Abstract
Two-dimensional crystalline bacterial surface layers (S-layers) are found in a broad range of bacteria and archaea as the outermost cell envelope component. The self-assembling properties of the S-layers permit them to recrystallize on solid substrates. Beyond their biological interest as S-layers, they are currently used in nanotechnology to build supramolecular structures. Here, the structure of S-layers and the interactions between them are studied through surface force techniques. Scanning force microscopy has been used to study the structure of recrystallized S-layers from Bacillus sphaericus on mica at different 1:1 electrolyte concentrations. They give evidence of the two-dimensional organization of the proteins and reveal small corrugations of the S-layers formed on mica. The lattice parameters of the S-layers were a = b = 14 nm, γ = 90° and did not depend on the electrolyte concentration. The interaction forces between recrystallized S-layers on mica were studied with the surface force apparatus as a function of electrolyte concentration. Force measurements show that electrostatic and steric interactions are dominant at long distances. When the S-layers are compressed they exhibit elastic behavior. No adhesion between recrystallized layers takes place. We report for the first time, to our knowledge, the value of the compressibility modulus of the S-layer (0.6 MPa). The compressibility modulus is independent on the electrolyte concentration, although loads of 20 mN m−1 damage the layer locally. Control experiments with denatured S-proteins show similar elastic properties under compression but they exhibit adhesion forces between proteins, which were not observed in recrystallized S-layers.
INTRODUCTION
Membrane interactions have been the subject of intense research in the past decades, not only to understand membrane adhesion or fusion, but also to control the properties of artificial membranes in relation with biomedical applications such as drug delivery systems. Several tools exist to probe through direct measurements the interactions either at the level of a single molecule, such as optical tweezers (1), biomembrane force probe (2), and atomic force microscopy (3–7), or at a more collective level with techniques such as surface force apparatus (SFA) (8), vesicle micromanipulation (9,10), and the thin film balance (11,12). The interactions of lipid layers are now fairly well understood (13–16), at least in their main features, which include van der Waals forces, double layer forces, entropic (undulation and protrusion) forces, and hydration forces. In contrast, the ones between more complex membranes, not to mention real membranes (17), remain insufficiently explored. Lipid bilayers can be decorated with functionalized lipids or protein receptors and ligands, which may give them various types of interactions, including forces related to molecular recognition. The use of lipids with large flexible headgroups may give steric repulsions with a range depending on the headgroup features (18), which can interestingly modulate the adhesion. Lipids functionalized with groups, which recognize each other, produce an adhesion-free energy in good agreement with theory (19). Receptors and ligands have, in the past decade, been the subject of numerous studies with various techniques (20–22). Real membranes are often covered by a very complex extracellular membrane matrix (23) made of various proteins distributed inhomogeneously along the membrane, which makes them difficult to use in a model study. Bacterial protein layers (surface layers, S-layers) are two-dimensional (2D) crystalline arrangements of proteins that are composed of a single (glyco)protein and constitute one of the most common outermost cell envelope components of the prokaryotic organisms bacteria and archaea (24,25). Their structure and morphology have been extensively studied (26): these proteins recrystallize through a self-assembly process on a wide range of substrates and exhibit oblique (p2), square (p4), or hexagonal (p3, p6) lattice symmetry, the center-to-center spacing between the morphological units ranging 3–30 nm (27). They have a typical thickness of 5–10 nm.
Fine control and characterization of the crystal structure is crucial to anchor molecular functional units such as streptavidin or antibodies in a regular array (28). This is the basis of multifunctional biological chips that can be used in the future as model systems to investigate in detail molecular interactions (29–31). The S-layers as the main part of the bacterial cell envelope play a role in the protection of the organism against hostile environmental conditions. They are of major importance for cell-cell interaction and cell growth and division and should withstand expansion to keep the cell shape in archaea (25,32,33). In this work, recrystallized bacterial S-layers are used as a model system to investigate the relation between structure, stability, and interaction forces between cell walls. This is studied with the scanning force microscope and the SFA. Recently, the correlation between these techniques has been successfully used to study the stability of supported lipid bilayers (7). As a consequence, the application of both techniques to measure physicochemical properties of bacterial S-layers such as thickness, elasticity, charge density, thermal stability, and interaction forces contribute to understand the behavior of these biological systems. We report the timescale of the recrystallization process of the S-protein SbpA on mica, investigated with scanning force microscopy (SFM). The morphology of the S-layer in water at different electrolyte concentrations of KNO3 was also studied via SFM. We have found that the 2D crystalline structure of the S-layer is independent of the electrolyte concentration. The thickness of the recrystallized S-layer was obtained with the SFM and found to be 13.5 nm, which corresponds to a protein bilayer (34). The interaction force between two S-layer surfaces studied by SFA in water under the same electrolyte conditions is repulsive. At separation distances between 30 and 100 nm, the interaction depends on the electrolyte concentration. Below 30 nm, the repulsive force does not depend on electrolyte concentration and the S-layers exhibit elastic behavior. No adhesion takes place between recrystallized S-layers. The mean value of the compressibility modulus of the S-layers is ∼0.6 MPa. Experiments with denatured S-layers show another repulsion regime, similar compressibility modulus, and the appearance of adhesion forces between them.
MATERIALS AND METHODS
S-layers
The bacterial cell surface layer protein SbpA used in this study was isolated from Bacillus sphaericus CCM 2177. Growth in continuous culture, cell wall preparation, extraction of S-layer protein with guanidine chloride (GHCl), dyalization, and further centrifugation was done according to Sleytr et al. (35). The concentration of SbpA was adjusted with Milli-Q water (Millipore, Bedford, MA) to 1 mg mL−1. This solution was used for all recrystallization experiments.
SbpA recrystallization
Recrystallization experiments were carried out in mini petri dishes (30 mm diameter, 5 mL volume). Mica slides of ∼1 cm2 area were immersed and kept for at least 1 h in buffer solution containing protein monomers (the minimum incubation time has been found to be 30 min, see Results). The protein/buffer volume ratio was 0.1:1 for every sample. The buffer consisted of 0.5 mM Tris-HCl buffer, pH 9, with added 10 mM CaCl2. The samples with recrystallized protein were washed with Milli-Q water before starting the experiments. KNO3 (Merck suprapure) was the electrolyte used in the experiments. The water used had a specific resistivity of 18.2 MΩ cm−1 (Elgastat Maxima for HPLC unit). The mica used was high quality Ruby Muscovite. Aqueous solutions adjusted to pH 3 were prepared with Millipore water, citric acid-monohydrate (Merck, Darmstadt, Germany), and hydrochloric acid (Sigma-Aldrich, St. Louis, MO) indistinctively.
Scanning force microscopy
SFM images have been recorded in contact mode in water, 10 mM and 100 mM KNO3 at room temperature using a Nanoscope III multimode (Veeco Instruments, Santa Barbara, CA). The multimode was equipped with a 12 × 12 μm2 scanner. Silicon nitride (Si3N4) cantilevers (NP-S, Veeco Instruments) with a spring constant of 0.1 N m−1 were used. The spring constants of the cantilevers were calculated using the thermal method (36) in water. The scan rate for imaging was ∼5 Hz (5 μm s−1 for a 500 × 500 nm2 scan area) under a constant force of 0.2–2.5 nN. The imaging force for protein resolution depends on the chosen medium: in water, forces of 1.5–2.5 nN allowed protein resolution; meanwhile for 10 mM and 100 mM KNO3, lower forces of 0.2–1 nN were necessary to get the crystalline structure of the film. The thickness of the studied S-layers was obtained using the “scratching” method. Scratching of the S-layer was carried out by applying a scanning force of 10–20 nN at 5 μm s−1 over a 250 × 250 nm2 area. Then, a larger region containing the damaged area was scanned under the nonwearing imaging conditions (see above). Thickness of the S-layer was computed by surface profile analysis on different positions and samples.
Surface force apparatus
Interactions between S-layers have been measured with a homemade SFA. This technique uses cleaved mica sheets (1–4 μm thick), with back surfaces silvered, which are glued to cylindrical glass lenses with curvature radius R (2 cm in this case). The two surfaces are in a crossed-cylinder geometry and they can be moved toward or away from each other by a micrometer shaft and a differential spring system. SFA measures the force F between the mica surfaces as a function of the actual distance D between them. Here, a leaf spring is used to measure the force with an uncertainty of 10−7 N, whereas the distance is obtained by white light multiple beam interferometry with Angstrom resolution. For a complete description of this technique, see Israelachvili and Adams (8).
The force at a particular distance between the surfaces is measured once thermal equilibrium is attained. Then, the surfaces are brought closer to each other and the procedure is repeated at the new distance. The experiment is completed when the load force between the surfaces in contact reaches a certain value (i.e., maximum load).
The distance between the S-protein layers was computed using the contact between the bare mica substrates as reference (i.e., zero distance). Two more parameters are important in this type of experiment: the delay time between two consecutive force measurements and an adequate maximum load. The former is determined by the time needed for equilibration of the SFA system at a particular distance, which is ∼45 s. Accordingly, intervals of 60 s were chosen for all experiments. To check whether the compression of S-layers can result in irreversible changes in their structure, various SFA experiments with different maximum loads (expressed as Fmax/R) were performed. The experimental protocol was the following: i), the surfaces were approached and pressed until Fmax/R = 2 mN m−1 before separation, ii), the same procedure was performed with an Fmax/R = 20 mN m−1, and iii), step i was repeated for comparison. The measurements in steps i and ii were carried out twice to check reproducibility. This protocol was performed on at least two different locations within the S-layers to check their homogeneity. The experiments were repeated on different samples and done in pure water (pH ≅ 5.5) and in aqueous solutions of KNO3 (10 mM and 100 mM).
RESULTS
Recrystallization and structure of an S-layer studied by SFM S-layer recrystallization on freshly cleaved mica was monitored online using contact SFM. The SFM tip was mounted in the enclosed liquid cell on the bare mica substrate, and the cell was filled with protein solution. After stabilization, the tip was continuously scanning the same area (2 μm2) on the mica substrate and images of the scanned surface were sequentially recorded. Fig. 1 shows the height images of the recrystallizing S-layer recorded at different times. Although after 4 min the mica seems to have some S-proteins on the surface, it is after 7 min when the first S-layer domains of size between 40 and 120 nm are clearly distinguished. The recrystallization process takes places continuously with time, and after 32 min the S-proteins cover the scanned area forming the S-layer. From this time on, the roughness (root mean square (RMS), Rq) of the S-layer remains approximately equal to 1 nm. No noticeable difference in the protein layer structure can be seen at longer times.
FIGURE 1.
(a–f) SFM height images obtained in contact mode on S-protein layers recrystallized on mica recorded at different self-assembling times: (a) mica, after 4 min; (b) after 7 min; (c) after 11 min; (d) after 21 min; (e) after 21 min; and (f) after 32 min. The S-layer covers the whole area after 32 min, and no significant differences between the images can be observed at later times.
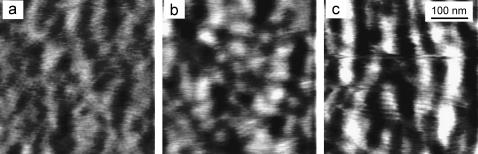
Fig. 2 shows 500 × 500 nm2 height images obtained with SFM in contact mode of S-protein layers crystallized on mica in water, 10 mM and 100 mM KNO3 aqueous solutions, respectively. Two main features can be seen: the typical crystalline structure of the SbpA layer, the p4 square lattice (26), and the different crystalline domains extending over areas of 50 × 50 – 150 × 150 nm2. This is typically seen when simultaneous nucleation points are formed in the early stages of the S-protein recrystallization on hydrophilic substrates (37). Fig. 3 shows deflection SFM images corresponding to the height images shown in Fig. 2, respectively, together with the 2D Fourier transforms on depicted regions of different areas within the images (white frame rectangles). The square lattice parameters obtained by 2D Fourier transformation on the periodical structures of at least two different domains within the scanned area are a = 14 ± 1 nm, b = 14 ± 2 nm for Fig. 3 a, a = 14 ± 2 nm, b = 14 ± 2 nm for Fig. 3 b, and a = 14 ± 1 nm, b = 14 ± 1 nm for Fig. 3 c. Within the experimental error, these values agree with those obtained for S-layers on bacteria (26). These values also show that the 2D crystalline structure of the S-layer does not depend on the electrolyte, i.e., the S-protein layer crystals are not altered by the presence of electrolyte concentration.
FIGURE 2.
(a–c) SFM height images obtained in contact mode on S-protein layers crystallized on mica. (a) pure water; (b) KNO3 10 mM; and (c) KNO3 100 mM. The vertical scale is 3 nm from black to white. The images show different positions of the same sample.
FIGURE 3.
(a–c) SFM deflection images obtained in contact mode on S-protein layers crystallized on mica. (a) pure water; (b) KNO3 10 mM; and (c) KNO3 100 mM. The vertical scale is 1 nm from dark brown to white. In lower panels, the 2D Fourier transformation corresponds to each domain within each image. Note the halo corresponding to the network periodicity.
The RMS roughness (Rq) of the protein layer was found to be within a range from 0.3 nm to 0.6 nm over different samples and positions (see Table 1). S-layers were scratched with an SFM tip and subsequently imaged over a larger area to obtain the layer thickness. Care was taken to scratch with a force high enough to remove the soft S-layer without damaging the mica substrate underneath. Wear studies on bare mica show that mica is damaged by the SFM tip when the scratch force is at least 90 nN (wear is visible, with a step height of 0.3 nm in Fig. 4 b). The mechanical properties of the mica do not depend on the electrolyte.
TABLE 1.
RMS roughness (Rq) of S-layers as a function of the KNO3 concentration obtained from SFM images; the values are averages from three different samples
| KNO3 concentration/M | Rq (RMS)/nm |
|---|---|
| Milli-Q water | 0.29 ± 0.05 |
| 0.01 | 0.6 ± 0.3 |
| 0.1 | 0.6 ± 0.3 |
FIGURE 4.
(a) Height SFM images in contact mode of scratched S-protein layers crystallized on mica (applied shear force, 10 nN). The scratched area is 250 × 250 nm2. The surface profile below corresponds to the depicted line on the image. A step height of 13.3 ± 0.1 nm is obtained from the profile. The vertical scale from black to white is 5 nm. (b) Height SFM image in contact mode of a bare mica surface, showing the scratched region when the shear force is 90 nN (almost an order of magnitude larger than the force necessary to scratch the S-layer). The step height between a worn and a nonworn mica region is 0.3 nm.
We have found that a shear force of 10 nN is enough to remove the S-layer from the mica substrate, and the obtained S-layer thickness does not show significant differences when the shear force is increased to 20 nN. Fig. 4 a shows the scratched area and the corresponding surface profile along the depicted line. It can be seen that crystalline structure is absent within the scratched area, whereas it is present in the remaining, nonscratched region. The step height of 13.3 ± 0.1 nm shows the figure corresponds, therefore, to the thickness of the S-layer. Averaging over surface profiles along different lines, different scratching areas, and different S-layer samples gives a value of 13.5 ± 0.9 nm. This value corresponds to one protein bilayer, and it is in agreement with the thickness of recrystallized SbpA on hydrophilic Si wafers (37) and on polyelectrolyte supports (34).
Forces between two S-layers studied by SFA in aqueous media
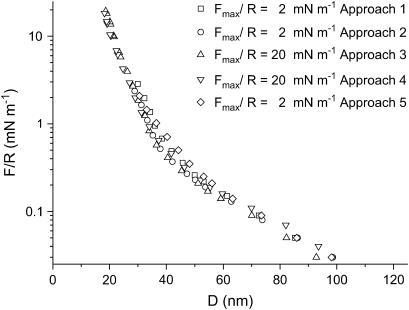
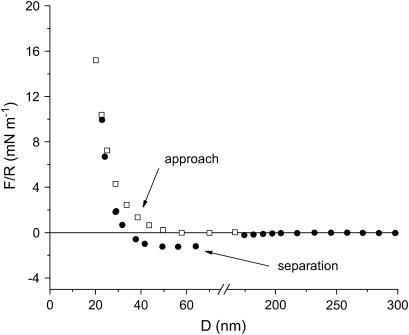
Forces between S-layers in water measured with the SFA are shown in Fig. 5. As can be seen, the interactions between the S-layers are similar after they have been pressed against each other with a load of 20 mN m−1. This shows that the layers are not damaged by the strongest applied compression. Upon separation, no adhesive force is measured and no hysteresis in the force is observed. Therefore, only approaching curves are shown. Another remarkable feature of these curves is the existence of several regimes as a function of the separation distance (D). At D > 100 nm, the force is zero. For distances in the interval 40 nm < D < 100 nm, the force varies exponentially with distance with a decay length of λW1 ∼ 22 ± 2 nm. Finally, the last regime of interaction observed appears at distances 20 nm < D < 40 nm, where the curves also show an exponential decreasing behavior but with a decay length smaller than that of the previous regime (λW2 = 5.5 ± 0.4 nm).
FIGURE 5.
SFA interaction measurements between two S-layers in aqueous media as a function of the separation distance D for two values of Fmax/R and different approaches. The force/distance profiles are reproducible even after the S-layers were brought to a force F/R of 20 mN m−1. The error of the measurement is 0.05 mN m−1, no error bars have been plotted to make the graph clearer.
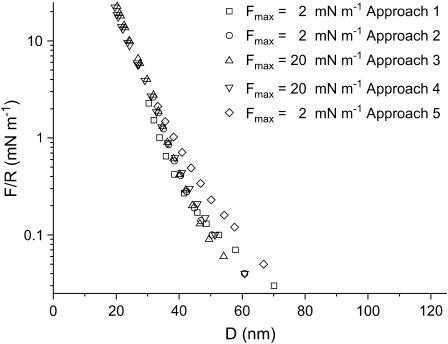
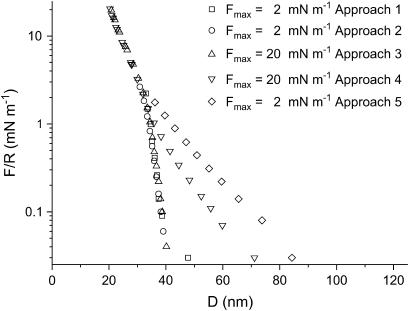
Fig. 6 shows the interaction curves in the presence of 10 mM KNO3 aqueous solution. The force is zero for D > 75 nm. Below this distance, the interaction force is repulsive showing a decay length of 9 ± 1 nm. Below 40 nm, a second exponential regime sets in. This regime is similar to the one observed in pure water, i.e., an exponential repulsion with a decay length of 5.5 nm. In the curves shown in Fig. 6 (10 mM KNO3) only the last one is shifted toward larger distances, indicating that the layer has been affected by the compression at 20 mN m−1 in contrast with S-layers in pure water. Like the results obtained in pure water, no adhesive force is measured upon separation of the surfaces and no hysteresis is observed. Fig. 7 shows the interaction between S-layers in 100 mM KNO3 aqueous solution. When the layers have not been subjected to a strong compression, a first exponential force regime sets in within the range 32 nm < D < 40 nm and a decay length of λw3 ∼ 1.7 ± 0.3 nm. At smaller distances, the same force regime is observed as found in 10 mM electrolyte and in water. Fig. 7 also shows that there are significant differences in the curves obtained after the layers have been subjected to a strong compression. This shows that compression at high electrolyte concentrations produces irreversible changes in the protein layer structure. As a result the repulsive interaction at distances higher than 32 nm is of longer range. Contrarily, the short-range repulsion at distances shorter than 32 nm is not affected. As in the preceding cases, no adhesion is observed upon separation.
FIGURE 6.
SFA interaction measurements between two S-layers in 10 mM KNO3 solution as a function of the separation distance D for two values of Fmax/R and different approaches. After the S-layers were brought to a force F/R of 20 mN m−1, the long-range force regime is different (error bar, 0.05 mN m−1).
FIGURE 7.
SFA interaction measurements between two S-layers in 100 mM KNO3 solution as a function of the separation distance for several values of Fmax/R and different approaches. After the S-layers were brought to a force F/R of 20 mN m−1, the forces are of longer range. However, the short-range regime is not affected (error bar, 0.05 mN m−1).
Control experiments with denatured S-layers were carried out. Table 2 shows the decay length of the repulsive force and the calculated elastic modulus when the denatured S-layers are approached and compressed, respectively. Fig. 8 shows the interaction between two denatured S-layer surfaces in 100 mM KNO3. It can be seen that after compression the denatured protein layers show adhesion forces, which could not be measured for recrystallized S-layers.
TABLE 2.
Decay length and compressibility modulus for denatured S-layers
| Water | 10 mM | 100 mM | |
|---|---|---|---|
| λ1 (nm) | 20 ± 4 | 6.0 ± 1.6 | 6.2 ± 1.4 |
| λ2 (nm) | 5.7 ± 0.6 | 6.0 ± 1.6 | 6.2 ± 1.4 |
| k (Mpa) | 0.60 ± 0.06 | 0.64 ± 0.15 | 0.7 ± 0.2 |
The value of the modulus is similar to the value obtained for recrystallized S-layers. Conversely, the decay lengths in the presence of KNO3 denote only one repulsive regime.
FIGURE 8.
SFA interaction measurement between two denatured S-layer surfaces. The decay of the repulsive force is different than for recrystallized S-layers. The main result is the appearance of adhesion forces, which are not detected when the protein layer is crystalline.
DISCUSSION
S-proteins recrystallize on mica, forming a protein bilayer with crystal domains of 50 × 50 – 150 × 150 nm2 in size and an RMS roughness of ∼1 nm. The Rq values are small relative to the variation of the forces with distance shown in Figs. 5–7, and they should not have any effect on these forces. The interaction between S-layers in water and in 1:1 electrolyte aqueous solution is characterized by two force regimes. At distances >32 nm, the repulsive force depends on electrolyte concentration, whereas at smaller distances, no influence of the electrolyte could be detected. At 100 mM KNO3, the protein layers appear to be modified after being pressed with a maximum load of 20 mN m−1.
At 10 mM KNO3 or in pure water, the protein layer is not affected by the stronger compression. In all cases, there is no adhesion. This is consistent with the SFM observation that once a bilayer is adsorbed, no more protein adsorbs on it.
Repulsive forces between recrystallized S-layers start at distances of 100 nm, 60 nm, and 40 nm in pure water, 10 mM electrolyte, and 100 mM electrolyte, respectively. The shorter range of the forces when electrolyte is present is consistent with the lower force needed in SFM to image the structure of the protein layer, in contrast with the force needed to image the S-layers in pure water. In the long-distance regime, the force is repulsive, with an exponential decay length that decreases when the electrolyte concentration is increased. One is, therefore, tempted to attribute this force to an electrostatic double-layer interaction. However, the decay lengths are not close to the values expected from a purely electrostatic interaction: in water, the decay length is 22 nm, much lower than the expected value of 170 nm; in 10 mM KNO3, one would expect 3 nm, but the experimental value is 9 nm; only in the case of 100 mM KNO3 is the decay length (1.7 nm) not far from the expected value (0.96 nm). This leads to the conclusion that in addition to electrostatic double-layer forces, there should be other forces contributing to the interaction.
One possible origin could be that after the layers are formed from a solution composed of 0.5 mM Tris-HCl and 10 mM CaCl2 and the surfaces transferred into the SFA, some CaCl2 would also be transferred in the apparatus. However, this is unlikely because the surfaces are first transferred from a 5 mL beaker into a 2 L beaker of ultrapure water to wash away the excess of proteins and ions of the solution before being mounted in the SFA. Dissimilarities between the decay length obtained experimentally and the Debye length in pure water have been previously found in other SFA experiments (38). Another possible origin of these small forces could be steric forces coming from a small quantity of polypeptide chains sticking out from the proteins of the S-layer. In fact, in our case, steric and double-layer interactions seem to act over the same distance range and are, therefore, mixed. These are generally called electrosteric interaction (39). The short-range force regime is the same in the three electrolyte concentrations studied. At distances <32 nm, the proteins are pressed against each other and the measured forces are likely to be the elastic response of the S-layers.
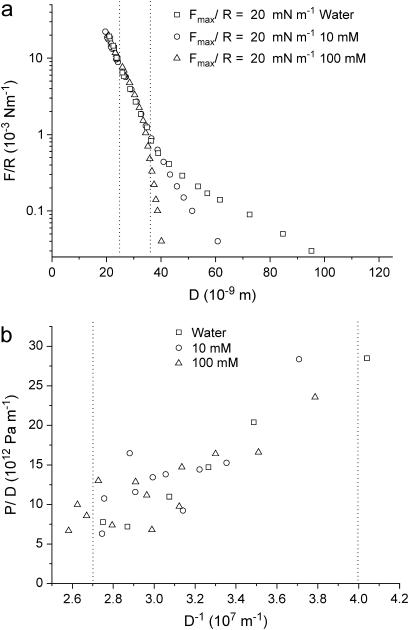
One can therefore conclude that the elastic compression of S-layers is a purely reversible and reproducible process within these experimental conditions. In the three electrolyte concentrations studied, the compression regime starts around 32 nm. This distance is close to two recrystallized S-layers on mica and is fairly consistent with the thickness (13.5 nm) measured by SFM. The compressibility of the S-layers can be quantified through the elastic modulus, k, which characterizes the deformation of a material (ΔD) when it is subjected to a change of pressure ΔP perpendicular to the surface: k = D dP/dD. The pressure P is obtained from Derjaguin's approximation (40), which relates the force F(D) between two curved surfaces of radius R to the energy per unit area E(D) between flat surfaces: F(D) = 2πRE(D). Taking the derivative relative to D of each member leads to the pressure P = (2πR)−1dF/dD. Therefore, from the values of F/R and D, one can calculate P versus D. The value of the compressibility modulus of the S-layers is given by the slope in the graph of Fig. 9 b. In this way, a value of k at the onset of the compression can be deduced from the points between the two dotted vertical lines in Fig. 9 a. This value, 0.6 ± 0.2 MPa, obtained for water, 10 mM, and 100 mM, is independent of electrolyte concentration. The same calculation has been carried out for the denatured S-protein layer (see Table 2), obtaining the same value within experimental error. This could be explained by considering that the denaturation process does not lead to protein desorption from the mica surface (41), leading to the compression of the same amount of polymer for a constant volume. However, the important feature of the control experiments with denatured proteins is the existence of adhesion forces. These results show that recrystallized S-layers avoids cell wall-cell wall fusion, meaning that another component of the bacterial cell wall should be responsible for such biological phenomenon. Even though Table 2 shows the decay of the repulsive force between denatured S-layers, no real output of the repulsive regime can be asserted. This is due to the fact that there are several ways of denaturating the S-layer leading to different surface topology (41) and therefore to a variety of force regimes. Mechanical properties of other bacterial surfaces have been reported in the literature. Thus the B. subtilis envelope was found to have a compressibility of 107 Pa (42), and the turgor pressure of the M. gryphiswaldense envelope was 105 Pa (43). A compressibility modulus of 2.5 × 107 Pa was reported for gram negative Murein sacculi (44).
FIGURE 9.
(a) Effect of the electrolyte concentration in the interaction forces between S-layers. (b) Rescaled graph showing the elastic compression of the S-layer calculated using Derjaguin's approximation. The points between the two dotted lines are used to calculate the elastic modulus of the S-layers from the equation dP/dD = k/D.
CONCLUSIONS
The recrystallization of the S-layer protein SbpA on mica has been monitored online with a scanning force microscope. The self-assembly process led to single crystal domains of areas ranging from 50 × 50 nm2 to 150 × 150 nm2 after approximately half an hour. The calculated lattice parameters on single crystal domains, a = b = 14 nm, γ = 90°, are in agreement with values obtained for bacteria. Neither the lattice parameters nor the RMS roughness of the protein layer showed any dependence on the used KNO3 concentrations. The threshold shear force necessary to disrupt the recrystallized SbpA protein layer on mica was 10 nN. A thickness of the adsorbed protein layer of 13.5 nm, which corresponds to a protein bilayer, was determined by the scratching method. This value is in agreement with the distance measured interferometrically with the SFA when two S-protein layers are brought into contact. The interaction forces between two S-layers in aqueous media are repulsive. The surfaces have been found to be nonadherent for recrystallized S-layers. Conversely, denaturation of the protein layer leads to adhesion behavior. These results show that recrystallized SbpA proteins do not promote cell-cell adhesion. The repulsive interaction between recrystallized S-layers follows two regimes. The long-range regime is exponential with a decay length that changes with ionic strength. However, electrostatic double-layer forces are not enough to explain the experimental results, and the effect of steric interactions cannot be ruled out. At high ionic strength and after the S-layers have been compressed, the interaction range changes, indicating that the surface of the protein layers has been modified. At smaller distances, below 40 nm, a short-range regime is observed, which does not depend strongly on the ionic strength. The exponential decay length is ∼5 nm. This force regime is not altered after applying a high load (20 mN m−1) upon the surfaces, in contrast to what was observed at longer distances illustrating the stability of the protein conformation. In this regime, the compression of the S-layer takes place. Although the core of the protein layer was damaged locally after several compression cycles at 20 mN m−1, a value of 0.6 Mpa of the compressibility modulus of the recrystallized S-layer could be obtained.
Acknowledgments
The authors thank J. Friedmann and R. Berger for technical support. R. v. Klitzing, K. Melzak, and S. Rodríguez are acknowledged for reading the manuscript.
This work was supported by the FP5 EU-RTN program “Nanocapsules” No. HPRN-CT-2000-00159 and AFOSR project F49620-03-1-0222. S.M.F. also thanks the Max Planck Society for financial support. J.L.T.H. is a Ramón y Cajal Senior Research Fellow (Program of the Spanish Ministry of Education and Science).
References
- 1.Askin, A. 1997. Optical trapping and manipulation of neutral particles using lasers. Proc. Natl. Acad. Sci. USA. 94:4853–4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans, E., K. Ritchie, and R. Merkel. 1995. Sensitive force technique to probe molecular adhesion and structural linkages at biological interfaces. Biophys. J. 68:2580–2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binnig, G., C. F. Quate, and Ch. Gerber. 1986. Atomic force microscope. Phys. Rev. Lett. 56:930–933. [DOI] [PubMed] [Google Scholar]
- 4.Butt, H.-J. 1991. Measuring electrostatic, van der Waals, and hydration forces in electrolyte solutions with an atomic force microscope. Biophys. J. 60:1438–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rief, M., F. Oesterheld, B. Heymann, and H. E. Gaub. 1997. Single molecule force spectroscopy on polysaccharides by atomic force microscopy. Science. 275:1295–1297. [DOI] [PubMed] [Google Scholar]
- 6.Williams, P. M., S. B. Fowler, R. B. Best, J. L. Toca-Herrera, K. Scott, A. Steward, and J. Clarke. 2003. Hidden complexity in the mechanical properties of titin. Nature. 442:446–449. [DOI] [PubMed] [Google Scholar]
- 7.Benz, M., T. Gutsmann, C. Nianhuan, R. Tadmor, and J. Israelachvili. 2004. Correlation of SFM and SFA measurements concerning the stability of supported lipid bilayers. Biophys. J. 86:870–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Israelachvili, J. N., and G. E. Adams. 1978. Measurement of forces between two mica surfaces in aqueous electrolyte solutions in the range 0–100 nm. J. Chem. Soc. Faraday Trans I. 4:975–1001. [Google Scholar]
- 9.Pincet, F., T. Le Bouar, Y. Zhang, J. Esnault, J. M. Mallet, E. Perez, and P. Sinay. 2001. Ultraweak sugar-sugar interactions for transcient cell adhesion. Biophys. J. 40:1354–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ly, H. V., D. E. Block, and M. L. Longo. 2002. Interfacial tension effect of ethanol on lipid bilayer rigidity, stability, and area/molecule: a micropipet aspiration approach. Langmuir. 18:8988–8995. [Google Scholar]
- 11.Bergeron, V., and P. M. Claesson. 2002. Structural forces reflecting polyelectrolyte organization from bulk solutions and within surface complexes. Adv. Colloid Interface Sci. 96:1–20. [DOI] [PubMed] [Google Scholar]
- 12.Klitzing, R. V., and H.-J. Müller. 2002. Film stability control. Curr. Opin. Colloid Interface Sci. 7:18–25. [Google Scholar]
- 13.Israelachvili, J. N., and H. Wennerstrom. 1992. Entropic forces between amphiphilic surfaces in liquids. J. Phys. Chem. 96:520–553. [Google Scholar]
- 14.Rand, R. P., and V. A. Parsegian. 1989. Hydration forces between phospholipid bilayers. Biochim. Biophys. Acta. 988:351–376. [Google Scholar]
- 15.Toca-Herrera, J. L., H. J. Müller, R. Krustev, T. Pfohl, and H. Möhwald. 1999. Influence of ethanol on the thickness and free energy of film formation of DMPC foam films. Colloids Surf. A. 152:357–365. [Google Scholar]
- 16.Toca-Herrera, J. L., H. J. Müller, R. Krustev, and H. Möhwald. 2000. Effect of charged lipid DMPG on the thickness and contact angle of foam films. J. Phys. Chem. B. 104:5486–5491. [Google Scholar]
- 17.Chu, Y. S., W. A. Thomas, O. Eder, F. Pincet, E. Perez, J.-P. Thiery, and S. Dufour. 2004. Quantification of separation forces in e-cadherin-mediated cell doublets reveals rapid adhesion strengthened by actin cytoskeleton remodeling through RAC and CDC42 but not RHO. J. Cell Biol. 167:1163–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhl, T. L., D. E. Leckband, D. D. Lasic, and J. N. Israelachvili. 1994. Modulation of interaction forces between bilayers exposing short-chained ethylene-oxide headgroups. Biophys. J. 66:1479–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pincet, F., E. Perez, J. C. Loudet, and L. Lebeau. 2001. Reversible adhesion with mobile sites: a test of the theory. Phys. Rev. Lett. 87:178101–178104. [DOI] [PubMed] [Google Scholar]
- 20.Helm, C. A., W. Knoll, and J. N. Israelachvili. 1991. Measurement of ligand receptor interactions. Proc. Natl. Acad. Sci. USA. 88:8169–8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sivasankar, S., W. Brieher, N. Lavrik, B. Gumbiner, and D. Leckband. 1999. Direct molecular force measurements of multiple adhesive interactions between cadherin domains. Proc. Natl. Acad. Sci. USA. 96:11820–11824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Merkel, R., P. Nassoy, A. Leung, K. Ritchie, and E. Evans. 1999. Energy landscapes of receptor-ligand bonds explored with dynamic force spectroscopy. Nature. 397:50–53. [DOI] [PubMed] [Google Scholar]
- 23.Sabri, S., M. Soler, C. Foa, A. Pierres, A. M. Benoliel, and P. Bongrand. 2000. Glycocalyx modulation is a physiological means of regulating cell adhesion. J. Cell Sci. 113:1589–1600. [DOI] [PubMed] [Google Scholar]
- 24.Sleytr, U. B., P. Messner, D. Pum, and M. Sára, editors. 1996. Crystalline Bacterial Cell Surface Proteins (Molecular Biology Intelligence Unit). Academic Press R. G. Handes, Austin, TX.
- 25.Sleytr, U. B., and T. J. Beveridge. 1999. Bacterial S-layers. Trends Microbiol. 7:253–260. [DOI] [PubMed] [Google Scholar]
- 26.Sleytr, U. B., P. Messner, D. Pum, and M. Sára. 1999. Crystalline bacterial cell surface layers (S-layers): from supramolecular cell structure to biomimetics and nanotechnology. Angew. Chem. Int. Ed. Engl. 38:1034–1054. [DOI] [PubMed] [Google Scholar]
- 27.Sleytr, U. B., M. Sára, D. Pum, B. Schuster, P. Messner, and C. Schaeffer. 2002. Self assembly protein systems: microbial S-layers. In Biopolymers, Vol. 7. A. Steinbuechel, S. Fahnenstock, editors. Wiley-VCH, Weinheim, Germany. 285–338.
- 28.Sleytr, U. B., M. Sára, D. Pum, and B. Schuster. 2005. Crystalline bacterial cell surface layers (S-layers): a versatile self-assembly system, In Supramolecular Polymers, 2nd ed. A. Ciferri, editor. CRC Press, Taylor & Francis Group, Boca Raton, FL. 583–613.
- 29.Moll, D., C. Huber, B. Schlegel, D. Pum, U. B. Sleytr, and M. Sára. 2002. S-layer-streptavidin fusion proteins as template for nanopatterned molecular arrays. Proc. Natl. Acad. Sci. USA. 99:14646–14651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ilk, N., S. Küpcü, G. Moncayo, S. Klimt, R. Ecker, R. Hofer-Warbinek, E. M. Egelseer, U. B. Sleytr, and M. Sára. 2004. Heterologous expression of a chimaeric S-layer/enhanced green fluorescent protein to follow the uptake of S-layer-coated liposomes into eukaryotic cells. Biochem. J. 370:441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Völlenkle, C., S. Weigert, N. Ilk, E. M. Egelseer, V. Weber, F. Loth, D. Falkenhagen, U. B. Sleytr, and M. Sára. 2004. Construction of a functional S-layer fusion protein comprising an IgG-binding domain for the development of specific adsorbents for extracorporeal blood purification. Appl. Environ. Microbiol. 70:1514–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sleytr, U. B., and M. Sára. 1997. Bacterial and archaeal S-layer proteins. Structure-function relationships and their biotechnological applications. Trends Biotechnol. 15:20–26. [DOI] [PubMed] [Google Scholar]
- 33.Seltmann, G., and O. Holst. 2002. The Bacterial Cell Wall. Springer, Heidelberg, Germany. 219–261.
- 34.Toca-Herrera, J. L., R. Krastev, V. Bosio, S. Küpcü, D. Pum, A. Fery, M. Sara, and U. B. Sleytr. 2005. Recrystallization of bacterial S-layers on flat polyelectrolyte surfaces and hollow polyelectrolyte capsules. small. 1:339–348. [DOI] [PubMed] [Google Scholar]
- 35.Sleytr, U. B., M. Sara, S. Küpcü, and P. Messner. 1986. Structural and chemical characterization of S-layers of selected strains of Bacillus stearothermophilus and Desulfotomaculum nigrificans. Arch. Microbiol. 146:19–24. [DOI] [PubMed] [Google Scholar]
- 36.Hutter, J., and J. Bechhoefer. 1993. Determination of the spring constants of probes for force microscopy/spectroscopy. J. Rev. Sci. Instrum. 64:1868–1873. [Google Scholar]
- 37.Gyorvary, E., O. Stein, D. Pum, and U. B. Sleytr. 2003. Self-assembly and recrystallization of bacterial S-layer proteins at silicon supports imaged in real time by atomic force microscopy. J. Microsc. 212:300–306. [DOI] [PubMed] [Google Scholar]
- 38.Argillier, J. F., R. Ramachandran, W. C. Harris, and M. Tirrell. 1991. Polymer surfactant interactions studied with the surface force apparatus. J. Coll. Interface Sci. 146:242–250. [Google Scholar]
- 39.Borget, P., F. Lafuma, and C. Bonnet-Gonnet. 2005. Characterizations and properties of hairy latex particles. J. Coll. Interface Sci. 285:136–145. [DOI] [PubMed] [Google Scholar]
- 40.Israelachvili, J. N. 1992. Intermolecular and Surface Forces. Academic Press, London.
- 41.Toca-Herrera, J. L., S. Moreno-Flores, J. Friedmann, D. Pum, and U. B. Sleytr. 2004. Chemical and thermal denaturation of crystalline bacterial S-layer proteins. Microsc. Res. Techniq. 65:226–234. [DOI] [PubMed] [Google Scholar]
- 42.Thwaites, J. J., U. C. Surana, and A. M. Jones. 1991. Mechanical properties of Bacillus subtilis cell walls: effect of ions and lysozyme. J. Bacteriol. 173:204–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arnoldi, M., M. Fritz, E. Bäuerlein, M. Radmacher, E. Sackmann, and A. Boulbitch. 2000. Bacterial turgor pressure can be measured by atomic force microscopy. Phys. Rev. E. 62:1034–1044. [DOI] [PubMed] [Google Scholar]
- 44.Dufrene, Y. F. 2002. Atomic force microscopy, a powerful tool in microbiology. J. Bacteriol. 184:5205–5213. [DOI] [PMC free article] [PubMed] [Google Scholar]