment (vs > 30,000 nm/s), viscous damping contributes a hidden viscous force fv to the total force. Therefore, the probe force at each speed versus detachment has to be corrected using f = fe + fv. The viscous force is given by fv = 0.00048 versus Evans et al. (24).
FIGURE 2.
(A) Biomembrane force probe. The upper panel shows the aspirated RBC with the attached bead and the probe bead held by the second micropipette. The probe bead is brought in and out of contact with the bead on the RBC. The lower panel is a cartoon of the bound cadherins at the bead-bead junction. (B–D) Representative force versus time profiles obtained with the steady-ramp (B), jump/ramp (C), and constant force (D) modes of the BFP.
The homophilic interactions between C-Cadherin fragments were tested with three different modes of the biomembrane force probe, which varied in their force-displacement trajectories. In all cases, the beads were brought into contact and held for 0.1 s with an impingement force of ∼10 pN. In the steady ramp mode (Fig. 2 B), the beads were separated at a constant loading rate rf = df/dt (pN/s). In the presence of a bond, this loading rate was maintained until the bond broke. With the steady-ramp mode, the bonds were probed at loading rates ranging from 50 to 50,000 pN/s. In the jump/ramp mode of the BFP (see Fig. 2 C), the bonds were first rapidly pulled at a loading rate of ∼5000 pN/s until the force reached a preset value (jump phase). The surviving bonds were then pulled at a lower loading rate until they broke (ramp phase). In the third BFP mode, the time to bond failure was determined while maintaining the bond under a constant, low force. Thus, a force was quickly applied at ∼800 pN/s up to a small value (∼40 pN), after which the force was held constant until the bond failed (see Fig. 2 D). This is a limiting case of the jump/ramp mode, in which loading rate during the ramp phase is zero. In both cases, the magnitude of the preset force is chosen to ensure the rupture of weaker bonds, so that the stronger bonds, the majority of which survive the jump, are then selectively probed in the subsequent ramp or force-clamp phase.
To ensure that these rupture events correspond to single bonds, we used three different criteria. In the most rigorous approach, apparent peaks in the tails at higher forces were compared with that expected if the force was shared between two (or more) bonds. The data were also fit to different probability distributions, to test whether multibond contacts could account for the high force tails. We also considered the data consistency between different measurements. Generally, the force tails vary between samples and with experimental variables such as the impingement force. They also vary with the number of measurements, since rare events are more prominent in larger data sets. Features that were not reproducible were thus discounted. By contrast, the principle peaks analyzed were reproducible. These analyses reflect peaks, which consistently appear in each data set, independent of the sample, the size of the data set, etc. Finally, the internal consistency of the three BFP approaches provided a further test of the analyses.
Two types of control measurements confirmed that the measured binding events are due to specific cadherin bonds. In one control, the beads were not functionalized with cadherin. In the second set of control measurements, EDTA added to the medium removed calcium and eliminated cadherin's adhesive function. The latter completely abrogates adhesion, whereas some residual binding can be detected at low calcium concentrations (18).
Lifetime of molecular bonds under force
When a molecular bond is subjected to an external bond, the dissociation rate (koff) increases exponentially with the external force as first suggested by Bell (25), according to
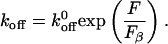 |
Here, 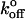 is the rate of dissociation in the absence of force and Fβ is the so-called thermal force, which is related to the thermally averaged projection of the transition state along the line of force, xβ, by (26),
is the rate of dissociation in the absence of force and Fβ is the so-called thermal force, which is related to the thermally averaged projection of the transition state along the line of force, xβ, by (26),
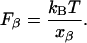 |
At applied forces greater than the thermal force, the rate of reassociation vanishes, and the likelihood S(t) of remaining in the bound state is given by
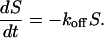 |
In the case of constant force, the normalized probability density for bond failure between times t and t + dt is given by
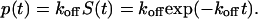 |
The probability of bonds breaking at times larger than t is therefore
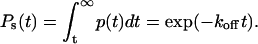 |
This is the expression for the survival probability at time t, Ps(t), which is used in the analysis of the constant force experiments.
Dynamic force spectroscopy
When a bond is subjected to a steadily increasing force, the rupture forces are distributed according to a probability function determined by the energy barriers to unbinding and the loading rate (rf). In the case of simple bonds, i.e., those confined by only one energy barrier, the rupture forces Fr are distributed according to the probability density p(f) (26),
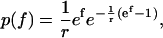 |
where
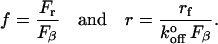 |
The most probable rupture force Fr* is derived from this probability function, and is
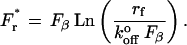 |
Thus, plots of Fr* versus the loge of the loading rate (Lnrf) will be linear functions that are scaled by Fβ and have an intercept that depends on 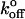 .
.
RESULTS
The CEC1-2 fragment exhibited two bound states
Homophilic CEC1-2 binding was tested initially using the steady ramp mode of the BFP. In these measurements, the probe and target beads are first brought into contact and then pulled at a constant loading rate df/dt until the bonds fail. Fig. 2 B shows a typical force profile for the steady ramp mode with a nominal loading rate of 1000 pN/s. We measured the forces necessary to break the bonds at four nominal loading rates (NLRs), namely: 100, 1000, 10,000, and 50,000 pN/s. For each case, the effective (or actual) loading rates (ELRs) were directly calculated from the force profiles (see Table 1).
TABLE 1.
Effective loading rates (ELRs) in the steady ramp experiments
| Effective loading rates (pN/s)
|
||
|---|---|---|
| NLR (pN/s) | CEC1-2 | CEC1-5 |
| 100 | 69 ± 6 | 68 ± 5 |
| 1000 | 682 ± 50 | 664 ± 54 |
| 10,000 | 7158 ± 837 | 6894 ± 750 |
| 50,000 | 31,519 ± 4719 | 29,672 ± 3991 |
Because of the stochastic nature of single bond rupture, the measured rupture forces follow a probability distribution that is defined by the energy landscape of the bond and by the loading rate. Fig. 3 A shows the distribution of the rupture forces obtained at a loading rate of 69 ± 6 pN/s. The majority of rupture forces (∼80%) forms one prominent peak, with the most probable force being 34.3 ± 1.5 pN. For a bond confined by a single barrier, the most probable rupture force Fr* varies with the logarithm of the loading rate rf according to
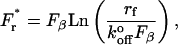 |
where 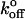 is the rate of dissociation in the absence of force and Fβ is the so-called thermal force which is determined by the dimensions of the energy barrier (26). For a bond confined by a single barrier, the plot of Fr* versus Ln(rf) yields a linear plot with a slope scaled by Fβ, and with an intercept determined by the unstressed dissociation rate
is the rate of dissociation in the absence of force and Fβ is the so-called thermal force which is determined by the dimensions of the energy barrier (26). For a bond confined by a single barrier, the plot of Fr* versus Ln(rf) yields a linear plot with a slope scaled by Fβ, and with an intercept determined by the unstressed dissociation rate 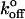 (see Materials and Methods).
(see Materials and Methods).
FIGURE 3.
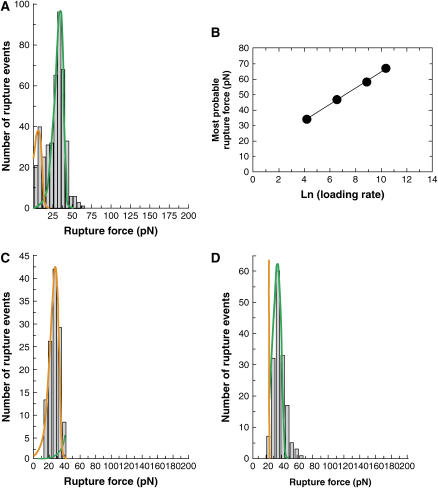
Measured strengths of trans-bonded CEC12 fragments. (A) Histogram obtained from a steady ramp test of CEC12 versus CEC12 at 69 ± 5 pN/s. The solid lines correspond to the probability distribution at 69 ± 5 pN/s. See text and Table 1 for corresponding bond parameters. (B) Most probable rupture force of the prominent peak (green) versus the logarithm of the loading rate. (C and D) Force histograms obtained with the jump/ramp mode of the BFP. (C) Shows the bonds that failed during the jump phase along with the probability distribution that best fit the data (orange curve). The majority of attachments (∼60%) survived the jump and failed during the ramp phase. (D) Shows the corresponding histogram of rupture forces measured in the ramp phase. The solid green curve is the probability distribution defined by the corresponding bond parameters for this bound state.
Between CEC1-2 fragments, the force histograms exhibit a prominent peak measured at each of the loading rates used. The corresponding most probable force Fr* increased linearly with the logarithm of the loading rate (see Fig. 3 B). The parameters of the corresponding bound state, determined from the above linear dependence, are Fβ = 5.27 ± 0.13 pN and 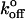 = 0.019 ± 0.004 s−1. The theoretical distribution associated with these parameters (main population) at a loading rate of 69 pN/s, is plotted along with the experimental distribution in Fig. 3 A.
= 0.019 ± 0.004 s−1. The theoretical distribution associated with these parameters (main population) at a loading rate of 69 pN/s, is plotted along with the experimental distribution in Fig. 3 A.
Fig. 3 A shows that, besides the main peak, there is a second population of smaller rupture forces (∼20%). However, it was not possible to unambiguously distinguish this population from the prominent peak with the steady ramp method. Instead, the jump/ramp mode of the BFP was used in order to separate the two populations. During the jump phase, the beads were separated at a nominal loading rate of 6000 pN/s. The corresponding effective loading rates are shown in Table 2. Approximately 40% of the bonds ruptured during this phase, and these rupture events correspond to the population of weakest homophilic bonds. Fig. 3 C shows the corresponding histogram along with the theoretical distribution that best fit the low rupture force data. The parameters of the corresponding bond, estimated from the nonlinear fitting, are Fβ = 5.23 ± 0.46 pN and 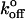 = 3.93 ± 1.54 s−1. The jump phase ended at a force of 24 ± 4 pN and the surviving bonds were then pulled at 78 ± 10 pN/s. The resulting distribution of rupture events during this latter ramp phase (Fig. 3 D) corresponds to the prominent peak in the steady ramp experiments in Fig. 3 A (Fβ = 5.27 ± 0.13 pN;
= 3.93 ± 1.54 s−1. The jump phase ended at a force of 24 ± 4 pN and the surviving bonds were then pulled at 78 ± 10 pN/s. The resulting distribution of rupture events during this latter ramp phase (Fig. 3 D) corresponds to the prominent peak in the steady ramp experiments in Fig. 3 A (Fβ = 5.27 ± 0.13 pN; 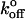 = 0.019 ± 0.004 s−1).
= 0.019 ± 0.004 s−1).
TABLE 2.
Effective loading rates (ELRs) in the jump/ramp experiments
| Jump phase
|
Ramp phase
|
||||
|---|---|---|---|---|---|
| Fragment | NLR (pN/s) | ELR preceding rupture (pN/s) | ELR preceding a ramp (pN/s) | NLR (pN/s) | ELR (pN/s) |
| CEC1-2 | 6000 | 4276 ± 565 | 4524 ± 764 | 100 | 78 ± 10 |
| CEC1-5 | 6000 | 4103 ± 591 | 4296 ± 494 | 10 | 7.8 ± 0.9 |
CEC1-5 forms four bound states
The homophilic adhesion by CEC1-5 was first investigated with the steady ramp mode at the same nominal loading rates as used in the CEC12 measurements (compare to Fig. 3 A). Consistent with the presence of additional domains, the distributions of rupture forces measured between CEC1-5 fragments were more complex than those measured between CEC1-2 fragments. This broader histogram could be interpreted as a superposition of the bound states exhibited by CEC1-2 plus two additional stronger bonds. Both steady ramp and jump/ramp BFP measurements confirmed this.
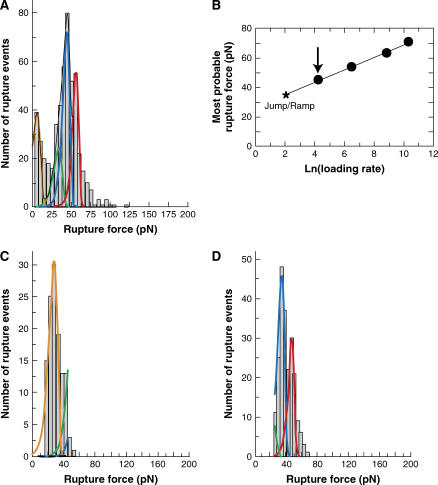
Fig. 4 A shows the probability distribution of the rupture forces obtained at a steady loading rate of 68 ± 5 pN/s. One of the new bonds clearly defined the prominent peak at each loading rate. Fig. 4 B shows that the most probable force defined by this peak occurs at higher forces than observed with CEC1-2. The most probable rupture force associated with this dominant peak varied linearly with the logarithm of the loading rate, and the corresponding bond parameters are Fβ = 4.2 ± 0.11 pN and 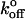 = 0.00039 ± 0.00012 s−1. The theoretical distribution at 68 pN/s defined by these parameters (blue curve) is superimposed on the histogram in Fig. 4 B. However, the width of the distribution exceeded the theoretical width based on the known uncertainty in the BFP measurements. This indicated that this single bound state was insufficient to describe the data. The presence of an additional population with slightly higher rupture forces than the dominant peak indicated the existence of an additional binding event. The latter bond could not be clearly discriminated at steady loading rates greater than 100 pN/s, but the two stronger bonds were clearly separated at a loading rate of ∼8 pN/s in jump/ramp experiments. This mode was used to separate the weaker bonds, exhibited by the CEC1-2 segments, from the additional stronger states. During the jump phase, the beads were separated at a nominal loading rate of 6000 pN/s (see Table 2 for the corresponding ELRs) until the force reached a value of 27 ± 4 pN. The surviving bonds were then pulled at 7.8 ± 0.9 pN/s. The rupture forces obtained during the latter ramp phase (60%) formed two clearly defined populations (Fig. 4 D). The peak at the lower force (Fig. 4 D, blue line) agrees with the distribution measured in the steady ramp experiments (Fig. 4 A). The parameters for the bound state that generated the second population are Fβ = 3.96 ± 0.18 pN and
= 0.00039 ± 0.00012 s−1. The theoretical distribution at 68 pN/s defined by these parameters (blue curve) is superimposed on the histogram in Fig. 4 B. However, the width of the distribution exceeded the theoretical width based on the known uncertainty in the BFP measurements. This indicated that this single bound state was insufficient to describe the data. The presence of an additional population with slightly higher rupture forces than the dominant peak indicated the existence of an additional binding event. The latter bond could not be clearly discriminated at steady loading rates greater than 100 pN/s, but the two stronger bonds were clearly separated at a loading rate of ∼8 pN/s in jump/ramp experiments. This mode was used to separate the weaker bonds, exhibited by the CEC1-2 segments, from the additional stronger states. During the jump phase, the beads were separated at a nominal loading rate of 6000 pN/s (see Table 2 for the corresponding ELRs) until the force reached a value of 27 ± 4 pN. The surviving bonds were then pulled at 7.8 ± 0.9 pN/s. The rupture forces obtained during the latter ramp phase (60%) formed two clearly defined populations (Fig. 4 D). The peak at the lower force (Fig. 4 D, blue line) agrees with the distribution measured in the steady ramp experiments (Fig. 4 A). The parameters for the bound state that generated the second population are Fβ = 3.96 ± 0.18 pN and 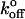 = 1.02 ± 0.16 × 10−5 s−1. The latter is particularly slow, and it is questionable whether this could be measured reliably by conventional binding techniques. However, it is important to note that these parameters and the associated errors correspond to the distribution that best fit the experimental histogram at a loading rate of 7.8 pN/s, as shown in Fig. 4 D. Table 3 summarizes the parameters for the different bound states.
= 1.02 ± 0.16 × 10−5 s−1. The latter is particularly slow, and it is questionable whether this could be measured reliably by conventional binding techniques. However, it is important to note that these parameters and the associated errors correspond to the distribution that best fit the experimental histogram at a loading rate of 7.8 pN/s, as shown in Fig. 4 D. Table 3 summarizes the parameters for the different bound states.
FIGURE 4.
Mechanical strengths of CEC1-5 bonds. (A) Histogram of CEC1-5 versus CEC1-5 rupture forces measured under a steady ramp of 68 ± 5 pN/s. The solid lines are the probability distributions for the small fraction (∼30%) of rupture events attributed to the bound states measured between CEC12 fragments: namely, the orange line corresponds to the bound state with Fβ = 5.2 and koff = 3.9 s−1, and the green line corresponds to the bond with Fβ = 5.3 and koff = 0.02 s−1. The blue and red solid curves correspond to the stronger bound states exhibited by CEC1-5. (B) The most probable rupture force corresponding to the prominent peak in the CEC1-5 force histograms versus the logarithm of the loading rates. (C) Force histogram measured during the jump phase (4103 ± 591 pN/s) in the jump/ramp measurements. The majority of bonds (∼60%) survived the jump and failed during the ramp phase (7.8 ± 0.9 pN/s) shown in D. The corresponding histogram shows two, resolved peaks. The peak described by the blue curve is the same as the peak exhibiting the linear dependence shown in A. The second peak was fit by the probability distribution shown by the red curve. The bond parameters pertaining to the red and blue distributions are given in the text and summarized in Table 1.
TABLE 3.
Summary of the parameters obtained from nonlinear fits of the force histograms to the probability distribution
| State (color code) | Fβ (pN) | koff (s−1) | to (s) | t40 (s) | t40* (s) |
|---|---|---|---|---|---|
| Orange | 5.23 ± 0.46 | 3.93 ± 1.54 | 0.25 ± 0.1 | 1.2 ± 1 × 10−4 | — |
| Green | 5.27 ± 0.13 | 0.019 ± 0.004 | 52.6 ± 11.1 | 0.026 ± 0.011 | 0.019 ± 0.001 |
| Blue | 4.2 ± 0.11 | 3.9 ± 0.7 × 10−4 | 2.5 ± 0.5 × 103 | 0.19 ± 0.08 | 0.22 ± 0.01 |
| Red | 3.96 ± 0.18 | 1.02 ± 0.16 × 10−5 | 9.8 ± 1.5 × 104 | 4.02 ± 2.48 | 3.13 ± 0.19 |
Control measurements demonstrated that these different bound states are all calcium-dependent. In the presence of EDTA, <5% of probe touches to the surface resulted in an adhesive event, compared with >15% in the presence of calcium. In addition, the peak in the latter histograms was at ∼15 pN, compared with the values of >25 pN measured with calcium. Absent protein on the beads, <3% of probe touches resulted in binding events. There were too few data to identify any obvious peaks, and the random forces ranged up to ∼70 pN. The results of both control measurements differed substantially from the data obtained with active protein on the probes, confirming that the features in Figs. 3 and 4 are due to cadherin adhesion.
Force-clamp measurements quantify CEC1-5 bond lifetimes
Based on the unstressed dissociation rates of the different cadherin bonds, as characterized by steady ramp and jump/ramp measurements, the lifetimes of the different bound states differ by at least two orders of magnitude. Thus, direct measurements of the lifetimes of CEC1-5 bonds would further confirm their existence, and verify the kinetic rates determined by force spectroscopy. The measured lifetimes of trans-bonded CEC1-5 complexes subjected to a constant force confirmed this prediction.
These constant force measurements provided separate verification of the multistate characteristics of homophilic CEC1-5 adhesion. In the lifetime measurements, following contact for 0.1 s, the beads were separated at loading rate of ∼800 pN/s until the force reached ∼40 pN. The bonds were then held at this force until they failed. The rupture times ranged from ∼0.001 s to ∼10 s. For the analysis, the normalized survival probability Psurv was computed according to
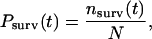 |
where nsurv (t) is the number of bonds that rupture at times bigger than t and N is the total number of bonds. The survival probability as a function of time is shown in Fig. 5.
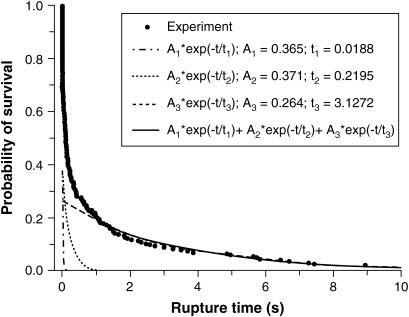
FIGURE 5.
The survival probability versus the rupture time for CEC1-5 bonds held at 40 pN. The circles are the experimentally measured survival probabilities. The solid line through the data is the superposition of the three exponential functions indicated by the dashed (A1), dotted (A2), and dash-dot curves (A1).
The time-dependence of Psurv(t) cannot be described by a single, first-order dissociation event. Instead, the data are best fit by a superposition of exponential decays, indicative of more than one homophilic CEC1-5 bond. The constant force of ∼40 pN ruptures the weakest bonds (Fig. 4 A, orange line), and all surviving bonds contribute to the decay curves. The survival time dependence of the probability curve is best fit by the superposition of three decaying exponentials, which correspond to three first-order dissociation events:
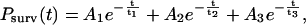 |
with
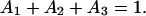 |
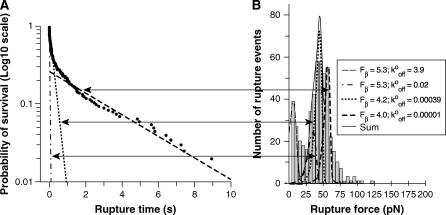
Here Ai is the fraction of bonds in state i, and ti is the corresponding lifetime. The curves in Fig. 5 correspond to the three exponential terms and their superposition. Fig. 6 shows the same data plotted on a log10 scale. Here, three linear regimes are clearly seen, and the arrows indicate their relationship to the peaks comprising the force histograms identified by force spectroscopy. An F-test justified the number of exponentials required to fit the decay curve (27). The use of three exponentials substantially improved the fit relative to a two-exponential function, but four exponentials did not significantly improve the fit (see Supplementary Material).
FIGURE 6.
(A) Logarithm10 of the survival probability Psurv versus rupture time for the CEC1-5 bonds held at 40 pN. The lines indicate the three exponential decay curves shown in Fig. 5. (B) Map between the exponential decay functions and the probability distributions describing the peaks in the CEC1-5 force histogram.
The lifetimes determined from the fit to the decay curve agree quantitatively with the kinetic parameters that define the green, blue, and red states determined from the steady ramp and jump/ramp measurements. This can be appreciated from the data in the last two columns of Table 3. The t40 column gives the predicted lifetime for the different bound states subject to an applied force of 40 pN, whereas the t40* column gives the values estimated from the distribution of survival times (Fig. 5). The values in the t40 column were calculated from
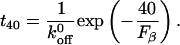 |
The weakest bond (orange line) did not contribute to the survival probability distribution because of its small lifetime of 0.1 ms at a force of 40 pN. Most of the rupture events associated with this state broke during the rapid jump phase.
DISCUSSION
These single molecule rupture measurements revealed the multistate nature of homophilic C-Cadherin bonds. The multiple binding events are directly related to the modular structure of the protein's extracellular domain. The CEC1-2 fragment interactions exhibited only two bound states with rapid kinetics whereas the CEC1-5 fragment exhibits four bound states, which include the same two observed with CEC1-2. The different bonds exhibit a hierarchy of kinetic rates, with dissociation rates ranging from 3.9 s−1 between the outer bonds to 0.00001 s−1 for the longest-lived, strongest bond between the full extracellular domains. On the other hand, the two states exhibited only by the full extracellular domain are long-lived, with lifetimes >103 s.
The hierarchy of mechanical strengths found for C-cadherin is qualitatively similar to that previously reported for human E-cadherin (20). There are, however, quantitative differences in the dissociation rates of at least an order of magnitude. The C-cadherin bonds have longer lifetimes (slower dissociation rates), with the exception of the strongest “red” state. The most remarkable finding is the CEC1-2 bond with a lifetime of 52 ± 11 s, which is 25-fold longer than the longest EEC1-2 bond lifetime of 1–2 s (20). The lifetime of the second CEC1-2 bond is twofold greater than the corresponding EEC1-2 bond. On the other hand, the “blue” state is 10-fold greater than the corresponding E-cadherin bond. The strongest “red” states are comparable, within error. In general, the C-cadherin bonds are stronger and rupture at higher forces than E-cadherin at comparable loading rates.
The short-lived states exhibited by the two outermost domains deserve special attention. Several studies support the involvement of domain EC1 in cadherin selectivity, but the structural features of trans EC1 association, suggested by the crystal structures, is the same for both C- and E-cadherin. Moreover, on account of their low apparent affinities, there are no prior reports of differences in the physical properties of the N-terminal bonds. That the lifetime of one of the two CEC1-2 bonds is 25-fold greater than that of the corresponding EEC1-2 bond suggests that kinetic discrimination may underlie cadherin selectivity. On the other hand, the similarly long-lived states associated with the full-length protein might be important for strong, stable adhesion after the brief recognition step. Indeed, Duguay et al. (28) showed that cadherin-dependent cell segregation in aggregation measurements depended on the agitation rates (shear stress) used. It is well known that cell or particle aggregation under shear depends on the association and dissociation rates relative to the shear rate (29–32). Thus, rapid kinetics may dictate which cells preferentially associate initially, while the stronger, slower forming bonds determine the final adhesion strength.
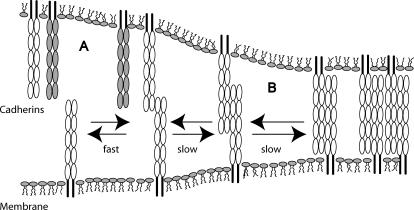
The hypothetical scenario described above is summarized in the model in Fig. 7. In studies of E-cadherin (20), the outer bonds form rapidly during initial intermolecular contact, but then convert to the strong, bound states within a few seconds. In light of prior surface force measurements, it is reasonable to assume that the strong, slow bonds correspond to interdigitated, opposing ectodomains. These BFP findings suggest that differences in the kinetics of the fast, outer bonds may underlie the cadherin discrimination. Once the appropriate bonds form, these weak, fast bonds transition to the more stable, strong bonds, which comprise the mature junctions.
FIGURE 7.
Proposed model for cadherin junction assembly. (A) Cadherins rapidly bind via their outer domains and kinetically discriminate between different cadherins (shaded and open). Differences in the binding kinetics of different classical cadherins may provide the kinetic proofreading that underlies selective cadherin binding. (B) Once the proteins engage, the cell membranes are held in close proximity sufficiently long for the slower, strong bonds to form between overlapping proteins. In this staged cadherin junction assembly, the sequence of binding events is governed by the hierarchy of kinetic rates.
Based on these and previous molecular adhesion measurements, the evidence shows that protein segments in addition to the EC1 domains contribute to adhesion. This contradicts a widely held view that only EC1 domains are involved in adhesion. For example, recent electron tomography images of desmosomal cadherin junctions suggested that EC1 contacts predominate, although there was a considerable number of different ectodomain configurations in the images (10). Both the quantified lifetimes and strengths of the C-cadherin (and E-cadherin) bonds show that the full-length ectodomains will form multiple bound states. The bond energies and the redistribution of the populations with time favor the stronger bonds, which require segments other than EC1-2 (20). The origin of the differences is unresolved, and further investigations are obviously needed to determine why desmosomal cadherin in fixed processed tissue exhibits apparently different behavior than the different classical cadherins in bulk water.
A consideration of other biophysical studies suggests qualitative and quantitative parallels with these BFP findings. Surface force measurements of C-cadherin interactions similarly exhibited modular interactions. The trans interactions between the full extracellular domains formed three energetically distinct trans-bonded associations that spanned three different membrane separations (18). The stronger associations involve the interdigitation of the cadherin domains, whereas the weakest interaction requires the EC1 domain. By analogy, these BFP findings suggest that the long-lived states, which are limited to the CEC1-5 fragment, involve overlapping, interdigitated EC domains. Similarly, the SFA experiments with the CEC1-2 fragment showed only one trans-bonded association involving domains EC1 of opposing molecules. These single molecule results show that the CEC1-2 fragment forms two substates, which could not be distinguished with SFA measurements. One of these could be due to cis dimer formation, while the other is due to adhesion between opposing domains. Because the BFP lacks the spatial information in the SFA measurements, we cannot distinguish between the two possibilities. Alternatively, steered molecular dynamics simulations of the rupture of a cadherin EC1-EC1 adhesive complex suggested a two-state dissociation of CEC1 domains (33). In these simulations, after extraction from the hydrophobic pocket, the conserved Trp2 was trapped in another substate before the bond finally broke completely.
Atomic force microscopy measurements with the full extracellular domain of vascular endothelial-cadherin, at loading rates ranging from 1.2 × 102 to 1.2 × 104 pN/s, also defined three different populations (peaks) with different bond strengths (34). The authors attributed this to cadherin oligomerization, and assumed the population at the lowest force was due to the single pairwise cadherin interaction. From the force spectra, they estimated the putative cadherin bond lifetime to be ∼0.55 s, which is the same order of magnitude as the fast bonds identified in these BFP measurements. Baumgartner et al. did not, however, investigate the additional peaks at higher rupture forces, so further comparisons are not possible. Additionally, flow chamber measurements quantified the lifetimes of human EEC1-2 bonds subjected to constant shear forces (1–4 pN) (35). The lifetime of the EEC1-2 bond thus measured was ∼2 s, which agreed quantitatively with one of the EEC1-2 peaks identified in BFP measurements (20). However, the prior investigations of cadherin dynamics do not individually provide as complete an analysis of the cadherin binding dynamics as the BFP measurements described here. Nevertheless, comparisons of the appropriate parameters with these results show good agreement with the force distributions and the bond kinetics.
In summary, these single bond rupture investigations demonstrate that cadherin forms multiple bound states that require different EC domains. Although this contradicts one view that only the N-terminal domains mediate adhesion, the measured bond lifetimes support the hypothesis that the EC1 domains confer binding selectivity. Both the bond rupture and lifetime measurements expose significant differences between homophilic C-cadherin and homophilic E-cadherin dissociation rates. Although the N-terminal strand dimer interfaces of both proteins are structurally very similar (8,11), these measurements reveal substantial kinetic differences that may underlie cadherin selectivity in vivo.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Supplementary Material
Acknowledgments
We thank Saiko Rosenberger for providing the protein samples used in this study. M.V.B. thanks Koji Kinoshita for his excellent technical assistance in the use of the BFP.
This work was supported by National Institutes of Health grants No. 5 RO1 GM51338 (to D.L.), and No. HL65333 and No. HL31579 (both to E.E.).
References
- 1.Takeichi, M. 1995. Morphogenetic roles of classic cadherins. Curr. Opin. Cell Biol. 7:619–627. [DOI] [PubMed] [Google Scholar]
- 2.Gumbiner, B. M. 1996. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 84:345–357. [DOI] [PubMed] [Google Scholar]
- 3.Takeichi, M. 1990. Cadherins: a molecular family important in selective cell-cell adhesion. Annu. Rev. Biochem. 59:237–252. [DOI] [PubMed] [Google Scholar]
- 4.Shapiro, L., A. M. Fannon, P. D. Kwong, A. Thompson, M. S. Lehmann, G. Grubel, J.-F. Legrand, J. Als-Nielsen, D. R. Colman, and W. A. Hendrickson. 1995. Structural basis of cell-cell adhesion by cadherins. Nature. 374:327–337. [DOI] [PubMed] [Google Scholar]
- 5.Nagar, B., M. Overduin, M. Ikura, and J. M. Rini. 1996. Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature. 380:360–364. [DOI] [PubMed] [Google Scholar]
- 6.Tamura, K., W.-S. Shan, W. A. Hendrickson, D. R. Colman, and L. Shapiro. 1998. Structure-function analysis of cell adhesion by neural (N-) cadherin. Neuron. 20:1153–1163. [DOI] [PubMed] [Google Scholar]
- 7.Pertz, O., D. Bozic, A. W. Koch, C. Fauser, A. Brancaccio, and J. Engel. 1999. A new crystal, Ca2+ dependence and mutational analysis reveal molecular details of E-cadherin homoassociation. EMBO J. 18:1738–1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boggon, T. J., J. Murray, S. Chappuis-Flament, E. Wong, B. M. Gumbiner, and L. Shapiro. 2002. C-Cadherin ectodomain structure and implications for cell adhesion mechanisms. Science. 296:1308–1313. [DOI] [PubMed] [Google Scholar]
- 9.Häussinger, D., T. Ahrens, H.-J. Sass, O. Pertz, J. Engel, and S. Grzesiek. 2002. Calcium-dependent homoassociation of E-cadherin by NMR spectroscopy: changes in mobility, conformation and mapping of contact regions. J. Mol. Biol. 324:823–839. [DOI] [PubMed] [Google Scholar]
- 10.He, W., P. Cowin, and D. L. Stokes. 2003. Untangling desmosomal knots with electron tomography. Science. 302:823–839. [DOI] [PubMed] [Google Scholar]
- 11.Häussinger, D., T. Ahrens, T. Aberle, J. Engel, J. Stetefeld, and S. Grzesiek. 2004. Proteolytic E-cadherin activation followed by solution NMR and x-ray crystallography. EMBO J. 23:1699–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrison, O. J., E. M. Corps, T. Berge, and P. J. Kilshaw. 2005. The mechanism of cell adhesion by classical cadherins: the role of domain 1. J. Cell Sci. 118:711–721. [DOI] [PubMed] [Google Scholar]
- 13.Takeichi, M. 1991. Cadherin cell adhesion receptors as a morphogenetic regulator. Science. 251:1451–1455. [DOI] [PubMed] [Google Scholar]
- 14.Nose, A., K. Tsuji, and M. Takeichi. 1990. Localization of specificity determining states in cadherin cell adhesion molecules. Cell. 61:147–155. [DOI] [PubMed] [Google Scholar]
- 15.Niessen, C. M., and B. M. Gumbiner. 2002. Cadherin-mediated cell sorting not determined by binding or adhesion specificity. J. Cell Biol. 156:389–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sivasankar, S., W. Brieher, N. Lavrik, B. Gumbiner, and D. Leckband. 1999. Direct molecular force measurements of multiple adhesive interactions between cadherin ectodomains. Proc. Natl. Acad. Sci. USA. 96:11820–11824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sivasankar, S., B. M. Gumbiner, and D. Leckband. 2001. Direct measurements of multiple adhesive alignments and unbinding trajectories between cadherin extracellular domains. Biophys. J. 80:1758–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu, B., S. Chappuis-Flament, E. Wong, I. E. Jensen, B. M. Gumbiner, and D. Leckband. 2003. Functional analysis of the structural basis of homophilic cadherin adhesion. Biophys. J. 84:4033–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chappuis-Flament, S., E. Wong, L. D. Hicks, C. M. Kay, and B. M. Gumbiner. 2001. Multiple cadherins extracellular repeats mediate homophilic binding and adhesion. J. Cell Biol. 154:231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perret, E., A. Leung, H. Feracci, and E. Evans. 2004. Trans-bonded pairs of E-cadherin exhibit a remarkable hierarchy of mechanical strengths. Proc. Natl. Acad. Sci. USA. 101:16472–16477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levi, G., D. Ginsberg, J. Girault, I. Sabanay, J. P. Thiery, and B. Geiger. 1991. EP-Cadherin in muscles and epithelia of Xenopus laevis embryos. Development. 113:1335–1344. [DOI] [PubMed] [Google Scholar]
- 22.Lee, C.-H., and B. Gumbiner. 1995. Disruption of gastrulation movements in Xenopus by a dominant-negative mutant for C-cadherin. Dev. Biol. 171:363–373. [DOI] [PubMed] [Google Scholar]
- 23.Evans, E., K. Ritchie, and R. Merkel. 1995. Sensitive force technique to probe molecular adhesion and structural linkages at biological interfaces. Biophys. J. 68:2580–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Evans, E., A. Leung, D. Hammer, and S. Simon. 2001. Chemically distinct transition states govern rapid dissociation of single L-selectin bonds under force. Proc. Natl. Acad. Sci. USA. 98:3784–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bell, G. I. 1978. Models for the specific adhesion of cells to cells. Science. 200:618–627. [DOI] [PubMed] [Google Scholar]
- 26.Evans, E. 2001. Probing the relation between force-lifetime and chemistry in single molecular bonds. Annu. Rev. Biophys. Biomol. Struct. 30:105–128. [DOI] [PubMed] [Google Scholar]
- 27.Bates, D. M., and D. G. Watts. 1988. Non-Linear Regression Analysis and its Applications. John Wiley and Sons, New York.
- 28.Duguay, D., R. A. Foty, and M. S. Steinberg. 2003. Cadherin-mediated cell adhesion and tissue segregation: qualitative and quantitative determinants. Dev. Biol. 253:309–323. [DOI] [PubMed] [Google Scholar]
- 29.Swift, D. L., S. Friedlander, and K. Friedlander. 1964. The coagulation of hydrosols by Brownian motion and laminar shear flow. J. Colloid Sci. 19:621–647. [Google Scholar]
- 30.Dembo, M., D. C. Torney, K. Saxman, and D. Hammer. 1988. The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proc. R. Soc. Lond. B Biol. Sci. 234:55–83. [DOI] [PubMed] [Google Scholar]
- 31.Hammer, D. A., and S. M. Apte. 1992. Simulation of cell rolling and adhesion on surfaces in shear flow: general results and analysis of selectin-mediated neutrophil adhesion. Biophys. J. 63:35–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goldsmith, H. L., T. A. Quinn, G. Drury, C. Spanos, F. A. McIntosh, and S. I. Simon. 2001. Dynamics of neutrophil aggregation in Couette flow revealed by videomicroscopy: effect of shear rate on two-body collision efficiency and doublet lifetime. Biophys. J. 81:2020–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bayas, M. V., K. Schulten, and D. Leckband. 2004. Forced dissociation of the strand dimmer interface between C-cadherin ectodomains. Mech. Chem. Biosys. 1:101–111. [PubMed] [Google Scholar]
- 34.Baumgartner, W., P. Hinterdorfer, W. Ness, A. Raab, D. Vestweber, H. Schindler, and D. Drenckhahn. 2000. Cadherin interaction probed by atomic force microscopy. Proc. Natl. Acad. Sci. USA. 97:4005–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perret, E., A.-M. Benoliel, P. Nassoy, A. Pierres, V. Delmas, J.-P. Thiery, P. Bongrand, and H. Feracci. 2002. Fast dissociation kinetics between individual E-cadherin fragments revealed by flow chamber analysis. EMBO J. 21:2537–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD—visual molecular dynamics. J. Mol. Graph. 14:33–38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.