Abstract
Infection of domestic rabbits with cottontail rabbit papillomavirus (CRPV) causes local papillomas which progress to carcinomas in more than 80% of cases. This animal model system therefore allows the identification of molecular mechanisms required for the induction and progression of epithelial tumors. The viral E2 protein stimulates both viral DNA replication and transcription, and these functions can be genetically separated. We introduced the respective mutations into CRPV E2 and found, in line with published data for other papillomavirus E2 proteins, that mutation of the highly conserved amino acid 37 or 73 resulted in replication-competent but transactivation-deficient E2 proteins, whereas E2 proteins with mutations at residue 39 were replication deficient and transactivation competent. The R37A, I73L, and I73A E2 mutants, showing a loss of transactivation function, and the R37K E2 mutant, which is still transactivation competent, were introduced into the whole genome of CRPV, which was then injected into the skin of rabbits. Strikingly, the ability to induce tumors within 6 weeks was abolished by each of the E2 mutations, in contrast to the tumor induction rate (93%) obtained with wild-type CRPV DNA. Two small papillomas induced by mutant E2 I73A CRPV DNA appeared as late as 12 or 24 weeks postinjection, were significantly smaller, and showed no further extension of growth. These data suggest that functionally conserved amino acids in the transactivation domain of E2 are also required for the induction and growth of epithelial tumors in rabbits infected with CRPV.
A suitable animal model in which to investigate molecular processes involved in tumor induction and progression by papillomaviruses is the New Zealand White rabbit, which develops local papillomas within 3 to 8 weeks after injection of cottontail rabbit papillomavirus (CRPV) DNA into the skin (45). In contrast to the productive virus infection observed in the cottontail rabbit, which is the natural host, infection of New Zealand White rabbits with CRPV causes an abortive infection (45). As many as 80% of the initially benign epithelial tumors progress to carcinomas within the next 6 to 14 months, and these finally metastasize without the need for cofactors (45). Previous studies using deletion or insertion mutations within the genome have shown that all open reading frames (ORF) of CRPV, except for E5 and L2, are required for tumor induction in rabbits (5, 6, 25, 26, 46). In contrast, tissue culture experiments have demonstrated that expression of CRPV LE6 and E7 is sufficient to induce anchorage-independent growth of NIH 3T3 cells and to induce tumors in nude mice injected with the respective cell lines (25). When viral transcripts from benign and malignant tumors were compared by a number of different techniques, no major quantitative or qualitative differences that would have been indicative of a prominent role of a single viral gene product could be observed (27, 29, 48).
A master regulator of early gene expression and viral DNA replication is the multifunctional E2 protein of papillomaviruses. The E2 protein forms dimers that specifically bind to the palindromic DNA sequence ACCN6GGT, which can be found in multiple copies in the upstream regions of early and late promoters of papillomaviruses (24). In addition, E2 is capable of stimulating transcription through multimerized E2 binding sites located upstream of artificial promoters in transient reporter assays (2, 10, 14, 19, 21, 28, 40, 42, 44). As subsequent studies revealed, several early bovine papillomavirus 1 (BPV1) promoters are activated through E2 in a binding-site-dependent manner as well (18, 20, 30, 39). In contrast, the early promoter P97, situated at the beginning of the E6 ORF of human papillomavirus 16 (HPV16), and the respective highly conserved promoters in HPV6, -11, -18, and -31 and CRPV have consistently been found to be repressed by binding of E2 to proximal E2-binding sites (4, 12, 14, 32, 34, 43). Besides the early promoters of BPV1, the only natural HPV promoter identified that is activated by E2 is the HPV6a E7 promoter (32).
During DNA replication, E2 forms a complex with the viral E1 protein and thereby increases the origin of replication recognition by E1 and subsequently replication efficiency (41). To separate the replicative function from the transactivation function, a single amino acid exchange in E2 of BPV1 or HPV11, -16, -18, or -31 was found to be sufficient (1, 7, 9, 13, 16, 17, 35, 42). Another activity described for the BPV1 E2 protein is mediation of the attachment of the episomal viral DNA to host cell chromosomes (22, 23, 38). In contrast to this well-described multifunctionality of the E2 protein, only a few data exist linking these activities to the carcinogenic activity of papillomaviruses. Only in the case of BPV1 were the transactivating and replicative functions shown to be essential for virus-mediated transformation of cells (7, 11, 31, 36). In contrast, the E2 gene of HPV16 and the transactivating function of E2 in the context of the whole genome of HPV31 were found to be dispensable for immortalization of primary keratinocytes (33, 42).
Our intention was to characterize functions of the E2 protein of CRPV in more detail in order to gain insight into the role of E2 in induction and progression of tumors in vivo. As has been described for other papillomaviruses, we were able to separate the transactivating and replicating functions of CRPV E2 by single amino acid exchanges in the N-terminal region, and we demonstrated for the first time that the transactivating ability of E2 is required for induction of papillomas in vivo.
MATERIALS AND METHODS
Recombinant plasmids.
The E2-dependent luciferase reporter plasmid p6xE2BS-luc has been described previously (42). The noncoding region (NCR) of CRPV (CRPV nucleotides [nt] 7339 to 151) was amplified by PCR using plasmid CRPV-pLAII, described previously (26), as a template, with an upstream primer containing an SstI restriction site (5′-CTAAACGCAAGAGCTCTACTTAATTG-3′) and a downstream primer with an NcoI restriction site (5′-GCAGTTCTCCATGGGCTCTAAGTTTC-3′). The PCR-generated fragment was cloned into the pGL3-Basic Vector (Promega), which contains a luciferase gene, and was designated CRPV-pGL3-NCR.
The coding region of CRPV E2 (CRPV nt 3111 to 4284) was amplified by PCR using plasmid CRPV-pLAII as a template, with the upstream primer P3096 (5′-GAAGACGAGGGATCCGATGGAGGCTCTC-3′) and the downstream primer P4305 (5′-GGGTTACATGCATACAGGATCCTAAAGCCC-3′), each containing a BamHI restriction site. For CRPV E1 (CRPV nt 1362 to 3171), an upstream primer containing an EcoRI restriction site (5′-CCCGGAGTGTTGTAAGAATTCATGGCTG-3′) and a downstream primer with a BamHI restriction site (5′-CCAAACTCGTGCTCGGATCCTCATAGAG-3′) were used. Both fragments were cloned into the pSG5 expression vector (Stratagene), and the resulting constructs were designated E1-pSG5 and E2-pSG5, respectively. Site-specific mutagenesis of E2-pSG5 was performed by PCR with the oligonucleotides shown in Table 1 and with P3096 and P4305 as other primers. Mutations of amino acids (aa) 37 and 73 of E2 within the context of the whole CRPV genome were performed by site-specific mutagenesis-PCR with CRPV-pLAII as a template, using the oligonucleotide primers of Table 1 and two other primers, one upstream of the mutations introduced (5′-CAGCAAGAGTCAAACATTTGTTG-3′) and one downstream (5′-GTTTTCGTTGCTTTGCCGGCCTTC-3′). The 2-kb large PCR amplicon that contained the respective mutation was subcloned back into CRPV-pLAII by using the original SacI and MluI restriction sites of the wild-type (wt) clone. The mutants within the whole CRPV genome were designated E2-R37K-CRPV, E2-R37A-CRPV, E2-I73L-CRPV, and E2-I73A-CRPV. All PCR-amplified fragments were confirmed by DNA sequence analysis. Additionally, the complete E2-I73A-CRPV genome was sequenced and found to contain no mutations in comparison to the wt sequence except for the mutated site introduced.
TABLE 1.
Oligonucleotide sequences used for the generation of mutated CRPV-E2-pSG5 expression constructs
| Oligonucleotide | Sequencea | Mutated aa (total nt) |
|---|---|---|
| wt R37 | CACTGGAACTTACTAAGAAAAGAACAGGTCC | |
| E2-R37K | CACTGGAACTTACTAAAAAAAGAACAGGTCC | 37 (3205-3235) |
| E2-R37A | CACTGGAACTTACTAGCAAAAGAACAGGTCC | 37 (3205-3235) |
| wt E39 | AACTTACTAAGAAAAGAACAGGTCCTTTTAC | |
| E2-E39Q | AACTTACTAAGAAAACAACAGGTCCTTTTAC | 39 (3211-3241) |
| E2-E39A | AACTTACTAAGAAAAGCACAGGTCCTTTTAC | 39 (3211-3241) |
| wt I73 | TGTGCAAAGCAAGCCATAGAAATGGTGCTGTAC | |
| E2-I73L | TGTGCAAAGCAAGCCTTAGAAATGGTGCTGTAC | 73 (3313-3345) |
| E2-I73A | TGTGCAAAGCAAGCCGCAGAAATGGTGCTGTAC | 73 (3313-3345) |
Sequences are shown in the 5′→3′ direction. Nucleotide changes resulting in amino acid exchanges are underlined. The corresponding wt sequences are printed in bold italics.
Cell culture.
The SCC13 cell line was maintained as described previously (42). The sf1Ep cell line (ATCC CCL68) is an epithelial cottontail rabbit cell line that is maintained in a medium consisting of a 1:1 mixture of M199 and F12 (both from Life Technologies) supplemented with 1 nM dexamethasone, 100 pM 17β-estradiol, 1 nM T3, 10 ng of PGF2α/ml, 5 mg of insulin/liter (all from Sigma-Aldrich), 100,000 IU of penicillin-streptomycin (Life Technologies)/liter, and 0.588 g of sodium bicarbonate/liter. In addition, 5% fetal calf serum (Seromed, Berlin, Germany), 10 μg of mouse epidermal growth factor (Sigma Aldrich)/liter, 50 mM HEPES, and 20 mM NaOH are added.
Western blot analysis.
Approximately 5 × 105 sf1Ep cells were seeded into 60-mm-diameter dishes. The next day, cells were transfected with 1 μg of expression vector DNA in the presence of 15 μl of Lipofectamine (Life Technologies) in OptiMEM (Life Technologies) in accordance with the manufacturer's recommendations. Cells were harvested 48 h after transfection, and crude nuclear extracts were prepared as described previously (42). Briefly, cells were washed once with cold phosphate-buffered saline (PBS), scraped in 1 ml of cold PBS into a microcentrifuge tube, and pelleted by centrifugation (at 20,000 × g for 30 s at 4°C). The cell pellet was incubated for 5 min on ice in 250 μl of lysis buffer (10 mM HEPES [pH 7.9], 300 mM saccharose, 50 mM NaCl, 1.5 mM MgCl2, 0.25 mM EGTA, 0.5% [vol/vol] Igepal CA 630 [Sigma-Aldrich], 1 mM EDTA, 1 mM dithiothreitol [DTT], 0.5 mM sodium orthovanadate, 50 mM NaF, and protease inhibitor cocktail [Sigma Aldrich]). Nuclei were pelleted at 3,000 × g and 4°C for 5 min. The nuclear pellet was extracted on ice for 30 min with 30 μl of elution buffer (20% [vol/vol] glycerol, 10 mM HEPES [pH 7.9], 500 mM NaCl, 1.5 mM MgCl2, 0.1 mM EGTA, 0.1 mM EDTA, 1 mM DTT, 50 mM NaF, 0.5 mM sodium orthovanadate, and protease inhibitor cocktail [Sigma Aldrich]). The supernatant representing the crude nuclear extract was recovered by centrifugation (at 16,000 × g for 5 min at 4°C) in a microcentrifuge. Aliquots were snap-frozen and stored at −80°C.
Protein sample buffer was added to aliquots of the crude nuclear extracts that were equal in volume, and the samples were heated to 95°C for 5 min and then separated by sodium dodecyl sulfate (SDS)-12% polyacrylamide gel electrophoresis. Proteins were transferred to a nitrocellulose membrane (Hybond ECL; Amersham Pharmacia) in a buffer containing 10 mM 3-(cyclohexylamino)-1-propanesulfonic acid (CAPS) (pH 10.3) and 10% methanol at 70 V for 2 h. The membrane was blocked with 5% nonfat dry milk in Tris-buffered saline-0.1% Tween 20 (MTBST) overnight. Then it was incubated for 2 h at room temperature with a primary antibody diluted 1:1,000 in MTBST. To detect bound antibody, a swine anti-rabbit antibody coupled to horseradish peroxidase (Jackson Immunochemicals) was added at a 1:1,500 dilution in MTBST and the membrane was incubated for another hour. E2 proteins were detected by using the chemiluminescence reagent ECL (Amersham Pharmacia) and were visualized by use of autoradiography film (Hyperfilm ECL; Amersham Pharmacia). The rabbit polyclonal antiserum against CRPV E2 was provided by F. O. Wettstein (3).
Electromobility shift assay.
Electromobility shift assays were carried out with 20,000 cpm of a 32P-end-labeled double-stranded oligonucleotide containing an E2 binding site (nt 45 to 70 of HPV31). Binding reactions were carried out for 15 min on ice in a final volume of 20 μl containing equal volumes of the crude nuclear extracts described in the preceding section, the labeled oligonucleotide, and final concentrations of 10 mM HEPES (pH 7.9), 20% glycerol, 100 mM KCl, 1.5 mM DTT, and 2 μg of poly(dI-dC) (Amersham Pharmacia). Complexes were separated in a native 7% polyacrylamide gel (55 parts acrylamide to 1 part bisacrylamide) containing 0.25× Tris-borate-EDTA (1× Tris-borate-EDTA is 89 mM Tris base, 89 mM boric acid, and 2 mM EDTA). Gels were run at 200 V, dried, autoradiographed with an intensifying screen or exposed to storage screens, and then visualized with a Fuji BAS 1800 phosphorimager.
Transient luciferase expression assay.
Approximately 105 SCC13 cells were seeded into 35-mm-diameter dishes. The next day, cells were cotransfected with 200 ng of a luciferase reporter plasmid and 10 ng of pSG5 or the respective E2-pSG5 expression vector DNA in the presence of 5 μl of Lipofectamine (Life Technologies). Luciferase assays were carried out 48 h after transfection. Cells were washed twice with cold PBS and then lysed by addition of 150 μl of cold luciferase extraction buffer (0.1 M potassium phosphate [pH 7.8], 1% Triton X-100, 1 mM DTT). Lysates were cleared by centrifugation (at 20,000 × g for 5 min at 4°C), and 20- to 40-μl aliquots of extract were subjected to luminometer analysis as described in the manufacturer's manual. Transient luciferase expression assays were repeated with different plasmid preparations three to five times, each with duplicate measurements.
Transient replication analysis.
Approximately 5 × 105 SCC13 cells were seeded into 60-mm-diameter dishes. The next day, cells were transfected with 1 μg of E1-pSG5, 500 ng of CRPV-pGL3-NCR, and 200 ng of the respective E2-pSG5 expression vector in the presence of 15 μl of Lipofectamine. Cells were harvested 48 h after transfection, and low-molecular-weight DNA was purified as described previously (41). DNA aliquots of equal volumes were incubated with the restriction enzymes DpnI (5 U) and HpaI (15 U) (both from New England Biolabs) and 10 μg of RNAse A at 37°C for 5 h. The DNAs were separated on a 0.8% agarose gel.
Southern blot analysis.
DNA was transferred onto a nylon membrane (GeneScreen Plus; NEN Life Science Products) by capillary transfer with 0.4 M NaOH as a transfer buffer for 5 h. Eighty nanograms of linearized CRPV-pGL3-NCR served as a hybridization probe; the probe was labeled with 50 μCi of [α-32P]dCTP (specific activity, 6,000 Ci/mmol; Amersham Pharmacia) and Ready-To-Go DNA labeling beads (without dCTP) (Amersham Pharmacia) and was purified by NucTrapProbe (Stratagene). Hybridization of the probe to the DNA on the membrane was carried out with 10 × 106 cpm of the probe in 10 ml of hybridization buffer (4× SSPE [1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA {pH 7.7}], 5× Denhardt solution, 50% formamide, 1% SDS, 10% dextran sulfate, and 0.2 mg of salmon sperm [Life Technologies]/ml) overnight at 42°C. Then the membrane was washed twice in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1%SDS and twice in 0.1× SSC-0.1% SDS at room temperature, and twice in 0.1× SSC-1% SDS at 50°C. Afterward the blot was exposed to storage screens and visualized with a Fuji BAS 1800 phosphorimager.
Animal system.
Female New Zealand White rabbits were anesthesized with 25 mg of ketamine hydrochloride/kg of body weight and 5 mg of 2% Xylazine/kg. The backs of the rabbits were shaved with electric clippers and a razor blade. All sites to be infected were marked with black tattoo ink. CRPV virions (4.8 × 106 particles/μl in PBS provided by F. O. Wettstein) were introduced in aliquots of 5 μl per infection site into the rabbit skin by use of a tattoo needle. For infection via the “helios gene gun” (Bio-Rad), 1 μg of DNA was precipitated on 0.5 mg of gold as described by the manufacturer. Each injection site on the rabbit skin was bombarded 10 times with 1 μg of DNA by use of the gene gun and and 350 to 400 lb of helium pressure gas/in2 (47). Papillomas were removed surgically 3 and 6 months postinjection, snap-frozen, and stored at −80°C. DNA was isolated after proteinase K digestion in melting buffer (0.1 M EDTA [pH 8.0], 0.05 M Tris [pH 8.0], 0.5% [vol/vol] SDS) by phenol-chloroform extraction and ethanol precipitation. The coding region of the E2 gene containing the mutation site was amplified by PCR (35 cycles) with the upstream primer 5′-CAGCAAGAGTCAAACATTTGTTG-3′ and the downstream primer 5′-GTTTTCGTTGCTTTGCCGGCCTTC-3′. The amplicon was sequenced by using an internal primer (5′-GACGAGGGTGACGATGGAGGCTC-3′) and the BigDye Terminator DNA sequencing kit (ABI PRISM) and was analyzed in an ABI PRISM 310 Genetic Analyzer.
RESULTS
Mutation analysis of CRPV E2.
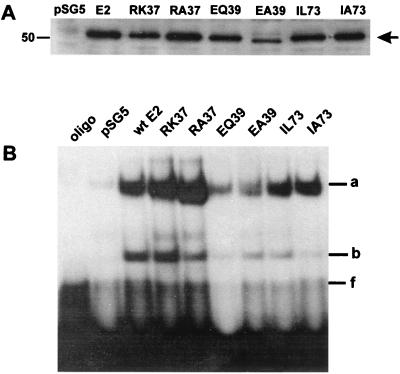
To determine the role of the transcription-modulating activities of the CRPV E2 protein in the development and/or progression of tumors in vivo, we first either mutated the amino acid residue at sequence position 37, 39, or 73 of the E2 gene to alanine (E2-R37A-pSG5, E2-E39A-pSG5, and E2-I73A-pSG5) or introduced a conservative change (E2-R37K-pSG5, E2-E39Q-pSG5, and E2-I73L-pSG5) in the background of the E2-pSG5 expression vector (Fig. 1; Table 1). To analyze the expression levels of mutant and wt E2 proteins, we transfected sf1Ep cells, a rabbit epithelial cell line, with 1 μg of wt CRPV E2-pSG5 or one of the mutant expression vectors, prepared crude nuclear extracts, and performed Western blot analysis. Using a polyclonal anti-CRPV E2 antiserum, we observed a single band, indicating the presence of E2 proteins, with a molecular size of approximately 50 kDa in transfected cells, which is in line with the calculated size of 45 kDa (Fig. 2A). This suggests that none of the mutations introduced into E2-pSG5 caused severe instability of the respective proteins. To investigate if the mutant E2 proteins are still able to bind DNA in a sequence-specific manner, we used the same nuclear extracts to perform electromobility shift assays with a 32P-labeled oligonucleotide that contains an E2 binding site. Two specific retarded complexes (Fig. 2B, bands a and b) were detected in extracts isolated from cells transfected with wt or mutant E2-pSG5 genes; these complexes most likely represent E2, since they were absent from vector-transfected cell extracts (Fig. 2B). The presence of two complexes rather than one could be due either to protein degradation or to posttranslational modifications of the E2 proteins. Taken together, these data suggest that mutation of amino acid 37, 39, or 73 of CRPV E2-pSG5 did not have a strong effect on protein stability or on the ability to bind DNA in a sequence-specific manner.
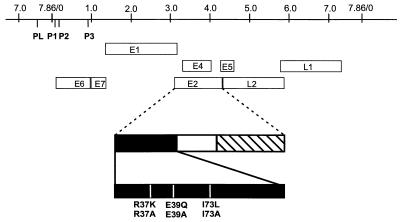
FIG. 1.
Linear map of CRPV. Open boxes, ORF. The early promoters P1, P2, and P3 and the late promoter PL are indicated. The structure of the CRPV E2 protein is diagrammed below. Solid rectangle, amino-terminal domain of E2, required for stimulation of DNA replication and transcription. Striped rectangle, carboxy-terminal domain, mediating sequence-specific DNA binding and dimerization of E2 proteins. Mutations resulting in amino acid changes of highly conserved residues (residues 37, 39, and 73) are marked by vertical white lines.
FIG. 2.
(A) Western blot analysis of nuclear extracts from sf1Ep cells transfected with CRPV wt E2-pSG5 or one of the E2 mutants R37K-, R37A-, E39Q-, E39A-, I73L-, and I73A-pSG5. The control lane contains an extract from cells transfected with the parental pSG5 vector. The position of the E2 proteins is indicated by an arrow. A molecular-size marker (in kilodaltons) is shown on the left. (B) Electromobility shift assay of CRPV wt and mutant E2 proteins present in nuclear extracts of transfected sf1Ep cells. Control lanes contained no protein extract (oligo) or an extract from cells transfected with the parental vector (pSG5). The retarded bands corresponding to the E2-DNA complex (a and b) and the unbound 32P-labeled oligonucleotide containing an E2 binding site (f) are indicated on the right.
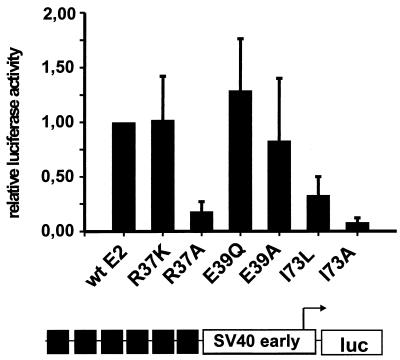
It has been shown that wt CRPV E2-pSG5 stimulates the transcriptional activity of reporter constructs containing E2 binding sites upstream of a minimal promoter element and of the CRPV regulatory region which contains multiple early and late promoters (14, 15). To investigate the transcription-modulating properties of the E2 mutants generated, SCC13 cells were cotransfected with the E2-responsive synthetic luciferase reporter plasmid p6xE2BS-luc and wt or mutant E2-pSG5 expression vectors. On average, the wt E2-pSG5 expression vector stimulated the basal luciferase activity of the reporter plasmid 60-fold over that with the empty expression vector. In contrast, cotransfection of the reporter plasmid with either the E2-R37A-pSG5 or the E2-I73A-pSG5 mutant stimulated luciferase activity only to 18 or 8% of the wt E2 level, respectively. Surprisingly, mutation of isoleucine at position 73 to leucine (E2-I73L-pSG5) reduced transactivation activity only to 33%, and the conservative change of arginine at position 37 to lysine (E2-R37K-pSG5) showed no effect at all (Fig. 3). In line with published data for other papillomavirus types (1, 7, 9, 13, 16, 17, 35, 42), mutation of amino acid 39 (E2-E39A-pSG5 and E2-E39Q-pSG5) had no significant effect on transactivation (Fig. 3).
FIG. 3.
Transient luciferase expression assays. SCC13 cells were transfected with expression vectors for CRPV wt E2 or the R37K, R37A, E39Q, E39A, I73L, or I73A E2 mutant protein and the E2-responsive reporter plasmid p6xE2BS-luc and were analyzed for luciferase activity. The reporter plasmid, diagrammed below the graph, consists of six E2-binding sites (solid boxes) upstream of the minimal simian virus 40 (SV40) early promoter, which drives the expression of the luciferase gene (luc). The luciferase activity obtained by cotransfection of each E2 mutant expression plasmid is given relative to the activity of wt E2-transfected cells, which was set to 1. Error bars, standard deviations.
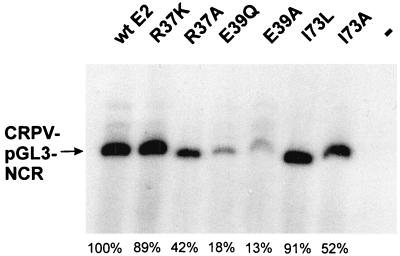
Next, we wanted to test if the E2-pSG5 mutants with impaired transactivation are capable of stimulating the E1-dependent replication of the CRPV origin. To this end, we performed a transient replication assay. SCC13 cells were cotransfected with a construct containing the complete NCR of CRPV (CRPV-pGL3-NCR), which includes the origin of replication mapped by Chiang et al. (8) together with an expression plasmid for the CRPV E1 protein (E1-pSG5) and with either wt E2-pSG5 or one of the E2 mutants E2-R37K-pSG5, E2-R37A-pSG5, E2-E39Q-pSG5, E2-E39A-pSG5, E2-I73L-pSG5, and E2-I73A-pSG5. Low-molecular-weight DNA was isolated, digested with the restriction enzyme DpnI in order to distinguish replicated DNA from transfected DNA, and analyzed by Southern blot analysis (Fig. 4). In agreement with earlier studies, replication of the CRPV origin was dependent on the presence of both E1 and E2 proteins. E2-pSG5 mutants E2-R37K-pSG5, E2-R37A-pSG5, E2-I73L-pSG5, and E2-I73A-pSG5 stimulated replication of the ori plasmid to 42 to 91% of the levels with wt E2-pSG5, whereas E2 mutant E2-E39Q-pSG5 or E2-E39A-pSG5 showed decreased abilities, stimulating replication to 18 or 13% of the E2-pSG5 wt levels, respectively (Fig. 4). In summary, these experiments demonstrated that the transactivation function of CRPV E2-pSG5 is highly dependent on residues R37 and I73 in the amino terminus and can be genetically separated from the replication function, as has been described for other papillomaviruses (1, 7, 9, 13, 16, 17, 35, 42).
FIG. 4.
Autoradiograph of a transient DNA replication assay. SCC-13 cells were transfected with plasmid CRPV-pGL3-NCR either alone (−) or together with an expression vector for CRPV E1 and an expression vector for CRPV wt E2 or the R37K, R37A, E39Q, E39A, I73L, or I73A mutant E2 protein. Low-molecular-weight DNA was extracted, digested with DpnI and HpaI, and analyzed by Southern blot hybridization. The position of DpnI-resistant CRPV-pGL3-NCR DNA is indicated by an arrow. Percentages given refer to the amounts of replicating plasmids of the respective E2 mutants in relation to the wt E2 protein (100%).
Transactivation by E2 is required for tumor induction in vivo.
Up to now, tissue culture experiments have provided contradictory results regarding the role of the E2 transactivation function in cell immortalization and transformation. Whereas immortalization of primary human keratinocytes by HPV31 was not impaired by the transactivation-negative HPV31 E2:IL-73 mutant, the corresponding BPV1 L73 mutant was unable to transform C127 cells (7, 42). To determine the role of the CRPV E2 transactivating function in tumor induction in the rabbit system, in a first experiment the I73A mutant, with highly impaired transactivation, was introduced into the context of the whole CRPV genome (E2-I73A-CRPV) and injected into the skin of four domestic rabbits. The skin on the back of each rabbit was divided into four rectangles, with eight injection sites within each rectangle. Two diagonal rectangles were injected either with wt CRPV-pLAII DNA or with whole genomic E2-I73A-CRPV DNA. Four additional sites on each rabbit were infected with CRPV particles. In contrast to wt CRPV-pLAII DNA, which induced papillomas in 85% of injected sites within 6 weeks, no papillomas developed at the sites injected with E2-I73A-CRPV DNA in the same time period. During follow-up, however, we observed the development of one small papilloma after 12 weeks and another after 24 weeks out of a total of 56 sites injected with E2-I73A-CRPV DNA (Table 2). In contrast to wt CRPV-induced tumors, no further significant increase in tumor size was observed for the two I73A-induced papillomas. The tumors were removed 6 months postinfection, DNA was isolated, and the presence of the I73A mutation was confirmed by direct sequencing after PCR amplification. A second experiment with a total of 40 infection sites each for I73A mutant DNA and wt DNA in five rabbits confirmed the inability of the I73A mutant to induce tumors even within 12 weeks (Table 2). To test for the tumor-inducing ability of the other E2 mutants, four more rabbits were injected with the DNAs of wt CRPV-pLAII, E2-I73L-CRPV, E2-R37A-CRPV, and E2-R37K-CRPV (Fig. 5). Again, 6 weeks after infection, no tumors had developed on the sites injected with the E2 mutants, whereas large papillomas were present at all sites injected with wt CRPV-pLAII DNA (Fig. 5 and Table 2).
TABLE 2.
Overview of tumor development in domestic rabbits after injection with CRPV virions, wt CRPV-pLAII DNA, or E2 mutant CRPV DNA
| No. of wks postinjection | Inoculum | No. of rabbits | No. of tumors/no. of sites (%) | Maximum avg diam (mm)a |
|---|---|---|---|---|
| 6 | ||||
| Expt 1 | Virus stock | 4 | 12/16 (75) | ND |
| wt DNA | 4 | 48/56 (85) | ND | |
| I73A mutant DNA | 4 | 0/56 (0) | ND | |
| Expt 2 | wt DNA | 5 | 40/40 (100) | ND |
| I73A mutant DNA | 5 | 0/40 (0) | ND | |
| Expt 3 | wt DNA | 4 | 32/32 (100) | ND |
| R37K mutant DNA | 4 | 0/32 (0) | ND | |
| R37A mutant DNA | 4 | 0/32 (0) | ND | |
| I73L mutant DNA | 4 | 0/32 (0) | ND | |
| 12 | ||||
| Expt 1 | Virus stock | 4 | 14/16 (88) | 14 |
| wt DNA | 4 | 42/56 (75)b | 23 | |
| I73A mutant DNA | 4 | 1/56 (1.8) | 2 | |
| Expt 2 | wt DNA | 5 | 40/40 (100) | 31 |
| I73A mutant DNA | 5 | 0/40 (0) | 0 | |
| 24 (expt 1) | Virus stock | 4 | 12/16 (75)b | Confluent |
| wt DNA | 4 | 42/56 (75) | Confluent | |
| I73A mutant DNA | 4 | 2/56 (3.6) | 3 |
ND, not determined. Confluent, tumor size could not be determined because tumors could not be separated from each other.
Decrease due to regression of papillomas in one rabbit.
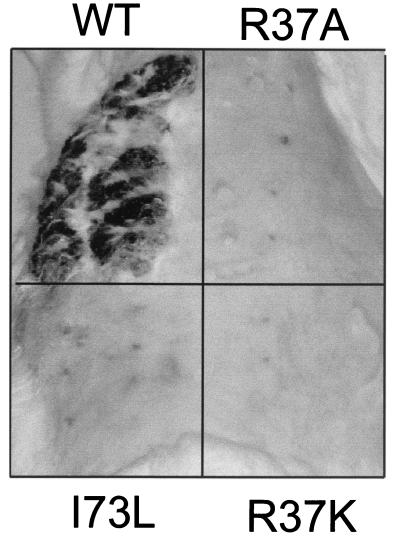
FIG. 5.
Photograph of the back of a rabbit 6 weeks after infection with CRPV-pLAII wt DNA (WT), E2-R37K-CRPV DNA (R37K), E2-R37A-CRPV DNA (R37A), or E2-I73L-CRPV DNA (I73L) by use of a gene gun. Papilloma development was visible only at sites injected with wt CRPV-pLAII DNA.
DISCUSSION
Previous work had suggested that all CRPV proteins, with the exception of E5 and L2, are required for tumor induction in the rabbit (5, 6, 26, 27, 46). In the case of E2, this finding was based on the fact that frameshift mutations resulted in truncated proteins lacking the dimerization and DNA binding domain, which is indispensable for almost all functions of E2 (24, 46). In this study we created single amino acid mutations aiming to knock out defined functions of E2. Following this strategy, we identified isoleucine at position 73 (I73) of CRPV E2 and arginine at position 37 (R37) as critical for transactivation of synthetic reporter constructs containing E2 binding sites, and we identified glutamic acid at position 39 (E39) as required for enhancing E1-dependent replication of plasmids containing a CRPV origin of replication. These results are in line with earlier findings described for the E2 proteins of other papillomaviruses (1, 7, 9, 13, 16, 17, 35, 42). Interestingly, the conservative change from arginine to lysine (R37K) had no effect on transcription stimulation by CRPV E2, as has been demonstrated for BPV1 E2, whereas the very same E2 mutants of HPV11 and -31 (7, 9, 42) were almost completely deficient in transactivation.
When we tested all transactivation-deficient E2 mutants embedded in the context of the whole genome of CRPV for tumor induction in rabbits, we noted, surprisingly, an almost complete loss of tumorigenic potential, even in the case of the transactivation-competent R37K mutant. Further studies are required to investigate the effect of the R37K E2 mutant on the homologous viral promoters in detail in order to better understand the behavior of this mutant in the animal.
Interestingly, a mutation in the context of the whole genome of BPV1 (BPV1 E2 L73) similar to our E2-I73L-CRPV causes a complete loss of transforming activity in C127 mouse fibroblast cells. This is most likely due, however, to diminished expression of the transforming proteins E6, E7, and E5, because the respective promoters P89 and P2443 are highly dependent on E2 stimulation (7, 18, 20, 30, 39). In contrast, the same mutation in the E2 gene of the HPV31 genome did not affect the immortalization of human keratinocytes (42). That may be explained by E2 C-terminus-mediated repression of the HPV31 P97 promoter, which is responsible for expression of the immortalizing viral oncogenes E6 and E7 and is not affected by mutations in the N terminus. However, to date it is not clear to what extent molecular events leading to cell transformation or cell immortalization in vitro contribute to the development of epithelial tumors in vivo. Based on previous findings, one would expect loss of the transactivating capability of E2 to coincide with deregulation of viral gene expression. Earlier findings demonstrated that shortly after infection of rabbits, high levels of E6 and E7 transcripts coincided with detectable levels of E2 transcripts in epithelial stem cells (37). High expression levels of E6 and E7 transcripts were also maintained in the process of tumor progression (48). Therefore, it is conceivable that loss of E2 transactivator functions may influence the efficiency of tumor induction and growth. However, no detailed information is yet available regarding the regulation of the four individual CRPV promoters by E2. So far, experiments have been carried out only with reporter constructs that carry multiple promoters. On the one hand, CRPV E2 repressed a reporter construct containing both the late promoter PL and the early promoter P1 (14). On the other hand, a reporter construct harboring the early promoter P2 in addition to PL and P1 was activated by E2 (15).
Interestingly, our animal experiments suggest that the transactivation domain of E2 is required not only for tumor induction but also for the extension of tumor growth, as indicated by the fact that the two papillomas induced by E2-I73A-CRPV showed highly retarded growth potential in addition to their late appearance.
In summary, we have shown here that the transactivation domain in the N terminus of CRPV E2, which functionally depends on amino acids R37 and I73, is essential for induction of papillomas in New Zealand White rabbits. This suggests that the ability of E2 to stimulate viral and/or cellular gene expression contributes to papillomavirus-mediated tumorigenesis.
Acknowledgments
This work was supported by a grant from the Wilhelm Sander-Stiftung (99.111.1) to T.I.
We thank Felix O. Wettstein for providing the CRPV stock and the anti-CRPV E2 antibody and Bettie Steinberg for technical advice regarding infection techniques.
REFERENCES
- 1.Abroi, A., R. Kurg, and M. Ustav. 1996. Transcriptional and replicational activation functions in the bovine papillomavirus type 1 E2 protein are encoded by different structural determinants. J. Virol. 70:6169-6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Androphy, E. J., D. R. Lowy, and J. T. Schiller. 1987. Bovine papillomavirus E2 trans-activating gene product binds to specific sites in papillomavirus DNA. Nature (London) 325:70-73. [DOI] [PubMed] [Google Scholar]
- 3.Barbosa, M. S., and F. O. Wettstein. 1988. E2 of cottontail rabbit papillomavirus is a nuclear phosphoprotein translated from an mRNA encoding multiple open reading frames. J. Virol. 62:3242-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernard, B. A., C. Bailly, M. C. Lenoir, M. Darmon, F. Thierry, and M. Yaniv. 1989. The human papillomavirus type 18 (HPV18) E2 gene product is a repressor of the HPV18 regulatory region in human keratinocytes. J. Virol. 63:4317-4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandsma, J. L., Z. H. Yang, S. W. Barthold, and E. A. Johnson. 1991. Use of a rapid, efficient inoculation method to induce papillomas by cottontail rabbit papillomavirus DNA shows that the E7 gene is required. Proc. Natl. Acad. Sci. USA 88:4816-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brandsma, J. L., Z. H. Yang, D. DiMaio, S. W. Barthold, E. Johnson, and W. Xiao. 1992. The putative E5 open reading frame of cottontail rabbit papillomavirus is dispensable for papilloma formation in domestic rabbits. J. Virol. 66:6204-6207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brokaw, J. L., M. Blanco, and A. A. McBride. 1996. Amino acids critical for the functions of the bovine papillomavirus type 1 E2 transactivator. J. Virol. 70:23-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiang, C. M., M. Ustav, A. Stenlund, T. F. Ho, T. R. Broker, and L. T. Chow. 1992. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc. Natl. Acad. Sci. USA 89:5799-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper, C. S., S. N. Upmeyer, and P. L. Winokur. 1998. Identification of single amino acids in the human papillomavirus 11 E2 protein critical for the transactivation or replication functions. Virology 241:312-322. [DOI] [PubMed] [Google Scholar]
- 10.Cripe, T. P., T. H. Haugen, J. P. Turk, F. Tabatabai, P. G. Schmid III, M. Durst, L. Gissmann, A. Roman, and L. P. Turek. 1987. Transcriptional regulation of the human papillomavirus-16 E6-E7 promoter by a keratinocyte-dependent enhancer, and by viral E2 trans-activator and repressor gene products: implications for cervical carcinogenesis. EMBO J. 6:3745-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiMaio, D., and J. Settleman. 1988. Bovine papillomavirus mutant temperature sensitive for transformation, replication and transactivation. EMBO J. 7:1197-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong, G., T. R. Broker, and L. T. Chow. 1994. Human papillomavirus type 11 E2 proteins repress the homologous E6 promoter by interfering with the binding of host transcription factors to adjacent elements. J. Virol. 68:1115-1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ferguson, M. K., and M. R. Botchan. 1996. Genetic analysis of the activation domain of bovine papillomavirus protein E2: its role in transcription and replication. J. Virol. 70:4193-4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujii, T., J. L. Brandsma, X. Peng, S. Srimatkandada, L. Li, A. Canaan, and A. B. Deisseroth. 2001. High and low levels of cottontail rabbit papillomavirus E2 protein generate opposite effects on gene expression. J. Biol. Chem. 276:867-874. [DOI] [PubMed] [Google Scholar]
- 15.Giri, I., and M. Yaniv. 1988. Study of the E2 gene product of the cottontail rabbit papillomavirus reveals a common mechanism of transactivation among papillomaviruses. J. Virol. 62:1573-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grossel, M. J., F. Sverdrup, D. E. Breiding, and E. J. Androphy. 1996. Transcriptional activation function is not required for stimulation of DNA replication by bovine papillomavirus type 1 E2. J. Virol. 70:7264-7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris, S. F., and M. R. Botchan. 1999. Crystal structure of the human papillomavirus type 18 E2 activation domain. Science 284:1673-1677. [DOI] [PubMed] [Google Scholar]
- 18.Haugen, T. H., T. P. Cripe, G. D. Ginder, M. Karin, and L. P. Turek. 1987. trans-Activation of an upstream early gene promoter of bovine papilloma virus-1 by a product of the viral E2 gene. EMBO J. 6:145-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hawley-Nelson, P., E. J. Androphy, D. R. Lowy, and J. T. Schiller. 1988. The specific DNA recognition sequence of the bovine papillomavirus E2 protein is an E2-dependent enhancer. EMBO J. 7:525-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hermonat, P. L., B. A. Spalholz, and P. M. Howley. 1988. The bovine papillomavirus P2443 promoter is E2 trans-responsive: evidence for E2 autoregulation. EMBO J. 7:2815-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirochika, H., T. R. Broker, and L. T. Chow. 1987. Enhancers and trans-acting E2 transcriptional factors of papillomaviruses. J. Virol. 61:599-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ilves, I., S. Kivi, and M. Ustav. 1999. Long-term episomal maintenance of bovine papillomavirus type 1 plasmids is determined by attachment to host chromosomes, which is mediated by the viral E2 protein and its binding sites. J. Virol. 73:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehman, C. W., and M. R. Botchan. 1998. Segregation of viral plasmids depends on tethering to chromosomes and is regulated by phosphorylation. Proc. Natl. Acad. Sci. USA 95:4338-4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McBride, A. A., H. Romanczuk, and P. M. Howley. 1991. The papillomavirus E2 regulatory proteins. J. Biol. Chem. 266:18411-18414. [PubMed] [Google Scholar]
- 25.Meyers, C., J. Harry, Y. L. Lin, and F. O. Wettstein. 1992. Identification of three transforming proteins encoded by cottontail rabbit papillomavirus. J. Virol. 66:1655-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nasseri, M., C. Meyers, and F. O. Wettstein. 1989. Genetic analysis of CRPV pathogenesis: the L1 open reading frame is dispensable for cellular transformation but is required for papilloma formation. Virology 170:321-325. [DOI] [PubMed] [Google Scholar]
- 27.Nasseri, M., and F. O. Wettstein. 1984. Differences exist between viral transcripts in cottontail rabbit papillomavirus-induced benign and malignant tumors as well as non-virus-producing and virus-producing tumors. J. Virol. 51:706-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Phelps, W. C., and P. M. Howley. 1987. Transcriptional trans-activation by the human papillomavirus type 16 E2 gene product. J. Virol. 61:1630-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phelps, W. C., S. L. Leary, and A. J. Faras. 1985. Shope papillomavirus transcription in benign and malignant rabbit tumors. Virology 146:120-129. [DOI] [PubMed] [Google Scholar]
- 30.Prakash, S. S., B. H. Horwitz, T. Zibello, J. Settleman, and D. DiMaio. 1988. Bovine papillomavirus E2 gene regulates expression of the viral E5 transforming gene. J. Virol. 62:3608-3613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rabson, M. S., C. Yee, Y. C. Yang, and P. M. Howley. 1986. Bovine papillomavirus type 1 3′ early region transformation and plasmid maintenance functions. J. Virol. 60:626-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rapp, B., A. Pawellek, F. Kraetzer, M. Schaefer, C. May, K. Purdie, K. Grassmann, and T. Iftner. 1997. Cell-type-specific separate regulation of the E6 and E7 promoters of human papillomavirus type 6a by the viral transcription factor E2. J. Virol. 71:6956-6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romanczuk, H., and P. M. Howley. 1992. Disruption of either the E1 or the E2 regulatory gene of human papillomavirus type 16 increases viral immortalization capacity. Proc. Natl. Acad. Sci. USA 89:3159-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romanczuk, H., F. Thierry, and P. M. Howley. 1990. Mutational analysis of cis elements involved in E2 modulation of human papillomavirus type 16 P97 and type 18 P105 promoters. J. Virol. 64:2849-2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakai, H., T. Yasugi, J. D. Benson, J. J. Dowhanick, and P. M. Howley. 1996. Targeted mutagenesis of the human papillomavirus type 16 E2 transactivation domain reveals separable transcriptional activation and DNA replication functions. J. Virol. 70:1602-1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarver, N., M. S. Rabson, Y. C. Yang, J. C. Byrne, and P. M. Howley. 1984. Localization and analysis of bovine papillomavirus type 1 transforming functions. J. Virol. 52:377-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmitt, A., A. Rochat, R. Zeltner, L. Borenstein, Y. Barrandon, F. O. Wettstein, and T. Iftner. 1996. The primary target cells of the high-risk cottontail rabbit papillomavirus colocalize with hair follicle stem cells. J. Virol. 70:1912-1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skiadopoulos, M. H., and A. A. McBride. 1998. Bovine papillomavirus type 1 genomes and the E2 transactivator protein are closely associated with mitotic chromatin. J. Virol. 72:2079-2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spalholz, B. A., P. F. Lambert, C. L. Yee, and P. M. Howley. 1987. Bovine papillomavirus transcriptional regulation: localization of the E2-responsive elements of the long control region. J. Virol. 61:2128-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spalholz, B. A., Y. C. Yang, and P. M. Howley. 1985. Transactivation of a bovine papilloma virus transcriptional regulatory element by the E2 gene product. Cell 42:183-191. [DOI] [PubMed] [Google Scholar]
- 41.Stenlund, A. 1996. Papillomavirus DNA replication, p. 679-698. In M. L. DePamphilis (ed.), DNA replication in eukaryotic cells. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Stubenrauch, F., A. M. Colbert, and L. A. Laimins. 1998. Transactivation by the E2 protein of oncogenic human papillomavirus type 31 is not essential for early and late viral functions. J. Virol. 72:8115-8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stubenrauch, F., T. Zobel, and T. Iftner. 2001. The E8 domain confers a novel long distance transcriptional repression activity on the E8∧E2C protein of high-risk human papillomavirus type 31. J. Virol. 75:4139-4149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ushikai, M., M. J. Lace, Y. Yamakawa, M. Kono, J. Anson, T. Ishiji, S. Parkkinen, N. Wicker, M. E. Valentine, I. Davidson, et al. 1994. trans-Activation by the full-length E2 proteins of human papillomavirus type 16 and bovine papillomavirus type 1 in vitro and in vivo: cooperation with activation domains of cellular transcription factors. J. Virol. 68:6655-6666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wettstein, F. O. 1987. Papillomaviruses and carcinogenic progression. I. Cottontail rabbit (Shope) papillomavirus, p. 167-186. In N. P. Salzman and P. M. Howley (ed.), The Papovaviridae, vol. 2. Plenum Publishing Corporation, New York, N.Y.
- 46.Wu, X., W. Xiao, and J. L. Brandsma. 1994. Papilloma formation by cottontail rabbit papillomavirus requires E1 and E2 regulatory genes in addition to E6 and E7 transforming genes. J. Virol. 68:6097-6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao, W., and J. L. Brandsma. 1996. High efficiency, long-term clinical expression of cottontail rabbit papillomavirus (CRPV) DNA in rabbit skin following particle-mediated DNA transfer. Nucleic Acids Res. 24:2620-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeltner, R., L. A. Borenstein, F. O. Wettstein, and T. Iftner. 1994. Changes in RNA expression pattern during the malignant progression of cottontail rabbit papillomavirus-induced tumors in rabbits. J. Virol. 68:3620-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]