Abstract
The final stages of budding and release of a retroviral particle from the cell require the late (L) domain of Gag. Recently, ubiquitin and ubiquitin ligases have been implicated in the late stages of retroviral budding. In a yeast two-hybrid screen of a T-cell cDNA library to identify cellular proteins that interact with human immunodeficiency virus type 2 (HIV-2) Gag polyprotein, we identified Tsg101, an inactive homologue of ubiquitin ligase E2. Tsg101 and HIV-2 Gag interact specifically in vitro and in vivo. The interaction requires the L domain PTAPP motif in the p6 domain of HIV-2 Gag and the N-terminal Ubc-conjugation homology domain of Tsg101. Tsg101 is incorporated into HIV-2 virions. Expression of the N-terminal Ubc-conjugation homology domain of Tsg101 inhibits the release of HIV-2 virus particles. Overexpression of Tsg101 results in an increase in the level of ubiquitination of HIV-2 Gag. Our results provide evidence for recruitment of the ubiquitination machinery of the cell during late stages of the viral life cycle, mediated by the viral Gag protein.
Retroviral Gag proteins play a crucial role in the virus life cycle, mediating virion assembly, association with viral genomic RNA, and virus release (47). The Gag protein is synthesized as a polyprotein precursor that, on activation of the viral protease during the budding process, is cleaved to yield the mature virion components: matrix (MA), capsid (CA), nucleocapsid (NC), p6, and two spacer peptides p2 and p1. Gag proteins contain conserved domains required for membrane targeting (M), interaction between Gag polyproteins (I), and late stages of virus particle assembly and release from the cell (L). Although the amino acid sequences of the L domains are not conserved among retroviruses, they are functionally equivalent and interchangeable between different members of the Retroviridae (36). The L domain is required for the final stages of virion release from the cell (14, 20, 54, 55) and has recently been reported to interact with the ubiquitination machinery (37, 43, 46, 52). Further evidence for a role of ubiquitin and ubiquitin ligases in retroviral budding has come from the observation that depletion of intracellular pools of free ubiquitin by treatment of cells with proteasome inhibitors results in inhibition of retroviral budding (37, 43). Ubiquitin is also found within retroviral particles (1, 33, 34, 40), and a small proportion of human immunodeficiency virus type 1 (HIV-1) p6 and Moloney murine leukemia virus (MoMLV) p12 (both of which contain the L domain) present in virions is monoubiquitinated (14, 56).
A number of cellular proteins interact with Gag. Cyclophilin A, a prolyl isomerase, interacts with HIV-1 Gag (30) and is incorporated into virions, enhancing virion infectivity (12). HO3, a histidyl-tRNA synthetase, binds to MA of HIV-1 (26). KIF4, a microtubule-associated motor protein, interacts with retroviral Gag proteins, including those of HIV-1 (48) and MoMLV (23). The cytoskeletal proteins actin, ezrin, moesin, and cofilin have been found in virions (35). Human translation factor elongation factor 1α (EF1α) interacts with HIV-1 Gag MA and NC domains and is incorporated into virions (5). A novel virion-associated protein, called VAN, binds to HIV-1 MA (16). The C-terminal fragment of nucleolin interacts with MoMLV Gag, inhibiting virion assembly (3).
HIV-1 and HIV-2 are both members of the lentivirus subfamily of retroviruses. While they have similar genetic organization, there is limited homology at the nucleotide and amino acid levels (17). HIV-2 differs from HIV-1 in the mechanism it uses to select its genomic RNA for encapsidation (21), suggesting that the Gag proteins of HIV-1 and HIV-2 may interact with different cellular proteins. In this report we describe the identification of Tsg101 as an HIV-2 Gag-interacting protein. Tsg101 interacts specifically with HIV-2 Gag in vitro and in vivo. The PTAPP motif in p6 of Gag and the N-terminal Ubc-homology domain of Tsg101 are required for the interaction. Furthermore, we demonstrate that Tsg101 is incorporated into HIV-2 particles. Overexpression of full-length Tsg101 causes an increased level of ubiquitination of Gag. Expression of the N-terminal Ubc-conjugation homology domain of Tsg101 (amino acid residues 1 to 167) inhibits the release of HIV-2 particles.
MATERIALS AND METHODS
Construction of plasmids for use in the yeast two-hybrid system.
Yeast expression plasmids pLexA (bait) and pB42AD (prey), carrying HIS3 and TRP1 selection markers, respectively, were obtained from Clontech. Gag coding sequences of HIV-1 HXB2 (11) and HIV-2 ROD (17) were cloned between the EcoRI and SalI restriction sites of pLexA and pB42AD from pGexGag1 and pGexGag2 (see below) to generate pLexAGag1, pLexAGag2, pB42ADGag1, and pB42ADGag2, respectively. Subdomains of the HIV-2 Gag gene were cloned into the EcoRI and SalI restriction sites of pLexA from pGex-based plasmids (see below) to generate pLexAMA2, pLexACA2, pLexANC2, pLexAGag2Δp2-p6, and pLexAGag2Δp6. pAMPTsg101, which contains human Tsg101 cDNA, was a kind gift from S. Cohen (Stanford). Tsg101 sequences from BglI (Klenow polymerase treated) to SalI (nucleotides 92 to 1494) were cloned from pAMPTsg101 into the EcoRV and SalI sites of Bluescript KSII, generating pKSTsg101. The Tsg101 coding sequence was then cloned as an EcoRI-XhoI fragment from pKSTsg101 into the EcoRI to XhoI sites of the yeast activation domain plasmid pYesTrp2 (Invitrogen) to generate pYesTrpTsg. Deletions of Tsg101 were constructed by removing sequences between the SphI and XhoI sites (592-1263), generating pYesTrpTsgΔSX, and between ClaI and XhoI (1030-1263) sites, generating pYesTrpTsgΔCX. An N-terminal truncation of Tsg101 was constructed by cloning an SphI fragment of Tsg101 (592-1263) from pYesTrpTsg into the SphI site of pYesTrp2, generating pYesTrpTsgΔUbc.
Construction of plasmids for in vitro binding assays.
DNA sequences encoding Gag proteins of HIV-1 HXB2 (11) and HIV-2 ROD (17) were PCR amplified using Pfu Turbo DNA polymerase (Stratagene) with IGAGF (5′-CATAGAATTCATGGGTGCGAGAGCGTCA-3′) and 1GAGR (5′-CATGTAGTCGACTTATTGTGACGAGGGGTC-3′) primers for HIV-1 Gag or 2GAGF (5′-CTATGAATTCATGGGCGCGAGAAACTCC-3′) and 2GAGR (5′-CACGTCGTCGACCTACTGGTCTTTTCCAAA-3′) primers for HIV-2 Gag. The resulting PCR products were digested with EcoRI and SalI and cloned into the EcoRI and SalI sites of pGex4T1 (Amersham Pharmacia Biotech) to generate pGexGag1 and pGexGag2, respectively. Bacterial expression vectors containing subdomains of HIV-2 Gag were generated by PCR amplification, as above, using the following primers: 2GAGF and 2MAR (5′-GATCGTCGACGTAATTTCCTCCCTTCTC-3′) for pGexMA2 (nucleotides 546 to 950), 2CAF (5′-GATCGAATTCCCAGTGCAACATGTAGGC-3′) and 2CAR (5′-GCGCGTCGACCATTAATCTAGCTTTCTG-3′) for pGexCA2 (nucleotides 951 to 1640), and 2NCF (5′-CTATGAATTCGCCCAGCAGAGAAAGGCA-3′) and 2NCR (5′-CTATTAGTCGACACCTGCCTGTCTATCTGG-3′) for pGexNC2 (nucleotides 1692 to 1838). pGexGag2Δp2-p6 was constructed by cloning an EcoRI-HindIII fragment (nucleotides 546 to 1458) from pGexGag2 into the EcoRI and HindIII sites of pGexCA2; the resulting plasmid contains HIV-2 nucleotides 546 to 1640 and lacks the p2, NC, p1, and p6 domains. Plasmid pGexGag2Δp6 was constructed by PCR amplification of HIV-2 sequences using primers 2CAF and 2NCR. The PCR product was digested with HindIII (nucleotide 1458) and SalI (nucleotide 1839) and cloned into the HindIII and SalI sites of pGexGag2. The resulting plasmid contains HIV-2 nucleotides 546 to 1839 and lacks the p1 and p6 domains. The PTAPP motif in the p6 domains of HIV-1 and HIV-2 Gag was deleted by PCR mutagenesis (Quik Change mutagenesis kit, Stratagene), using pGexGag1 and oligonucleotides 1DPTAPPs (5′-CAGAGCAGACCAGAGGAAGAGAGCTTCAGG-3′) and 1DPTAPPas (5′-CCTGAAGCTCTCTTCCTCTGGTCTGCTCTG-3′) for HIV-1 Gag and pGexGag2 and 2DPTAPPs (5′-CCGCAGGGGCTGACAGTGGATCCAGCAGTG-3′) and 2DPTAPPas (5′-CACTGCTGGATCCACTGTCAGCCCCTGCGG-3′) for HIV-2 Gag. The mutations were confirmed by DNA sequencing.
Construction of mammalian expression plasmids.
pSVR is an infectious proviral clone of HIV-2 ROD containing a simian virus 40 origin of replication (32). Restriction site positions are given with respect to the first nucleotide of the viral RNA. pSVRΔNB, which has a 550-bp deletion in the env gene between NsiI and BstXI (nucleotides 6369 to 6919), has been previously described (15). pSVRΔPTAPP was constructed by replacing an EcoRV fragment of pSVRΔNB (nucleotides 1101 to 2939) with an EcoRV fragment from pKS2EV-ΔPTAPP, which contains a deletion of the p6 PTAPP motif. pKS2EV-ΔPTAPP was constructed by oligonucleotide-directed mutagenesis (25) of pKS2EV−, a plasmid based on Bluescript KSIII containing an EcoRV fragment (nucleotides 1101 to 2939) of HIV-2, and the mutagenic oligonucleotide 2DPTAPPs. The mutations were confirmed by DNA sequencing. A proviral clone with deletions in the pol and env genes was constructed by cloning the Gag open reading frame from pGexGag2 as an EcoRI-SalI (nucleotides 546 to 2114) fragment into the EcoRI and XhoI sites in the polylinker of pcDNA3 (Invitrogen), generating plasmid pcDNAGag2. An EcoRI (nucleotide 546)-XbaI fragment from pcDNAGag2 (using an XbaI site in the polylinker immediately 3′ to the XhoI site) was then cloned into the EcoRV (nucleotide 1101) and XbaI (nucleotide 5067) sites of pSVRΔNB. The resulting plasmid, pSVRGagΔNB, lacks the pol gene (nucleotides 2114 to 5067) and therefore expresses uncleaved Gag polyprotein.
Tsg101 was amplified by PCR using primers ETSGF (5′-CATAGAATTCTATGGCGGTGTCGGAGAGC-3′) and XTSGR (5′-CATACTCGAGGTAGAGGTCACTGAGACCGGCAG-3′) and cloned as an EcoRI-XhoI fragment into pTriEx1.1 (Novagen). The resulting plasmid, pTriExTsg, contains the Tsg101 coding sequence (nucleotides 91 to 1260), with an in-frame polyhistidine tag at the C terminus. For use in the in vivo ubiquitination assay, the polyhistidine tag was removed from pTriEx1.1 by digestion with XhoI and DraIII, followed by treatment with Klenow polymerase and religation, generating plasmid pTriEx1.1ΔHis. Tsg101 was cloned into pTriEx1.1ΔHis as an EcoRI-XhoI fragment (nucleotides 91 to 1260), to produce plasmid pTriExTsgΔHis. Constructs expressing 5′ (nucleotides 91 to 591) and 3′ (nucleotides 592 to 1260) regions of Tsg101 were generated by PCR using primers ETSGF and TSG591R (5′-GAATCTCGAGCATGCCTGGCATGTAGG-3′) for the 5′ region of Tsg101 or TSG592F (5′-GATCGAATTCTCCAGGTGGAATCTCTCC-3′) and XTSGR for the 3′ region of Tsg101 and cloned between the EcoRI and XhoI sites in pTriEx1.1ΔHis. The resulting plasmids were pTriEx5′TsgΔHis and pTriEx3′TsgΔHis respectively. The integrity of the PCR-amplified sequences was confirmed by DNA sequencing.
pH6M-Ub, which expresses yeast ubiquitin tagged with histidine and myc, was a kind gift from R. Kopito (53).
All HIV-2-based plasmids were propagated in TOPF10′ (Invitrogen) at 30°C to minimize recombination. All other plasmids were propagated in DH5α.
Yeast two-hybrid library screening.
The Matchmaker LexA two-hybrid system and Jurkat cDNA library used in this study were obtained from Clontech. This system identifies potential bait-interacting proteins by selection for leucine prototrophy and a screen for β-galactosidase activity in Saccharomyces cerevisiae EGY48 (ura3 his3 trp1 LexAop6-LEU2). A total of 2 × 106 colonies were screened according to the manufacturers instructions. The putative bait-interacting clones were amplified by PCR using primers B42ADF (5′-TGACTGGCTGAAATCGAATGG-3′) and B42ADR (5′-GCAAGGTAGACAAGCCGACAA-3′). The PCR products were purified using QIAquick PCR purification columns (Qiagen), and the sequence of each library insert was determined and compared with entries in the GenBank database.
Yeast β-galactosidase activity assay.
The β-galactosidase activity of double transformants was measured quantitatively by assaying for enzyme activity using the chromogenic substrate o-nitrophenyl-β-d-galactopyranoside (ONPG) as specified by the manufacturer (Yeast Protocols Manual; Clontech). The enzymatic activity represents at least three separate assays on independent transformants.
Preparation of yeast cell lysates.
Yeast cell lysates were prepared for Western blotting as follows. Overnight cultures of transformed yeast were inoculated into 50 ml of yeast induction medium (SD Gal/Raf-His-Trp-Ura; Clontech), and incubated at 30°C until the optical density at 600 nm (OD600) was 0.4 to 0.6. The cells were harvested by centrifugation at 1,000 × g for 5 min at 4°C, washed with ice-cold water, and centrifuged again. Cells were resuspended in 100 μl/7.5 OD600 units of prewarmed (60°C) cracking buffer (7 M urea, 4.4% [wt/vol] sodium dodecyl sulfate [SDS], 35.4 mM Tris-HCl [pH 6.8], 88 μM EDTA, 1.24 mg of bromophenol blue per ml, 127 mM β-mercaptoethanol, 9 μM pepstatin A, 1.9 μM leupeptin, 9 mM benzamidine, 5.5 μM aproptinin, 4.4 mM phenylmethylsulfonyl fluoride) incubated at 70°C for 10 min. The cells were disrupted by vortexing with glass beads (425 to 600 μm, in diameter; Sigma) (80 μl/7.5 OD600 units). Lysates were cleared by centrifugation for 5 min in a microcentrifuge, and supernatants were boiled briefly before being loaded onto SDS-polyacrylamide gels for Western blot analysis.
In vitro binding assays.
Glutathione S-transferase (GST) fusion proteins were prepared essentially as described by Smith and Johnson (44), except that bacteria were cultured for 3 h and then induced with isopropyl-β-d-thiogalactopyranoside for a further 3 h. In vitro transcription-translation or coupled transcriptions-translations (Promega) were used to generate [35S]methionine-labeled proteins as described by the manufacturer. GST fusion proteins (500 ng) on beads were preincubated with bovine serum albumin at room temperature at a final concentration of 0.2 mg of bovine serum albumin per ml for 5 min and then rotated for 1 h at room temperature with 2 to 5 μl of in vitro-translated test protein in a final volume of 200 μl of EBC buffer (140 mM NaCl, 0.5% Nonidet P-40, 100 mM NaF, 200 μM orthovanadate, 50 mM Tris-HCl [pH 8.0], 0.5 mM phenylmethylsulfonyl fluoride). The beads were washed three times in 1 ml of NETN buffer (100 mM NaCl, 1 mM EDTA, 0.5% Nonidet P-40, 20 mM Tris-HCl [pH 8.0], 0.5 mM phenylmethylsulfonyl fluoride), centrifuged at 2,000 rpm in a microcentrifuge for 2 min, and boiled in 4× SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer. Bound proteins were resolved on SDS-polyacrylamide gels and detected by autoradiography.
Cell culture and transfections.
293T cells were maintained in Dulbecco's modified Eagle's medium (Gibco-BRL) supplemented with 10% fetal calf serum, penicillin, and streptomycin. Transient transfection of 293T cells was performed by the calcium phosphate method. The cells were harvested 48 h later. Tissue culture supernatant was cleared by filtration through a 0.45-μm-pore-size filter. Particles released from the transfected cells were pelleted by polyethylene glycol precipitation by the addition of 0.5 volume of 30% polyethylene glycol 8000-0.4 M NaCl for 16 h at 4°C. The precipitate was collected by centrifugation at 2,000 rpm in an MSE 43124-129 rotor at 4°C for 45 min and resuspended in 0.5 ml of TNE (10 mM Tris-Cl, 150 mM NaCl, 1 mM EDTA [pH 7.5]). This material was layered over an equal volume of TNE containing 20% sucrose and centrifuged at 98,000 × g for 2 h at 4°C. Virus particles were resuspended in SDS-PAGE loading buffer and analyzed by Western blotting. Where required, virus particle release was assessed using a reverse transcriptase assay (39).
Western blotting.
Western blotting analyses were performed using 12.5% polyacrylamide gels and Hybond-C extra transfer membrane (Amersham Pharmacia Biotech) as specified by the manufacturer protocol. Monoclonal antibody to HIV-2 p26 (Chemicon) and monoclonal antibody to Tsg101 (GeneTex) were used at a 1:1,000 dilution, monoclonal antibody to V5 epitope (Clontech) was used at a 1:5,000 dilution, monoclonal antibody to C-terminal polyhistidine epitope (Invitrogen) was used at a 1:5,000 dilution, and monoclonal antibody to LexA (Clontech) was used at a 1:2,000 dilution. Horseradish peroxidase-conjugated donkey anti-mouse immunoglobulin G secondary antibody (Chemicon) was used at a 1:5,000 dilution and detected by staining with enhanced chemiluminescence Western blotting detection reagent (Amersham Pharmacia Biotech) as specified by the manufacturer.
In vivo ubiquitination of HIV-2 Gag.
pH6M-Ub was introduced into 293T cells by cotransfection with plasmids expressing HIV-2 Gag and Tsg101. At 24 h later, the transfected cultures were treated with MG132 (2 μM) for an additional 16 h. The cells were harvested, and 10% of the cells were resuspended in SDS-PAGE loading buffer and analyzed by Western blotting, using anti-26 monoclonal antibody, to confirm that equivalent levels of Gag protein were expressed in each sample. The remaining cells were resuspended in lysis buffer (6 M guanidine hydrochloride, 100 mM NaH2PO4, 20 mM imidazole, 10 mM Tris-Cl [pH 8]), sonicated for 1 min on ice, and then rotated at room temperature for 1 h. The His-tagged proteins were purified on Ni-nitrotriacetate (NTA) spin columns (Qiagen), washed four times with 600 μl of wash buffer (8 M urea, 100 mM NaH2PO4, 20 mM imidazole, 10 mM Tris-Cl [pH 6.2]), and eluted once with 200 μl of elution buffer (8 M urea, 100 mM NaH2PO4, 20 mM imidazole, 10 mM Tris-Cl [pH 4.0]) and once with 200 μl of elution buffer containing 250 mM imidazole. The eluted samples were pooled, precipitated with 10% trichloroacetic acid, and analyzed by Western blotting using anti-p26 monoclonal antibody.
RESULTS
Identification of an interaction between HIV-2 Gag and Tsg101.
HIV-2 Gag was used as bait in a yeast two-hybrid screen to identify interacting proteins encoded by a Jurkat cDNA library, as described in Materials and Methods. Uncleaved Gag polyprotein was used rather than one of its mature cleavage products, because this is the functional intracellular form of Gag. Approximately 2 × 106 transformants were screened, and several positive clones were identified. Comparison of the cDNA sequences with the GenBank database revealed that one of the clones was human EF1α and another was human Tsg101, an inactive homologue of ubiquitin ligase E2, both of which have been reported to interact with HIV-1 Gag (5, 13, 51).
Tsg101 interacts specifically with HIV-2 Gag.
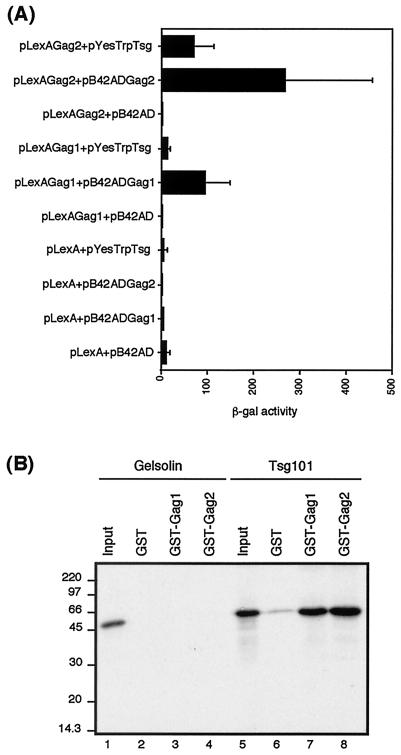
To confirm the specificity of the interaction between HIV-2 Gag and Tsg101, we cotransformed yeast with plasmids expressing either HIV-1 or HIV-2 Gag as DNA-binding domain fusion proteins and Tsg101 as an activation domain fusion and determined the strength of the interaction by quantitative β-galactosidase assays. Western blotting (data not shown) confirmed the level of expression of bait and prey fusion proteins. The results are shown in Fig. 1A. Tsg101 interacted specifically with HIV-2 Gag and also with HIV-1 Gag, as previously reported (13, 51). The interaction between HIV-2 Gag and Tsg101 appears to be stronger than that between HIV-1 Gag and Tsg101, despite similar levels of both bait proteins being expressed in the transformed yeast (data not shown).
FIG. 1.
Tsg101 binds specifically to HIV-2 Gag. (A) Analysis of the interaction between Tsg101 and HIV-2 Gag in the yeast two-hybrid system. Yeast were transformed with different combinations of plasmids encoding the bait and prey fusion proteins, as indicated. Reporter gene activation was measured by β-galactosidase assay of the yeast lysates (as described in Materials and Methods). The data represent the mean β-galactosidase (β-gal) activity from at least three separate assays on independent transformants. (B) In vitro binding of Tsg101 to HIV-1 and HIV-2 Gag proteins. In vitro-transcribed and translated Tsg101 (lanes 6 to 8) or gelsolin (lanes 2 to 4) was analyzed by GST fusion pull-down assays for the ability to bind GST, GST-Gag1, or GST-Gag2 immobilized on glutathione-Sepharose beads. Inputs were 10% of the amount of Tsg101 (lane 5) or gelsolin (lane 1) used in each assay. The positions of marker proteins are indicated in kilodaltons.
Further evidence of the specificity of the interaction was obtained by in vitro binding assays using GST-Gag fusion proteins and in vitro-translated Tsg101. Figure 1B shows that Tsg101 interacts specifically with both HIV-1 and HIV-2 Gag. There was no binding of Tsg101 to GST alone, confirming the specificity of the interaction between HIV-2 Gag and Tsg101. The cellular protein gelsolin, used as a nonspecific control, did not bind to GST, GST-Gag1, or GST-Gag2. The apparent discrepancy between the results obtained using yeast two-hybrid assays and GST fusion-protein binding assays may reflect difference in protein expression and folding when HIV-2 Gag and Tsg101 are produced in different expression systems.
Deletion of the p6 domain of HIV-2 Gag abolishes the interaction between Tsg101 and HIV-2 Gag.
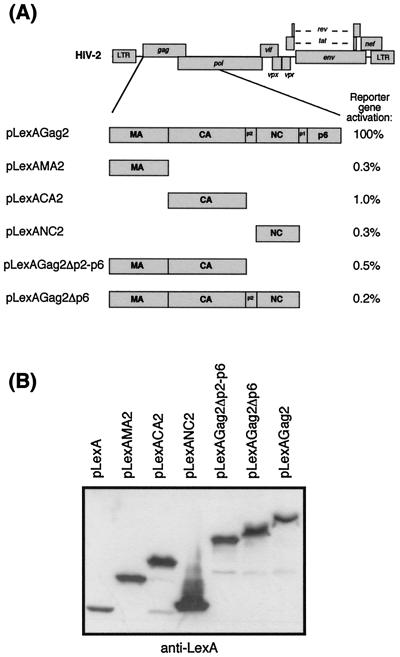
To map the region of the HIV-2 Gag polyprotein required for interaction with Tsg101, plasmids containing N- and C-terminally deleted HIV-2 Gag fused to the LexA DNA-binding domain were assayed for lacZ reporter gene activation using the two-hybrid assay. The LexA fusion constructs are shown diagrammatically in Fig. 2A and are described in Materials and Methods. The results of the yeast two-hybrid assays are shown in Fig. 2A. None of the Gag deletion mutants interacted with Tsg101: the reporter gene activation for the Gag truncations was reduced to 1% or less of that of full-length Gag in all cases, despite the expression of similar levels of the truncated Gag proteins (Fig. 2B). Deletion of the C-terminal p6 domain alone abolished the interaction with Tsg101, suggesting that the ability of HIV-2 Gag to interact with Tsg101 was determined by elements in the p6 domain of Gag.
FIG. 2.
Determination of the region of HIV-2 Gag involved in binding Tsg101. (A) Schematic diagram of HIV-2 Gag domains expressed in yeast as C-terminal fusion proteins with the LexA DNA-binding domain. Yeast were transformed with full-length Tsg101 (pYesTrpTsg) and either full-length or truncated Gag fusion proteins, and reporter gene activity was measured by the β-galactosidase assay. The results are expressed as a percentage of the β-galactosidase activity observed with Tsg101 and full-length Gag (117 units). The data were derived from the mean β-galactosidase activity from at least three separate assays on independent transformants. (B) Duplicate lysates of transformed yeast described in panel A were analyzed by Western blotting using anti-LexA monoclonal antibody to confirm that the various Gag fusion proteins were expressed to similar levels.
The PTAPP motif in the late assembly domain of HIV-2 Gag is required for interaction between Tsg101 and HIV-2 Gag.
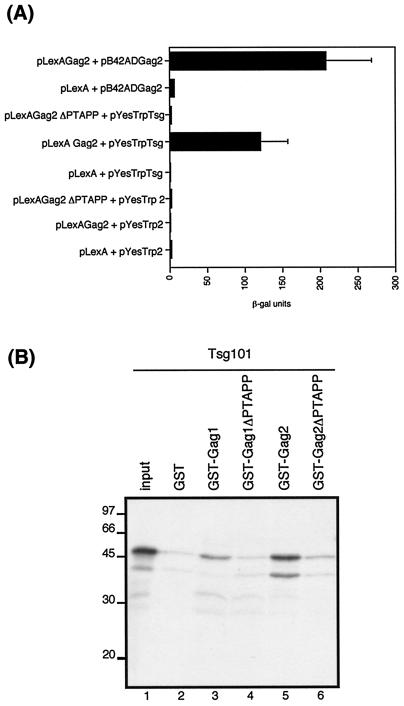
The C-terminal p6 region of HIV-2 Gag contains a PTAPP motif which is conserved among lentiviruses. This motif is required for efficient release of virions from the cell (14, 20) and has been reported to interact with the host cell ubiquitination machinery (46). It was therefore of interest to determine what role Tsg101, an inactive E2 ubiquitin conjugase homologue, may be playing in the retroviral life cycle. We generated an HIV-2 Gag expression plasmid in which the PTAPP motif was deleted. The effect of this deletion was determined by quantitative yeast two-hybrid assays following transformation of Tsg101 with either wild-type or PTAPP deletion mutant plasmids. Comparable levels of wild-type and PTAPP deletion proteins were expressed in the transformed yeast, as confirmed by Western blotting (data not shown). As shown in Fig. 3A, deletion of the PTAPP motif abolished the interaction of HIV-2 Gag with Tsg101. Further evidence for the requirement for the PTAPP motif in the interaction with Tsg101 was obtained from in vitro binding assays. The PTAPP deletion was cloned into HIV-2 Gag in the context of a GST fusion construct, and the ability of wild-type and mutant Gag proteins to interact with in vitro-translated Tsg101 was determined; the results are shown in Fig. 3B. As shown above (in Fig. 1B), wild-type HIV-2 Gag interacted specifically with Tsg101; however, deletion of the PTAPP motif resulted in a marked reduction in the level of binding to Tsg101. We included wild-type HIV-1 Gag and the homologous PTAPP deletion mutant in these binding assays; as expected, deletion of the PTAPP motif abolished the interaction of HIV-1 Gag with Tsg101.
FIG. 3.
The PTAPP late assembly domain is required for interaction of HIV-2 Gag with Tsg101. (A) Yeast two-hybrid analysis of the interaction between either wild-type or ΔPTAPP mutant Gag and Tsg101. Yeast were transformed with different combinations of plasmids encoding the bait and prey fusion proteins, as indicated. The data represent the mean β-galactosidase (β-gal) activity from at least three separate assays on independent transformants. (B) In vitro binding of Tsg101 to HIV-1 and HIV-2 Gag proteins. In vitro-transcribed and translated Tsg101 (lanes 2 to 6) was analyzed by GST fusion pull-down assays for the ability to bind GST, GST-Gag1, GST-Gag1ΔPTAPP, GST-Gag2, or GST-Gag2ΔPTAPP immobilized on glutathione sepharose beads. Input (lane 1) was 10% of the amount of Tsg101 used in each assay. The positions of marker proteins are indicated in kilodaltons.
The Ubc homology domain at the N terminus of Tsg101 is required for interaction with HIV-2 Gag.
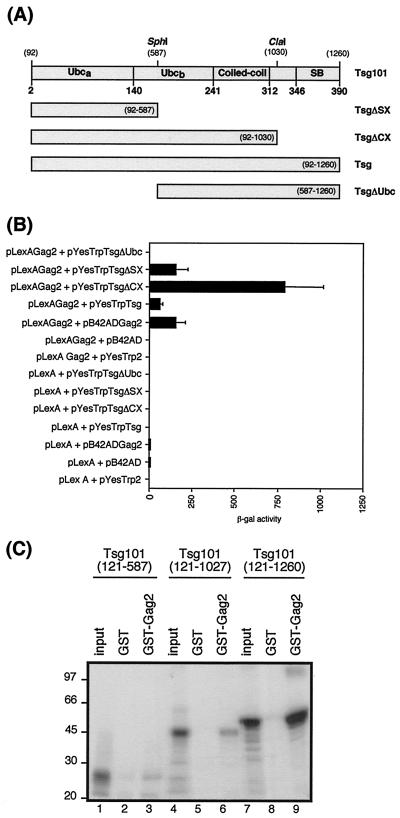
Tsg101 protein contains a region of homology to ubiquitin-conjugating enzyme E2 at the N terminus, a central coiled-coil domain, and a C-terminal steadiness box (10). To map the region of Tsg101 required for interaction with HIV-2 Gag, we constructed C-terminal and N-terminal deletions of Tsg101, shown in Fig. 4A. The ability of the Tsg101 truncations to interact with HIV-2 Gag polyprotein was assayed by quantitative β-galactosidase assays (Fig. 4B). The Tsg101 mutants with C-terminal deletions (TsgΔSX and TsgΔCX) interacted with HIV-2 Gag to a greater extent than did full-length Tsg101. The relative levels of expression of the truncated Tsg101 fusion constructs in the transformed yeast were analyzed by Western blotting and found to be similar to those of full-length Tsg101 (data not shown). Therefore, the increased interaction between HIV-2 Gag and TsgΔCX and TsgΔSX was not due to increased levels of the truncated proteins in the transformed yeast but may instead be due to conformational changes in Tsg101 when C-terminal sequences are removed. Deletion of the N-terminal Ubc-homology domain in TsgΔUbc abolished the interaction with HIV-2 Gag, indicating the importance of this region for the interaction.
FIG. 4.
Identification of the region of Tsg101 required for interaction with HIV-2 Gag. (A) Schematic diagram of Tsg101 mutants expressed in yeast as fusion proteins with the B42 activation domain. Numbers in parentheses indicate the nucleotide positions. Amino acid residues are shown in bold. Ubc, ubiquitin conjugase domain; SB, steadiness box. (B) Yeast two-hybrid analysis of the interaction between HIV-2 Gag and Tsg101 truncations. Yeast were transformed with different combinations of plasmids encoding the bait and prey fusion proteins, as indicated. The data represent the mean β-galactosidase (β-gal) activity from at least three separate assays on independent transformants. (C) In vitro binding of Tsg101 truncations to HIV-2 Gag protein. 35S-labeled Tsg101 truncations were prepared by in vitro transcription of template DNA linearized with SphI (nucleotide 587), ClaI (nucleotide 1030), or XhoI (nucleotide 1260), followed by in vitro translation. The Tsg101 truncations were analyzed by GST fusion pull-down assays for the ability to bind GST or GST-Gag2 immobilized on glutathione-Sepharose beads. Inputs (lanes 1, 4, and 7) were 10% of the amount of the relevant 35S-labeled Tsg101 protein used in each assay. The positions of marker proteins are indicated in kilodaltons.
We also tested the ability of the C-terminal truncation mutants of Tsg101 to interact with HIV-2 Gag using in vitro GST fusion protein-binding assays. The results are shown in Fig. 4C. Full-length Tsg101 bound specifically to GST-Gag2, as we had previously observed, and the C-terminal truncation mutants of Tsg101 (TsgΔCX and TsgΔSX) also bound to HIV-2 Gag, albeit to a lesser extent than did full-length Tsg101.
The results of the yeast two-hybrid β-galactosidase assays confirm that the N-terminal Ubc-homology domain is required for the interaction with HIV-2 Gag. The apparent discrepancies in the levels of interaction of the C-terminally truncated Tsg101 proteins with HIV-2 Gag protein detected using yeast two-hybrid β-galactosidase assays compared to GST fusion protein-binding assays may be caused by differences in protein synthesis and folding when Tsg101 is produced in different expression systems.
Tsg101 is incorporated into virions.
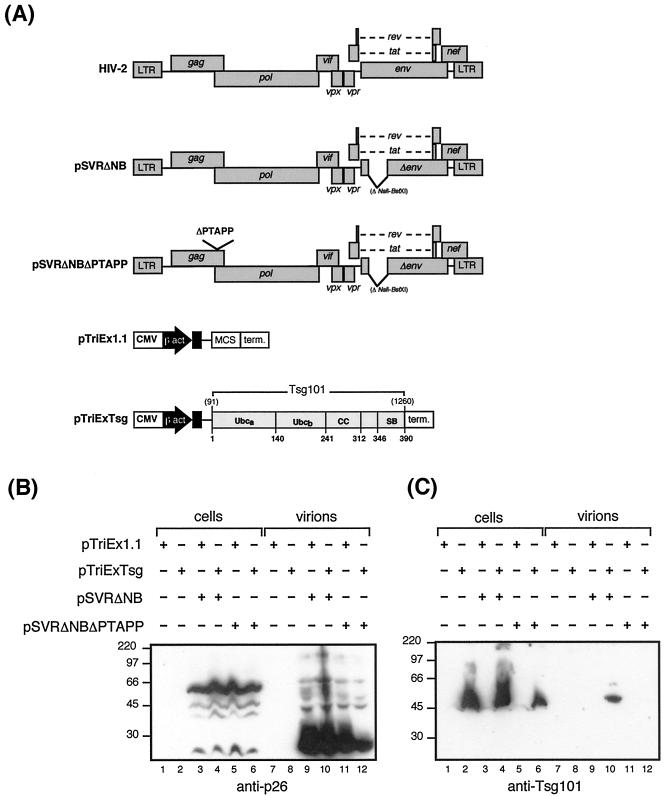
Having shown a specific interaction between Tsg101 and HIV-2 Gag, we asked whether Tsg101 was incorporated into HIV-2 particles. 293T cells were cotransfected with either wild-type Gag or PTAPP-deleted Gag protein and either control or Tsg101 expression constructs. The expression constructs used in this experiment are shown in Fig. 5A. Cell and virion samples were analyzed by Western blotting (Fig. 5B and C). Figure 5B shows Gag polyprotein in the cells and mature p26 capsid in the virions. In Fig. 5C, a 45-kDa Tsg101 protein was detected in the transfected cells. Tsg101 was detected in virions released from cells transfected with wild-type Gag expression construct. However, incorporation of Tsg101 into virus particles was not observed when the HIV-2 Gag PTAPP motif was deleted. The failure to detect Tsg101 in virus particles released from cells transfected with the PTAPP-deleted Gag expressor could reflect the slightly lower levels of p26 released in virus particles from cells transfected with HIV-2 Gag ΔPTAPP and Tsg101; however, exposure of the Western blots for longer periods did not reveal the presence of Tsg101 in Fig. 5C (lane track 12) (data not shown).
FIG. 5.
Tsg101 is incorporated into HIV-2 particles. (A) Schematic diagram of HIV-2 Gag and Tsg101 expression constructs. CMV, Cytomegalovirus immediate-early enhancer; β act., chicken β-actin promoter; MCS, multicloning site; term., rabbit β-globin terminator; CC, coiled-coil domain; SB, steadiness box. (B and C) 293T cells were transfected with Tsg101 expression plasmid (pTriExTsg) or control plasmid (pTriEx1.1) and either an env-deleted HIV-2 provirus (pSVRΔNB) or a PTAPP deletion mutant (pSVRΔNBΔPTAPP), as indicated. Cell and virus particle samples were prepared as described in Materials and Methods and subjected to analysis by Western blotting using monoclonal antibodies to either p26 (HIV-2 capsid) (B) or Tsg101 (C). The positions of marker proteins are indicated in kilodaltons.
Previous studies of the role of the PTAPP motif in the p6 domain of HIV-1 Gag in HeLa cells have reported that deletion of the motif inhibits virus particle release (14, 20). We did not observe this in our system, and this could be due to the cell type used in our study. Recently, Demirov and colleagues reported that the late domain of HIV-1 p6 functions in a cell type-dependent manner (7). They observed that while deletion of the PTAPP motif caused a severe reduction in the release of virus from HeLa cells, a much milder reduction was seen when 293T cells were used. This could explain the detection of p26 protein in virions produced from cells transfected with HIV-2 Gag lacking the PTAPP motif.
Tsg101 promotes ubiquitination of HIV-2 Gag.
The identification of Tsg101 as an inactive homologue of E2 ubiquitin conjugase enzymes has led to speculation that Tsg101 may act as a dominant negative regulator of ubiquitination (24, 38). In fact, Li et al. recently demonstrated that Tsg101 acts as a dominant negative regulator of ubiquitination of the oncoprotein Mdm2 (29).
To test whether overexpression of Tsg101 affected the level of ubiquitination of HIV-2 Gag protein, we adapted an in vivo ubiquitination assay from a method described by Treier and colleagues (49), as described in Materials and Methods. Briefly, the ubiquitination assay involves cotransfection of a hexahistidine (His6)-tagged ubiquitin expressor with an HIV-2 Gag expression construct in the presence or absence of a Tsg101 expression construct. The transfected cells are treated with proteasome inhibitor MG132 to prevent degradation of the ubiquitinated Gag proteins prior to lysis in guanidine hydrochloride. These denaturing conditions prevent degradation or deubiquitination of any protein-ubiquitin conjugates as well as any noncovalent protein-protein interactions. The His6-tagged ubiquitin and any His6-tagged ubiquitinated proteins are then purified by Ni-NTA chromatography. The eluted ubiquitinated proteins are then analyzed by Western blotting using an HIV-2 Gag-specific antibody.
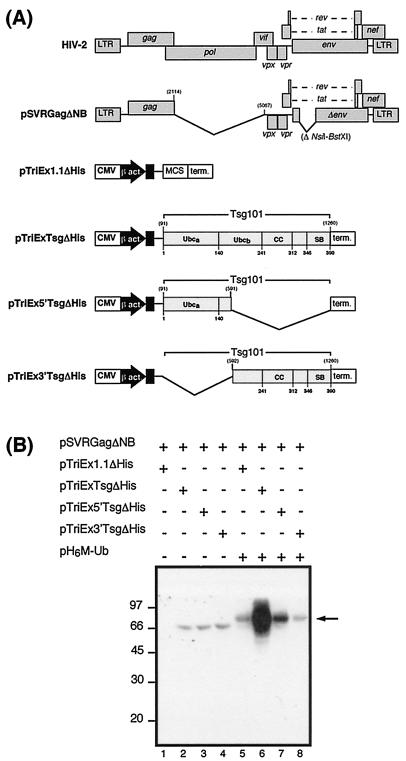
The HIV-2 Gag and Tsg101 expression constructs used in this experiment are shown in Fig. 6A. The relative expression levels of the full-length and truncated Tsg101 proteins were determined by Western blotting of lysates of 293T cells transfected with full-length Tsg101 (pTriExTsg), the N-terminal portion of Tsg101 (pTriEx5′Tsg), or the C-terminal portion of Tsg101 (pTriEx3′Tsg) using an antibody specific for C-terminal hexahistidine epitope-tagged proteins. The truncated Tsg101 proteins were expressed at comparable levels to full-length Tsg101 (data not shown). The hexahistidine epitope tag was deleted from the Tsg101 expression constructs used in the ubiquitination assay to prevent these proteins binding directly to the Ni-NTA columns. The results of the ubiquitination assay are shown in Fig. 6B. Instead of acting as an inhibitor of ubiquitination as predicted, overexpression of full-length Tsg101 resulted in a greater than 10-fold increase in the level of ubiquitinated Gag (lane 6) compared to the level of ubiquitination observed when the control vector was cotransfected (lane 5), as determined by densitometry. Overexpression of the N-terminal portion of Tsg101 (amino acid residues 1 to 167) resulted in a two- to three-fold increase in the level of Gag ubiquitination (lane 7), whereas overexpression of the C-terminal portion of Tsg101 (residues 168 to 390) did not increase ubiquitination levels (lane 8). The possible significance of these observations is discussed below.
FIG. 6.
Effect of Tsg101 expression on ubiquitination of HIV-2 Gag. (A) Schematic diagram of HIV-2 Gag and Tsg101 expression constructs. Abbreviations are as in Fig. 5. (B) 293T cells were transfected with constructs encoding a protease-negative, env-deleted HIV-2 provirus (pSVRGagΔNB) and either full-length Tsg101 (pTriExTsgΔHis), the N-terminal portion of Tsg101 (pTriEx5′TsgΔHis), the C-terminal portion of Tsg101 (pTriEx3′TsgΔHis), or control plasmid (pTriEx1.1ΔHis) in the presence or absence of a His-Myc-tagged ubiquitin expression construct (pH6M-Ub). At 48 h after transfection, protein extracts were applied to Ni-NTA columns, and ubiquitin-labeled HIV-2 Gag protein was eluted and detected by Western blotting with anti-p26 antibody. The positions of marker proteins are indicated in kilodaltons.
Overexpression of the N-terminal portion of Tsg101 inhibits particle budding.
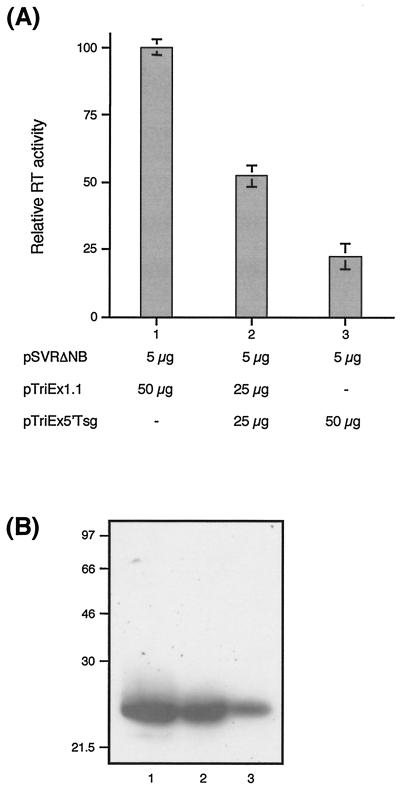
It has recently been reported that overexpression of the N-terminal portion of Tsg101 inhibits HIV-1 particle budding (6). Having demonstrated that HIV-2 Gag interacts with Tsg101, we investigated whether budding of HIV-2 particles would also be inhibited by overexpression of the N-terminal portion of Tsg101. We cotransfected 293T cells with 5 μg of env-deleted HIV-2 proviral construct pSVRΔNB (shown in Fig. 5A) and increasing amounts of pTriEx5′Tsg (pSVRΔNB-to-pTriEx5′Tsg DNA ratios, 1:5 and 1:10). The amount of plasmid DNA in each transfection was adjusted to a total of 55 μg with empty vector (pTriEx1.1). Expression of HIV-2 Gag protein in the transfected cells was studied by Western blotting (data not shown) and found to be equivalent at all DNA ratios. Virus particle production was measured by the reverse transcriptase assay. The results (Fig. 7A) show that expression of the N-terminal portion of Tsg101 inhibited virus particle release in a dose-responsive manner. Virus particle release was reduced by 47% at a DNA ratio of 1:5 and by 77% at a DNA ratio of 1:10, compared to control vector. Figure 7B shows a representative Western blot of virus particles released from the transfected cells and confirms the observation in Fig. 7A that overexpression of 5′Tsg inhibits virus release. These data are in agreement with the observation that HIV-1 budding can be inhibited by expression of the N-terminal portion of Tsg101 (6) and extend the evidence for a functional interaction between Tsg101 and the Gag protein of another member of the lentivirus family.
FIG. 7.
Overexpression of the N-terminal portion of Tsg101 inhibits virus particle release. (A) 293T cells were transfected with 5 μg of pSVRΔNB and either 50 μg of pTriEx1.1 (empty vector; lane 1), 25 μg of pTriEx5′Tsg plus 25 μg pTriEx1.1 (1:5 DNA ratio of pSVRΔNB to pTriEx5′Tsg; lane 2), or 50 μg of pTriEx5′Tsg (1:10 DNA ratio of pSVRΔNB to pTriEx5′Tsg; lane 3). At 48 h after transfection, virus particle samples were prepared as described in Materials and Methods and virus release was quantified by a reverse transcriptase assay. The data are the mean of three independent transfections. The error bars represent standard deviation. (B) Virus particle samples were also subjected to analysis by Western blotting using monoclonal antibody to HIV-2 p26. Lane numbering is as in panel A. The positions of marker proteins are indicated in kilodaltons.
DISCUSSION
In this report, we describe the identification of an interaction between HIV-2 Gag and human Tsg101 in a yeast two-hybrid screen of a Jurkat cDNA library. We have confirmed the specificity of the interaction in vitro and in vivo. While we were carrying out this work, there have been reports of an interaction between HIV-1 Gag and Tsg101 (13, 31, 51). We demonstrated that the PTAPP motif in the p6 domain of HIV-2 Gag is required for interaction with Tsg101. Since the PTAPP motif is conserved amongst the p6 Gag domains of all known primate lentiviruses, it is not surprising that both HIV-1 and HIV-2 Gag proteins interact with Tsg101 via this motif. In agreement with studies on the interaction of Tsg101 with HIV-1 Gag (51), we have demonstrated that the N-terminal Ubc-conjugation homology domain of Tsg101 is required for interaction with HIV-2 Gag and that overexpression of this portion of Tsg101 (amino acid residues 1 to 167) inhibits the release of HIV-2 particles. In addition, we have shown that Tsg101 is incorporated into HIV-2 particles via an interaction with HIV-2 Gag. We have also demonstrated that overexpression of Tsg101 leads to an increase in the levels of ubiquitinated Gag protein.
Tsg101 was originally identified as a tumor susceptibility protein since its deficiency causes neoplastic transformation in murine NIH 3T3 cells and tumorigenesis in nude mice (28). Subsequent analysis of the amino acid sequence of Tsg101 revealed that the N-terminal portion of Tsg101 had homology to ubiquitin E2 enzyme variants (UEVs) (24, 38). UEVs have homology to ubiquitin-conjugating enzymes; however, they lack an essential cysteine residue in the active site of the enzyme. The identification of Tsg101 as an inactive homologue of E2 enzymes led to speculation that Tsg101 may act as a negative regulator of ubiquitination (24, 38). Subsequently, it was reported that Tsg101 inhibits the ubiquitin-mediated degradation of Mdm2 by acting as an inhibitor of ubiquitination, resulting in down-regulation of p53 levels (29, 42). The process of ubiquitination involves the sequential action of three enzymes: a ubiquitin activation enzyme (E1), a ubiquitin-conjugating enzyme (E2), and a ubiquitin protein ligase (E3). The process results in the formation of an isopeptide bond between the C-terminal glycine residue of the ubiquitin molecule and a lysine residue in the substrate protein. Successive rounds of addition of ubiquitin molecules can lead to the formation of a polyubiquitin tail, which is recognized by the 28S proteasome and targets the ubiquitinated protein for degradation.
In addition to the well-established role of ubiquitin in protein degradation, it is becoming apparent that ubiquitination also serves noncanonical functions, such as controlling protein trafficking within the cell (9, 18). Whereas polyubiquitination directs proteins for degradation by the proteasome, monoubiquitination can serve as a signal for sorting to multivesicular bodies (MVB). The vacuolar protein-sorting (Vps) pathway sorts membrane-bound proteins for eventual degradation in the lysosome (27). Proteins can enter the Vps pathway from the Golgi or via endocytosis from the plasma membrane. Vesicles carrying proteins from either source fuse with endosomes, which mature to form MVB. The MVB fuse with the lysosome, and the endocytosed proteins are degraded by vacuolar proteases. The budding of enveloped viruses has been likened to budding of vesicles into endosomes (37). In both cases, membranes invaginate away from, rather than into, the cytoplasm. Tsg101 functions in the Vps pathway (2, 4). Garrus et al. demonstrated that the Vps pathway was essential for HIV-1 budding (13). Interestingly, the mammalian protein Nedd4, which is a member of the ubiquitin ligase E3 family, is involved in the down-regulation of several channel proteins (41). Nedd4 also interacts with the L domain of Rous sarcoma virus Gag (22). Taken together, these observations provide an intriguing link between the Vps pathway and the process of virus budding and may explain the observed requirement for ubiquitination in the budding process (37, 43, 46).
We observed an increase in the level of ubiquitinated HIV-2 Gag when Tsg101 was overexpressed. Thus, instead of acting as a dominant negative inhibitor of ubiquitination, as in the regulation of Mdm2 (29), Tsg101 is actually enhancing the level of HIV-2 Gag ubiquitination. A possible explanation of this could be that Tsg101, which itself is an inactive homologue of ubiquitin ligase E2 enzymes, is recruiting an active ubiquitin ligase to the site of budding. The formation of a heterodimer between an inactive UEV and an active E2 enzyme Ubc13 is required for postreplicative DNA repair in yeast (19, 45), and a related human UEV1a/Ubc heterodimer is required for IκBα kinase activation (8). While the requirement for the UEV in formation of the active heterodimer is clear, the role of the UEV subunit in catalysis and specificity remains to be established. Both human and yeast UEV/Ubc heterodimers interact with a RING domain protein, which may be either a cognate E3 enzyme or a substrate of the E2/UEV heterodimer (8, 50). Similarly, it is possible that Tsg101 may form a heterodimer with an active ubiquitin ligase, which results in the ubiquitination of HIV-2 Gag protein, which is bound to Tsg101 via an interaction between the Ubc domain of Tsg101 and the PTAPP domain of Gag. We are currently attempting to identify which Ubc enzyme is responsible for catalyzing the ubiquitination of HIV-2 Gag.
Acknowledgments
This work was funded by the Wellcome Trust (grant 057224).
We thank Andrew Lever for helpful discussions and for critical reading of the manuscript.
REFERENCES
- 1.Arthur, L. O., J. W. Bess, Jr., R. C. Sowder, Jr., R. E. Benveniste, D. L. Mann, J. C. Chermann, and L. E. Henderson. 1992. Cellular proteins bound to immunodeficiency viruses: implications for pathogenesis and vaccines. Science 258:1935-1938. [DOI] [PubMed] [Google Scholar]
- 2.Babst, M., G. Odorizzi, E. J. Estepa, and S. D. Emr. 2000. Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic 1:248-258. [DOI] [PubMed] [Google Scholar]
- 3.Bacharach, E., J. Gonsky, K. Alin, M. Orlova, and S. P. Goff. 2000. The carboxy-terminal fragment of nucleolin interacts with the nucleocapsid domain of retroviral gag proteins and inhibits virion assembly. J Virol. 74:11027-11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bishop, N., and P. Woodman. 2001. Tsg101/mammalian vps23 and mammalian vps28 interact directly and are recruited to vps4-induced endosomes. J. Biol. Chem. 276:11735-11742. [DOI] [PubMed] [Google Scholar]
- 5.Cimarelli, A., and J. Luban. 1999. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J. Virol. 73:5388-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demirov, D. G., A. Ono, J. M. Orenstein, and E. O. Freed. 2002. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc. Natl. Acad. Sci. USA 99:955-960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Demirov, D. G., J. M. Orenstein, and E. O. Freed. 2002. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 76:105-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deng, L., C. Wang, E. Spencer, L. Yang, A. Braun, J. You, C. Slaughter, C. Pickart, and Z. J. Chen. 2000. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103:351-361. [DOI] [PubMed] [Google Scholar]
- 9.Dupre, S., C. Volland, and R. Haguenauer-Tsapis. 2001. Membrane transport: ubiquitylation in endosomal sorting. Curr. Biol. 11:R932-R934. [DOI] [PubMed] [Google Scholar]
- 10.Feng, G. H., C. J. Lih, and S. N. Cohen. 2000. TSG101 protein steady-state level is regulated posttranslationally by an evolutionarily conserved COOH-terminal sequence. Cancer Res. 60:1736-1741. [PubMed] [Google Scholar]
- 11.Fisher, A. G., E. Collalti, L. Ratner, R. C. Gallo, and F. Wong Staal. 1985. A molecular clone of HTLV-III with biological activity. Nature 316:262-265. [DOI] [PubMed] [Google Scholar]
- 12.Franke, E. K., H. E. Yuan, and J. Luban. 1994. Specific incorporation of cyclophilin A into HIV-1 virions. Nature 372:359-362. [DOI] [PubMed] [Google Scholar]
- 13.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 14.Gottlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffin, S. D., J. F. Allen, and A. M. Lever. 2001. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of Gag protein confer specificity. J. Virol. 75:12058-12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta, K., D. Ott, T. J. Hope, R. F. Siliciano, and J. D. Boeke. 2000. A human nuclear shuttling protein that interacts with human immunodeficiency virus type 1 matrix is packaged into virions. J. Virol. 74:11811-11824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guyader, M., M. Emerman, P. Sonigo, F. Clavel, L. Montagnier, and M. Alizon. 1987. Genome organization and transactivation of the human immunodeficiency virus type 2. Nature 326:662-669. [DOI] [PubMed] [Google Scholar]
- 18.Hicke, L. 2001. A new ticket for entry into budding vesicles-ubiquitin. Cell 106:527-530. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann, R. M., and C. M. Pickart. 1999. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96:645-653. [DOI] [PubMed] [Google Scholar]
- 20.Huang, M., J. M. Orenstein, M. A. Martin, and E. O. Freed. 1995. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 69:6810-6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaye, J. F., and A. M. Lever. 1999. Human immunodeficiency virus types 1 and 2 differ in the predominant mechanism used for selection of genomic RNA for encapsidation. J. Virol. 73:3023-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kikonyogo, A., F. Bouamr, M. L. Vana, Y. Xiang, A. Aiyar, C. Carter, and J. Leis. 2001. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc. Natl. Acad. Sci. USA 98:11199-11204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim, W., Y. Tang, Y. Okada, T. A. Torrey, S. K. Chattopadhyay, M. Pfleiderer, F. G. Falkner, F. Dorner, W. Choi, N. Hirokawa, and H. C. Morse, III. 1998. Binding of murine leukemia virus Gag polyproteins to KIF4, a microtubule-based motor protein. J. Virol. 72:6898-6901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koonin, E. V., and R. A. Abagyan. 1997. TSG101 may be the prototype of a class of dominant negative ubiquitin regulators. Nat. Genet. 16:330-331. [DOI] [PubMed] [Google Scholar]
- 25.Kunkel, T. A., J. D. Roberts, and R. A. Zakour. 1987. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 154:367-382. [DOI] [PubMed] [Google Scholar]
- 26.Lama, J., and D. Trono. 1998. Human immunodeficiency virus type 1 matrix protein interacts with cellular protein HO3. J. Virol. 72:1671-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemmon, S. K., and L. M. Traub. 2000. Sorting in the endosomal system in yeast and animal cells. Curr. Opin. Cell Biol. 12:457-466. [DOI] [PubMed] [Google Scholar]
- 28.Li, L., and S. N. Cohen. 1996. Tsg101: a novel tumor susceptibility gene isolated by controlled homozygous functional knockout of allelic loci in mammalian cells. Cell 85:319-329. [DOI] [PubMed] [Google Scholar]
- 29.Li, L., J. Liao, J. Ruland, T. W. Mak, and S. N. Cohen. 2001. A TSG101/MDM2 regulatory loop modulates MDM2 degradation and MDM2/p53 feedback control. Proc. Natl. Acad. Sci. USA 98:1619-1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luban, J., K. L. Bossolt, E. K. Franke, G. V. Kalpana, and S. P. Goff. 1993. Human immunodeficiency virus type 1 Gag protein binds to cyclophilins A and B. Cell 73:1067-1078. [DOI] [PubMed] [Google Scholar]
- 31.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 32.McCann, E. M., and A. M. Lever. 1997. Location of cis-acting signals important for RNA encapsidation in the leader sequence of human immunodeficiency virus type 2. J. Virol. 71:4133-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ott, D. E., L. V. Coren, E. N. Chertova, T. D. Gagliardi, and U. Schubert. 2000. Ubiquitination of HIV-1 and MuLV Gag. Virology 278:111-121. [DOI] [PubMed] [Google Scholar]
- 34.Ott, D. E., L. V. Coren, T. D. Copeland, B. P. Kane, D. G. Johnson, R. C. Sowder, Jr., Y. Yoshinaka, S. Oroszlan, L. O. Arthur, and L. E. Henderson. 1998. Ubiquitin is covalently attached to the p6Gag proteins of human immunodeficiency virus type 1 and simian immunodeficiency virus and to the p12Gag protein of Moloney murine leukemia virus. J. Virol. 72:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ott, D. E., L. V. Coren, B. P. Kane, L. K. Busch, D. G. Johnson, R. C. Sowder, Jr., E. N. Chertova, L. O. Arthur, and L. E. Henderson. 1996. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J. Virol. 70:7734-7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Parent, L. J., R. P. Bennett, R. C. Craven, T. D. Nelle, N. K. Krishna, J. B. Bowzard, C. B. Wilson, B. A. Puffer, R. C. Montelaro, and J. W. Wills. 1995. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J. Virol. 69:5455-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patnaik, A., V. Chau, and J. W. Wills. 2000. Ubiquitin is part of the retrovirus budding machinery. Proc. Natl. Acad. Sci. USA 97:13069-13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ponting, C. P., Y. D. Cai, and P. Bork. 1997. The breast cancer gene product TSG101: a regulator of ubiquitination? J. Mol. Med. 75:467-469. [PubMed] [Google Scholar]
- 39.Potts, B. J. 1990. "Mini' reverse transcriptase (RT) assay, p. 102-106. In A. Aldovini and B. D. Walker (ed.), Techniques in HIV research. Stockton Press, New York, N.Y.
- 40.Putterman, D., R. B. Pepinsky, and V. M. Vogt. 1990. Ubiquitin in avian leukosis virus particles. Virology 176:633-637. [DOI] [PubMed] [Google Scholar]
- 41.Rotin, D., O. Staub, and R. Haguenauer-Tsapis. 2000. Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. J. Membr. Biol. 176:1-17. [DOI] [PubMed] [Google Scholar]
- 42.Ruland, J., C. Sirard, A. Elia, D. MacPherson, A. Wakeham, L. Li, J. L. de la Pompa, S. N. Cohen, and T. W. Mak. 2001. p53 accumulation, defective cell proliferation, and early embryonic lethality in mice lacking tsg101. Proc. Natl. Acad. Sci. USA 98:1859-1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schubert, U., D. E. Ott, E. N. Chertova, R. Welker, U. Tessmer, M. F. Princiotta, J. R. Bennink, H. G. Krausslich, and J. W. Yewdell. 2000. Proteasome inhibition interferes with gag polyprotein processing, release, and maturation of HIV-1 and HIV-2. Proc. Natl. Acad. Sci. USA 97:13057-13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smith, D. B., and K. S. Johnson. 1988. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 67:31-40. [DOI] [PubMed] [Google Scholar]
- 45.Spence, J., S. Sadis, A. L. Haas, and D. Finley. 1995. A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol. Cell. Biol. 15:1265-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Strack, B., A. Calistri, M. A. Accola, G. Palu, and H. G. Gottlinger. 2000. A role for ubiquitin ligase recruitment in retrovirus release. Proc. Natl. Acad. Sci. USA 97:13063-13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swanstrom, R., and J. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. Coffin and S. Hughes and H. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 48.Tang, Y., U. Winkler, E. O. Freed, T. A. Torrey, W. Kim, H. Li, S. P. Goff, and H. C. Morse III. 1999. Cellular motor protein KIF-4 associates with retroviral Gag. J. Virol. 73:10508-10513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Treier, M., L. M. Staszewski, and D. Bohmann. 1994. Ubiquitin-dependent c-Jun degradation in vivo is mediated by the delta domain. Cell 78:787-798. [DOI] [PubMed] [Google Scholar]
- 50.Ulrich, H. D., and S. Jentsch. 2000. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 19:3388-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vogt, V. M. 2000. Ubiquitin in retrovirus assembly: actor or bystander? Proc. Natl. Acad. Sci. USA 97:12945-12947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ward, C. L., S. Omura, and R. R. Kopito. 1995. Degradation of CFTR by the ubiquitin-proteasome pathway. Cell 83:121-127. [DOI] [PubMed] [Google Scholar]
- 54.Wills, J. W., C. E. Cameron, C. B. Wilson, Y. Xiang, R. P. Bennett, and J. Leis. 1994. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J. Virol. 68:6605-6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiang, Y., C. E. Cameron, J. W. Wills, and J. Leis. 1996. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J. Virol. 70:5695-5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan, B., X. Li, and S. P. Goff. 1999. Mutations altering the moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 18:4700-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]