Abstract
The Gag protein of retroviruses alone can polymerize into regular virus-like particles (VLPs) both in vitro and in vivo. In most circumstances the capsid (CA) and nucleocapsid (NC) domains of Gag as well as some form of nucleic acid are required for this process. The mechanism by which NC-nucleic acid interaction promotes assembly has remained obscure. We show here that while deletion of the NC domain of Rous sarcoma virus Gag abolishes formation and budding of VLPs at the plasma membranes of baculovirus-infected insect cells, replacement of NC with a dimer-forming leucine zipper domain restores budding of spherical particles morphologically similar to wild-type VLPs. The positioning of the dimerization domain appears to be critical for proper assembly, as the insertion of a 5-amino-acid flexible linker upstream of the zipper domain leads to budding of tubular rather than spherical particles. Similar tubular particles are formed when the same linker is inserted upstream of NC. The tubes are morphologically distinct from tubes formed when the p10 domain upstream of CA is deleted. The fact that a foreign dimerization domain can functionally mimic NC suggests that the role of nucleic acid in retroviral assembly is not to serve as a scaffold but rather to promote the formation of Gag dimers, which are critical intermediates in the polymerization of the Gag shell.
All retroviruses encode a single structural protein, Gag, that alone can assemble into spherical virus-like particles (VLPs) and bud from a host cell membrane. The Gag polyprotein invariably contains a membrane binding (MA), a capsid (CA), and a nucleic acid binding (NC) domain, as well as additional domains which vary from genus to genus. In the formation of nascent virions, wild-type retroviral Gag proteins polymerize into spherical shells composed of about 1,500 (Rous sarcoma virus [RSV] [34, 40]), 2,100 (Mason-Pfizer monkey virus [29]), or more than 4,000 (human immunodeficiency virus [HIV] [unpublished data]) protein molecules. After maturation, which is induced by proteolytic cleavage of Gag by the viral protease (PR), the virion core forms from the liberated CA domain. Unlike that of the immature particle, the morphology of this core may be that of an irregular polyhedron, a tube, or a cone, depending on the virus genus (2, 20, 21, 36, 39). At least in some cases, purified CA protein can polymerize into structures resembling a mature core. For example, HIV type 1 (HIV-1) CA forms tubes or cones at high salt concentrations (14), a process driven exclusively by CA-CA interactions (14, 35). The molecular interactions utilized by HIV-1 CA have been extensively studied (9, 10, 24). By contrast, the molecular interactions that drive assembly of full-length Gag are poorly understood.
In vitro, purified Gag proteins readily polymerize into regular spherical particles or other higher-order structures under select conditions (4-7, 10, 13-15, 24, 26, 35, 43). Several generalizations can be made about assembly from in vitro assembly studies. First, the CA domain is absolutely required for the formation of regular higher-order structures. Second, the MA domain is dispensable for assembly (6, 7, 14, 15), although the sequence immediately upstream of CA plays an important role in shape determination (13, 18, 35). Third, at physiological salt concentrations both the NC domain and nucleic acid are required for polymerization to occur (6, 7, 14). Finally, spherical particles that closely mimic wild-type VLPs can form from Gag constructs containing NC (37, 40).
In vivo, Gag protein is released from cells into the medium in a membrane-enclosed, particulate form, a process referred to as budding. Budding is not synonymous with assembly, which we define as the polymerization of protein into a regular higher-order structure. The process of budding requires the presence of three functional domains: a membrane binding domain (the N-terminal portion of MA), an “interaction,” or I, domain (within NC), and a late domain (38). Of these, only the I domain is required for assembly. The process of assembly has the same requirements in vivo as in vitro. Thus, MA is dispensable for assembly (12, 30), although proteins with no membrane binding domain can assemble only intracellularly (12). The region upstream of CA can play an important role in shape determination (41), core assembly does not occur without CA, and NC is generally required for polymerization into a higher-order structure. Under some conditions HIV-1 is an exception to the latter rule. Although under most circumstances at least a portion of NC is required for both assembly and budding (32), in overexpression systems HIV-1 Gag can assemble into various structures, including slightly irregular spheres, in the absence of NC (11, 12, 16, 19, 27). Perhaps related is the fact that HIV-1 is also the only retrovirus reported to contain a dimerization domain near the C terminus of CA (9). Deletion of this domain abolishes NC-independent assembly (11).
Two models for Gag assembly that could explain the function of the NC-nucleic acid interaction in retroviral assembly have been proposed. The first model suggests that nucleic acid serves as a scaffold holding a particle together and that the function of NC is that of an adhesive, to attach Gag to the scaffold (6, 28). The second model suggests that NC-nucleic acid interactions serve to drive Gag multimerization (42) and that Gag-Gag interactions form the scaffold that holds the particle together. Three lines of evidence support the latter theory. First, it has been shown that in vitro assembly occurs efficiently in the presence of very short DNA oligonucleotides (40), which could serve as a scaffold only if they were somehow bridged together. Second, the minimum-size oligonucleotide that supports efficient assembly in vitro is exactly twice the size of the binding site of the NC domain (25). Third, two groups have shown that in transfected cells, replacement of the HIV-1 NC domain by a leucine zipper protein dimerization (or trimerization) domain allows the release of Gag protein in some particulate form into the medium (3, 42). These groups addressed only whether Gag could exit the cell and did not examine whether the zipper-containing proteins could assemble into spherical VLPs.
Using the RSV system and high-level expression of Gag by means of baculovirus vectors, we have found that a leucine zipper dimerization domain substituted for the NC domain allows the polymerization of Gag and extrusion of membrane-enclosed particles in a manner similar to that of wild type Gag. Surprisingly, the exact placement of the dimerization domain relative to CA determines if the chimeric Gag particles are spherical or tubular. This finding that a protein dimerization domain promotes regular polymerization of Gag in vivo implies that dimerization is of fundamental importance in retrovirus assembly.
MATERIALS AND METHODS
Viruses and cells.
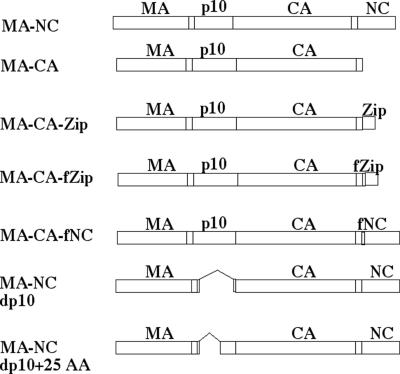
All baculovirus infections were performed with Sf9 spinner culture cells grown in Sf-900 II serum-free medium (Life Technologies). The baculovirus construct RSV MA-NC (RSV ΔPR) has been described previously (17). The vectors expressing MA-NC dp10 and MA-NC dp10 + 25 AA were constructed by replacing the p10 region of vector pFB MA-NC (17) with the p10 region from constructs Δp10.52 (6) and Min0 (18), respectively. The vector expressing MA-CA was constructed by replacing the CA-NC junction of vector pFB-Myr0 (17) with the equivalent region from construct pET-3xc MA-CA (40), resulting in an open reading frame that ends immediately after the CA-NC spacer peptide SP (amino acids 1 to 488). The vector expressing MA-CA-Zip was constructed by synthesizing an oligonucleotide linker that added the leucine zipper domain of CREB1 (amino acids 284 to 327; HUMCREB accession no. M27691) to the C terminus of vector pFB MA-CA. The vector construct MA-CA-fZip was engineered the same way as MA-CA-Zip except that the 5-amino-acid sequence GSGSG was added to the N terminus of the leucine zipper. The vector expressing MA-CA-fNC was constructed by engineering the identical linker sequence (GSGSG) between CA-SP and NC of RSV ΔPR. All of the vector constructs were shuttled into baculoviruses according to the Bac-to-Bac baculovirus expression system protocol (Life Technologies).
TEM and SEM.
Cells to be viewed by transmission electron microscopy (TEM) and scanning electron microscopy (SEM) were infected with the recombinant baculovirus at a low multiplicity and harvested 3 days later. Cells were fixed in 2.5% glutaraldehyde, postfixed with 1% osmium tetroxide, and dehydrated through a graded series of ethanol washes. Samples for TEM analysis were embedded in Spurr resin (33), thin sectioned, and viewed on a Morgani 268 TEM. Samples for SEM analysis were critical point dried, mounted onto aluminum stubs, sputter coated with gold-palladium, and viewed on a Hitachi 4500 SEM.
Particle solubility and density.
For VLP analysis, cells were removed from VLP-containing medium by centrifugation at 1,000 × g for 5 min. For solubility experiments, 1 ml of VLP-containing medium was either left unaltered or adjusted to 0.5% Triton X-100, incubated for 20 min at room temperature, and spun in a microcentrifuge at 20,000 × g for 60 min. The VLP pellets were resuspended in 1× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) buffer and separated by SDS-PAGE. Proteins were either stained with Coomassie blue or transferred to a polyvinylidene difluoride membrane and probed with either a polyclonal rabbit anti-RSV CA antibody or a monoclonal mouse anti-baculovirus gp64 antibody (kindly provided by Gary Blissard). For density experiments, VLPs were first pelleted by centrifugation for 30 min at 45,000 rpm in a 50.2 Ti rotor (Beckman). The pelleted VLPs were then resuspended in 200 μl of STE (100 mM NaCl, 10 mM Tris [pH 7.5], 1 mM EDTA) and layered onto a 4-ml 10-to-60% (wt/wt) step sucrose gradient prepared in STE. Gradients were spun for 2 h at 50,000 rpm in an SW60 rotor (Beckman). The resulting gradients were collected in 10 fractions of 420 μl. A sample of each fraction was separated by SDS-PAGE, transferred onto a polyvinylidene difluoride membrane, and probed with a polyclonal rabbit anti-CA antibody. VLP-containing fractions were pooled, diluted with STE, and spun in a microcentrifuge at 20,000 × g for 60 min. VLP pellets were processed and viewed by TEM as described above. The density of each collected fraction was determined by refractometry.
RESULTS
Replacement of NC with a leucine zipper.
Two laboratories have shown that the HIV-1 NC domain can be replaced with a dimer-forming leucine zipper without compromising budding efficiency (3, 42), but neither of these groups tested whether these chimeric proteins assemble correctly. In order to examine if dimerization is sufficient to promote VLP assembly, we substituted the leucine zipper domain from the human cyclic AMP response element binding protein (HUMCREB) for the NC domain of RSV Gag (Fig. 1). In the first construct, the zipper was positioned immediately after the 12-amino-acid spacer segment (SP) between CA and NC, thereby resulting in an exact replacement of NC (MA-CA-Zip). In a second construct, the dimerization domain was separated from SP by an artificial 5-amino-acid flexible linker, GSGSG (MA-CA-fZip). These chimeric proteins, as well as the Gag proteins MA-NC and MA-CA, used as positive and negative controls, respectively, were expressed in insect cells by means of baculovirus vectors, and the cells were examined by SEM and TEM of thin sections (Fig. 2). Because the PR domain of RSV inhibits proper VLP assembly in insect cells (17), all of the constructs described here lack a PR domain.
FIG. 1.
Schematic representation of expressed proteins. MA-NC corresponds to the RSV Gag protein lacking its PR domain at the C terminus.
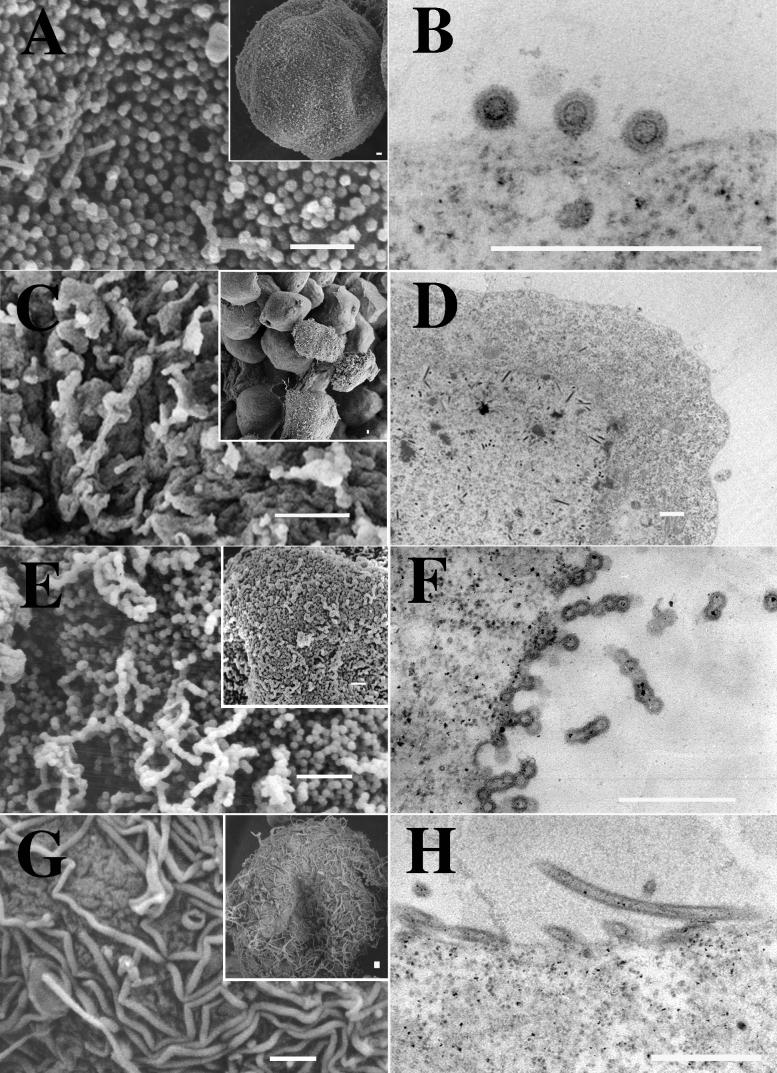
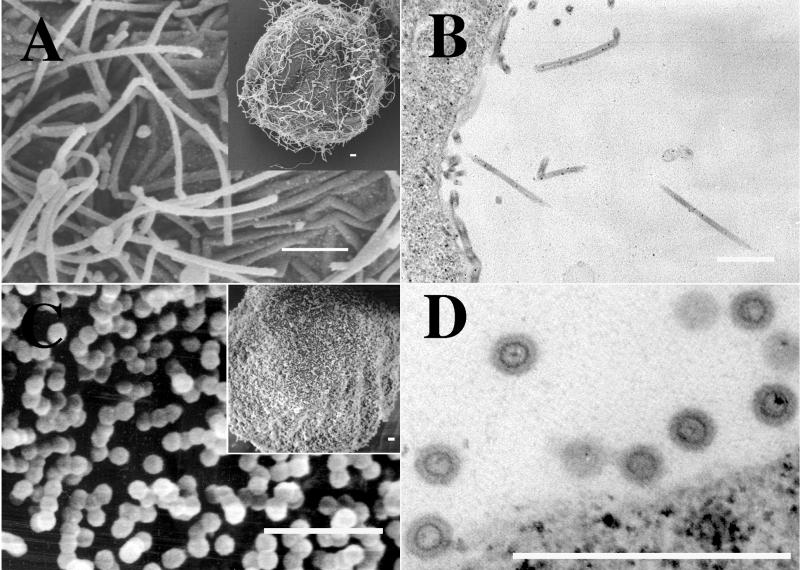
FIG. 2.
EM images of budding structures. Cells were infected with baculovirus vectors expressing either MA-NC (A and B), MA-CA (C and D), MA-CA-Zip (E and F), or MA-NC-fZip (G and H). Panels A, C, E, and G are SEM images (insets are low-magnification images), and panels B, D, F, and H are thin-section TEM images. Bars, 500 nm.
MA-NC was found to produce regular spherical particles covering the surfaces of infected cells (Fig. 2A), as described previously (17). Thin sections of these cells revealed the circular outlines of spherical particles with a single predominant ring or shell near the particle center, presumably representing the CA domain (Fig. 2B). Deletion of the NC domain (MA-CA) abolished assembly as detected by this visual criterion. In a field of infected cells (Fig. 2C, inset), most cells expressing MA-CA were bald, lacking any surface structures. A few cells displayed some surface texture, but when they were viewed at a higher magnification (Fig. 2C), the texture appeared to be irregular wavy protuberances, a morphology commonly seen in baculovirus-infected cells. Thin-section electron microscopy (EM) confirmed the absence of recognizable viral structures (Fig. 2D). No electron-dense aggregates, characteristic of retrovirus Gag protein in the process of assembly, appeared beneath the plasma membrane. The cigar-shaped baculovirus particles visible in the nucleus verified that cells had indeed been infected.
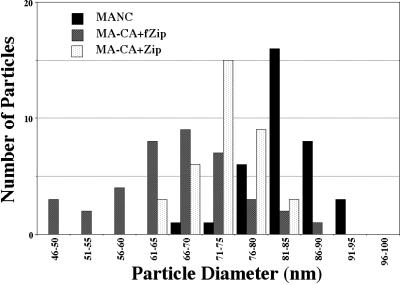
The two chimeric Gag-zipper proteins formed regular budding structures at the plasma membrane. Insect cells expressing the MA-CA-Zip protein showed spherical particles coating their surfaces (Fig. 2E). While they were similar overall to VLPs formed by MA-NC, the MA-CA-Zip particles differed in that many appeared to be fused into long chains and aggregates of merged spheres. Thin-section TEM confirmed this conclusion (Fig. 2F), revealing that, unlike MA-NC VLPS, most of the MA-CA-Zip VLPs had incompletely closed shells. Chains of particles in which each VLP seemed to have formed out of the incomplete VLP that came before it were frequently observed. In contrast to these nearly spherical particles, the budding structures on cells expressing MA-CA-fZip were of an entirely different morphology (Fig. 2G and H). These cells displayed flexible tubular structures on their surfaces. The tubes were not homogeneous in width and frequently had kinks. The average diameters ± standard deviations of MA-NC, MA-CA-Zip, and MA-CA-fZip particles were 83 ± 6, 73 ± 5, and 66 ± 10 nm (Fig. 3). The average diameter of the dark inner ring of MA-NC was 46 ± 3 nm. Although the zipper particles also contained dark inner rings, these were not distinct enough for accurate measurements to be taken. The 5-to-10%-smaller diameter of the MA-CA-Zip particles compared with the MA-NC particles might be explained by the hypothesis that the unclosed spheres are more easily stretched or crushed. Indeed, the MA-CA-Zip structures often appeared slightly ovular.
FIG. 3.
Size distribution of MA-NC, MA-CA-Zip, and MA-CA-fZip. Diameters of spherical (MA-NC and MA-CA-Zip) or tubular (MA-CA-fZip) particles were measured in TEM thin sections.
We were surprised that the addition of 5 amino acids upstream of the leucine zipper had such a dramatic effect on assembly. In order to determine whether this insert could exert the same effect in the context of NC, we introduced the same linker sequence at the junction between CA-SP and NC (MA-CA-fNC) (Fig. 1). SEM and TEM images of insect cells expressing this construct showed tubular particles morphologically identical to those from the equivalent construct containing the zipper (Fig. 4). Although with an average diameter of 79 ± 9 nm these tubes were somewhat larger than the MA-CA-fZip tubes, they appeared similar in their apparent flexibility and heterogeneity in width.
FIG. 4.
EM images of budding structures of MA-CA-fNC. (Top) SEM; (Bottom) TEM. Bar, 500 nm.
In summary, we have shown that Gag protein containing a foreign dimerization domain in place of NC is capable of assembling into regular spherical particles. Further, addition of a 5-amino-acid flexible linker (GSGSG) placed upstream of the dimerization domain, or of NC itself, resulted in the formation of tubular particles. These data suggest that the primary role of NC in assembly is to promote Gag dimerization and that the positioning of the dimerization domain has a strong influence on viral morphology.
MA-NC dp10 tubes are distinct.
The tubes that formed upon addition of 5 amino acids upstream of the NC or of the zipper domain represent the first example in which an RSV Gag protein has been observed to be extruded through the membrane in vivo in a tubular shape. Polymerization of retroviral Gag proteins into tubes is not without precedent, however. Moloney murine leukemia virus (M-MuLV) Gag buds as tubes when its p12 domain is deleted (41), HIV-1 Gag buds as tubes in an overexpression system when sequences downstream of CA are deleted (11, 12, 16, 19, 27), and both HIV-1 and RSV assemble into tubes in vitro when sequences upstream of CA are deleted (5, 6, 10, 13-15, 18, 24). Furthermore, in vitro CA itself assembles into tubes, and for some larger Gag proteins a change in pH can shift the mode of polymerization between spherical and tubular (15). A mechanistic understanding of the rules governing spherical or tubular assembly remains elusive.
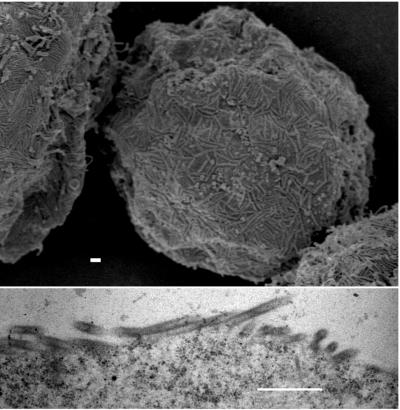
We considered the possibility that the tubular budding of MA-CA-fZip and MA-CA-fNC represented a default assembly pathway. If this were correct, then all tubes assembled with Gag constructs might be expected to have the same morphological properties. To address this question, we expressed an RSV Gag construct with a wild-type CA-NC junction but with 52 of the 62 amino acids of its p10 domain deleted (MA-NC dp10). This protein is known to assemble into regular tubular particles in vitro, as well as in Escherichia coli, in the absence of the N-terminal membrane binding domain, which had been removed to improve solubility (6, 18). Baculovirus-mediated expression of MA-NC dp10 led to the budding of tubular particles from the plasma membrane, but these tubes were distinct from those formed by the two linker insertion mutants described above (Fig. 5A and B). They were long (up to several micrometers) and narrow, appeared rigid, and contained a distinct dark inner ring that was most evident in cross sections. In summary, genetic alterations at the N and C termini of Gag can result in the assembly of tubes, but this is not a default assembly pathway, since the two kinds of tubes are morphologically distinct.
FIG. 5.
EM images of p10 budding structures. Cells were infected with baculovirus vectors expressing either MA-NC dp10 (A and B) or MA-NC dp10 + 25 AA (C and D). (A and C) SEM images; (B and D) thin-section TEM images. Bars, 500 nm.
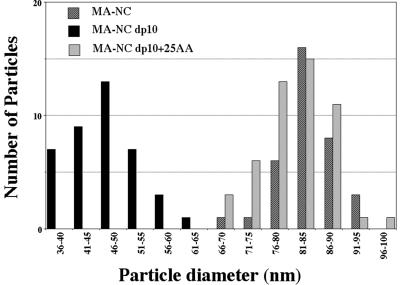
In vitro and in E. coli, the stretch of 25 amino acids at the C terminus of p10, i.e., just upstream of the CA domain, is sufficient to restore the spherical assembly phenotype to a Gag protein with the dp10 deletion (18). To examine if these amino acids also function similarly in vivo, we created a baculovirus vector encoding the protein MA-NC dp10 + 25 AA (Fig. 1). The VLPs budding from insect cells expressing this protein were spherical and indistinguishable from MA-NC VLPs (Fig. 5C and D). The distributions of diameters of the MA-NC dp10 + 25 AA spheres and the MA-NC spheres were almost identical, but that of the MA-NC dp10 tubes showed considerably smaller diameters (Fig. 6). The average diameters ± standard deviations of MA-NC, MA-NC dp10, and MA-NC dp10 + 25 AA VLPs were 83 ± 6, 47 ± 4, and 82 ± 6 nm, respectively. The average diameters ± standard deviations of the dark inner rings of MA-NC, MA-NC dp10, and MA-NC dp10 + 25 AA VLPs were 46 ± 3, 25 ± 4, and 50 ± 4 nm, respectively. In summary, the assembly properties of RSV Gag appear the same in vitro and in vivo; Gag constructs lacking the p10 domain assemble into tubes, but 25 amino acids of p10 are sufficient to convert the assembly process to one that yields spheres.
FIG. 6.
Size distribution of MA-NC, MA-NC dp10, and MA-NC dp10 + 25 AA particles. Diameters of spherical (MA-NC and MA-NC dp10 + 25 AA) or tubular (MA-NC dp10) particles were measured in TEM thin sections.
Biochemical characterization of baculovirus-produced particles.
To complement the qualitative data obtained by EM, we examined the biochemical properties of several types of Gag particles collected after budding from recombinant-baculovirus-infected insect cells. Initially, in order to characterize budding rates, cells were pulse-labeled with [35S]methionine and Gag protein released into the medium was analyzed at time points after a period of chase with excess cold methionine. In addition to the constructs already described, we also examined a mutant lacking its late domain (dp2b, with the sequence TASAPPPPYVG deleted). All the proteins, including the negative controls MA-CA and dp2b, were released into the medium at similar rates (data not shown). This finding was unexpected because it has been well established that both NC and p2b are required for efficient budding of RSV at normal expression levels in mammalian and avian cells, which is consistent with our observation that proteins lacking NC are incapable of forming viral particles visible by EM analysis. We therefore believe that some of these biochemical measurements of protein in the growth medium reflect an artifact of baculovirus overexpression and not true budding. The expression level achieved in the baculovirus expression system is likely to saturate the plasma membrane, which could have the effect of slowing the release of regular particles while permitting the release of some nonparticulate protein, perhaps through cellular blebbing (31; data not shown).
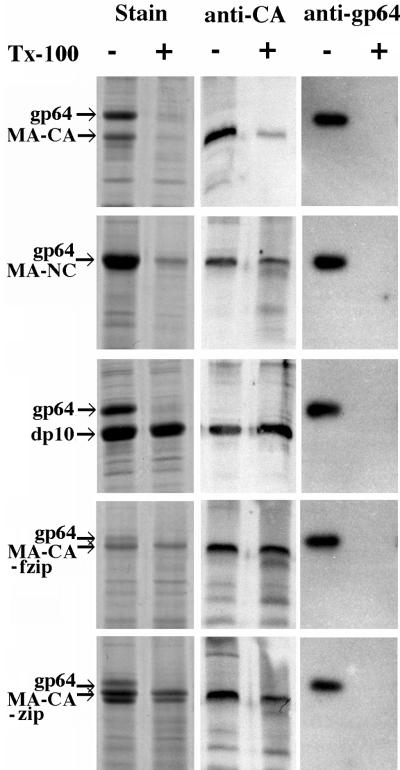
Next, we sought to characterize the particles biochemically. In order to determine if they were detergent resistant, samples of medium were treated with 0.5% Triton X-100 for 20 min before centrifugation (Fig. 7). Detergent treatment had little effect on the recovery of MA-NC, MA-NC dp10, or the zipper proteins in the pellet fractions, as would be expected for stable immature retrovirus particles. For MA-NC this is most evident in the Western blot, because the baculovirus gp64 protein comigrates with MA-NC in SDS-PAGE (Fig. 7, left panel). In contrast, very little of the MA-CA protein was recovered after detergent treatment, which suggests that, as was observed by EM (Fig. 2C and D), MA-CA does not form into true viral structures. Thin-section EM of each of the detergent-treated pellets (except MA-CA, which did not have a substantial pellet) revealed that the morphology of the particles was the same as that observed at the plasma membrane (data not shown). These thin sections also verified that the detergent-treated pellets contained stable baculovirus cores. As an internal control we used the same samples to assess the effect of detergent treatment on the baculovirus gp64 membrane protein. gp64 became completely soluble after detergent treatment, providing verification that the detergent conditions were sufficient to disrupt viral membranes. Because the baculovirus cores contained nucleic acid but could not be separated from the retroviral particles, we were unable to determine the nucleic acid contents of the particles formed from the various Gag proteins. Taken together, these results demonstrate that each of the baculovirus-produced VLPs has a detergent-stable core but that the nonparticulate MA-CA protein that exited the cell did so through cellular blebbing or some other means not involving formation of a stable core.
FIG. 7.
Detergent stability of particles. Particles were pelleted after incubation with or without Triton X-100 (Tx-100) as described in Materials and Methods. Pellets were dissolved in SDS, and the proteins were separated by SDS-PAGE and then either stained with Coomassie blue, probed with an antibody against RSV CA, or probed with an antibody against baculovirus gp64.
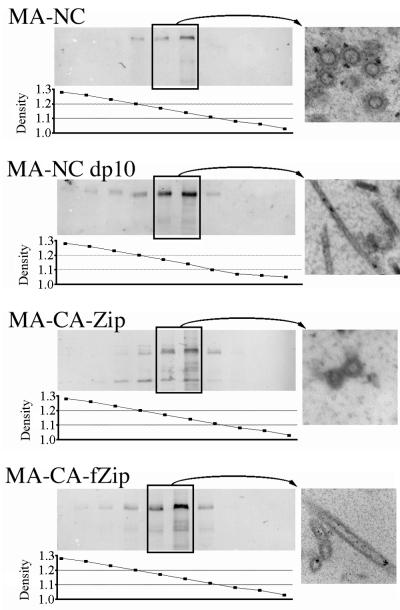
To further characterize the baculovirus-produced VLPs, we compared the densities of the zipper- and NC-containing particles. VLPs from the medium of cells expressing MA-NC, MA-NC dp10, MA-CA-Zip, or MA-CA-fZip were concentrated and centrifuged to equilibrium in 10-to-60% (wt/wt) sucrose gradients. Each gradient was divided into 10 fractions, and a portion of each fraction was analyzed either by refractometry to determine the density or by Western blotting with an anti-RSV CA antiserum to determine the location of Gag within the gradient (Fig. 8). The VLPs from all four constructs migrated to a density of approximately 1.14 to 1.17 g/cm3, consistent with the normal density of retroviruses in sucrose. Particles from the equilibrium gradients were pooled, pelleted, thin sectioned, and viewed by TEM. In each case the morphology of recovered VLPs matched the morphology observed at the plasma membranes of cells expressing the corresponding construct (Fig. 8). In multiple repeats of this gradient fractionation, a portion of the zipper-containing proteins frequently migrated to a higher density (around 1.2 g/cm3) in the sucrose gradient. Thin sections of the higher-density particles revealed no difference in particle morphology. We predict that these higher-density particles, when they appear, are particles that have had all or part of their membranes stripped, thus altering their density in sucrose. In summary, all four types of particles analyzed in this study are regular VLPs with normal retroviral densities in sucrose, regardless of whether they have a zipper or an NC domain.
FIG. 8.
Density of particles. Pelleted particles from each expression construct were resuspended and then centrifuged to equilibrium in 10-to-60% (wt/wt) sucrose gradients. A portion of each fraction was separated by SDS-PAGE and probed with an antibody against RSV CA. The density of each fraction was determined by refractometry. Fractions from the boxed lanes were pooled, diluted to reduce the sucrose concentration, and then centrifuged again to collect the particles. The pellets were then viewed by thin-section TEM and are shown on the right.
DISCUSSION
We have shown that the function of the NC domain of RSV Gag in promoting assembly of VLPs can be replaced by a small protein dimerization domain that neither interacts with nucleic acid nor by itself polymerizes into higher-order structures. Depending on the exact positioning of the leucine zipper, the VLPs either are morphologically very similar to classic immature retrovirus particles or are distinct tubular particles. It has been reported previously that a dimer-forming or a trimer-forming leucine zipper could replace NC in budding assays (3, 42), but the ultrastucture of the particles released into the medium was not determined. In particular, it was not known whether chimeric Gag-leucine zipper particles were regular in shape, and thus were built on the same principles as wild-type virions, or, alternatively, whether they were produced by blebbing of the plasma membrane. The results presented in this paper are important because they strongly suggest that the mechanism by which the NC domain promotes assembly is induction of the formation of Gag dimers, which in turn are essential intermediates in the formation of immature virions. It cannot be determined from these data whether every Gag protein is required to dimerize, or whether dimerization is simply required for initiation of the assembly process. We did not test whether a trimerization domain could replace NC in promoting regular assembly, but because a Gag dimer could theoretically still be induced by a trimer-forming leucine zipper, results from expression of such a construct would likely be ambiguous.
The spherical particles formed by MA-CA-Zip appear similar, but not identical, to wild-type immature particles. In particular, the MA-CA-Zip particles are spherical and of nearly the same diameter, but they are not closed. Two possible explanations might account for this difference. First, RNA might act as a secondary scaffolding in the assembly of Gag proteins with an NC domain. That is, the Gag-Gag interactions could drive polymerization, but RNA could play some role in the final shaping of the core. However, the fact that assembly can occur in vitro with very short oligonucleotides argues against this explanation. A second and more likely explanation for the incomplete assembly is simply that a specific positioning of the dimerization domain relative to CA is required for the proteins to achieve the correct conformation to yield a complete sphere. In support of this notion, we found that addition of a 5-amino-acid flexible linker between CA and NC or between CA and the leucine zipper had a dramatic effect on viral morphology. In both cases, spherical assembly was abolished and the resulting particles appeared as fat tubes.
In vitro assembly systems have provided key insights into the mechanism of retrovirus assembly. Purified Gag proteins spontaneously polymerize into spherical or tubular particles (4, 6, 7, 10, 13-15, 24, 35). Unlike the situation with in vivo studies, complications due to protein-membrane interactions do not arise in vitro and the role of nucleic acid can be assessed independently. With few exceptions, formation of regular structures requires a Gag protein with intact CA and NC domains (or a portion of NC), as well as nucleic acid. Based on their ultrastructure, spherical particles accurately mimic immature virions that have budded from cells, except that they lack a membrane (15, 37, 40). Tubular particles appear to represent the mature retrovirus core, which has a conical, tubular, or irregular polyhedral shape in infectious virions, depending on the virus genus. This inference is indirect and is based in part on the observation that HIV-1 Gag proteins capable of forming tubes also form cones (10, 15, 24). Reconstruction of HIV-1 tubes from cryo-EM images has led to a molecular model for the cone (24). For RSV, the sequence immediately N-terminal to the CA domain plays a specific shape-determining role. Deletion or replacement of the 25 amino acids immediately upstream of RSV CA leads to the formation of regular tubes (18). Although an extensive collection of mutations in this 25-amino-acid region has been scored for spherical or tubular particle assembly (18), it is still unclear how this domain acts as an effector for particle morphology.
The results presented here show that the same 25-amino-acid sequence upstream of CA that is essential for spherical polymerization of RSV Gag proteins in vitro is also essential in the baculovirus system, where assembly and envelopment occur at the plasma membrane. The tubular structures that form when the p10 domain is deleted appear to match the equivalent in vitro structures in both size and morphology, with the exception that the in vivo particles contain a lipid envelope. This concordance further supports the faithfulness of the in vitro assembly system. In a previous study of particles released from transfected COS cells, it was shown that the dp10 deletion in RSV Gag results in particles that migrate more slowly in rate zonal sedimentation assays than wild-type RSV VLPs (23). This study did not examine particle morphology. The slower migration was previously interpreted to be the result of smaller particles, but in light of the tubular morphology observed here, we reinterpret the slower migration as due to the higher friction coefficient of tubes compared with spheres.
The observation that RSV requires a specific sequence upstream of CA for spherical assembly supports the idea that HIV and RSV Gag proteins have somewhat different assembly requirements. For HIV-1 there is no evidence that a specific sequence in the upstream MA domain promotes spherical-particle formation. In vivo, deletion of most of HIV-1 MA does not inhibit spherical assembly (30). It has been proposed that by rupturing the salt bridge between Pro1 and Asp51, any N-terminal extension of CA would lead to refolding of part of CA, and that this refolding would favor a spherical polymerization mode (35). In contrast to HIV-1, M-MuLV Gag appears to behave in the same manner as RSV Gag. M-MuLV Gag with a deletion in the p12 domain between MA and CA forms tubular particles when expressed in 293T cells (41), suggesting that a specific amino acid sequence in p12 is required for spherical assembly. No structural information is available for N-terminally extended CA protein from RSV or M-MuLV, and it is not known how these domains direct assembly.
The function of NC in assembly, both in vivo and in vitro, has remained obscure. The interaction domain that has been mapped to NC could be envisioned to bring Gag molecules together directly, via protein-protein contacts in NC, or indirectly, by acting as a binding surface to collect Gag molecules on a long RNA molecule. Biochemical experiments have argued for the latter (8). Indeed, it has been proposed that RNA is itself a scaffold holding the particle together (28). The presence as well as the precise positioning of NC in Gag appears to be of critical importance in assembly. The stretch of 12 amino acids (or 14 in HIV-1) comprising the spacer between CA and NC is very sensitive to mutation, and as described above, deletion of part or all of this segment of Gag leads to grossly aberrantly assembled particles budding from the plasma membrane (1, 22). Quantitative studies on RSV Gag assembly in vitro with oligonucleotides of defined length have provided insight into the role of NC in assembly. Briefly, it was found that the minimum-size oligonucleotide capable of promoting efficient assembly was exactly twice the size of the binding site of NC (25). These results have been interpreted to mean that upon binding to nucleic acid, the NC domain promotes dimerization of Gag and that this dimerization is critical for assembly. The finding reported here, that a protein dimerization domain can replace NC in vivo, strongly supports models in which a Gag dimer is an essential intermediate in assembly.
Acknowledgments
We thank Gary Blissard for providing the antibody to gp64 and John Wills for providing the Δp10.52 vector.
This work was supported by NIH grant CA20081.
REFERENCES
- 1.Accola, M. A., S. Höglund, and H. G. Göttlinger. 1998. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J. Virol. 72:2072-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Accola, M. A., A. Ohagen, and H. G. Göttlinger. 2000. Isolation of human immunodeficiency virus type 1 cores: retention of Vpr in the absence of p6gag. J. Virol. 74:6198-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Accola, M. A., B. Strack, and H. G. Göttlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell, S., R. J. Fisher, E. M. Towler, S. Fox, H. J. Issaq, T. Wolfe, L. R. Phillips, and A. Rein. 2001. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc. Natl. Acad. Sci. USA 98:10875-10879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campbell, S., and A. Rein. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, S., and V. M. Vogt. 1997. In vitro assembly of virus-like particles with Rous sarcoma virus Gag deletion mutants: identification of the p10 domain as a morphological determinant in the formation of spherical particles. J. Virol. 71:4425-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cimarelli, A., S. Sandin, S. Höglund, and J. Luban. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74:3046-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gamble, T. R., S. Yoo, F. F. Vajdos, U. K. von Schwedler, D. K. Worthylake, H. Wang, J. P. McCutcheon, W. I. Sundquist, and C. P. Hill. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 10.Ganser, B. K., S. Li, V. Y. Klishko, J. T. Finch, and W. I. Sundquist. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80-83. [DOI] [PubMed] [Google Scholar]
- 11.Gay, B., J. Tournier, N. Chazal, C. Carriere, and P. Boulanger. 1998. Morphopoietic determinants of HIV-1 Gag particles assembled in baculovirus-infected cells. Virology 247:160-169. [DOI] [PubMed] [Google Scholar]
- 12.Gheysen, D., E. Jacobs, F. de Foresta, C. Thiriart, M. Francotte, D. Thines, and M. De Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 13.Gross, I., H. Hohenberg, C. Huckhagel, and H. G. Kräusslich. 1998. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J. Virol. 72:4798-4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gross, I., H. Hohenberg, and H. G. Kräusslich. 1997. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur. J. Biochem. 249:592-600. [DOI] [PubMed] [Google Scholar]
- 15.Gross, I., H. Hohenberg, T. Wilk, K. Wiegers, M. Grättinger, B. Müller, S. Fuller, and H. G. Kräusslich. 2000. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 19:103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hockley, D. J., M. V. Nermut, C. Grief, J. B. Jowett, and I. M. Jones. 1994. Comparative morphology of Gag protein structures produced by mutants of the gag gene of human immunodeficiency virus type 1. J. Gen. Virol. 75:2985-2997. [DOI] [PubMed] [Google Scholar]
- 17.Johnson, M. C., H. M. Scobie, and V. M. Vogt. 2001. PR domain of Rous sarcoma virus Gag causes an assembly/budding defect in insect cells. J. Virol. 75:4407-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joshi, S. M., and V. M. Vogt. 2000. Role of the Rous sarcoma virus p10 domain in shape determination of Gag virus-like particles assembled in vitro and within Escherichia coli. J. Virol. 74:10260-10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jowett, J. B., D. J. Hockley, M. V. Nermut, and I. M. Jones. 1992. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. J. Gen. Virol. 73:3079-3086. [DOI] [PubMed] [Google Scholar]
- 20.Kingston, R. L., N. H. Olson, and V. M. Vogt. 2001. The organization of mature Rous sarcoma virus as studied by cryo-electron microscopy. J. Struct. Biol. 136:67-80. [DOI] [PubMed] [Google Scholar]
- 21.Kotov, A., J. Zhou, P. Flicker, and C. Aiken. 1999. Association of Nef with the human immunodeficiency virus type 1 core. J. Virol. 73:8824-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kräusslich, H. G., M. Facke, A. M. Heuser, J. Konvalinka, and H. Zentgraf. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krishna, N. K., S. Campbell, V. M. Vogt, and J. W. Wills. 1998. Genetic determinants of Rous sarcoma virus particle size. J. Virol. 72:564-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li, S., C. P. Hill, W. I. Sundquist, and J. T. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409-413. [DOI] [PubMed] [Google Scholar]
- 25.Ma, Y. M., and V. M. Vogt. 2002. Rous sarcoma virus Gag protein-oligonucleotide interaction suggests a critical role for protein dimer formation in assembly. J. Virol. 76:5452-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morikawa, Y., T. Goto, and K. Sano. 1999. In vitro assembly of human immunodeficiency virus type 1 Gag protein. J. Biol. Chem. 274:27997-28002. [DOI] [PubMed] [Google Scholar]
- 27.Morikawa, Y., D. J. Hockley, M. V. Nermut, and I. M. Jones. 2000. Roles of matrix, p2, and N-terminal myristoylation in human immunodeficiency virus type 1 Gag assembly. J. Virol. 74:16-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parker, S. D., J. S. Wall, and E. Hunter. 2001. Analysis of Mason-Pfizer monkey virus Gag particles by scanning transmission microscopy. J. Virol. 75:9543-9548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Göttlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Royer, M., S. S. Hong, B. Gay, M. Cerutti, and P. Boulanger. 1992. Expression and extracellular release of human immunodeficiency virus type 1 Gag precursors by recombinant baculovirus-infected cells. J. Virol. 66:3230-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sandefur, S., R. M. Smith, V. Varthakavi, and P. Spearman. 2000. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 74:7238-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spurr, A. R. 1969. A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26:31-43. [DOI] [PubMed] [Google Scholar]
- 34.Vogt, V. M., and M. N. Simon. 1999. Mass determination of Rous sarcoma virus virions by scanning transmission electron microscopy. J. Virol. 73:7050-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.von Schwedler, U. K., T. L. Stemmler, V. Y. Klishko, S. Li, K. H. Albertine, D. R. Davis, and W. I. Sundquist. 1998. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 17:1555-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Welker, R., H. Hohenberg, U. Tessmer, C. Huckhagel, and H. G. Kräusslich. 2000. Biochemical and structural analysis of isolated mature cores of human immunodeficiency virus type 1. J. Virol. 74:1168-1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilk, T., I. Gross, B. E. Gowen, T. Rutten, F. de Haas, R. Welker, H. G. Kräusslich, P. Boulanger, and S. D. Fuller. 2001. Organization of immature human immunodeficiency virus type 1. J. Virol. 75:759-771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wills, J. W., and R. C. Craven. 1991. Form, function, and use of retroviral gag proteins. AIDS 5:639-654. [DOI] [PubMed] [Google Scholar]
- 39.Yeager, M., E. M. Wilson-Kubalek, S. G. Weiner, P. O. Brown, and A. Rein. 1998. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc. Natl. Acad. Sci. USA 95:7299-7304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu, F., S. M. Joshi, Y. M. Ma, R. L. Kingston, M. N. Simon, and V. M. Vogt. 2001. Characterization of Rous sarcoma virus Gag particles assembled in vitro. J. Virol. 75:2753-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yuan, B., S. Campbell, E. Bacharach, A. Rein, and S. P. Goff. 2000. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J. Virol. 74:7250-7260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang, Y., H. Qian, Z. Love, and E. Barklis. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 72:1782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuber, G., J. McDermott, S. Karanjia, W. Zhao, M. F. Schmid, and E. Barklis. 2000. Assembly of retrovirus capsid-nucleocapsid proteins in the presence of membranes or RNA. J. Virol. 74:7431-7441. [DOI] [PMC free article] [PubMed] [Google Scholar]