Abstract
The highly conserved Vpr protein mediates cell cycle arrest, transcriptional transactivation, and nuclear import of the preintegration complex in human immunodeficiency virus type 1. To identify functional domains in simian immunodeficiency virus (SIV) mac239 Vpr, we mutagenized selected motifs within an α-helical region and two C-terminal HxRxG motifs. All Vpr mutants located to the nucleus. Substitution of four amino acids in the α-helical domain did not interfere with cell cycle arrest, while a single substitution abolished cell cycle arrest function. Mutation of the first HxRxG motif to AxAxA also resulted in loss of cell cycle arrest, while mutation of the second motif had no effect. Interestingly, both Vpr mutants impaired in cell cycle arrest function also showed reduced transactivation of the SIV long terminal repeat, suggesting that arrest of cells at G2/M mediates or contributes to transactivation by Vpr.
The vpr gene is conserved in all primate immunodeficiency viruses and codes for a virion-associated protein that localizes to the nucleus of infected cells (2, 11, 26, 27). A number of functions have been attributed to the Vpr protein, but its function in viral pathogenicity is not yet understood. One conserved feature of primate immunodeficiency virus Vpr is the induction of a cell cycle arrest in the G2/M phase in mammalian cells which appears to be partially dependent on the species of the employed cell system (9, 19, 23). The ability of Vpr to act as a weak transcriptional transactivator of the viral long terminal repeat (LTR) and a variety of cellular promoters has been described previously (3, 18), and for human immunodeficiency virus type 1 (HIV-1) Vpr, a correlation between cell cycle arrest and transactivation functions has been proposed (5). Upon HIV-1 infection of nondividing cells or macrophages, Vpr also contributes to the nuclear import of the preintegration complex (7, 24). The abilities of Vpr to mediate the nuclear import of the preintegration complex and cell cycle G2 arrest are two genetically separable and independent functions in HIV-1 (25). Interestingly, these two functions are distributed on two genes, vpr and vpx, in the HIV-2-simian immunodeficiency virus sm (SIVsm) group (4). Animal studies using vpr and vpx deletion variants of SIVmac239 showed that the deletion of either gene still allows AIDS progression, whereas in the absence of both genes, no pathogenicity was observed (6, 8). However, a selection advantage for a functional vpr gene was observed in infected rhesus macaques (10). Taken together, these results suggest that vpr plays an important role in viral pathogenicity.
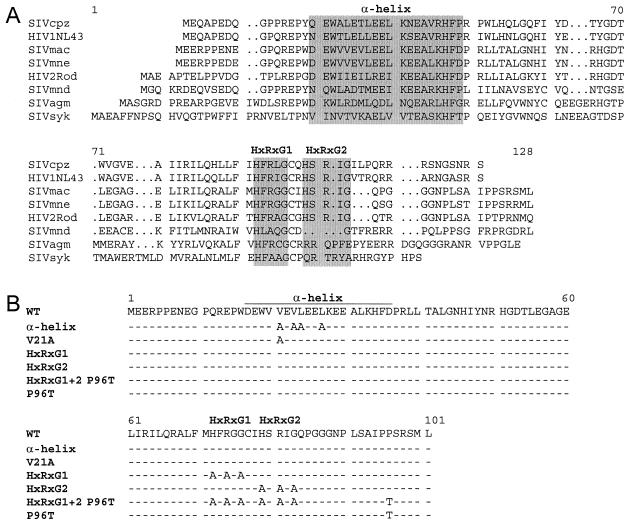
The aim of this study was to define the motifs in Vpr of SIVmac239 that are important for cell cycle arrest in the G2/M phase and for transactivation of the homologous LTR. For transient expression studies, the vpr open reading frame of SIVmac239 was amplified by PCR using a set of primers (CGGGATCCGAAGAAAGACCTCCAGAAA and CGGAATTCATAGCATGCTTCTAGAGGG) and cloned into the expression vector pCMV6Myc (22) by using the restriction sites for BamHI and EcoRI introduced by the PCR primers (underlined). The correct sequence of wild-type Vpr was verified. The obtained plasmid, pCMV6M-239Vpr, resulted in the expression of an N-terminal Myc epitope-tagged (EQKLISEEDL) Vpr protein missing only the first methionine residue of wild-type Vpr. Two conserved domains in the Vpr protein of SIVmac239 were targeted for mutagenesis using spliced overlap extension PCR with pCMV6M-239Vpr as the template (Fig. 1B). Substitutions were carefully selected to potentially affect only one of the analyzed Vpr functions rather than to produce complete loss of function in the mutant protein. In the N-terminal α-helix, the introduced mutations were not intended to break the helical structure since this most likely causes functional defect. Substitutions of valine residues at positions 21 and 23 and leucine residues at positions 24 and 27 in the conserved N-terminal amphipathic α-helix were selected, as well as the single mutation of valine 21 to alanine. In the Saccharomyces cerevisiae system, deletion of the conserved C-terminal HxRxG motifs (HxRxG1 and HxRxG2) resulted in loss of cell cycle arrest in G2 (12). To identify which of the two motifs is important for arrest in G2 in the mammalian system, both motifs were mutated individually and in combination. This resulted in the mutation of histidine, arginine, and glycine residues in the two HxRxG motifs at positions 72, 74, and 76 and at positions 79, 81, and 83 to AxAxA individually and in combination. Another Vpr mutation arose by accident and resulted in the replacement of proline at position 96 by threonine. The correct sequences of all mutants were verified. In addition, the vpr open reading frame of the HIV-1 NL4-3 isolate was cloned into the same expression vector to produce the N-terminal Myc-tagged fusion protein.
FIG. 1.
Amino acid sequences of primate immunodeficiency virus Vpr proteins and mutants used in this study. (A) Alignment of the amino acid sequences of Vpr proteins from SIVcpz, HIV-1 NL4-3, SIVmac, SIVmne, HIV-2 Rod, SIVmnd, SIVagm, and SIVsyk. Sequences were obtained from the Los Alamos database. Gaps ( · ) were introduced to optimize the alignment. The conserved amphipathic α-helix and HxRxG motifs are highlighted with gray boxes. (B) Schematic representation of mutated amino acids in SIVmac239 Vpr. The consensus sequence is shown on top. Identical amino acids are indicated by dashes, and mutated amino acids are shown in capital letters.
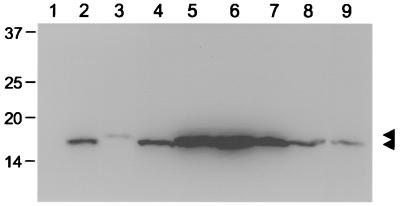
For analysis of the expression and stability of wild-type and mutant Vpr proteins, COS cells were transiently transfected with pCMV6Myc expression constructs by using a DEAE-dextran protocol (20). Wild-type HIV-1 NL4-3 Vpr was used as a control in all experiments. The cells were maintained in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum. Whole-cell lysates were prepared 48 h posttransfection, and the expression of Vpr proteins was determined by Western blotting with antibodies against the N-terminal Myc epitope (Fig. 2). The expression levels of Vpr mutant proteins were comparable to those of wild-type SIVmac239 and HIV-1 NL4-3 Vpr proteins, except that the α-helix mutant showed expression at low levels. A similar stabilizing effect has been observed previously for the α-helix in HIV-1 Vpr (14).
FIG. 2.
Expression and stability of SIVmac239 Vpr protein and derived mutants. COS cells were transfected with 3 μg of plasmid DNA of the empty expression vector (lane 1) or expression constructs for N-terminal Myc-tagged SIVmac239 Vpr (lane 2), HIV-1 NL4-3 Vpr (lane 9), and the SIVmac239 α-helix (lane 3), V21A (lane 4), HxRxG1 (lane 5), HxRxG2 (lane 6), HxRxG1+2 P96T (lane 7), and P96T (lane 8) mutant Vpr proteins. Whole-cell lysates were prepared 48 h after transfection and subsequently separated by electrophoresis. After transfer of proteins to a membrane filter, Vpr proteins were detected by immunoblotting with the Myc monoclonal antibody 9E10 and an enhanced chemiluminescence protocol. The positions of Vpr proteins are indicted by arrowheads, and molecular weight markers are indicated on the left.
Localization of the Vpr mutants within the cell was analyzed by indirect immunofluorescence, taking advantage of the N-terminal Myc epitope. For this purpose, COS cells were transfected with the pCMV6Myc expression constructs by using a Lipofectamine (GibcoBRL, Rockville, Md.) transfection protocol. After 36 h, the cells were fixed with 3% paraformaldehyde for 30 min at room temperature and Vpr proteins were detected by using Cy3-conjugated anti-c-Myc antibodies (Sigma, St. Louis, Mo.). Nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole [3 μg/ml]; Roche, Mannheim, Germany) before embedding. Immunofluorescence images were analyzed with an Axioplan 2 microscope (Zeiss), and the two images for anti-Myc and DAPI staining were merged using Metaview software (Universal Imaging Corp., Brandywine, Pa.). No fluorescence was observed in cells transfected with the vector only (data not shown). Wild-type SIVmac239 Vpr protein was located predominantly in the nucleus, while the nucleoli were excluded (Fig. 3). Comparable localization was observed for all Vpr mutants (Fig. 3). HIV-1 Vpr also localized to the nucleus, but the exclusion of nucleoli was less pronounced. In addition, wild-type and mutant vpr reading frames were cloned in pEGFP-C1 (Clontech, Palo Alto, Calif.) to create Vpr fusion proteins with green fluorescent protein (GFP). The GFP-Vpr fusion proteins were localized evenly throughout the cells at 30 h posttransfection, and nuclear staining was observed only after 72 h (data not shown). In addition, fusion to GFP severely impaired the ability of Vpr proteins to induce G2/M arrest (data not shown), indicating that fusion to GFP either interferes with SIVmac239 Vpr conformation or results in aberrant localization, and was therefore not studied further.
FIG.3.
Subcellular localization of SIVmac239 Vpr and its mutant variants. Approximately 2 μg of pCMV6Myc expression plasmids for wild-type SIVmac239 Vpr or the derived α-helix, V21A, HxRxG1, HxRxG2, HxRxG1+2 P96T, or P96T mutant, as well as the empty vector, was introduced into COS cells. After 36 h, Myc epitope-tagged Vpr proteins were detected by using Cy3-conjugated anti-Myc antibodies and nuclei were stained with DAPI and visualized by fluorescence microscopy. Fluorescence pictures were processed, and anti-Myc staining was overlaid with DAPI (merge).
To determine the amino acid motifs of SIVmac239 Vpr that are important for the induction of G2/M cell cycle arrest, HeLa cells were transfected with pCMV6Myc expression constructs for wild-type and mutant Vpr proteins. To allow specific analysis of transfected cells, a plasmid expressing farnesylated GFP was cotransfected. The Vpx protein of SIVmac239 was included as an additional negative control for cell cycle analyses. For this purpose, the vpx open reading frame was amplified by PCR and cloned into the pCMV6Myc expression vector and the correct sequence was confirmed. At 36 to 48 h posttransfection, cells were harvested and stained with propidium iodide. The DNA content of the cells was analyzed by flow cytometry, and the cell cycle profiles of cells expressing GFP were compared (Fig. 4A). The ratios of the numbers of cells in the G2/M phase to those in G1 (G2/M:G1 ratios) were calculated. For easy comparison, the ratio for wild-type Vpr was set as 1 and the relative values for Vpr mutants were deduced (Fig. 4B). As expected, the expression of SIVmac239 or HIV-1 NL4-3 vpr caused an increase in the number of cells in the G2 or M phase of the cell cycle. Cells transfected with the HxRxG1 and HxRxG1+2 P96T vpr mutants showed no effect on the cell cycle profiles, whereas transfection with the HxRxG2 or P96T vpr mutant still increased the percentage of cells in the G2/M phase. These findings indicate that the loss of function is due to mutations in the first HxRxG motif of SIVmac239 Vpr. For HIV-1 Vpr, similar data have been published previously showing the importance of the histidine residue at position 71 in the first HxRxG motif for G2/M arrest (1, 12, 13, 21). Expression of the Vpr α-helix mutant still induced the cell cycle arrest, but the single V21A mutation rendered the Vpr variant incapable of arresting the cells in G2/M. From these data, we cannot conclude whether the defect is due to the altered conformation and/or stability of the protein or whether the mutation in the α-helix specifically disrupts another domain in Vpr which might be important for the induction of G2/M arrest. While induction of apoptosis has been described previously for HIV-1 Vpr (15-17), no influence on apoptosis was detected in cells expressing wild-type SIVmac239 Vpr or any of the mutants analyzed (data not shown). These studies included analyses of nuclear fragmentation and detection of nucleosomes in the cytoplasm by enzyme-linked immunosorbent assay (Roche).
FIG. 4.
Induction of cell cycle arrest by SIVmac239 Vpr and its mutant variants. HeLa cells were transfected with 15 μg of pCMV6Myc expression constructs and 10 μg of an expression plasmid for farnesylated GFP by using a calcium phosphate precipitate protocol. At 36 to 48 h after transfection, cells were harvested, fixed with 70% ethanol for at least 2 h on ice, and stained with propidium iodide (0.1% Triton X-100, 20 μg of propidium iodide/ml, 5 μg of RNase A/ml) for 30 min at 37°C. The DNA content of 30,000 cells was analyzed on a FACSCalibur (Becton Dickinson, Franklin Lakes, N.J.). Data acquisition and analysis were performed with CellQuest (Becton Dickinson) and ModFit (Verity Software House, Topsham, Maine) software with the doublet discrimination module activated. Samples were gated for GFP expression and exclusion of cellular debris. (A) Representative cell cycle profiles for the mutants in this series. The left peaks (M1) indicate cells in G1, and the right peaks (M2) indicate cells in G2/M; cells in S phase are shown between the G1 and G2 peaks. The percentages of cells in either G1 or G2/M were calculated by the ModFit program and are shown in the upper right of each graph. (B) Mean ratios of the numbers of cells in the G2/M phase to those in G1 (G2/M:G1 ratios) were calculated from three to six independent experiments, and standard deviations were deduced. For easy comparison, the ratio for wild-type Vpr was set as 1 and the relative values for Vpr mutants were deduced.
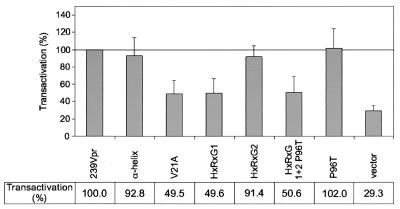
Next, we tested the ability of the Vpr mutants to transactivate the SIVmac239 LTR. For this purpose, a fragment of the SIVmac239 LTR (nucleotides 9751 to 10320) was cloned into the vector pUC19luc. COS cells were transfected with the SIVmac239 LTR luciferase reporter construct, a β-galactosidase reporter construct as an internal control (pSV-β-galactosidase; Promega, Madison, Wis.), and one of the corresponding Vpr expression plasmids (Fig. 5) following a DEAE-dextran protocol. Cells were harvested 60 to 72 h posttransfection. Luciferase and β-galactosidase assays were carried out in accordance with the protocols of the luciferase assay system provided by Promega and were analyzed in a luminometer (Chiron, Emeryville, Calif.). After normalization to β-galactosidase activity, the values for wild-type SIVmac239 Vpr were set as 100% and those for the Vpr mutants and vector control were calculated in relation to them. The mean values for luciferase activity and standard deviations were calculated from six independent experiments (Fig. 5). A 4- to 10-fold activation was observed for wild-type SIVmac239 Vpr compared with that for the vector control. Mutation of four amino acid residues in the α-helical domain, mutation of the second HxRxG motif, and replacement of proline at position 96 by threonine had no influence on the ability of Vpr to transactivate the SIV LTR. However, mutation of the single valine residue at position 21, mutation of the first HxRxG motif, and mutation of both HxRxG motifs reduced the transactivation potential of the respective Vpr mutants compared with that of wild-type Vpr. In these studies with an SIV LTR reporter construct, HIV-1 NL4-3 Vpr showed low levels of transactivation (data not shown).
FIG. 5.
Transactivation of SIVmac239 Vpr and its derived mutants. COS cells were transfected in 10-cm-diameter dishes with 7 μg of the SIVmac239 LTR luciferase reporter construct, 1 μg of a β-galactosidase reporter construct, and 15 μg of the corresponding Vpr expression plasmids. Transfected cells were harvested 60 to 72 h after transfection, washed once with phosphate-buffered saline, and lysed in 1 ml of cell culture lysis reagent (Promega). Luciferase and β-galactosidase assays were performed, and luciferase values were normalized for β-galactosidase activity. Corrected luciferase values for wild-type SIVmac239 Vpr were set as 100%, and transactivation values for the Vpr mutants and the vector control were calculated in relation to them. Mean values and standard deviations were calculated from six independent experiments.
Since all mutants analyzed in this study still localized to the nucleus, we cannot conclude whether nuclear localization of SIVmac239 Vpr is required for cell cycle arrest and transactivation of the LTR. Results obtained with mutations in the α-helical domain of Vpr showed a somewhat more complex pattern. Mutation of four amino acids had no effect on cell cycle arrest or transactivation, even though protein expression was decreased. In contrast, the single amino acid substitution V21A abolished both functions. As all substitutions were conservative, we assume that the single amino acid substitution may interfere with the correct protein conformation. Interestingly, the mutation of the first C-terminal HxRxG motif impaired both the cell cycle arrest and transactivation functions, whereas the mutation of the second motif had no influence on either Vpr function. These data suggest a functional linkage between cell cycle arrest and transactivation by SIVmac239 Vpr similar to that previously proposed for HIV-1 Vpr (5). So far, the role of cell cycle arrest in the progression to AIDS is still unclear and needs further investigation. Introducing mutations into SIVmac239 proviral DNA that destroy the Vpr cell cycle arrest function and testing these virus variants for their pathogenesis potential in the rhesus model may provide new insight into the role of Vpr in viral load and persistence in the host.
Acknowledgments
We thank Briggite Besieger for critical reading and Bernhard Fleckenstein for continuous support.
This work was supported by German BMBF grant no. 01 Kl 9766/6, a fellowship from the German BMBF (S.M.L.), and Deutsche Forschungsgemeinschaft grant no. SFB 466.
REFERENCES
- 1.Berglez, J. M., L. A. Castelli, S. A. Sankovich, S. C. Smith, C. C. Curtain, and I. G. Macreadie. 1999. Residues within the HFRIGC sequence of HIV-1 Vpr involved in growth arrest activities. Biochem. Biophys. Res. Commun. 264:287-290. [DOI] [PubMed] [Google Scholar]
- 2.Cohen, E. A., G. Dehni, J. G. Sodroski, and W. A. Haseltine. 1990. Human immunodeficiency virus vpr product is a virion-associated regulatory protein. J. Virol. 64:3097-3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cohen, E. A., E. F. Terwilliger, Y. Jalinoos, J. Proulx, J. G. Sodroski, and W. A. Haseltine. 1990. Identification of HIV-1 vpr product and function. J. Acquir. Immune Defic. Syndr. 3:11-18. [PubMed] [Google Scholar]
- 4.Fletcher, T. M., III, B. Brichacek, N. Sharova, M. A. Newman, G. Stivahtis, P. M. Sharp, M. Emerman, B. H. Hahn, and M. Stevenson. 1996. Nuclear import and cell cycle arrest functions of the HIV-1 Vpr protein are encoded by two separate genes in HIV-2/SIV(SM). EMBO J. 15:6155-6165. [PMC free article] [PubMed] [Google Scholar]
- 5.Forget, J., X. J. Yao, J. Mercier, and E. A. Cohen. 1998. Human immunodeficiency virus type 1 Vpr protein transactivation function: mechanism and identification of domains involved. J. Mol. Biol. 284:915-923. [DOI] [PubMed] [Google Scholar]
- 6.Gibbs, J. S., A. A. Lackner, S. M. Lang, M. A. Simon, P. K. Sehgal, M. D. Daniel, and R. C. Desrosiers. 1995. Progression to AIDS in the absence of a gene for vpr or vpx. J. Virol. 69:2378-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heinzinger, N. K., M. I. Bukinsky, S. A. Haggerty, A. M. Ragland, V. KewalRamani, M. A. Lee, H. E. Gendelman, L. Ratner, M. Stevenson, and M. Emerman. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc. Natl. Acad. Sci. USA 91:7311-7315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoch, J., S. M. Lang, M. Weeger, C. Stahl-Hennig, C. Coulibaly, U. Dittmer, G. Hunsmann, D. Fuchs, J. Muller, and S. Sopper. 1995. vpr deletion mutant of simian immunodeficiency virus induces AIDS in rhesus monkeys. J. Virol. 69:4807-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jowett, J. B., V. Planelles, B. Poon, N. P. Shah, M. L. Chen, and I. S. Chen. 1995. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J. Virol. 69:6304-6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lang, S. M., M. Weeger, C. Stahl-Hennig, C. Coulibaly, G. Hunsmann, J. Muller, H. Muller-Hermelink, D. Fuchs, H. Wachter, and M. M. Daniel. 1993. Importance of vpr for infection of rhesus monkeys with simian immunodeficiency virus. J. Virol. 67:902-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu, Y. L., P. Spearman, and L. Ratner. 1993. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J. Virol. 67:6542-6550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Macreadie, I. G., L. A. Castelli, D. R. Hewish, A. Kirkpatrick, A. C. Ward, and A. A. Azad. 1995. A domain of human immunodeficiency virus type 1 Vpr containing repeated H(S/F)RIG amino acid motifs causes cell growth arrest and structural defects. Proc. Natl. Acad. Sci. USA 92:2770-2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahalingam, S., V. Ayyavoo, M. Patel, T. Kieber-Emmons, and D. B. Weiner. 1997. Nuclear import, virion incorporation, and cell cycle arrest/differentiation are mediated by distinct functional domains of human immunodeficiency virus type 1 Vpr. J. Virol. 71:6339-6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mahalingam, S., S. A. Khan, R. Murali, M. A. Jabbar, C. E. Monken, R. G. Collman, and A. Srinivasan. 1995. Mutagenesis of the putative alpha-helical domain of the Vpr protein of human immunodeficiency virus type 1: effect on stability and virion incorporation. Proc. Natl. Acad. Sci. USA 92:3794-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nishizawa, M., M. Kamata, R. Katsumata, and Y. Aida. 2000. A carboxy-terminally truncated form of the human immunodeficiency virus type 1 Vpr protein induces apoptosis via G1 cell cycle arrest. J. Virol. 74:6058-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishizawa, M., M. Kamata, T. Mojin, Y. Nakai, and Y. Aida. 2000. Induction of apoptosis by the Vpr protein of human immunodeficiency virus type 1 occurs independently of G2 arrest of the cell cycle. Virology 276:16-26. [DOI] [PubMed] [Google Scholar]
- 17.Patel, C. A., M. Mukhtar, and R. J. Pomerantz. 2000. Human immunodeficiency virus type 1 vpr induces apoptosis in human neuronal cells. J. Virol. 74:9717-9726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Philippon, V., Z. Matsuda, and M. Essex. 1999. Transactivation is a conserved function among primate lentivirus Vpr proteins but is not shared by Vpx. J. Hum. Virol. 2:167-174. [PubMed] [Google Scholar]
- 19.Planelles, V., J. B. Jowett, Q. X. Li, Y. Xie, B. Hahn, and I. S. Chen. 1996. Vpr-induced cell cycle arrest is conserved among primate lentiviruses. J. Virol. 70:2516-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seed, B., and A. Aruffo. 1987. Molecular cloning of the CD2 antigen, the T-cell erythrocyte receptor, by a rapid immunoselection procedure. Proc. Natl. Acad. Sci. USA 84:3365-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selig, L., S. Benichou, M. E. Rogel, L. I. Wu, M. A. Vodicka, J. Sire, R. Benarous, and M. Emerman. 1997. Uracil DNA glycosylase specifically interacts with Vpr of both human immunodeficiency virus type 1 and simian immunodeficiency virus of sooty mangabeys, but binding does not correlate with cell cycle arrest. J. Virol. 71:4842-4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sells, M. A., and J. Chernoff. 1995. Epitope-tag vectors for eukaryotic protein production. Gene 152:187-189. [DOI] [PubMed] [Google Scholar]
- 23.Stivahtis, G. L., M. A. Soares, M. A. Vodicka, B. H. Hahn, and M. Emerman. 1997. Conservation and host specificity of Vpr-mediated cell cycle arrest suggest a fundamental role in primate lentivirus evolution and biology. J. Virol. 71:4331-4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subbramanian, R. A., X. J. Yao, H. Dilhuydy, N. Rougeau, D. Bergeron, Y. Robitaille, and E. A. Cohen. 1998. Human immunodeficiency virus type 1 Vpr localization: nuclear transport of a viral protein modulated by a putative amphipathic helical structure and its relevance to biological activity. J. Mol. Biol. 278:13-30. [DOI] [PubMed] [Google Scholar]
- 25.Vodicka, M. A., D. M. Koepp, P. A. Silver, and M. Emerman. 1998. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev. 12:175-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu, X. F., M. Matsuda, M. Essex, and T. H. Lee. 1990. Open reading frame vpr of simian immunodeficiency virus encodes a virion-associated protein. J. Virol. 64:5688-5693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan, X., Z. Matsuda, M. Matsuda, M. Essex, and T. H. Lee. 1990. Human immunodeficiency virus vpr gene encodes a virion-associated protein. AIDS Res. Hum. Retrovir. 6:1265-1271. [DOI] [PubMed] [Google Scholar]