Abstract
Infectious bursal disease virus (IBDV) is a double-stranded RNA (dsRNA) virus of the Birnaviridae family. Its two genome segments are encapsidated together with multiple copies of the viral RNA-dependent RNA polymerase, VP1, in a single-shell capsid that is composed of VP2 and VP3. In this study we identified the domains responsible for the interaction between VP3 and VP1. Using the yeast two-hybrid system we found that VP1 binds to VP3 through an internal domain, while VP3 interacts with VP1 solely by its carboxy-terminal 10 amino acids. These results were confirmed by using a reverse-genetics system that allowed us to analyze the interaction of carboxy-terminally truncated VP3 molecules with VP1 in infected cells. Coimmunoprecipitations with VP1- and VP3-specific antibodies revealed that the interaction is extremely sensitive to truncation of VP3. The mere deletion of the C-terminal residue reduced coprecipitation almost completely and also fully abolished production of infectious virions. Surprisingly, these experiments additionally revealed that VP3 also binds to RNA. RNase treatments and reverse transcription-PCR analyses of the immunoprecipitates demonstrated that VP3 interacts with dsRNA of both viral genome segments. This interaction is not mediated by the carboxy-terminal domain of VP3 since C-terminal truncations of 1, 5, or 10 residues did not prevent formation of the VP3-dsRNA complexes. VP3 seems to be the key organizer of birnavirus structure, as it maintains critical interactions with all components of the viral particle: itself, VP2, VP1, and the two genomic dsRNAs.
Infectious bursal disease virus (IBDV) is the causative agent of a highly contagious disease of young chickens. IBDV multiplies rapidly in developing B lymphocytes in the bursa of Fabricius, leading to immunosuppression and increased susceptibility to other diseases. Of the two IBDV serotypes, only serotype 1 is pathogenic to chickens (23). IBDV belongs to the family Birnaviridae (17). Members of this family are characterized by a double-stranded RNA (dsRNA) genome consisting of two segments (A and B) that are packaged within a single-shell icosahedral capsid of 60 nm.
The smaller genome segment, B, of IBDV (approximately 2.9 kb) encodes a single protein, viral protein 1 (VP1; 91 kDa). This protein is the putative RNA-dependent RNA polymerase (12). It has the intriguing feature of occurring in virions in two forms: a form in which it is covalently bound to the 5′ ends of the genomic dsRNA segments (viral protein genome-linked [VPg]) (16) and a free-polypeptide form. The larger dsRNA segment, A (approximately 3.3 kb), contains two partly overlapping open reading frames (ORFs). The first ORF encodes nonstructural protein VP5 (17 kDa). This protein proved to be nonessential for viral replication and infection (33, 41). The second ORF encodes a 110-kDa polyprotein, which is autocatalytically cleaved to give pVP2 (48 kDa), VP4 (28 kDa), and VP3 (32 kDa). The viral protease VP4, through a catalytic site belonging to the Lon protease family, is responsible for this self-processing of the polyprotein (5, 27, 35). During virus maturation, the precursor pVP2 is further processed into mature VP2 (40 kDa), probably as a result of a site-specific cleavage of pVP2 by VP4 protease activity (25, 27).
VP2 and VP3 are the major viral structural proteins. They form the proteinaceous capsid, the structure of which was shown by cryoelectron microscopy and image reconstruction to exhibit a T=13 lattice (10, 13). The capsid shell is composed of homotrimeric subunits of VP2 and VP3; 260 VP2 trimers constitute the outer surface, while the 200 Y-shaped VP3 trimers line the inner surface. Consistently, VP2 carries the major neutralizing epitopes (2, 4), while it has been suggested that VP3, by virtue of its basic carboxy-terminal domain, interacts with the packaged dsRNA (10, 22).
While these ultrastructural studies already provided important insight into the general architecture of the virus, essentially nothing is yet known about the specific interactions between the different viral components and how these give rise to the formation of viral particles. We and others recently reported that VP3 is able to associate with VP1 (30, 36). We showed that VP1-VP3 complexes are formed in the cytoplasm of IBDV-infected cells and eventually released into the cell culture medium, suggesting that the viral polymerase is incorporated into virions through interactions with the inner capsid protein (36). Likewise, Lombardo et al. (30) showed that VP1 is efficiently incorporated into IBDV virus-like particles produced in mammalian cells coexpressing the IBDV polyprotein and VP1.
In the present study we have followed up on these supposedly essential interactions. By using the yeast two-hybrid system as well as by mutagenesis of an infectious cDNA clone of the virus we mapped the domain in VP3 interacting with VP1 to the extreme carboxy-terminal tail of the polypeptide. This interaction appeared not to be required for negative-strand RNA synthesis but appeared to be crucial for the production of infectious progeny virus as deletion of just the carboxy-terminal residue was sufficient to abolish virus replication. These investigations also revealed that VP3 additionally binds to viral dsRNA, both of the A segment and of the B segment, and that it does so through a domain distinct from that binding to VP1.
MATERIALS AND METHODS
Cells, viruses, plasmids, and antisera.
QM5 cells (1) were cultured in QT35 medium (Gibco-BRL) supplemented with 5% fetal calf serum and 2% antibiotic solution ABII (1,000 U of penicillin [Yamanouchi], 1 mg of streptomycin [Radiumfarma], 20 μg of amphotericin B [Fungizone], 500 μg of polymixin B, and 10 mg of kanamycin/ml) in a CO2 (5%) incubator at 37°C. The classical IBDV isolate CEF94 is a derivate of PV1 which has been adapted for growth in cell cultures (8, 34). Recombinant fowlpox virus expressing the T7 polymerase gene (FPV-T7) (11) was received from the laboratory of M. Skinner (Compton Laboratory, Berks, United Kingdom). The preparation of the plasmids pHB36W (A segment) and pHB34Z (B segment), which contain full-length genomic cDNA of IBDV strain CEF94, has been described (8). Polyclonal rabbit antisera against VP1 and VP3 were produced by injecting rabbits with purified recombinant VP1 or VP3 (7, 36). A monoclonal antibody (MAb) directed against VP3 (MAb C3) was kindly provided by H. Müller (University of Leipzig, Leipzig, Germany).
Construction of two-hybrid expression plasmids.
Copy DNA encoding the full-length sequences of VP1 and VP3 of IBDV strain CEF94 or defined parts thereof (see Fig. 1) were amplified by PCR by using the Expand high-fidelity PCR system (Boehringer Mannheim). Plasmid pHB36W was used as the DNA template for preparing the VP3 constructs, and plasmid pHB34Z was used for the VP1 constructs. The sets of primers used were designed to introduce an EcoRI site at the upstream (5′) end and a stop codon plus either a SalI site (VP3 constructs) or an XhoI site (VP1 constructs) at the downstream (3′) end of each coding sequence (Table 1). PCR products were precipitated, digested with EcoRI and SalI (VP3 PCR fragments) or with EcoRI and XhoI (VP1 PCR fragments), gel purified by the QIAEX-II method (Qiagen), and ligated with T4 DNA ligase (New England BioLabs) into the yeast expression vectors pLexABD and pB42AD (Clontech). These vectors had previously been digested with EcoRI and XhoI. The ligation mixture was transformed into Escherichia coli DH5-α cells (Life Technologies), which were subsequently grown under ampicillin selection. Plasmid DNA prepared from several independent transformants was screened for the presence of the insert, and plasmids from positive clones were sequenced at the fusion junction by cycle sequencing with an ABI 310 sequencer (Perkin-Elmer) to ensure correct reading frames.
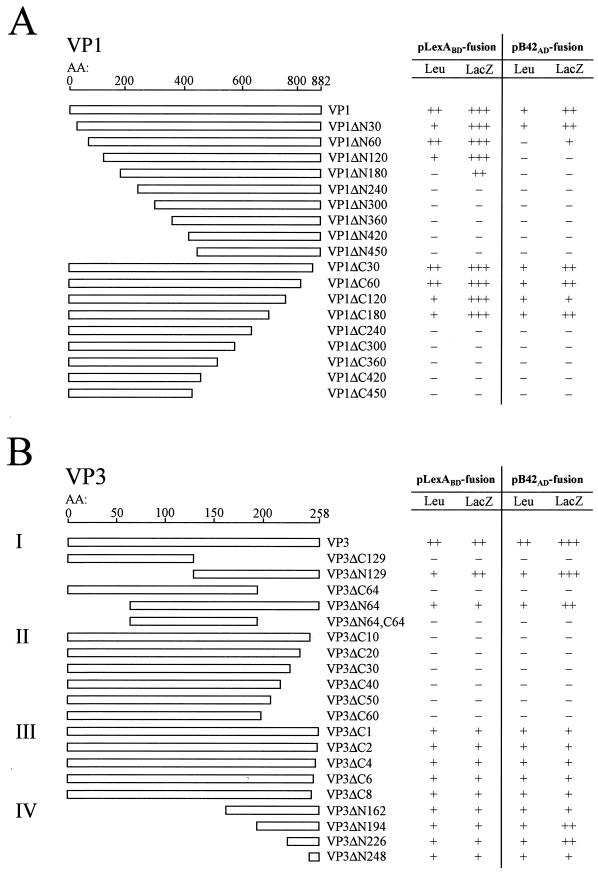
FIG. 1.
Mapping of the VP1-VP3 binding domain(s) by using deletion mutagenesis and the yeast two-hybrid system. Schematic representations of the deletion mutations in VP1 and VP3 are presented. VP1 and VP3 deletion mutants were cloned into pLexABD and pB42AD yeast expression vectors and tested for their ability to interact with full-length VP3 and full-length VP1, respectively. Interactions were assayed for leucine autotrophy (Leu) and for β-Gal activity (LacZ). For Leu, symbols are as follows: ++, clear growth (strong interaction); +, growth (interaction); −, no growth (no interaction); for LacZ, symbols are as follows: +++, deep blue colonies (very strong interaction); ++, blue colonies (strong interaction); +, light blue colonies (interaction); −, white colonies (no interaction). All results shown are representative of those for at least seven independent transformants. AA, amino acids.
TABLE 1.
Nucleotide sequences and positions of the primers used for VP1 and VP3 deletion mutagenesis, for the generation of mutant segment A cDNA constructs, and for RT-PCR analysis
| Primer | Nucleotide sequencea | Positionb | Purposec |
|---|---|---|---|
| p009 | ccgGAATTCATGAGTGACATTTTCAACAGTCCAC | B+ (nt 112) | VP1 and VP1ΔC derivates |
| p110 | cgtCTCGAGTCATGGCTGTTGGCGGCTCTCC | B− (nt 2754) | VP1 and VP1ΔN derivates |
| p075 | ccgGAATTCCTCTTGATCCCTAAAGTTTGGGTG | B+ (nt 202) | VP1ΔN30 |
| p076 | ccgGAATTCGTTTTGCAGCCACGGTCTCTGC | B+ (nt 292) | VP1ΔN60 |
| p078 | ccgGAATTCCCCAATGCGTACCCGCCAGAC | B+ (nt 472) | VP1ΔN120 |
| p080 | ccgGAATTCGAGGTCGCCACTGGAAGAAACC | B+ (nt 652) | VP1ΔN180 |
| p082 | ccgGAATTCGGCGACTTTGAGGTTGAAGATTAC | B+ (nt 832) | VP1ΔN240 |
| p084 | ccgGAATTCAAGAAGCTACTCAGCATGTTAAGTG | B+ (nt 1012) | VP1ΔN300 |
| p086 | ccgGAATTCAATAACGTGTTGAACATTGAAGGGTG | B+ (nt 1192) | VP1ΔN360 |
| p088 | ccgGAATTCGAGGCAAACTGCACTCGCCAAC | B+ (nt 1372) | VP1ΔN420 |
| p089 | ccgGAATTCCAAACATGGGCCACCTTTGCCATG | B+ (nt 1462) | VP1ΔN450 |
| p104 | cgtCTCGAGTCATCTGCTCGTTCCTGCTCCGAG | B− (nt 2664) | VP1ΔC30 |
| p103 | cgtCTCGAGTCACTGTGGGTTCTTGACTTCTGGG | B− (nt 2574) | VP1ΔC60 |
| p101 | cgtCTCGAGTCACTTGGACTTGTGGAGTTTCTCGG | B− (nt 2394) | VP1ΔC120 |
| p099 | cgtCTCGAGTCAGGGCTTGGGGGGTACTGGCTTG | B− (nt 2214) | VP1ΔC180 |
| p097 | cgtCTCGAGTCAGCAGGCTTTGTTCAGGAGTGGG | B− (nt 2034) | VP1ΔC240 |
| p095 | cgtCTCGAGTCAGAGATCTTTGCTGTATGTAGCTGAC | B− (nt 1854) | VP1ΔC300 |
| p093 | cgtCTCGAGTCACTCAATTGATTTGAACTCCTCGCTG | B− (nt 1674) | VP1ΔC360 |
| p091 | cgtCTCGAGTCAGGCAATGTTCATGGCAAAGGTGG | B− (nt 1494) | VP1ΔC420 |
| p090 | cgtCTCGAGTCAGGCTTGCATGTGTTGGCGAGTG | B− (nt 1404) | VP1ΔC450 |
| p057 | tcGAATTCGCTTCAGAGTTCAAAGAGACCCCC | A+ (nt 2396) | VP3 and VP3ΔC derivates |
| p006 | gatcGTCGACTCACTCAAGGTCCTCATCAGAGAC | A− (nt 3166) | VP3 and VP3ΔN derivates |
| p061 | aagtGTCGACTCAGTCGACTCAGCTTGGCCCTCGGTGCCCATTG | A− (nt 2782) | VP3ΔC129 |
| p060 | ccgGAATTCGGCCAGCTAAAGTACTGGCAGAAC | A+ (nt 2786) | VP3ΔN129 |
| p063 | aagtGTCGACTCAATAGACTTTGGCAACTTCGTCTATG | A− (nt 2977) | VP3ΔC64 and VP3ΔN64,C64 |
| p062 | ccgGAATTCAACGCACCACAAGCAGGCAGCAAG | A+ (nt 2588) | VP3ΔN64 |
| p064 | gatcGTCGACTCACCAGCGGCCCAGCCGACCAG | A− (nt 3136) | VP3ΔC10 |
| p065 | gatcGTCGACTCATGTTGGAGCATTGGGTTTTGGCTTG | A− (nt 3106) | VP3ΔC20 |
| p066 | gatcGTCGACTCATGGTAGAGCCCGCCTGGGATTGCG | A− (nt 3076) | VP3ΔC30 |
| p067 | gatcGTCGACTCACAGCTCCATCGCAGTCAAGAGCAGATC | A− (nt 3046) | VP3ΔC40 |
| p068 | gatcGTCGACTCACATCTGTTCTTGGTTTGGGCCACGTC | A− (nt 3016) | VP3ΔC50 |
| p069 | gatcGTCGACTCAGTTGATTTCATAGACTTTGGCAACTTCG | A− (nt 2986) | VP3ΔC60 |
| p106 | gatcGTCGACTCAAAGGTCCTCATCAGAGACGGTCC | A− (nt 3163) | VP3ΔC1 |
| p107 | gatcGTCGACTCAGTCCTCATCAGAGACGGTCCTG | A− (nt 3160) | VP3ΔC2 |
| p108 | gatcGTCGACTCAATCAGAGACGGTCCTGATCCAGC | A− (nt 3154) | VP3ΔC4 |
| p109 | gatcGTCGACTCAGACGGTCCTGATCCAGCGGCC | A− (nt 3148) | VP3ΔC6 |
| p110 | gatcGTCGACTCACCTGATCCAGCGGCCCAGCC | A− (nt 3142) | VP3ΔC8 |
| p111 | tcGAATTCGAAGAACAAATCCTAAGGGCAGCTAC | A+ (nt 2882) | VP3ΔN162 |
| p112 | tcGAATTCGAAATCAACCATGGACGTGGCCC | A+ (nt 2978) | VP3ΔN194 |
| p113 | tcGAATTCCCAAAGCCCAAGCCAAAACCCAATG | A+ (nt 3074) | VP3ΔN226 |
| p114 | tcGAATTCATCAGGACCGTCTCTGATGAGGAC | A+ (nt 3137) | VP3ΔN248 |
| p138 | CTCAAAGAAGATGGAGACC | A+ (nt 2704) | pHB36W derivates |
| p139 | CAGGTACCTCACTAAAGGTC | A− (nt 3177) | pHB36W-VP3Δ1(tag) |
| p140 | CAGGTACCTCATCAAAGGTCC | A− (nt 3177) | pHB36W-VP3Δ1(tga) |
| p141 | CAGGTACCTCACTCAAGGTCCTCTCAAGAGAC | A− (nt 3177) | pHB36W-VP3Δ5(tga) |
| p142 | CAGGTACCTCACTCAAGGTCCTCATCAGAGACGGTCCTTCACCAGC | A− (nt 3177) | pHB36W-VP3Δ10(tga) |
| pA(+) | CCCTGACCCTGTGTCCCCCACAGTC | A− (nt 295) | RT, (+)RNA segment A |
| pAC1 | GATCGGTCTGACCCCGGGGGAG | A+ (nt 3) | PCR, segment A specific |
| pANC1 | TAGGTCGAGGTCTCTGACCTGAGAG | A− (nt 264) | PCR, segment A specific |
| pA(−) | GTGCATGCAGAGAAGAGCCGGTTGGC | A+ (nt 2852) | RT, (−)RNA segment A |
| pANC2 | ACCCGCGAACGGATCCAATTTGGG | A− (nt 3256) | PCR, segment A specific |
| pAC2 | ATCTACGGGGCTCCAGGACAGGC | A+ (nt 2912) | PCR, segment A specific |
| pB(+) | GGGCGATGTGTTGGGTAGTACTTTGGG | B− (nt 458) | RT, (+)RNA segment B |
| pBC1 | ATACGATGGGTCTGACCCTCTGGG | B+ (nt 3) | PCR, segment B specific |
| pBNC1 | GGAAGTACTCCTGATCTCCAATAGGG | B− (nt 433) | PCR, segment B specific |
| pB(−) | GGGTTCCCACTCGACGAGTTCCTAGC | B+ (nt 2077) | RT, (−)RNA segment B |
| pBNC2 | CCCCCGCAGGCGAAGGCCGGG | B− (nt 2822) | PCR, segment B specific |
| pBC2 | TGAGCTGTCAGAGTTCGGTGAGGC | B+ (nt 2112) | PCR, segment B specific |
The primer sequences are listed 5′ to 3′. Restriction sites are in italics. Additional nucleotides 5′ of a restriction site are in lowercase. Boldface letters indicate an in-frame stop codon. Mutated nucleotides are underlined.
A+ and B+, A and B segment plus strands, respectively; A− and B−, A and B segment minus strands, respectively.
(+)RNA, plus strand RNA; (−)RNA, minus strand RNA.
Two-hybrid analysis.
All two-hybrid media, buffers, and protocols were prepared and used as described by Clontech (manual for the Matchmaker LexA two-hybrid system and Yeast Protocols Handbook). The yeast strain Saccharomyces cerevisiae EGY48 (Clontech) was transformed by using the lithium acetate method with the URA3+ plasmid p8op-lacZ (Clontech), which has a lacZ reporter gene preceded by an upstream LexA binding domain. Transformed cells were amplified and subsequently cotransformed with pLexABD (HIS3+) and pB42AD (TRP1+) constructs carrying the VP1 cDNA (or fragments thereof) and the VP3 cDNA (or segments thereof). Control plasmids were pLexABD-bicoid (pRHFM1; OriGene) and pB42AD-empty. Transformants were plated onto glucose-supplemented synthetic dropout medium (SD) lacking His, Ura, and Trp (SD/Glu/−His/−Ura/−Trp medium). His+ Ura+ Trp+ colonies from each transformation were subsequently plated onto SD/Gal/raffinose (Raf)/−His/−Ura/−Trp/−Leu medium to assess the transcriptional activation of the LEU2 reporter gene and onto SD/Gal/Raf/5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)/−His/−Ura/−Trp medium to assess the transcriptional activation of the lacZ reporter gene. Transformants containing plasmids pLexABD-53 (Clontech) and pB42AD-SV40 T (Clontech), in which 53 codes for the murine p53 protein, known to physically associate strongly with simian virus 40 large T antigen (14, 28), served as a positive control to assess assay conditions.
Modification of the full-length cDNA clone of IBDV segment A.
Plasmid pHB36W was used for oligonucleotide-directed mutagenesis to introduce stop codons in the 3′-terminal region of the coding strand of VP3 by PCR. Primers p139, p140, p141, and p142 (Table 1), each containing one or more nucleotide substitutions to create these stop codons, were used in combination with primer p138 (Table 1).
The resulting PCR fragments were digested with BglII and KpnI, purified with an agarose gel (Qiaex gel extraction kit), and ligated (Rapid ligation kit; Boehringer Mannheim) into the full-length segment A clone (pHB36W), which had been digested with the same restriction enzymes. The ligation mixture was subsequently used to transform E. coli DH5-α cells (Life Technologies). To confirm the correct nucleotide sequences of the pHB36W derivates, designated pHB36W-VP3Δ1(tag), pHB36W-VP3Δ1(tga), pHB36W-VP3Δ5(tga), and pHB36W-VP3Δ10(tga), we determined the nucleotide sequence of the exchanged part (BglII/KpnI fragment). Two independently obtained plasmids of each pHB36W derivate were used for in vitro transcription and translation and for transfection of QM5 cells.
In vitro transcription and translation.
In vitro T7 polymerase-driven expression was carried out by using the TNT Quick coupled transcription-translation system (Promega) as described by the manufacturer, except that reactions were performed in a final volume of 2.5 μl. Plasmid pHB36W or its derivatives (0.4 μg) were used as the template. The resulting viral proteins were resolved in 12% separating gels by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by autoradiography.
Transfection of QM5 cells.
QM5 cells were grown to 80% confluence in 60-mm-diameter dishes and infected with FPV-T7 (multiplicity of infection = 3). After 1 h, the cells were washed once with 5 ml of QT35 medium and covered with 5 ml of Optimem 1 (Gibco-BRL). In the meantime, 2.0 μg of DNA was mixed with 25 μl of Lipofectamine (Gibco-BRL) in 0.5 ml of Optimem 1 and kept at room temperature for at least 30 min. The QM5 cells were subsequently covered with 4 ml of fresh Optimem 1, and the DNA-Lipofectamine mixture was added. The transfection was performed overnight (18 h) in a 37°C incubator (5.0% CO2). The transfected monolayer was rinsed once with QT35 medium, and fresh QT35 medium supplemented with 5% fetal calf serum and 2% ABII was added. The plates were further incubated for another 24 h.
Radiolabeling of transfected cells, immunoprecipitation, and gel electrophoresis.
At 48 h posttransfection, cells were starved for 1 h in methionine-free Eagle's minimal essential medium (EMEM) (Gibco-BRL). Cells were then labeled for 3 h with 20 μCi of [35S]methionine (Amersham)/ml in methionine-free EMEM. At the end of the labeling, the cell cultures were lysed on ice in 1× phosphate-buffered saline (PBS)-TDS lysis buffer by using a 5× PBS-Triton-sodium deoxycholate-SDS (TDS) lysis buffer stock solution (5% Triton X-100, 2.5% sodium deoxycholate, 0.5% SDS, 0.7 M NaCl, 14 mM KCl, 50 mM Na2HPO4, 7.5 mM KH2PO4). Cell debris was removed by centrifugation at 4°C for 20 min at 13,000 × g. All lysates were pretreated with protein A-Sepharose (Amersham) prior to being immunoprecipitated with polyclonal anti-VP1 serum or monoclonal anti-VP3 (MAb C3) serum. Protein A-Sepharose-bound immune complexes were washed three times in 1× PBS-TDS lysis buffer and eluted in 30 μl of SDS sample buffer (60 mM Tris-HCl [pH 6.8], 2.5% SDS, 5% β-mercaptoethanol, 10% glycerol, 0.1% bromophenol blue). Proteins were resolved in 12% separating gels by SDS-PAGE and visualized by autoradiography.
RNase treatment of immunoprecipitates.
Before SDS-PAGE the protein A-Sepharose-bound immune complexes were resuspended in 10 μl of 10 mM Tris-HCl (pH 7.5) and treated as follows. For RNaseONE treatment, the immunoprecipitates were incubated for 45 min at 37°C with 1,000 U of RNaseONE (Promega)/ml in 10 mM Tris-HCl (pH 7.5)-5 mM EDTA-200 mM sodium acetate. RNaseONE, an RNase able to cleave a phosphodiester bond between any two ribonucleotides, catalyzes the degradation of single-stranded RNA (ssRNA) to cyclic nucleotide monophosphate intermediates (Promega). For RNase A treatment, the immunoprecipitates were incubated for 45 min at 37°C with RNase A at a concentration of 100 μg/ml in 10 mM MgCl2. RNase A is often considered an ssRNA-specific enzyme, though at a low salt concentration (i.e., 10 mM MgCl2) it degrades both ssRNA and dsRNA (29). At the end of the RNase treatments the Sepharose beads were collected and washed three times with 1× PBS-TDS lysis buffer, after which the immune complexes were eluted in 30 μl of SDS-sample buffer and subjected to SDS-PAGE.
RT-PCR analysis of immunoprecipitates.
For the negative- and positive-strand-specific reverse transcription-PCR (RT-PCR) of segments A and B, 1 μl of the respective immunoprecipitates was used. To prime cDNA synthesis on the genomic positive and negative strands of segment A, we used oligonucleotides pA(+) and pA(−), which hybridized to nucleotides (nt) 271 to 295 and 2852 to 2877 of the plus and minus strands, respectively (Table 1). To prime cDNA synthesis on the genomic positive and negative strands of segment B, we used oligonucleotides pB(+) and pB(−), which hybridized to nt 432 to 458 and 2077 to 2102 of the plus and minus strands, respectively (Table 1). For RT, samples containing 1 μl of the immunoprecipitate and 2 pmol of the primer in a total volume of 10 μl were incubated at 98°C for 2 min and immediately chilled on ice. Subsequently, 10 μl of an RT mixture containing 2× Superscript II first-strand-synthesis buffer (Gibco-BRL), 20 mM dithiothreitol, 1 mM (each) deoxynucleoside triphosphate, and 200 U of Superscript II (Gibco-BRL) was added. For the negative-control reaction, the addition of Superscript II enzyme was omitted. The RT reaction was carried out at 50°C for 60 min, after which the incubation was continued at 70°C for 15 min to inactivate the RT. The reaction products were amplified by PCR using primer pairs pAC1-pANC1 for the plus strand of segment A (yielding a PCR product of 262 bp corresponding to nt 3 to 264), pAC2-pANC2 for the minus strand of segment A (yielding a 345-bp PCR product corresponding to nt 2912 to 3256), pBC1-pBNC1 (yielding a 431-bp PCR product corresponding to nt 3 to 433) for the plus strand of segment B, and pBC2-pBNC2 for the minus strand of segment B (yielding a 711-bp product corresponding to nt 2112 to 2822) (Table 1). The PCR consisted of 25 cycles, each comprising 15 s of denaturation at 94°C, 30 s of annealing at 68°C, and 45 s of elongation at 72°C. The 25 cycles were followed by a 90-s incubation at 72°C.
Detection of infectious rescued IBDV.
To rescue infectious virus, transfected cells and culture fluid were freeze-thawed three times and the supernatant was filtered through a 200-nm-pore-size filter (Acrodisc; Gelman Sciences) to remove the FPV-T7 and cellular debris. The cleared supernatant was either stored at −20°C or used directly for further analysis. Infectious rescued IBDV was detected by inoculating a subconfluent monolayer of QM5 cells with part of the cleared supernatant. After incubation for 48 h at 37°C the cells were washed with PBS, dried, and stored at −20°C until an immunoperoxidase monolayer assay was performed by using a polyclonal antiserum against VP3 (39).
RESULTS
A carboxy-terminal binding site in VP3 is responsible for VP1-VP3 interaction.
The yeast two-hybrid system (20, 21, 38) is an increasingly popular tool for the detection of protein-protein interactions. Here we used the system to map the domains responsible for the VP1-VP3 interaction. Various truncated versions of VP1 and VP3 were inserted into the plexABD (“bait”) and pB42AD (“prey”) yeast expression plasmids. An interaction between the bait and prey fusion proteins allows yeast cells containing both plasmids to grow on medium lacking leucine and to produce β-galactosidase (β-Gal), giving rise to blue colonies on X-Gal-containing medium. The strength of the protein-protein interaction can be judged by the intensity of the blue phenotype, allowing a semiquantitative evaluation of the interaction between the candidate proteins (18, 28, 40). As positive controls, a combination of plasmids pLexABD-VP1 and pB42AD-VP3 and the reciprocal combination pLexABD-VP3 and pB42AD-VP1, each one containing a complete gene, were tested, and these produced the expected phenotypes. Individually, or in combination with the respective control plasmids pLexABD-bicoid and pB42AD-empty, these constructs were negative in both assays, confirming the specificity of the test system.
To map the interacting domain of VP3, five different deletion mutants were initially generated: VP3ΔC129 and VP3ΔN129, in which the carboxy-terminal and amino-terminal halves of VP3 were deleted, VP3ΔC64 and VP3ΔN64, lacking 64 amino acids at either end, and VP3ΔN64,C64, missing 64 amino acids from both termini of the protein (Fig. 1B, I). Similarly, a large series of amino- and carboxy-terminal-deletion mutants were generated for VP1 (Fig. 1A). The coding region of each of the deletion mutants was expressed by both pLexABD and pB42AD expression vectors, and the products were tested for their capability to interact with wild-type VP1 or VP3. To rule out the possibility of nonspecific transactivation of the reporter genes, all constructs were additionally assayed for reporter gene activation when expressed either alone or together with the respective control plasmids pLexABD-bicoid and pB42AD-empty. Since all constructs proved to be negative in these tests (data not shown), all interactions between the LexABD and B42AD fusion proteins were regarded as specific. The results of the assays for the VP1-VP3 interaction are presented in Fig. 1. Of the VP1 deletion mutants, both carboxy- and amino-terminally truncated fusion proteins scored positive in the test assessing production of β-Gal and growth on medium lacking Leu, except for the mutants with an amino- or carboxy-terminal truncation of 240 amino acids or larger (Fig. 1A). These results suggest that the internal core domain of VP1 is essential for the VP3 interaction. For the domain mapping of VP3 the results showed that a deletion of 64 amino acids or more at the carboxy terminus completely abolished binding to VP1, whereas the deletions at the amino terminus had no effect on this interaction (Fig. 1B, I). This indicated that the carboxyl terminus of VP3 is critical for the VP1-VP3 interaction.
To determine the interacting domain in VP3 more precisely, we constructed and tested six progressive-carboxy-terminal-deletion mutants, lacking 10, 20, 30, 40, 50, and 60 amino acids. None of these mutants showed reporter gene activation, suggesting that the binding domain of VP3 is located within the last 10 amino acids of the carboxyl terminus (Fig. 1B, II). We therefore constructed and tested five additional carboxy-terminal-deletion mutants lacking 1, 2, 4, 6, and 8 amino acids. Each of these deletion mutants had VP1 binding activity, albeit activity weaker than that of wild-type VP3 (Fig. 1B, III). To confirm these findings, we also tested a series of VP3 proteins with amino-terminal deletions by removing up to 248 of the 258 amino acids of the protein. All these truncated proteins, even the mutant VP3ΔN248, consisting of just the 10-residue carboxy-terminal domain of VP3, conferred growth on Leu− medium and gave blue colonies on X-Gal-containing medium (Fig. 1B, IV).
Collectively, these yeast two-hybrid experiments consistently identified the 10-amino-acid carboxy-terminal peptide of VP3 as the VP1 interaction domain. To independently confirm this conclusion and to study the function of the VP1-VP3 interaction, we made use of our reverse-genetics system (9) to generate viral VP3 carboxy-terminal-deletion mutants.
In vivo interaction between VP1 and carboxy-terminally truncated VP3.
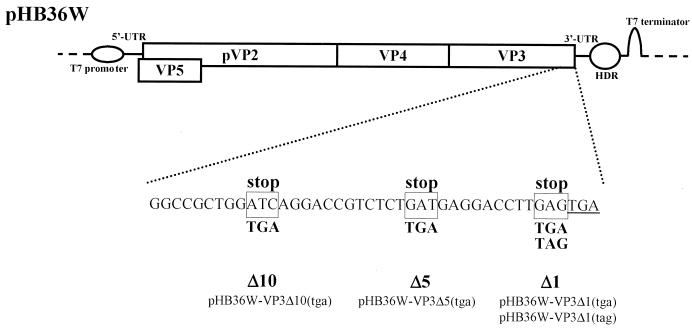
To test for a functional role for the VP1-VP3 interaction during IBDV replication, mutant IBDV cDNA clones of the full-length segment A were constructed by introducing stop codons in the 3′ terminus of the coding strand to generate carboxy-terminally truncated VP3 mutants. cDNA clones pHB36W-VP3Δ10(tga), pHB36W-VP3Δ5(tga), pHB36W-VP3Δ1(tga), and pHB36W-VP3Δ1(tag), containing a stop at codon positions 10, 5, or 1 preceding the natural stop codon of the VP3 gene, were prepared (Fig. 2). In all mutants the original codon was replaced by a TGA stop codon except in clone pHB36W-VP3Δ1(tag), where a TAG stop codon, which required only a single point mutation (GAG to TAG), was used.
FIG. 2.
Schematic representation of mutagenesis of the segment A cDNA clone. Plasmid pHB36W, containing the full-length cDNA sequence of segment A preceded by a T7 promoter sequence and followed by the autocatalytic hepatitis delta virus ribozyme (HDR) and a T7 terminator, is shown at the top. The nucleotide sequence of the 3′ terminus of the ORF for the polyprotein is depicted below. The TGA stop codon of VP3 is underlined. Mutated nucleotides are boxed, and introduced stop codons are indicated below in boldface. The generated plasmids are pHB36W-VP3Δ10(tga), pHB36W-VP3Δ5(tga), pHB36W-VP3Δ1(tga), and pHB36W-VP3Δ10(tag), according to the position and kind of stop codon in the 3′ terminus of the coding sequence of VP3. UTR, untranslated region.
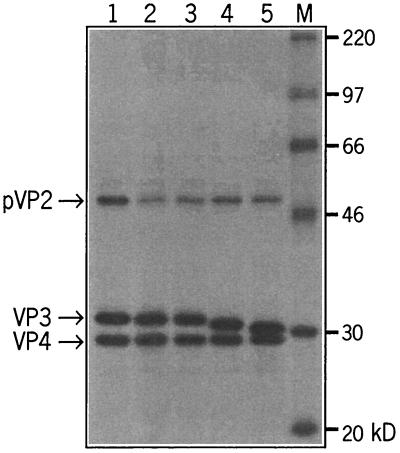
To analyze the expression and processing of the viral proteins of the mutated A segments, in vitro transcriptions and translations were performed by using a rabbit reticulocyte system. The radiolabeled translation products were subsequently analyzed by SDS-PAGE and autoradiography (Fig. 3). In all cases the viral polyprotein was processed normally into pVP2, VP3, and VP4. As predicted, the apparent molecular mass of VP3 decreased with increasing deletions: the removal of 5 or 10 carboxy-terminal amino acids gave rise to an apparent reduction in molecular mass of some 1 to 2 kDa.
FIG. 3.
Autoradiogram of an SDS-PAGE analysis of a coupled in vitro transcription-translation reaction. Plasmids containing the full-length A segment, pHB36W (lane 1), pHB36W-VP3Δ1(tag) (lane 2), pHB36W-VP3Δ1(tga) (lane 3), pHB36W-VP3Δ5(tga) (lane 4), and pHB36W-VP3Δ10(tga) (lane 5), were used as the DNA template to produce the viral proteins. The positions of the viral proteins and the sizes of the marker proteins (lane M) are indicated.
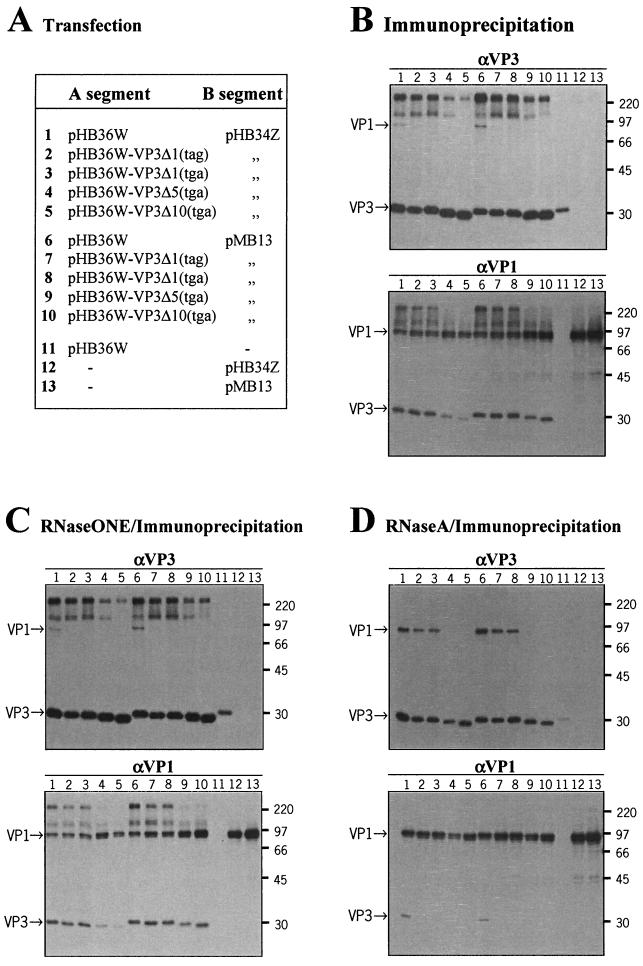
To study more directly the role of the VP3 carboxy-terminal domain in the interaction with VP1, we coexpressed the proteins in cells using the cotransfection system described previously (9). The wild-type and mutated full-length segment A cDNA clones, pHB36W, pHB36W-VP3Δ1(tag), pHB36W-VP3Δ1(tga), pHB36W-VP3Δ5(tga), and pHB36W-VP3Δ10(tga), were each transfected into QM5 cells together with either the wild-type full-length segment B cDNA clone (pHB34Z) or a mutant full-length segment B cDNA clone named pMB13 (Fig. 4A). The last clone was used as a negative control for virus replication. In this clone, the stem-loop structure in the 3′ terminus of the coding strand predicted by Boot et al. (8) was changed into a more stable stem-loop structure by the replacement of three adjacent cytosine residues (nt 2802 to 2804) by three guanines, which resulted in a construct that failed to yield virus in combination with wild-type segment A cDNA (6). Forty-eight hours posttransfection, cells were metabolically labeled for 3 h with [35S]methionine and subsequently subjected to immunoprecipitation with either anti-VP1 or anti-VP3 serum. The immunoprecipitates obtained were analyzed by SDS-PAGE, and the labeled proteins were visualized by autoradiography. As shown in Fig. 4B, top, when the anti-VP3 serum was used, comparable amounts of the full-length and the truncated VP3 proteins were detected in the transfected QM5 cells. VP1 was clearly coprecipitated with full-length VP3 but only in minor amounts with the carboxy-terminally truncated VP3Δ1 (lanes 1 to 3). The immunoprecipitations of VP3Δ5 and VP3Δ10 (lanes 4 to 5) completely failed to coprecipitate detectable amounts of VP1. This is in contrast with the anti-VP1 immunoprecipitations, where VP1 and VP3 were both present in all immunoprecipitates (lanes 1 to 5). The same results were found with the pMB13-cotransfected QM5 cells (lanes 6 to 10), indicating that productive viral replication is not required for VP1-VP3 binding. For IBDV strain CEF94 we recently showed that the free VP1 protein occurs in two forms with molecular masses of 90 and 95 kDa (36). In IBDV-infected cells both these forms appeared to interact with VP3 initially after their translation, though the interaction with the 90-kDa form gradually became undetectable thereafter (36). These observations explain the presence of the two discrete bands of free VP1 in the 90- to 95-kDa size range seen in the anti-VP1 immunoprecipitates (Fig. 4B to D), while only the 95-kDa band appears in the anti-VP3 immunoprecipitates; apparently only the 95-kDa form of VP1 interacted with VP3 and therefore was coprecipitated when the anti-VP3 serum was used.
FIG. 4.
Radioimmunoprecipitation analysis of VP1-VP3 interaction in transfected QM5 cells. (A) QM5 cells were (co)transfected with the indicated plasmids containing either the wild-type or a mutated cDNA of the A segment and/or B segment. At 48 h posttransfection cells were metabolically labeled for 3 h with [35S]methionine. Subsequently, cells were lysed and immunoprecipitated with anti-VP3 serum (αVP3) or with anti-VP1 serum (αVP1) followed by SDS-PAGE, either directly (B) or after an RNaseONE (C) or RNase A treatment (D). Positions of the viral proteins and sizes of marker proteins (in kilodaltons) are indicated.
Furthermore, with the exception of the controls of the transfected A segment or B segment alone (Fig. 4B to D, lanes 11 to 13), all immunoprecipitations revealed the presence of several bands in the high-molecular-mass region of >97 kDa (lanes 1 to 10). Our previous studies have shown that these bands are VP1 specific and represent VP1-RNA complexes, as ascertained by Western blot analysis using VP1-specific sera and RNase treatment (36). It was obvious that these VP1-RNA complexes were present in both the VP1-specific and the VP3-specific immunoprecipitates. Since, in the latter case, neither VP3Δ5 nor VP3Δ10 was able to coprecipitate VP1, we speculated that the presence of these VP1-RNA complexes in the VP3-specific immunoprecipitates might be a consequence of a direct VP3-RNA interaction rather than of a VP1-VP3 interaction. This assumption would also explain the unexpected coprecipitation of VP3Δ5 and VP3Δ10 proteins when the VP1 antibodies were used. The observation that the amount of coprecipitated VP3 correlated with the amount of precipitated VP1-RNA complex rather than with the amount of precipitated VP1 (lanes 1 to 5 and 6 to 10) also supports this interpretation. We therefore analyzed the sensitivity of the immune complexes to RNase.
VP3 interacts both with VP1 and with IBDV-specific dsRNA.
To study the VP1-VP3 interaction per se, i.e., without the interference of RNA-mediated coimmunoprecipitation, we performed an RNase treatment on all the VP1-specific and VP3-specific immunoprecipitates. To distinguish between ssRNA and dsRNA, we performed RNaseONE and RNase A treatment, thereby degrading specifically ssRNA or both ssRNA and dsRNA, respectively. The RNaseONE treatment failed to degrade the VP1-RNA complexes (Fig. 4C). The electrophoretic patterns of both the VP1- and VP3-specific coimmunoprecipitates were indistinguishable from those presented in Fig. 4B, indicating that the integrity of the VP1-RNA complexes is not dependent on ssRNA. In contrast, in the RNase A-treated samples, no high-molecular-mass (>97-kDa) complexes were present (Fig. 4D; lanes 1 to 13). Hence, the VP1-RNA complexes were maintained by dsRNA. Furthermore, in the VP3-specific immunoprecipitations, VP1 was coprecipitated with full-length VP3 and, to a lesser extent, with VP3Δ1 but not with VP3Δ5 and VP3Δ10. The reciprocal immunoprecipitations fully confirmed these results. The VP1 antiserum coprecipitated full-length VP3 and, again in much smaller amounts, VP3Δ1 (clearly seen after a prolonged exposure of the autoradiograph; data not shown). Obviously, VP3Δ5 and VP3Δ10 were no longer coprecipitated in the absence of dsRNA. Apparently, VP3 is able to associate with the VP1-dsRNA complexes through direct VP3-dsRNA interaction.
It was therefore of interest to determine whether the dsRNA in these complexes represented IBDV genome segment A- and/or B-specific sequences. To this end, we performed an RT-PCR analysis. The untreated and RNaseONE- and RNase A-treated VP3-specific immunoprecipitates were analyzed for the presence of positive- and negative-strand genomic RNA. As expected, RT-PCR products specific for the 5′ ends of both strands of both segments were obtained from the untreated and RNaseONE-treated samples but not from the RNase A-treated samples (Table 2; data for the RNaseONE-treated samples are not shown). The finding that no product was obtained when reverse transcriptase was omitted from the reaction mixture before PCR indicates that the PCR products were derived from RNA, not from contaminating DNA. In cells transfected with either pHB36W or pHB34Z alone, no RT-PCR products were obtained, confirming the absence of IBDV-specific VP1-dsRNA complexes in these immunoprecipitates. Plasmids pHB36W and pMB13, which are known to be incapable of generating infectious progeny virus when cotransfected into cells, appeared to be RNA replication competent as shown by the RT-PCR products obtained (Table 2). Hence, the RNaseONE-resistant but RNase A-sensitive high-molecular-mass material in the radioimmunoprecipitation analysis of this combination (Fig. 4B to D, lanes 6 to 10) was of IBDV-specific dsRNA origin. Infection induced in cells by using pMB13 and pHB36W is probably affected at the level of particle assembly or the particles produced are somehow not infectious.
TABLE 2.
RT-PCR analysis of the anti-VP3 immunoprecipitation reactions of Fig. 4B and D
| Transfected plasmid for segment:
|
Result of strand-specific RT-PCR with indicated primers for samplesa:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Untreated
|
RNase A treated
|
||||||||
| A | B | A+ | A− | B+ | B− | A+ | A− | B+ | B− |
| pHB36W | pHB34Z | + | + | + | + | − | − | − | − |
| pHB36W-VP3Δ1(tag) | pHB34Z | + | + | + | + | − | − | − | − |
| pHB36W-VP3Δ1(tga) | pHB34Z | + | + | + | + | − | − | − | − |
| pHB36W-VP3Δ5(tga) | pHB34Z | + | + | + | + | − | − | − | − |
| pHB36W-VP3Δ10(tga) | pHB34Z | + | + | + | + | − | − | − | − |
| pHB36W | − | − | − | − | − | − | − | − | |
| pHB34Z | − | − | − | − | − | − | − | − | |
| pHB36W | pMB13 | + | + | + | + | − | − | − | − |
Specific positive-sense and negative-sense primers of segments A and B (A+, A−, B+, and B−, respectively) were used for RT. Following RT, the reaction products were amplified by PCR using specific primer pairs based on 5′ terminal sequences of both the positive and negative strands of segments A and B (for details, see Materials and Methods). The plus and minus signs indicate the presence or absence of a specific PCR product. Controls, in which reverse transcriptase was omitted from the reaction, were negative.
As the RT-PCR analysis was performed with primers designed to amplify 5′-end sequences from plus as well as minus strand RNA, the results indicate that full-length dsRNA was present in the immunoprecipitates. Consequently, the higher-molecular-mass material of ≥220 kDa, which had barely entered the gel in the immunoprecipitation assays, most likely represents the full-length dsRNA-VPg complexes. The smear below it suggests the presence of incomplete dsRNA molecules such as VPg linked to partial dsRNA molecules, consisting of one full-length strand and one partial, complementary strand; this material runs faster than the full-length dsRNA-VPg complexes and appears as a smear due to its heterogeneity (15, 26). When full-length dsRNA-VPg complexes from another birnavirus, infectious pancreatic necrosis virus, were treated with RNase by Magyar et al. (31), VPg molecules carrying oligonucleotides that had survived the enzymatic treatment, supposedly due to steric hindrance by the unusually large VPg, were obtained. The two discrete bands seen migrating at ∼110 to 150 kDa might therefore represent two forms of VPg linked to short oligonucleotides that were inaccessible to RNase. Interestingly, these short VPg-linked oligonucleotides were not resistant to RNase A treatment.
Altogether, these observations supported the results of the yeast two-hybrid screen with the truncated VP3 deletion mutants. In addition, they revealed that the (co)precipitated VP1-RNA complexes contained genomic dsRNA and that VP3 is able to associate with these complexes by interacting directly with the dsRNA.
VP1-VP3 interaction is essential for the generation of infectious progeny.
To analyze the truncated-VP3 viral mutants, VP3Δ1(tag), VP3Δ1(tga), VP3Δ5(tga), and VP3Δ10(tga), for their ability to produce infectious virus, we examined the transfection supernatants of these mutants for the presence of recombinant IBDV. Aliquots of the supernatants were inoculated onto fresh monolayers of QM5 cells, which were subsequently incubated for 48 h and fixed. An IBDV-specific antibody assay was used to analyze whether infection had occurred. In none of the assays could IBDV proteins be detected in the QM5 monolayer except for the wild-type situation, where unmodified segments A and B had originally been transfected (data not shown). These results indicate that all mutant viruses were unable to generate infectious progeny.
DISCUSSION
Little is yet known about the nature and significance of the interactions between the viral components during the IBDV life cycle. Here we focused on the viral capsid protein VP3. We demonstrated that its interaction with VP1 is critically mediated by its carboxy-terminal domain. The removal of just the terminal residue was sufficient to affect its association with VP1 and inhibited the production of infectious progeny completely. The VP3 protein also appeared to interact with both segments of the viral dsRNA. Our results thus point to an essential role in virus morphogenesis for VP3, which is involved not only in the formation of the capsid shell but also in the incorporation of the genome and the viral RNA polymerase.
The VP3 domain interacting with VP1 was unambiguously mapped to the carboxy-terminal tail by two independent approaches. In yeast two-hybrid analyses the ability to associate with VP1 was lost when 10 or more terminal residues of VP3 were removed while, conversely, the corresponding decapeptide by itself was able to establish an interaction. These assays additionally demonstrated that the VP1 protein interacts with VP3 through its more internal domain, but further mapping thereof was not pursued. Site-directed mutagenesis of our infectious cDNA clone revealed that in IBDV infection the interaction between the two proteins is even more sensitive to changes in the VP3 carboxy-terminal tail. The mere deletion of the ultimate residue dramatically affected the strength of their association, as judged by the almost complete loss of their interaction in the coimmunoprecipitation assay and the complete loss of infectious-virus production.
These studies unexpectedly led us to the detection of the VP3-dsRNA interactions. The key observation here was that higher-molecular-mass material, which, as we observed earlier, contains VPg-linked RNA (36), was immunoprecipitated with anti-VP3 serum from lysates of cells undergoing infection with VP3Δ5 and VP3Δ10 mutant virus (Fig. 4B). Because this material was unlikely to have been precipitated through interaction of the mutant VP3 molecules with VPg (free VP1 was clearly not coprecipitated from these lysates), a direct VP3-RNA interaction seemed most plausible. RNase treatments subsequently confirmed this interpretation, demonstrating in addition that the interaction was double strand specific: RNase A but not the single-strand-specific RNaseONE degraded the immune complexes. Furthermore, RT-PCR analysis substantiated that the RNA was indeed of IBDV origin and that it contained both positive and negative strands of each of the two genome segments. As the RT primers were designed to amplify the 5′ end sequence from the plus or minus RNA strand, the results indicated that full-length dsRNA was produced. Less dsRNA-linked VP1 was detected in cells expressing the C-terminally truncated proteins VP3Δ5 and VP3Δ10 than in cells expressing wild-type VP3 or VP3Δ1, suggesting that the VP3 truncations either affect the capacity of VP3 to associate with dsRNA or impair dsRNA synthesis by affecting the capacity of VP1 to function as a replicase, transcriptase, or primase. Wild-type VP3 was found predominantly in association with the dsRNA-linked form of VP1, relatively little being associated with free VP1. This may indicate that not all free VP1 is bound to VP3 during infection. Alternatively, the observation might have been brought about by the experimental conditions used: there may have been differences in the efficiencies with which the different complexes (VP3-VP1 versus VP3-VPg-dsRNA) were immunoprecipitated, or the amount of anti-VP3 antibodies may have been limiting. VP3 is probably synthesized in excess of VP1 (VP3 constitutes 40% of the virion protein, and VP1 constitutes only 3% [17). Limiting VP3 antibodies would be expected to favor the precipitation of dsRNA-VP1-VP3 complexes over free VP1-VP3 complexes as the former probably carry relatively more VP3. Finally it is of note that the same autoradiographic patterns were obtained in different radioimmunoprecipitation assays, both with transfected and with infected cells, by using different labeling periods and different labeling times (data not shown), indicating that the results obtained in the present coimmunoprecipitation analysis were not skewed by unlabeled VP1 and VP3 formed prior to metabolic labeling.
Our results finally provide experimental evidence for the long-anticipated role of VP3 in binding to viral nucleic acids. In 1986 Hudson et al. (22) proposed such a role on the basis of the polypeptide's primary sequence. These investigators noticed that the predicted carboxy-terminal domain, with 12 positively charged residues (and 8 prolines) among its last 40 residues, is highly basic in nature. Interestingly, the terminal stretch of this domain is, in contrast, very acidic, with four negatively charged amino acids being present within the last five residues. This suggests that VP3 might possess two functional domains within its carboxy terminus: the extreme terminal part, which binds to VP1, and the positively charged domain immediately upstream thereof, which binds the viral dsRNA. Future mutation analyses will be required to experimentally confirm the role of the latter domain.
Knowledge about the morphogenesis of birnaviruses is still very limited. Most of what we know came from studies in which IBDV cDNAs were expressed in cells using vaccinia virus or baculovirus systems (13, 19, 30, 32, 37). These studies revealed that virus-like particles indistinguishable from authentic virions can be assembled just from the segment A-derived polyprotein precursor (19, 30). It appears that correct particle assembly is not dependent on the presence of viral RNA or VP1 or of the small protein VP5, also specified by segment A, but rather that it merely requires the proteins pVP2 and VP3, the latter acting as an internal scaffold (32). VP1 was coincorporated into such particles to an extent similar to that to which it was coincorporated into virions when its cDNA was coexpressed with segment A cDNA (30). These investigations in addition pointed to an important role of the carboxy-terminal tail of pVP2 in the assembly of the T=13 capsid, this terminal region being crucially involved in the control of the interactions among VP2 trimers and between VP2 trimers and VP3 trimers (13).
The results demonstrate that the VP3 protein is a key organizer in birnavirus morphogenesis. By its interactions with all the structural components in the virion it appears to create the interior architecture that is required for the proper execution of the replication and transcription activities that are the hallmark of dsRNA viruses. It is now clear that VP3 entertains interactions with itself, with VP2, with free VP1 molecules, most likely also with genome-bound VP1, and with dsRNA segments A and B. Different domains in the protein are responsible for these different interactions. The carboxy-terminal domains binding to the dsRNAs and to VP1 are, for instance, distinct from the region involved in the homotypic interactions giving rise to the VP3 trimers that have been observed in viral particles by ultrastructural analyses (10, 13; M. G. J. Tacken et al., unpublished data). We have not yet explored the domain(s) in the homotrimeric VP3 complex interacting with the homotrimeric VP2 complexes that constitute the outer surface of the viral capsid.
Of particular interest are the interactions of VP3 with VP1 and VPg. These interactions are obviously of critical importance, as the various activities embodied in the polymerase protein are essential for the virus life cycle. Though we did not provide direct evidence for it, we assume that VP3 interacts with VPg and that the interacting domains in VP1 and VPg are the same. Free VP1 molecules present in virions have been shown to carry covalently bound, short stretches of viral RNA, presumably 5′-terminal sequences (24). Their association with VP3 suggests that VP1 does not bind through the domain that contains the serine residue by which these oligonucleotides are attached and also by which VPg is linked to genomic RNA. It is therefore more likely that VP1 and VPg both bind through another, but identical, domain. It will be interesting to find out if and how the virus regulates the numbers of free and genome-bound VP1 molecules assembled in each viral particle. Higher-resolution structural analyses may shed light on this question and establish whether free VP1 and VPg assume distinct and specific positions within the internal virion cavity. Precise spatial arrangements are likely to be required within the virion for the expeditious functioning of this replication and transcription machine.
dsRNA viruses of higher eukaryotes invariably contain multiple genome segments. It is still an unresolved issue how these segments are correctly assorted during particle assembly. Among these viruses the birnaviruses are exceptional because of their genome-linked polymerase protein VPg, the other viruses all having a cap structure associated with the 5′end of the positive RNA strand of each segment. A role for the VPg moiety in the selection of the genome segments is therefore conceivable. Alternatively, this selection might be achieved by VP3 through its capacity to bind dsRNA. This capacity might of course also assist in the proper positioning of the two dsRNA segments within the virion. We have not investigated whether the dsRNA binding activity of VP3 is sequence specific and, if so, whether the sequences recognized are segment specific or occur in the common terminal genomic regions. If this dsRNA binding activity indeed operates at the level of segment selection during particle assembly, we assume that VP3 binds to specific double-stranded regions within the viral mRNAs, as all dsRNA viruses are assumed to synthesize their negative RNA strand only after the packaging of their mRNAs. The specific encapsidation of IBDV mRNAs might thus resemble the mechanism by which the hepatitis B virus assembles its pregenomic RNA. Here the viral polymerase binds to a specific stem-loop structure present in viral mRNAs, which subsequently leads to the polymerase-dependent encapsidation of the viral nucleic acid (3, 42). Much work will be required to resolve these issues, not only for IBDV but also for the other dsRNA viruses. It is expected that continued molecular-virology studies in combination with the ultrastructural analyses in progress will allow us to answer the many outstanding questions concerning the biology of these viruses.
Acknowledgments
We thank Hermann Müller (University of Leipzig, Leipzig, Germany) for providing monoclonal antiserum against VP3 and Patricia A. J. van den Beuken and Sylvia B. E. Verschuren for technical assistance.
REFERENCES
- 1.Antin, P. B., and C. P. Ordahl. 1991. Isolation and characterization of an avian myogenic cell line. Dev. Biol. 143:111-121. [DOI] [PubMed] [Google Scholar]
- 2.Azad, A. A., M. N. Jagadish, M. A. Brown, and P. J. Hudson. 1987. Deletion mapping and expression in Escherichia coli of the large genomic segment of a birnavirus. Virology 161:145-152. [DOI] [PubMed] [Google Scholar]
- 3.Bartenschlager, R., and H. Schaller. 1992. Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 11:3413-3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becht, H., H. Muller, and H. K. Muller. 1988. Comparative studies on structural and antigenic properties of two serotypes of infectious bursal disease virus. J. Gen. Virol. 69:631-640. [DOI] [PubMed] [Google Scholar]
- 5.Birghan, C., E. Mundt, and A. E. Gorbalenya. 2000. A non-canonical lon proteinase lacking the ATPase domain employs the Ser-Lys catalytic dyad to exercise broad control over the life cycle of a double-stranded RNA virus. EMBO J 19:114-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boot, H. J., K. Dokic, and B. P. Peeters. 2001. Comparison of RNA and cDNA transfection methods for rescue of infectious bursal disease virus. J. Virol. Methods 97:67-76. [DOI] [PubMed] [Google Scholar]
- 7.Boot, H. J., A. A. ter Huurne, A. J. Hoekman, B. P. Peeters, and A. L. Gielkens. 2000. Rescue of very virulent and mosaic infectious bursal disease virus from cloned cDNA: VP2 is not the sole determinant of the very virulent phenotype. J. Virol. 74:6701-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boot, H. J., A. A. ter Huurne, B. P. Peeters, and A. L. Gielkens. 1999. Efficient rescue of infectious bursal disease virus from cloned cDNA: evidence for involvement of the 3′-terminal sequence in genome replication. Virology 265:330-341. [DOI] [PubMed] [Google Scholar]
- 9.Boot, H. J., A. H. ter Huurne, and B. P. Peeters. 2000. Generation of full-length cDNA of the two genomic dsRNA segments of infectious bursal disease virus. J. Virol. Methods 84:49-58. [DOI] [PubMed] [Google Scholar]
- 10.Bottcher, B., N. A. Kiselev, V. Y. Stel'Mashchuk, N. A. Perevozchikova, A. V. Borisov, and R. A. Crowther. 1997. Three-dimensional structure of infectious bursal disease virus determined by electron cryomicroscopy. J. Virol. 71:325-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Britton, P., P. Green, S. Kottier, K. L. Mawditt, Z. Penzes, D. Cavanagh, and M. A. Skinner. 1996. Expression of bacteriophage T7 RNA polymerase in avian and mammalian cells by a recombinant fowlpox virus. J. Gen. Virol. 77:963-967. [DOI] [PubMed] [Google Scholar]
- 12.Bruenn, J. A. 1991. Relationships among the positive strand and double-strand RNA viruses as viewed through their RNA-dependent RNA polymerases. Nucleic Acids Res. 19:217-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caston, J. R., J. L. Martinez-Torrecuadrada, A. Maraver, E. Lombardo, J. F. Rodriguez, J. I. Casal, and J. L. Carrascosa. 2001. C terminus of infectious bursal disease virus major capsid protein VP2 is involved in definition of the T number for capsid assembly. J. Virol. 75:10815-10828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chien, C. T., P. L. Bartel, R. Sternglanz, and S. Fields. 1991. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. USA 88:9578-9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho, M. W., O. C. Richards, T. M. Dmitrieva, V. Agol, and E. Ehrenfeld. 1993. RNA duplex unwinding activity of poliovirus RNA-dependent RNA polymerase 3Dpol. J. Virol. 67:3010-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobos, P. 1993. In vitro guanylylation of infectious pancreatic necrosis virus polypeptide VP1. Virology 193:403-413. [DOI] [PubMed] [Google Scholar]
- 17.Dobos, P., B. J. Hill, R. Hallett, D. T. Kells, H. Becht, and D. Teninges. 1979. Biophysical and biochemical characterization of five animal viruses with bisegmented double-stranded RNA genomes. J. Virol. 32:593-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Estojak, J., R. Brent, and E. A. Golemis. 1995. Correlation of two-hybrid affinity data with in vitro measurements. Mol. Cell. Biol. 15:5820-5829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fernandez-Arias, A., C. Risco, S. Martinez, J. P. Albar, and J. F. Rodriguez. 1998. Expression of ORF A1 of infectious bursal disease virus results in the formation of virus-like particles. J. Gen. Virol. 79:1047-1054. [DOI] [PubMed] [Google Scholar]
- 20.Fields, S., and O. Song. 1989. A novel genetic system to detect protein-protein interactions. Nature 340:245-246. [DOI] [PubMed] [Google Scholar]
- 21.Fields, S., and R. Sternglanz. 1994. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 10:286-292. [DOI] [PubMed] [Google Scholar]
- 22.Hudson, P. J., N. M. McKern, B. E. Power, and A. A. Azad. 1986. Genomic structure of the large RNA segment of infectious bursal disease virus. Nucleic Acids Res. 14:5001-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jackwood, D. J., Y. M. Saif, and J. H. Hughes. 1982. Characteristics and serologic studies of two serotypes of infectious bursal disease virus in turkeys. Avian Dis. 26:871-882. [PubMed] [Google Scholar]
- 24.Kibenge, F. S., and V. Dhama. 1997. Evidence that virion-associated VP1 of avibirnaviruses contains viral RNA sequences. Arch. Virol. 142:1227-1236. [DOI] [PubMed] [Google Scholar]
- 25.Kibenge, F. S., B. Qian, J. R. Cleghorn, and C. K. Martin. 1997. Infectious bursal disease virus polyprotein processing does not involve cellular proteases. Arch. Virol. 142:2401-2419. [DOI] [PubMed] [Google Scholar]
- 26.Kordyban, S., G. Magyar, H. K. Chung, and P. Dobos. 1997. Incomplete dsRNA genomes in purified infectious pancreatic necrosis virus. Virology 239:62-70. [DOI] [PubMed] [Google Scholar]
- 27.Lejal, N., B. Da Costa, J. C. Huet, and B. Delmas. 2000. Role of Ser-652 and Lys-692 in the protease activity of infectious bursal disease virus VP4 and identification of its substrate cleavage sites. J. Gen. Virol. 81:983-992. [DOI] [PubMed] [Google Scholar]
- 28.Li, B., and S. Fields. 1993. Identification of mutations in p53 that affect its binding to SV40 large T antigen by using the yeast two-hybrid system. FASEB J. 7:957-963. [DOI] [PubMed] [Google Scholar]
- 29.Libonati, M., and S. Sorrentino. 2001. Degradation of double-stranded RNA by mammalian pancreatic-type ribonucleases. Methods Enzymol. 341:234-248. [DOI] [PubMed] [Google Scholar]
- 30.Lombardo, E., A. Maraver, J. R. Caston, J. Rivera, A. Fernandez-Arias, A. Serrano, J. L. Carrascosa, and J. F. Rodriguez. 1999. VP1, the putative RNA-dependent RNA polymerase of infectious bursal disease virus, forms complexes with the capsid protein VP3, leading to efficient encapsidation into virus-like particles. J. Virol. 73:6973-6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Magyar, G., H. K. Chung, and P. Dobos. 1998. Conversion of VP1 to VPg in cells infected by infectious pancreatic necrosis virus. Virology 245:142-150. [DOI] [PubMed] [Google Scholar]
- 32.Martinez-Torrecuadrada, J. L., J. R. Caston, M. Castro, J. L. Carrascosa, J. F. Rodriguez, and J. I. Casal. 2000. Different architectures in the assembly of infectious bursal disease virus capsid proteins expressed in insect cells. Virology 278:322-331. [DOI] [PubMed] [Google Scholar]
- 33.Mundt, E., B. Kollner, and D. Kretzschmar. 1997. VP5 of infectious bursal disease virus is not essential for viral replication in cell culture. J. Virol. 71:5647-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petek, M., P. N. D'Aprile, and F. Cancellotti. 1973. Biological and physico-chemical properties of the infectious bursal disease virus (IBDV). Avian Pathol. 2:135-152. [DOI] [PubMed] [Google Scholar]
- 35.Sanchez, A. B., and J. F. Rodriguez. 1999. Proteolytic processing in infectious bursal disease virus: identification of the polyprotein cleavage sites by site-directed mutagenesis. Virology 262:190-199. [DOI] [PubMed] [Google Scholar]
- 36.Tacken, M. G., P. J. Rottier, A. L. Gielkens, and B. P. Peeters. 2000. Interactions in vivo between the proteins of infectious bursal disease virus: capsid protein VP3 interacts with the RNA-dependent RNA polymerase, VP1. J. Gen. Virol. 81:209-218. [DOI] [PubMed] [Google Scholar]
- 37.Vakharia, V. N. 1997. Development of recombinant vaccines against infectious bursal disease. Biotech. Annu. Rev. 3:151-168. [Google Scholar]
- 38.Warbrick, E. 1997. Two's company, three's a crowd: the yeast two hybrid system for mapping molecular interactions. Structure 5:13-17. [DOI] [PubMed] [Google Scholar]
- 39.Wensvoort, G., C. Terpstra, J. Boonstra, M. Bloemraad, and D. Van Zaane. 1986. Production of monoclonal antibodies against swine fever virus and their use in laboratory diagnosis. Vet. Microbiol. 12:101-108. [DOI] [PubMed] [Google Scholar]
- 40.Yang, X., E. J. Hubbard, and M. Carlson. 1992. A protein kinase substrate identified by the two-hybrid system. Science 257:680-682. [DOI] [PubMed] [Google Scholar]
- 41.Yao, K., and V. N. Vakharia. 1998. Generation of infectious pancreatic necrosis virus from cloned cDNA. J. Virol. 72:8913-8920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ziermann, R., and D. Ganem. 1996. Homologous and heterologous complementation of HBV and WHV capsid and polymerase functions in RNA encapsidation. Virology 219:350-356. [DOI] [PubMed] [Google Scholar]