Abstract
The full-length human immunodeficiency virus type 1 (HIV-1) mRNA encodes two precursor polyproteins, Gag and GagProPol. An infrequent ribosomal frameshifting event allows these proteins to be synthesized from the same mRNA in a predetermined ratio of 20 Gag proteins for each GagProPol. The RNA frameshift signal consists of a slippery sequence and a hairpin stem-loop whose thermodynamic stability has been shown in in vitro translation systems to be critical to frameshifting efficiency. In this study we examined the frameshift region of HIV-1, investigating the effects of altering stem-loop stability in the context of the complete viral genome and assessing the role of the Gag spacer peptide p1 and the GagProPol transframe (TF) protein that are encoded in this region. By creating a series of frameshift region mutants that systematically altered the stability of the frameshift stem-loop and the protein sequences of the p1 spacer peptide and TF protein, we have demonstrated the importance of stem-loop thermodynamic stability in frameshifting efficiency and viral infectivity. Multiple changes to the amino acid sequence of p1 resulted in altered protein processing, reduced genomic RNA dimer stability, and abolished viral infectivity. The role of the two highly conserved proline residues in p1 (position 7 and 13) was also investigated. Replacement of the two proline residues by leucines resulted in mutants with altered protein processing and reduced genomic RNA dimer stability that were also noninfectious. The unique ability of proline to confer conformational constraints on a peptide suggests that the correct folding of p1 may be important for viral function.
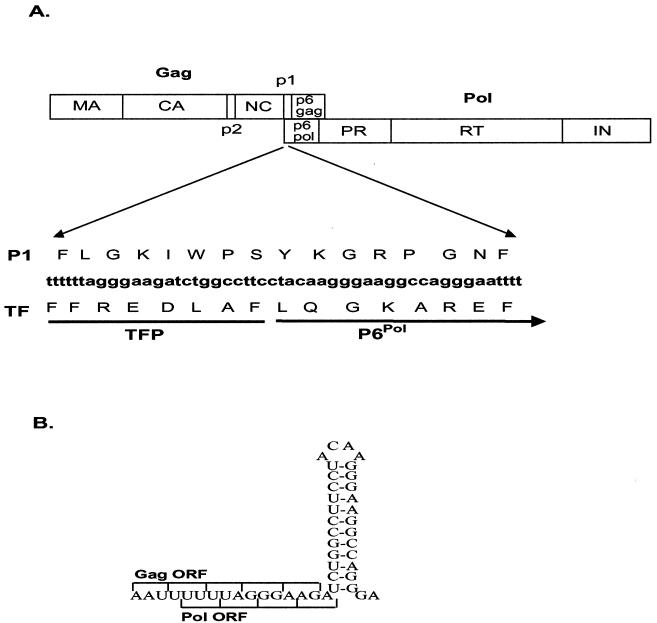
Human immunodeficiency virus type 1 (HIV-1) uses a −1 ribosomal frameshifting event that enables the precursor polyproteins Gag and Pol to be synthesized from the same genomic mRNA (for reviews, see references 19 and 24). The gag gene has its own distinct initiation and termination codons. The pol gene, however, lacks an initiation codon. Consequently, since pol partially overlaps and is in the −1 reading frame with respect to the gag gene, it is synthesized only as part of a GagProPol fusion protein (19, 20, 33, 42) (Fig. 1A). The frameshift required for the translation of GagProPol is promoted by an RNA signal comprising a “slippery” sequence (U UUU UUA), a 7-nucleotide spacer region, and a hairpin stem-loop (Fig. 1B). The stem-loop has been described as a simple RNA loop structure (19, 24, 30); however, recent work by Dinman et al (10) led them to predict that the RNA secondary structure required for efficient frameshifting may be a more complex triple-helix stem-loop. The −1 ribosomal frameshifting utilized by HIV-1 is thought to occur because ribosomes traveling along the mRNA translating gag interact with the stem-loop and as a result are temporarily stalled on the slippery sequence (19, 22, 38). While paused, approximately 1 in 20 ribosomes slip back 1 nucleotide, ensuring that Gag and GagProPol are synthesized in a strict ratio of 20 Gag molecules to each GagProPol (20). The ratio of Gag to GagProPol is critical for viral assembly and replication (18, 23, 29, 37).
FIG. 1.
(A) Schematic representation of the Gag and GagProPol polyproteins, showing the major domains and the stem-loop, p1, and TF overlapping region. (B) Wild-type frameshift coding region from HXB2-BH10.
The kinetics of ribosomal pausing at the frameshift signal are very precise. While the slippery sequence alone mediates a low level of frameshifting, the presence of the stem-loop increases the ribosomal frameshifting efficiency three- to fivefold (3, 5, 6, 30, 34). Bidou et al. (3) showed that mutations which decreased stem-loop thermodynamic stability also decreased the frequency of frameshifting events while increased stem-loop stability had a negative effect on frameshifting efficiency. Rather than using a stem-loop that maximizes frameshifting efficiency, it appears that the virus favors a stem-loop that supports the correct ratio of Gag to GagProPol synthesis. This is reflected in the great natural variation in the stability of the frameshift stem-loop among 76 gag nucleotide sequences from various strains and subtypes of HIV-1 examined by Chang et al. (7). For example, the average estimated thermodynamic stability (ΔG○) of the frameshift stem-loops from 29 subtype B viruses was −21.3 kcal/mol while that of 9 subtype A stem-loops was −17 kcal/mol and 5 of 7 subtype F sequences had an estimated stability of less than or equal to −15 kcal/mol (7).
The critical RNA secondary structure of the frameshift signal and the open reading frames for two proteins (p1 from Gag and the transframe protein [TF] from GagProPol) overlap (Fig. 1A). Accordingly, when mutations are introduced into the stem-loop RNA structure of the frameshift signal, the protein sequences of p1 and TF are also altered. The spacer peptide p1 is a 16-amino-acid protein to which no function has yet been ascribed. TF consists of two domains: a conserved N-terminal octapeptide (TFP) and a 48- to 60-amino-acid variable region (P6Pol) that are separated by a cleavage site (4, 25). While there is abundant evidence to suggest that TF is involved in the regulation of protease (PR) activity (1, 25, 32, 41, 43, 44), the mechanism of regulation remains unclear. TF-deleted mutants have enhanced polyprotein processing in vitro (25, 31), suggesting that the autocatalytic release of PR from TF acts as a triggering event for proteolytic processing (26, 31). Furthermore, cleavage of TF from PR is crucial for the stabilization of the PR dimeric structure (27), and Paulus et al. (32) have demonstrated that recombinant TF inhibits mature PR activity in vitro, suggesting an additional function for TF as a negative regulator of PR.
We have examined the frameshift region of HIV-1 to assess the effects of altering stem-loop stability in the context of the complete viral genome and to determine the roles of p1 and TF in HIV-1 replication. To isolate the RNA frameshift signal from p1 and TF, we have used a mutagenesis strategy which alters stem-loop stability and the amino acid sequences of p1 and TF independently. Mutants with a large decrease in stem-loop stability were found to be noninfectious, demonstrating the importance of stem-loop stability and frameshifting efficiency to viral replication. Mutations altering only TF, while preserving p1 sequence and stem-loop stability, did not affect virion function. However, changes to the amino acid sequence of p1, which retained the TF sequence and stem-loop stability, altered the virion and intracellular HIV-1 protein profiles and reduced the stability of genomic RNA dimers. In addition, these virions were noninfectious in peripheral blood mononuclear cells (PBMCs). Two additional p1 mutants were created to further examine the importance of p1 for HIV-1 replication. Replacement of the two highly conserved proline residues (positions 7 and 13) in p1 with leucines (P1:pro−), destroyed infectivity, altered protein profiles, and reduced genomic RNA dimer stability. The P1:pro+ mutant, which was designed to emulate the SLP1∗TF mutations while keeping the two proline residues intact (P1:pro+), displayed reduced infectivity but had protein profiles similar to wild-type profiles and had unchanged genomic RNA dimer stability. This study provides the first evidence that p1 and its two highly conserved proline residues are important for HIV-1 replication.
MATERIALS AND METHODS
Construction of DNA plasmids.
The HIV-1 DNA constructs used in this study were derived from the full-length wild-type HIV-1 plasmid HXB2-BH10 (40). Mutations were introduced into the region between DNA nucleotides nt 2096 and 2128 of HXB2-BH10 (5′-GATCTGGCCTTCCTACAAGGGAAGGCCAGGGAA -3′) which includes the frameshift stem-loop (SL) and encodes the p1 and TF proteins (Fig. 1A). Four initial frameshift region mutants have been named such that an asterisk indicates when an alteration has been made to SL, p1, or TF, with the wild-type being SLP1TF. Two additional p1 mutants were also created, P1:pro− and P1:pro+. All mutants were created using PCR stitch mutagenesis to introduce mutations via the PCR primers, as previously described (17), and subsequent cloning into HXB2-BH10 via the restriction sites ApaI and BclI. The resultant mutants are as depicted in Table 1, with the changes indicated in italics. The thermodynamic stability of the predicted stem-loop structures for each of the mutants was determined with the mfold program by M. Zuker of Washington University School of Medicine (12, 21, 35) using the server located at The Macfarlane Burnet Institute for Medical Research and Public Health (http://mfold.burnet.edu.au/) (Table 1). The SLP1TF mutations were also introduced into the protease-defective SVC21 P(−) plasmid (16, 28) and are referred to as P−/SLP1∗TF, P−/SLP1TF∗, P−/SL∗P1∗TF, and P−/SL∗P1TF∗. All constructs were sequenced to confirm the presence of the desired mutations and the absence of spontaneous mutations introduced via PCR mutagenesis.
TABLE 1.
Mutations within the frameshift region of wild-type HIV-1 (HXB2-BH10)
| Construct | DNA and protein sequence changesa | Energy (ΔG○, kcal/mol)b |
|---|---|---|
| Ile Trp Pro Ser Tyr LysGly Arg Pro Gly | ||
| SLP1TF (WT) | GATCTGGCCTTCCTACAAGGGAAGGCCAGGGAA | −21.4 |
| Asp Leu Ala Phe Leu Gln Gly Lys Ala Arg Glu | ||
| Thr Trp Leu Phe Tyr Lys Glu Lys Leu Gly | ||
| SLP1∗TF | GACCTGGCTTTTCTACAAGGAAAAGCTAGGGAA | −17.4 |
| Asp Leu Ala Phe Leu Gln Gly Lys Ala Arg Glu | ||
| Ile Trp Pro Ser Tyr Lys Gly Arg Pro Gly | ||
| SLP1TF∗ | GATTTGGCCGTCTTACAAAGGGCGGCCGGGGAA | −16.2 |
| Asp Leu Ala Val Leu Gln Arg Ala Ala Gly Glu | ||
| Thr Ser Arg Ser Tyr Lys Ala Lys Pro Ala | ||
| SL∗P1∗TF | GACCTCGCGTTCTTACAAGGCAAAGCCCGCGAA | −6.4 |
| Asp Leu Ala Phe Leu Gln Gly Lys Ala Arg Glu | ||
| Ile Trp Pro Ser Tyr Lys Gly Arg Pro Gly | ||
| SL∗P1TF∗ | GATATGGCCCAGTTACAAAGGGCGCCCAGGGAA | −7.1 |
| Asp Met Ala Gln Leu Arg Ala Pro Arg Glu | ||
| Thr Trp Pro Phe Tyr Lys Glu Gly Pro Gly | ||
| P1:pro+ | GACCTGGCCTTTCTACAAGGAAGGGCCAGGGAA | −21.2 |
| Asp Leu Ala Phe Leu Gln Gly Arg Ala Arg Glu | ||
| Ile Trp Leu Ser Tyr Lys Gly Arg Leu Gly | ||
| P1:pro− | GATCTGGCTTTCCTACAAGGGAAGGCTAGGGAA | −16.9 |
| Asp Leu Ala Phe Leu Gln Gly Lys Ala Arg Glu |
The upper amino acid sequence is that of p1, while the lower is that of TF. Mutated bases or amino acids are shown in italics; the stem region (Fig. 1) is underlined.
Virus production.
The calcium phosphate coprecipitation method was used for the transient transfection of 293T cells. DNA (10 μg) from each of the HIV-1 constructs was routinely used for transfection. The enhanced green fluorescent protein (EGFP; Clonetech) reporter plasmid (2 μg) was added to the DNA mixture to determine the transfection efficiency. Supernatants and cells were collected at 36 h posttransfection and separated by centrifugation for 30 min at 3,000 rpm (Beckman model GS-6 centrifuge). The reverse transcriptase (RT) activity of cell culture supernatants was measured by a micro RT assay as previously described (15).
Protein isolation and Western blot analysis.
Intracellular viral protein was isolated from transfected 293T cells by washing the cells twice with 1× Tris-buffered saline (TBS) and then lysing them with 2× TBS lysis buffer containing 1% Nonidet P-40, 20 mM phenylmethylsulfonyl fluoride, 1 μM pepstatin, and 1 μM leupeptin. Cell lysates were freeze-thawed in liquid nitrogen and then clarified by centrifugation for 20 min at 14,000 rpm (Eppendorf 5417R centrifuge). The transfection efficiency of the samples was monitored by measuring EGFP production from the reporter plasmid with a Bio Imaging Analyzer (Fuji Photo Film Co.). Virion protein was obtained by ultracentrifugation (Beckman model L-90, SW 41 rotor) of the transfection supernatants at 35,000 rpm for 1 h at 4°C through a 20% sucrose cushion. The viral pellets were resuspended in 50 μl of 2× TBS lysis buffer. Western blot analysis was carried out as previously described (37), using pooled sera from HIV-1-infected patients to detect total HIV-1 viral proteins.
Immunoprecipitation of protease-negative samples.
Intracellular viral protein was isolated from transfected 293T cells as described above, with lysis by RIPA buffer (300 mM NaCl, 50 mM Tris [pH 7.4], 0.5% [vol/vol] Triton x-100, 1 μM pepstatin, 1 μM leupeptin, 1 μM Pefabloc). The cellular protein was normalized on the basis of EGFP expression. Pooled sera from HIV-1-infected patients was added for incubation overnight at 4°C. Sepharose-G beads (Amersham Pharmacia Biotech) were then added, and incubation was continued for a further 6 h. The beads were washed five times with 500 μl of RIPA buffer, resuspended in 15 μl of sample buffer (100 mM Tris [pH 6.8], 3% sodium dodecyl sulfate [SDS], 33% glycerol, 0.03% bromophenol blue), and heated at 95°C for 10 min for Western blot analysis using an anti-p24 monoclonal antibody (NEN).
Analysis of virion RNA dimerization.
Viral supernatants were purified and concentrated by ultracentrifugation as described above. Virion pellets were resuspended in 500 μl of dimerization buffer (10 mM Tris [pH 7.5], 1 mM EDTA, 1% SDS, 50 mM NaCl), phenol-chloroform extracted and isolated for melting-curve analysis as previously described (13, 14). Similar amounts of genomic RNA were used to analyze the stability of the virion RNA dimer as previously described (37). Briefly, RNA samples that had been heat denatured for 10 min at the indicated temperatures were separated by electrophoresis in a 1% native agarose gel in 0.5× Tris-borate-EDTA buffer. Samples were transferred onto a Hybond N membrane (Amersham Pharmacia Biotech). Dimeric and monomeric RNAs were hybridized with a radioactive riboprobe (pGEM7zHIV-1), which is complementary to the 5′ end of the HIV-1 genomic RNA.
Infectivity assays.
Viral infectivity was measured using the 50% tissue culture infective dose (TCID 50) method (8). Briefly, PBMCs were isolated from HIV-seronegative buffy coats (supplied by the Red Cross Blood Bank, Melbourne, Victoria, Australia) as previously described (8). The PBMCs were stimulated with 10 μg of phytohemagglutinin (Murex Diagnostics) per ml and maintained for 3 days in RPMI 1640 medium (Gibco) containing 10% fetal bovine serum, gentamicin, glutamine, and 5% interleukin-2 (Boehringer). Viral supernatants, which were normalised for RT activity, were then mixed with 10 5 PBMCs in a 96-well tissue culture plate. Eight 10-fold dilutions of each virus were tested in triplicate. Viral infectivity was assessed by monitoring RT activity, as described above, with supernatants collected on days 3, 7, 10, and 14 postinfection.
RESULTS
Mutation of the frameshift region of HIV-1.
The critical RNA secondary structure of the frameshift signal and the open reading frames for the p1 and TF proteins overlap (Fig. 1A). We have created two classes of mutations, those that have a conservative effect on stem-loop stability (SL) and those that have a dramatic effect on stem-loop stability (SL∗). Within these classes are two mutants, one that alters the protein sequence of p1 (P1∗) and one that alters the protein sequence of TF (TF∗) (Table 1).
Three main factors influence the stability, and thus the frameshifting efficiency, of the stem-loop: A-U as opposed to G-C base pairing, alteration of the total number of base pairs, and alteration of the sequence of the actual loop (3). For each of the mutants described here, the 4-bp loop sequence remains wild type (Fig. 1; Table 1). The wild-type (HXB2-BH10) stem-loop has seven G-C and three A-U pairs (estimated ΔG○ = −21.1 kcal/mol−1). For the two mutants with a conservative effect on stem-loop stability, SLP1∗TF (estimated ΔG○ = −17.4 kcal/mol−1) and SLP1TF∗ (estimated ΔG○ = −16.2 kcal/mol−1), only the number of A-U versus G-C pairs was altered and the estimated ΔG○ was only 4 to 5 kcal/mol−1 more than that of the wild type. For the two SL∗ mutants, SL∗P1∗TF (estimated ΔG○ = −6.4 kcal/mol−1) and SL∗P1TF∗ (estimated ΔG○ = −7.1 kcal/mol−1), stem-loop stability was altered to an estimated ΔG○ of 14 to 15 kcal/mol−1 more than that of the wild type by reducing base pairing.
The thermodynamic stability of the frameshift stem-loop is important for GagProPol expression.
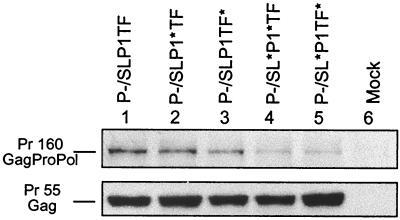
Protease-defective constructs that lack the ability to process the precursor proteins into their components were used to evaluate the relationship between the altered stem-loop stability of the mutants and frameshifting efficiency, as measured by Pr160-GagProPol synthesis. Comparing the amount of GagProPol synthesized relative to Pr55-Gag, the mutants with conservative alterations to stem-loop stability, P−/SLP1∗TF and P−/SLP1TF∗, synthesized GagProPol at a level comparable to that of the wild type (Fig. 2, lanes 2 and 3). As expected, the synthesis of GagProPol in the two mutants with major alterations to stem-loop stability, P−/SL∗P1∗TF and P−/SL∗P1TF∗, was greatly decreased compared to that in the wild type (Fig. 2, lanes 4 and 5).
FIG. 2.
Impact of frameshift region mutations on HIV-1 GagProPol expression. Cell lysates from transfections of 293T cells with protease-defective constructs were immunoprecipitated with sera from HIV-1-infected patients and then assayed by Western blot analysis using an anti-P24 antibody. Lane 1 shows the wild-type (P−/SLP1TF) profile.
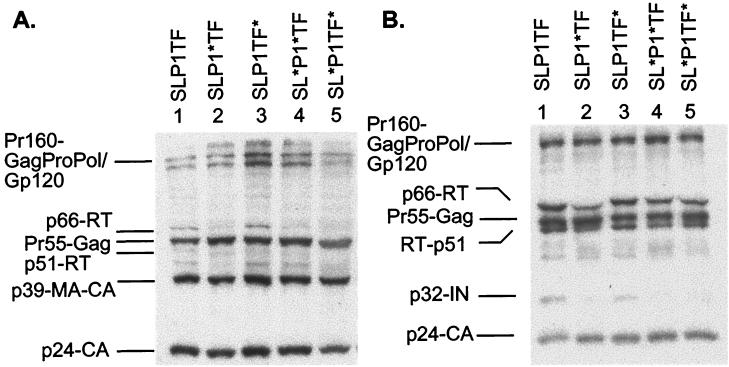
Mutants with reduced stem-loop stability or altered p1 amino acid sequences have defective cellular and viral protein patterns.
The HIV-1 proteins detected in cell lysates for each of the mutants were very similar to those of wild-type virus, with Pr55-Gag, p24-CA and Pr160-GagProPol/GP160-Env all being readily identified (Fig. 3A). In most cases, the levels of proteins synthesized from the mutant virion were comparable to those obtained from cells transfected with wild-type DNA (Fig. 3A, lane 1). The most obvious difference was a decrease in the p66-RT protein levels present in SLP1∗TF and the two stem-loop mutants SL∗P1∗TF and SL∗P1TF∗ (lanes 2, 4, and 5 respectively). Purified viral particles derived from cotransfected cells were also analyzed by Western blotting. For similar levels of p24-CA, the protein profile of SLP1TF∗ was comparable to that of the wild type whereas SLP1∗TF, SL∗P1∗TF, and SL∗P1TF∗ showed decreased levels of p66-RT, p51-RT, and p32-IN when compared to those in the wild type (Fig. 3B).
FIG. 3.
Impact of frameshift region mutations on HIV-1 protein profiles as determined by Western blot analysis. Cell (A) and virion (B) lysates were produced from transfections in 293T cells, resolved by SDS-polyacrylamide gel electrophoresis (10% polyacrylamide), and probed with sera from HIV-1-infected individuals. Lanes 1 show the wild-type (SLP1TF) HIV-1 protein profiles.
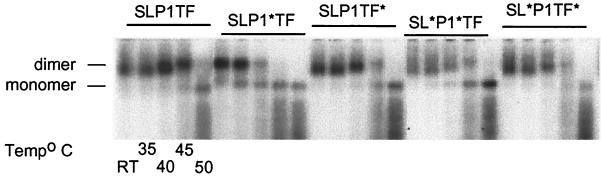
Mutants with altered p1 amino acid sequences display decreased genomic RNA dimer stability.
Dimeric RNA was present in all viruses (Fig. 4). Wild-type genomic virion RNA started dissociating from dimeric to monomeric RNA at 45°C. The stability of the SLP1∗TF dimeric RNA was dramatically reduced (Fig. 4), with some monomeric RNA being detectable even without heat treatment and only monomeric RNA present at 45°C. SL∗P1∗TF dimer RNA stability was also compromised, with monomeric RNA being clearly visible at 35°C (Fig 4). RNA isolated from SLP1TF∗ and SL∗P1TF∗ more closely resembled wild-type dimeric RNA stability.
FIG. 4.
Effect of frameshift region mutants on virion RNA dimerization. The impact of the frameshift region mutants on genomic RNA dimerization was determined using melting-curve and electrophoretic analysis of wild-type and mutant RNA. Virion RNA was resuspended in RNA dimerization buffer and heat denatured for 10 min at the indicated temperatures. Dimers and monomers were electrophoresed in a 1% native agarose gel and probed with an HIV-1 riboprobe. RT, room temperature.
Mutants with reduced stem-loop stability or altered p1 amino acid sequences are noninfectious in PBMCs.
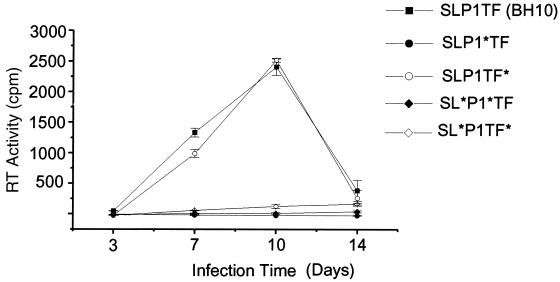
Wild-type and mutant virions derived from transfected 293T cells were normalized for RT activity and assayed for infectivity in PBMCs. Infectivity was determined in four different PBMC donors, and Fig. 5 is representative of the replication kinetics observed in these assays. The mutant with alterations only to the TF protein sequence (SLP1TF∗) replicated at a level similar to that of wild type, while the p1 mutant (SLP1∗TF) and the two mutants with large decreases in stem-loop stability (SL∗P1∗TF and SL∗P1TF∗) were noninfectious (Fig. 5).
FIG. 5.
Replication of the frameshift region mutants in PBMCs. Freshly isolated PBMCs were phytohemagglutinin stimulated for 3 days and then infected with either wild-type (SLP1TF) or mutant virion. Infectivity was monitored as RT activity in the culture supernatants, which were collected 3, 7, 10, and 14 days postinfection. The results shown represent the mean and standard deviation of duplicate samples.
Proline residues in p1 are important for HIV-1 function.
P1 has two proline residues (positions 7 and 13), which are both altered in SLP1∗TF. We chose to investigate these residues because proline has a cyclic structure capable of strongly influencing protein architecture and these proline residues are highly conserved among different strains of HIV-1. In addition, the p1-P6 Gag region is rich in this amino acid, which is surprising in a virus where proline residues are relatively uncommon (11). Two additional p1 mutants were created. The first mutant, P1:pro−, has the two proline residues of p1 (positions 7 and 13) replaced by leucines. The predicted stem-loop stability of this mutant of −21.2 kcal/mol was very similar to that of the wild type (−21.4 kcal/mol) (Table 1). The second mutant, P1:pro+, was designed to closely emulate the SLP1∗TF mutations while keeping the two proline residues intact. To maintain the functional stability of the frameshift stem-loop, P1:pro+ has an arginine-to-glycine mutation instead of the arginine-to-lysine mutation used in SLP1∗TF. The estimated stability of the P1:pro+ stem-loop (−16.9 kcal/mol) was similar to that of SLP1TF∗ (−16.2 kcal/mol), which replicates at wild-type levels. In addition, one residue (lysine 12) of TF was altered; since this residue is also altered in SLP1TF∗, it is unlikely that this will influence viral function.
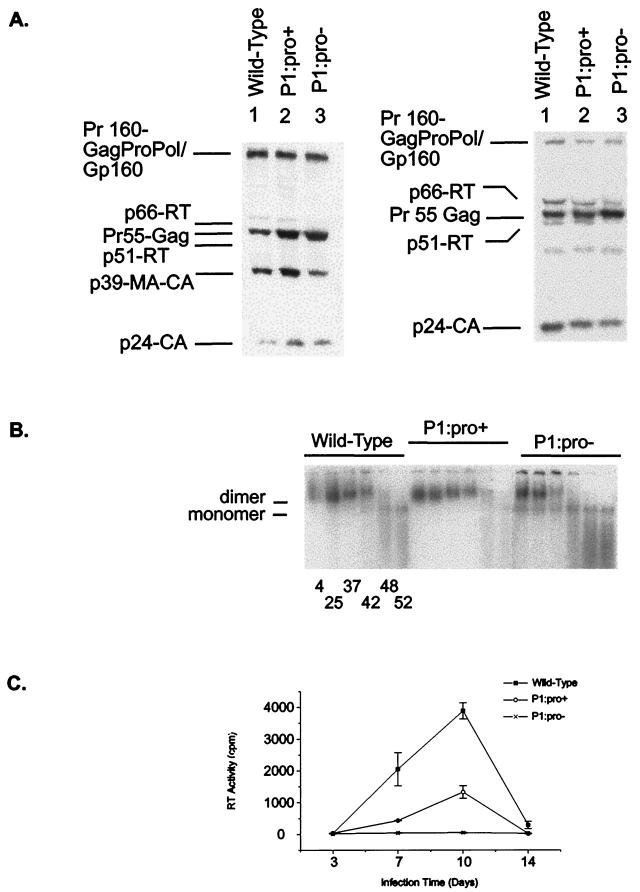
The cellular protein pattern of P1:pro+ was similar to that of the wild type (Fig. 6A), while P1:pro− showed a large decrease in the level of p66-RT (Fig. 6A), similar to that seen in SLP1∗TF (Fig. 3A). Purified viral particles were also analyzed by Western blotting. Compared to the wild-type virion profile, the P1:pro+ mutant displayed a slight increase in the level of Pr-55 Gag while the P1:pro− mutant displayed large decreases in the levels of p66-RT and p51-RT and an increased level of p55-Gag relative to the level of p24-CA (Fig. 6B). Again these differences in protein profile for P1:pro− were consistent with those observed for SLP1∗TF (Fig 3B). Virion genomic RNA analyses demonstrated that RNA was present in both mutants and that the stability of the P1:pro+ dimeric RNA was comparable to that of the wild type, while the P1:pro− dimeric RNA stability was dramatically reduced (Fig. 6C), resembling the stability of RNA dimers observed for SLP1∗TF (Fig. 4). The P1:pro+ mutant replicate in PBMCs at a low level compared to wild-type, while the P1:pro− mutant was noninfectious (Fig. 6D).
FIG. 6.
(A) Impact of frameshift region mutations on HIV-1 protein profiles as determined by Western blot analysis for cell (left) and virion lysates (right) that were probed with sera from HIV-1-infected individuals. (B) Effect of frameshift region mutants on virion RNA dimerization. (C) Replication of the p1 mutants in PBMCs. Infectivity was monitored as RT activity in the culture supernatants, which were collected 3, 7, 10, and 14 days postinfection. The results shown represent the mean and standard deviation of duplicate samples.
DISCUSSION
Within the frameshift region of HIV-1, the critical RNA secondary structures of the frameshift stem-loop, the Gag spacer peptide p1, and the GagProPol protein TF overlap. To characterize the frameshift region of HIV-1, we have created a series of mutants that systematically alter the stem-loop, p1, and TF. Changes to the stability of the frameshift stem-loop have demonstrated the importance of stem-loop thermodynamic stability in maintaining the optimal frameshifting efficiency required for viral infectivity. In addition, we provide the first evidence that the HIV-1 spacer peptide p1, and in particular the two proline residues of p1, are important for HIV-1 replication.
Two types of mutants were initially utilized in this study, those with very slight changes in stem-loop stability, which aimed to maintain frameshifting efficiency at wild-type levels, and those with a more dramatic decrease in stem-loop stability. Reduction of stem-loop stability by an estimated ΔG○ of 4-5 kcal/mol−1 (SLP1∗TF and SLP1TF∗) did not discernibly affect the translation efficiency of GagProPol, since the levels of GagProPol produced in the protease-negative constructs were comparable to those in the wild type. Moreover, one of the two mutants, SLP1TF∗, replicated as well as did wild-type virus in PBMCs. Taken together, these results most probably reflect the ability of the HIV-1 frameshift stem-loop to accommodate minor alterations to stability and maintain the balance between structural proteins and enzymes that is required for the production of infectious virion. This observation is consistent with the variability in frameshift stem-loop stability found in different strains and subtypes of HIV-1 (7) and with the results of a recent study by Telenti et al. demonstrating that natural variations in thermodynamic stability that resulted in decreased frameshifting and impaired GagProPol maturation were still able to support wild-type replication (39). Large reductions in stem-loop stability (an estimated ΔG○ of 14 to 15 kcal/mol−1) in mutants SL∗P1∗TF and SL∗P1TF∗ resulted in a marked decrease in GagProPol synthesis. The reduced level of GagProPol in SL∗P1∗TF and SL∗P1TF∗ was, however, sufficient to support protease activity, since processed products were observed in virion particles. Mutants with major disruptions to stem-loop stability (SL∗P1∗TF and SL∗P1TF∗) were found to be noninfectious in PBMCs. The decreased stem-loop stability of the SL∗ mutants did not affect the ability of these virions to package dimeric RNA and had little effect on genomic RNA dimer stability, with SL∗P1TF∗ having a dimer stability similar to that of the wild type.
TF is important for the activation and regulation of PR activity (1, 25, 30, 31, 41, 43, 44). In this study, mutations to the amino acid sequence of TF (SLP1TF∗) generated virions similar in profile to the wild-type virus. TF has two domains, TFP and P6Pol (4, 25). TFP is a highly conserved octapeptide that is a specific competitive inhibitor of the mature PR (25). In SLP1TF∗, only one amino acid in the TFP region was altered (phenylalanine to valine at position 8), which may not be sufficient to disrupt TFP function. The remaining changes were in the amino-acid-variable region of P6Pol, which has been suggested to be an unstructured protein whose importance lies in its ability to create a stable space between nucleocapsid (NC) and PR (2).
This study provides the first evidence that the spacer peptide p1 sequence is important for HIV-1 replication. The SLP1∗TF mutant, which includes six amino acid changes in the 16-amino-acid p1 sequence, had altered protein processing, reduced RNA dimer stability, and was noninfectious in PBMCs. Two additional p1 mutants were analyzed: P1:pro− has the two highly conserved proline residues of p1 replaced by leucines, while P1:pro+ was designed to closely emulate the SLP1∗TF mutations while keeping the two proline residues intact. Analysis of the P1:pro− mutant revealed defects in protein processing and RNA dimer stability similar to those observed for SLP1∗TF. The P1:pro+ mutant, however, showed protein patterns comparable to those of the wild type and also showed unchanged genomic RNA dimer stability. In addition, while P1:pro− was noninfectious in PBMCs, P1:pro+ displayed a low level of PBMC infection. These results further demonstrate that p1 is important for viral replication. Since the rigid cyclic structure of proline can confer unique conformational constraints on a peptide, dramatically affecting protein architecture, these results suggest that the proper folding of the p1 protein may be critical for HIV-1 replication. The fact that the P1:pro+ mutant was less infectious than the wild type suggests that p1 amino acids other than proline may also contribute to HIV-1 replication.
Western analysis of SLP1∗TF and P1:pro− showed an alteration to the wild-type cellular and virion protein patterns that most dramatically included a reduction in the amount of RT protein. In addition, the virion profile of P1:pro− displayed a large increase in the level of unprocessed Pr55-Gag. Taken together, these results suggest that the conformation of p1 may influence protein processing. Genomic RNA dimer stability and defects in proteolytic processing appear to be linked, since RNA dimers from PR-defective HIV-1 are less heat stable than dimers from wild-type HIV-1 (13). Stabilization of genomic RNA dimers is under the control of the mature NC, and it has recently been shown that p2/NC cleavage site mutants disrupt genomic RNA dimer stability and that cleavage of P2/NC in Gag is more important than in GagProPol for maintaining dimer stability (36). Even subtle changes to Gag processing in SLP1∗TF and P1:pro− that delay the cleavage of p2/NC may be sufficient to result in the decrease of RNA dimer stability observed in these p1 mutants. Interestingly, SL∗P1∗TF genomic RNA dimers were stable at temperatures up to 5°C higher than for those of SLP1∗TF. This result could represent a difference in the nature of the p1 amino acid mutations, or, alternatively, it may be that the effect of the stem-loop mutations override any functions specific to p1.
Any structural requirement for the p1 proline residues is likely to be transient, possibly influencing the arrangement of the Pr55-Gag precursor protein to yield the correct positioning for initiating or mediating proteolytic processing during HIV assembly. Since p1 forms part of p15-NC (NC-P1-P6), which specifically binds RNA and is tightly linked to the dimeric RNA genome and RT to form the ribonucleoprotein complex (9), modification of the protein configuration as a result of the proline mutations in SLP1∗TF and P1:pro− could alter the role of p15-NC, affecting genomic RNA dimerization and core formation. In addition, the importance of p1 proline residues in HIV replication may be due in part to their role in the overall protein folding in the proline-rich P1-P6 C terminus of Gag. Further investigation is required to explore whether p1 conformation is important for the function of the p55-Gag precursor protein or an intermediate such as p15-NC or if in fact p1-p6 acts as a single domain during the process of HIV-1 assembly.
Acknowledgments
We thank John Mills for critical review of the manuscript.
This study was funded by grants from the National Health and Medical Research Council (NHMRC) and the Macfarlane Burnet Institute Research Fund. Melissa Hill is a recipient of a Burnet Centenary fellowship and an amfAR fellowship. Miranda Shehu-Xhilaga is a recipient of an NHMRC Dora Lush Ph.D scholarship and an NHMRC C. J. Martin fellowship. Suzanne Crowe is supported by the Australian National Centre in HIV Virology Research (NCHVR), a grant from the Australian National Council of HIV/AIDS and Related Diseases, and the Macfarlane Burnet Institute Research Fund. Johnson Mak is the recipient of an NHMRC Peter Doherty postdoctoral fellowship.
REFERENCES
- 1.Almog, N., R. Roller, G. Arad, L. Passi-Even, M. A. Wainberg, and M. Kotler. 1996. A p6Pol-protease fusion protein is present in mature particles of human immunodeficiency virus type 1. J Virol. 70:7228-7232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beissinger, M., C. Paulus, P. Bayer, H. Wolf, P. Rosch, and R. Wagner. 1996. Sequence-specific resonance assignments of the 1H-NMR spectra and structural characterization in solution of the HIV-1 transframe protein p6. Eur. J. Biochem. 237:383-392. [DOI] [PubMed] [Google Scholar]
- 3.Bidou, L., G. Stahl, B. Grima, H. Liu, M. Cassan, and J. P. Rousset. 1997. In vivo HIV-1 frameshifting efficiency is directly related to the stability of the stem-loop stimulatory signal. RNA. 3:1153-1158. [PMC free article] [PubMed] [Google Scholar]
- 4.Candotti, D., C. Chappey, M. Rosenheim, P. M'Pele, J. M. Huraux, and H. Agut. 1994. High variability of the gag/pol transframe region among HIV-1 isolates. C. R. Acad. Sci. III. 317:183-189. [PubMed] [Google Scholar]
- 5.Cassan, M., V. Berteaux, P. O. Angrand, and J. P. Rousset. 1990. Expression vectors for quantitating in vivo translational ambiguity: their potential use to analyse frameshifting at the HIV gag-pol junction. Res. Virol. 141:597-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cassan, M., N. Delaunay, C. Vaquero, and J. P. Rousset. 1994. Translational frameshifting at the gag-pol junction of human immunodeficiency virus type 1 is not increased in infected T-lymphoid cells. J Virol. 68:1501-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, S. Y., R. Sutthent, P. Auewarakul, C. Apichartpiyakul, M. Essex, and T. H. Lee. 1999. Differential stability of the mRNA secondary structures in the frameshift site of various HIV type 1 viruses. AIDS Res. Hum. Retroviruses 15:1591-1596. [DOI] [PubMed] [Google Scholar]
- 8.Crowe, S. M., N. J. Vardaxis, S. J. Kent, A. L. Maerz, M. J. Hewish, M. S. McGrath, and J. Mills. 1994. HIV infection of monocyte-derived macrophages in vitro reduces phagocytosis of Candida albicans. J. Leukoc. Biol. 56:318-327. [DOI] [PubMed] [Google Scholar]
- 9.Darlix, J. L., G. Cristofari, M. Rau, C. Pechoux, L. Berthoux, and B. Roques. 2000. Nucleocapsid protein of human immunodeficiency virus as a model protein with chaperoning functions and as a target for antiviral drugs. Adv. Pharmacol. 48:345-372. [DOI] [PubMed] [Google Scholar]
- 10.Dinman, J. D., S. Richter, E. P. Plant, R. C. Taylor, A. B. Hammell, and T. M. Rana. 2002. The frameshift signal of HIV-1 involves a potential intramolecular triplex RNA structure. Proc. Natl. Acad. Sci. USA 99:5331-5336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitzgerald, D. J., E. C. Bronson, and J. N. Anderson. 1996. Compositional similarities between the human immunodeficiency virus and surface antigens of pathogens. AIDS Res. Hum. Retroviruses 12:99-106. [DOI] [PubMed] [Google Scholar]
- 12.Freier, S. M., R. Kierzek, J. A. Jaeger, N. Sugimoto, M. H. Caruthers, T. Neilson, and D. H. Turner. 1986. Improved free-energy parameters for predictions of RNA duplex stability. Proc. Natl. Acad. Sci. USA 83:9373-9377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu, W., R. J. Gorelick, and A. Rein. 1994. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 68:5013-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu, W., and A. Rein. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 67:5443-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goff, S., P. Traktman, and D. Baltimore. 1981. Isoltation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J. Virol. 1:239-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higuchi, R. 1990. Recombinant PCR, p. 177-183. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., New York, N.Y.
- 18.Hung, M., P. Patel, S. Davis, and S. R. Green. 1998. Importance of ribosomal frameshifting for human immunodeficiency virus type 1 particle assembly and replication. J. Virol. 72:4819-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacks, T. 1990. Translational suppression in gene expression in retroviruses and retrotransposons. Curr. Top. Microbiol. Immunol. 157:93-124. [DOI] [PubMed] [Google Scholar]
- 20.Jacks, T., M. D. Power, F. R. Masiarz, P. A. Luciw, P. J. Barr, and H. E. Varmus. 1988. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331:280-283. [DOI] [PubMed] [Google Scholar]
- 21.Jaeger, J. A., D. H. Turner, and M. Zuker. 1989. Improved predictions of secondary structures for RNA. Proc. Natl. Acad. Sci. USA 86:7706-7710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang, H. 1998. Direct structural evidence for formation of a stem-loop structure involved in ribosomal frameshifting in human immunodeficiency virus type 1. Biochim. Biophys. Acta 1397:73-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karacostas, V., E. J. Wolffe, K. Nagashima, M. A. Gonda, and B. Moss. 1993. Overexpression of the HIV-1 gag-pol polyprotein results in intracellular activation of HIV-1 protease and inhibition of assembly and budding of virus-like particles. Virology 193:661-671. [DOI] [PubMed] [Google Scholar]
- 24.Kollmus, H., M. W. Hentze, and H. Hauser. 1996. Regulated ribosomal frameshifting by an RNA-protein interaction. RNA 2:316-323. [PMC free article] [PubMed] [Google Scholar]
- 25.Louis, J. M., F. Dyda, N. T. Nashed, A. R. Kimmel, and D. R. Davies. 1998. Hydrophilic peptides derived from the transframe region of Gag-Pol inhibit the HIV-1 protease. Biochemistry 37:2105-2110. [DOI] [PubMed] [Google Scholar]
- 26.Louis, J. M., N. T. Nashed, K. D. Parris, A. R. Kimmel, and D. M. Jerina. 1994. Kinetics and mechanism of autoprocessing of human immunodeficiency virus type 1 protease from an analog of the Gag-Pol polyprotein. Proc. Natl. Acad. Sci. USA 91:7970-7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louis, J. M., E. M. Wondrak, A. R. Kimmel, P. T. Wingfield, and N. T. Nashed. 1999. Proteolytic processing of HIV-1 protease precursor, kinetics and mechanism. J. Biol. Chem. 274:23437-23442. [DOI] [PubMed] [Google Scholar]
- 28.Mak, J., M. Jiang, M. A. Wainberg, M. L. Hammarskjold, D. Rekosh, and L. Kleiman. 1994. Role of Pr160gag-pol in mediating the selective incorporation of tRNA(Lys) into human immunodeficiency virus type 1 particles. J. Virol. 68:2065-2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park, F., K. Ohashi, W. Chiu, L. Naldini, and M. A. Kay. 2000. Efficient lentiviral transduction of liver requires cell cycling in vivo. Nat. Genet. 24:49-52. [DOI] [PubMed] [Google Scholar]
- 30.Parkin, N. T., M. Chamorro, and H. E. Varmus. 1992. Human immunodeficiency virus type 1 gag-pol frameshifting is dependent on downstream mRNA secondary structure: demonstration by expression in vivo. J. Virol. 66:5147-5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Partin, K., G. Zybarth, L. Ehrlich, M. DeCrombrugghe, E. Wimmer, and C. Carter. 1991. Deletion of sequences upstream of the proteinase improves the proteolytic processing of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 88:4776-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paulus, C., S. Hellebrand, U. Tessmer, H. Wolf, H. G. Krausslich, and R. Wagner. 1999. Competitive inhibition of human immunodeficiency virus type-1 protease by the Gag-Pol transframe protein. J. Biol. Chem. 274:21539-21543. [DOI] [PubMed] [Google Scholar]
- 33.Ratner, L., W. Haseltine, R. Patarca, K. J. Livak, B. Starcich, S. F. Josephs, E. R. Doran, J. A. Rafalski, E. A. Whitehorn, K. Baumeister, et al. 1985. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature 313:277-284. [DOI] [PubMed] [Google Scholar]
- 34.Reil, H., H. Kollmus, U. H. Weidle, and H. Hauser. 1993. A heptanucleotide sequence mediates ribosomal frameshifting in mammalian cells. J. Virol. 67:5579-5584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serra, M. J., and D. H. Turner. 1995. Predicting thermodynamic properties of RNA. Methods Enzymol. 259:242-261. [DOI] [PubMed] [Google Scholar]
- 36.Shehu-Xhilaga, M. S. Pettit, R. Swanstrom, H.-G. Krausslich, J. Marshall, J.-Y. Lee, S. Crowe, and J. Mak. 2001. Proteolytic processing of the P2/nucleocapsid cleavage site is critical for human immunodeficiency virus type 1 RNA dimer maturation. J. Virol. 75:9156-9164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shehu-Xhilaga, M., S. M. Crowe, and J. Mak. 2001. Maintenance of the Gag/Gag-Pol ratio is important for human immunodeficiency virus type 1 RNA dimerization and viral infectivity. J. Virol. 75:1834-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Somogyi, P., A. J. Jenner, I. Brierley, and S. C. Inglis. 1993. Ribosomal pausing during translation of an RNA pseudoknot. Mol. Cell. Biol. 13:6931-6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Telenti, A., R. Martinez, M. Munoz, G. Bleiber, G. Greub, D. Sanglard, and S. Peters. 2002. Analysis of natural variants of the human immunodeficiency virus type 1 gag-pol frameshift stem-loop structure. J. Virol. 76:7868-7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Terwilliger, E., J. G. Sodroski, C. A. Rosen, and W. A. Haseltine. 1986. Effects of mutations within the 3′ orf open reading frame region of human T-cell lymphotropic virus type III (HTLV-III/LAV) on replication and cytopathogenicity. J. Virol. 60:754-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tessmer, U., and H. G. Krausslich. 1998. Cleavage of human immunodeficiency virus type 1 proteinase from the N-terminally adjacent p6∗ protein is essential for efficient Gag polyprotein processing and viral infectivity. J. Virol. 72:3459-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wain-Hobson, S., P. Sonigo, O. Danos, S. Cole, and M. Alizon. 1985. Nucleotide sequence of the AIDS virus, LAV. Cell 40:9-17. [DOI] [PubMed] [Google Scholar]
- 43.Zybarth, G., and C. Carter. 1995. Domains upstream of the protease (PR) in human immunodeficiency virus type 1 Gag-Pol influence PR autoprocessing. J. Virol. 69:3878-3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zybarth, G., H. G. Krausslich, K. Partin, and C. Carter. 1994. Proteolytic activity of novel human immunodeficiency virus type 1 proteinase proteins from a precursor with a blocking mutation at the N terminus of the PR domain. J. Virol. 68:240-250. [DOI] [PMC free article] [PubMed] [Google Scholar]