Abstract
The DNA sequences of the Oka varicella vaccine virus (V-Oka) and its parental virus (P-Oka) were completed. Comparison of the sequences revealed 42 base substitutions, which led to 20 amino acid conversions and length differences in tandem repeat regions (R1, R3, and R4) and in an origin of DNA replication. Amino acid substitutions existed in open reading frames (ORFs) 6, 9A, 10, 21, 31, 39, 50, 52, 55, 59, 62, and 64. Of these, 15 base substitutions, leading to eight amino acid substitutions, were in the gene 62 region alone. Further DNA sequence analysis showed that these substitutions were specific for V-Oka and were not present in nine clinical isolates. The immediate-early gene 62 product (IE62) of P-Oka had stronger transactivational activity than the mutant IE62 contained in V-Oka in 293 and CV-1 cells. An infectious center assay of a plaque-purified clone (S7-01) from the V-Oka with 8 amino acid substitutions in ORF 62 showed smaller plaque formation and less-efficient virus-spreading activity than did P-Oka in human embryonic lung cells. Another clone (S-13) with only five substitutions in ORF 62 spread slightly faster than S7-01 but not as effectively as P-Oka. Moreover, transient luciferase assay in 293 cells showed that transactivational activities of IE62s of S7-01 and S7-13 were lower than that of P-Oka. Based on these results, it appears that amino acid substitutions in ORF 62 are responsible for virus growth and spreading from infected to uninfected cells. Furthermore, the Oka vaccine virus was completely distinguishable from P-Oka and 54 clinical isolates by seven restriction-enzyme fragment length polymorphisms that detected differences in the DNA sequence.
Varicella-zoster virus (VZV) is a human herpesvirus that causes chickenpox (varicella) and shingles (herpes zoster). A live attenuated varicella vaccine, the Oka strain was originally developed by Takahashi et al. in Japan (56) and is routinely used in children in Japan and other countries, including the United States. Clinical symptoms caused by this live vaccine are very rare in healthy children. Although the Oka vaccine virus (V-Oka) is an avirulent virus, its parental virus (P-Oka), isolated from a patient with typical varicella, is thought to be virulent in vivo. It has not been clarified which gene(s) is involved in the pathogenicity of VZV infection. Thus, comparison of the complete genomes of V-Oka and P-Oka should reveal correlations between DNA sequence and virulence.
The complete DNA sequence of the VZV Dumas strain was first determined by Davison and Scott (9). The genome is a linear double-stranded DNA of ca. 125,000 bp and consists of unique long regions (ULs) flanked by terminal repeat long (TRL) and internal repeat long (IRL) inverted repeat regions, and a unique short (US) region flanked by internal repeat short (IRS) and terminal repeat short (TRS) inverted repeat regions. The genome contains ca. 70 open reading frames (ORFs), three of which exist in both IRS and TRS regions; genes 62 through 64 correspond to genes 69 through 71.
We previously determined the sequences of several genes from V-Oka and P-Oka and found differences between them (15, 16). As many as 15 base substitutions, representing 8 amino acid differences, were found in gene 62, located within the IRS region (or in gene 71 located within the TRS region) and its flanking regions, although no mutations were found in genes encoding other transactivators, such as genes 4, 10, 61, and 63. In addition to our report, another author reported that the DNA sequences of the P-Oka virus and a cosmid clone from the vaccine virus were different in gene 62 (2). The immediate-early gene 62 product (IE62) is an immediate-early (IE) protein consisting of 1,310 amino acids and is functionally conserved with herpes simplex virus type 1 (HSV-1) ICP4 (13). High levels of IE62 are associated with viral tegument (24) and a recombinant HSV-1 with both copies of ICP4 replaced by VZV IE62 has been constructed (11). IE62 can transactivate all three putative kinetic classes of VZV gene promoters—i.e., the IE, early, and late gene promoters (5, 10, 15, 16, 22, 38, 47, 52)—and can transrepress its own promoter (5, 12, 49). Production of infectious VZV generated by the transfection of purified viral DNA is strongly increased by the addition of an IE62-expressing plasmid, and it has been hypothesized that virion-associated IE62 may play a crucial role in stimulating the IE events upon infection (44). In our previous study, the transcriptional activity of the P-Oka IE62 was higher than that of V-Oka IE62, which possessed all eight amino acid substitutions, on all classes of VZV gene promoters, leading us to hypothesize that IE62 might play an important role in the VZV replicative cycle and, moreover, in the attenuation of VZV (15, 16). However, it is possible that other substitution(s) are responsible for the pathogenicity of VZV, since the DNA sequence comparisons between the Oka viruses have only been done for several genes, but not for the complete genome. To resolve this issue, the complete DNA sequence of both viruses was needed.
We determined here the complete genome sequences of the Oka viruses and found remarkable differences in some of the VZV genes. The present study provides the first step toward investigating the genes involved in VZV attenuation. Furthermore, new methods for distinguishing V-Oka from clinical isolates, including P-Oka, were established, based on the newly identified substitutions. These methods can be used for typing VZV strains and for quality control of the Oka varicella vaccine.
MATERIALS AND METHODS
Cells and viruses.
P-Oka was isolated in Japan from a patient with varicella and was propagated only in human embryonic lung (HEL) cells. A live, attenuated varicella vaccine (BIKEN, lot V-65), manufactured by the Research Foundation for Microbial Diseases of Osaka University, was used as V-Oka. Virus strains used as clinical isolates in the present study were isolated in Japan from patients with varicella (Kawaguchi, Inoue, RT-283, RT-342, and OZ-4) or zoster (Z-4, Z-5, Z-8, or RT-345) who had never been vaccinated. Strain RT-342 was isolated from the vesicles of a patient 10 days after vaccination, and strain RT-345 was isolated from the vesicles of a patient with shingles 19 months after vaccination. Clinical samples were isolated by the inoculation of HEL cells with vesicular fluids from patients and were propagated by three to five passages in HEL cells or MRC-5 cells.
PCR.
Genomic DNA was extracted from the infected cells as described previously (42). Approximately 500 primers (25 bp in length) were designed by referring to the nucleotide sequence of the Dumas strain (9). Almost all regions covering each genome were amplified as 43 overlapping pieces by PCR. The reaction mixtures for PCR contained 2.5 U of Ex Taq (Takara Shuzo Co., Kyoto, Japan), 200 μM concentrations of each deoxynucleoside triphosphate, 0.3 mM concentrations of each primer, and an extremely low amount of template DNA in 50 μl of Ex Taq buffer. Thirty cycles of amplification were performed, in which each cycle consisted of a denaturation step at 94°C for 1 min, an annealing step at 55°C for 1.5 min, and an extension step at 72°C for an appropriate period (1 min/kbp) in a thermal cycler (Takara Shuzo). For only the regions containing the R3 reiteration region and gene 62, the PCR conditions were as described above except that the denaturation time and the annealing time were each changed to 30 s. For an origin of replication region only, the amplified DNA fragments were obtained by shuttle PCR by using 2.5 U of LA Taq polymerase (Takara Shuzo), 200 μM concentrations of each deoxynucleoside triphosphate, 0.3 μM concentrations of each primer (32 bp in length), and an extremely low amount of template DNA in 50 μl of LA Taq buffer. Shuttle PCR conditions were follows: an incubation at 98°C for 2 min for the hot start and 30 cycles of 98°C for 30 s and 68°C for 2.5 min. All amplified products were purified by using a QIA PCR purification kit (Qiagen) to remove the primers.
Cloning of PCR products.
Determinations of the DNA sequences at both ends of the genomes, the flanking regions between the internal repeat long and IRS regions, and the R3 repeat were difficult to obtain by direct sequencing of the PCR products as described below, implying that various clones containing different sequences existed. Thus, PCR products containing the regions were individually inserted into a pCR2.1 vector and then transformed into competent Escherichia coli TOP10 5′ cells by the TA cloning method (Invitrogen). The plasmid DNAs were purified from cultured cells by using the QIA Prep Spin Kit (Qiagen).
Sequencing.
Direct sequencing of purified PCR products and plasmids was performed by using the BigDye-terminator cycle sequencing kit and a Genetic Analyzer 310 (both from Perkin-Elmer/Applied Biosystems, Chiba, Japan). The viral sequences were compiled by using the DNASIS computer programs (Hitachi Computer Engineering, Tokyo, Japan).
Plasmids.
Two effector plasmids were constructed for a transient luciferase assay to analyze the transcriptional activity of ORF 10 protein containing an amino acid substitution, which was caused by a partial base substitution within the HindIII site. A DNA fragment containing ORF 10 from V-Oka was amplified by PCR under the conditions described above. The amplified product was inserted into the pCR2.1 vector by the TA cloning method and then checked for the direction of the inserted fragment and whether the HindIII site was present or not. Plasmids pCG10(−) and pCG10(+) were constructed by inserting the KpnI-XhoI fragment from clones obtained by TA cloning into pCDNA3.1 (Invitrogen) containing the cytomegalovirus IE promoter. The HindIII site existed in pCG10(−), which bore the ORF 10 from P-Oka, but not in pCG10(+), which contained the ORF 10 present in V-Oka.
Five kinds of effector plasmids expressing IE62s were constructed. pPar-G62 and pVac-G62, which had eight amino acids that were different from each other, were selected from the clones and used for our previous study (15). After pPar-G62 and pVac-G62 were digested by NcoI and PstI (Takara Shuzo), pPV-G62Nco and pVP-G62Nco were constructed by connecting the small fragment from the digested pPar-G62 and pVac-G62 and the large fragment from the digested pVac-G62 and pPar-G62, respectively. pVmut4 was also selected from the clones from V-Oka; this clone had four amino acid differences in ORF 62 compared to pPar-G62. The reporter plasmid pPol-Luc contained ca. 750 bases upstream from the initiation codon of gene 29. This was constructed by transferring the NheI-BglII fragment from plasmid pPol-CAT as described previously (15) to the region upstream of the firefly luciferase gene in pGL3-Basic (Promega).
Transfection.
To analyze the transactivational activity of the ORF 10 proteins, CV1 cells, which were cultured in 35-mm plastic dishes at 105 cells/dish, were transfected by lipofection with SuperFect (Qiagen). Each reaction mixture for transfection consisted of 0.5 μg of the reporter plasmid, various amounts (0 to 2 μg) of either effector plasmid pCG10(+) or pCG10(−), and 0.25 μg of β-galactosidase-expressing plasmid pCH110 (Amersham Pharmacia) for normalizing the transfection efficiency between different dishes. The total amount of plasmid DNA in each transfection experiment was kept constant at 2.75 μg by adding the vector pCDNA3.1.
To analyze the transactivational activity of the different IE62s, 293 cells and guinea pig embryonic (GPE) cells (derived from the fourth week of pregnant guinea pigs), which were cultured in 35-mm plastic dishes at 105 cells/dish, were transfected with the SuperFect and Lipofectamine Plus reagents (Gibco-BRL), respectively. Each reaction mixture for transfection consisted of 0.25 mg of the reporter plasmid, the same amount of one of five effector plasmids that express IE62s, and 2 mg of the vector pUC19. All experiments were repeated at least three times independently.
Luciferase and β-galactosidase assays.
At 24 h after transfection, the cells were washed twice with phosphate-buffered saline. The cells were then lysed by the addition of 100 μl of reporter lysis buffer (Promega), followed by mixing for 15 min at room temperature. The lysate was collected in a new microcentrifuge tube, and the cell debris was removed by centrifugation at 12,000 rpm for 2 min. The luciferase activity of 10 μl of supernatant was determined by using a Berthold luminometer, model LB9507 (Berthold Japan, Tokyo, Japan), after the addition of 50 μl of substrate buffer (Promega). To normalize this transient assay for ORF 10 proteins to the transfection efficiency, the β-galactosidase activity of 10 μl of supernatant was also determined by using the luminometer, after the addition of 100 μl of the reaction buffer of the luminescent β-galactosidase detection kit II (Clontech) and incubation of the mixture at room temperature for 1 h. In another transient assay for various IE62s, the luciferase activity in each lysate was standardized to the protein concentration determined with the Bio-Rad protein reagent, because all of the internal plasmids used to normalize the transfection efficiency were stimulated by the IE62s.
Comparison of plaque sizes and infectious center assay.
HEL cells were infected with cell-free V-Oka, and virus clones were plaque purified twice. Two clones, S7-01 and S7-13, were obtained after DNA sequencing for use in further experiments. These clones were then propagated in HEL cells, and cell-free viruses were harvested by sonication as described previously (15). HEL or GPE cells were infected with the cell-free cloned viruses, P-Oka, and V-Oka, and their plaque sizes were compared after fixation and staining with methylene blue. An infectious center assay was also performed to examine the spreading of the viruses. HEL or GPE cells in 35-mm dishes were infected with cell-free viruses (ca. 50 PFU/plate). At 0 to 5 days postinfection, they were washed with phosphate-buffered saline (37°C) and treated with trypsin. The trypsin-treated cells were diluted and transferred onto monolayers of HEL cells in 35-mm dishes, and the numbers of infected cells were assessed by counting the plaques 7 days after the inoculation.
Restriction fragment length polymorphism (RFLP) analysis.
Six parts of the genome were amplified by PCR with six sets of primer pairs as shown in Table 1. The conditions for the PCR were as described above. Portions (3 μl) of the amplified products with three primer pairs—01-N12 and 01-R13, 50-N15 and 50-R17, or 60-N11 and 60-R12—were digested with 4 U of the restriction endonucleases AluI (Takara Shuzo), BstXI (Takara Shuzo), or BsrI (New England Biolabs), respectively. The same volumes of the products obtained by using primer pair 60-N06 and 60-R06 and primer pair 60-N26 and 60-R28 were digested independently with 4 U of the enzymes SfaNI (New England Biolabs)-AccII (Takara Shuzo) and SacII (New England Biolabs)-SmaI (New England Biolabs), respectively. Samples were digested for 1 h at 25°C for SmaI; 37°C for AluI, SfaNI, and SacII; 45°C for BstXI; 60°C for AccII; and 65°C for BsrI. The digested products were analyzed by electrophoresis in a 4% agarose gel (NuSieve 3.1; FMC BioProducts Co., Rockland, Maine) with ethidium bromide staining and then visualized with short-wave UV illumination of the gel.
TABLE 1.
Primers used in the RFLP analysis in this studya
| Name | Sequence | Position (nucleotide range) |
|---|---|---|
| 01-N12 | 5′-ATTGTATGCATGCGATTGCTATCGC-3′ | 5372-5396 |
| 01-R13 | 5′-GGTCTTCCACTTTAAAGGGGTTTGC-3′ | 6134-6110 |
| 50-N15 | 5′-CGATCACGTCGCTCACATCCAACCC-3′ | 93685-93709 |
| 50-R17 | 5′-ATGGCAGAAGAAACACGTATTGCCG-3′ | 94457-94433 |
| 60-N06 | 5′-GAGGACAACAGCTCCACCTTGACCG-3′ | 105277-105301 |
| 60-R06 | 5′-GAGTAATGTGGCCGCCCGGTTTTGG-3′ | 105613-105589 |
| 60-N26 | 5′-CCAAAACCGGGCGGCCACATTACTC-3′ | 105589-105613 |
| 60-R28 | 5′-ATTACTGTCGACCCGAGACCTGGCC-3′ | 106380-106356 |
| 60-N11 | 5′-AAGGGCTTCCGTCGGGCATCATGAG-3′ | 107729-107753 |
| 60-R12 | 5′-TCGGGTAAAAAGCCGGGCGATGAGC-3′ | 108497-108473 |
These primers were designed by referring to the nucleotide sequence of the Dumas strain. Nucleotide positions refer to those in the sequence of the Dumas strain.
RESULTS
Comparison of the complete DNA sequences of V-Oka and P-Oka. (i) Substitutions of bases and amino acids.
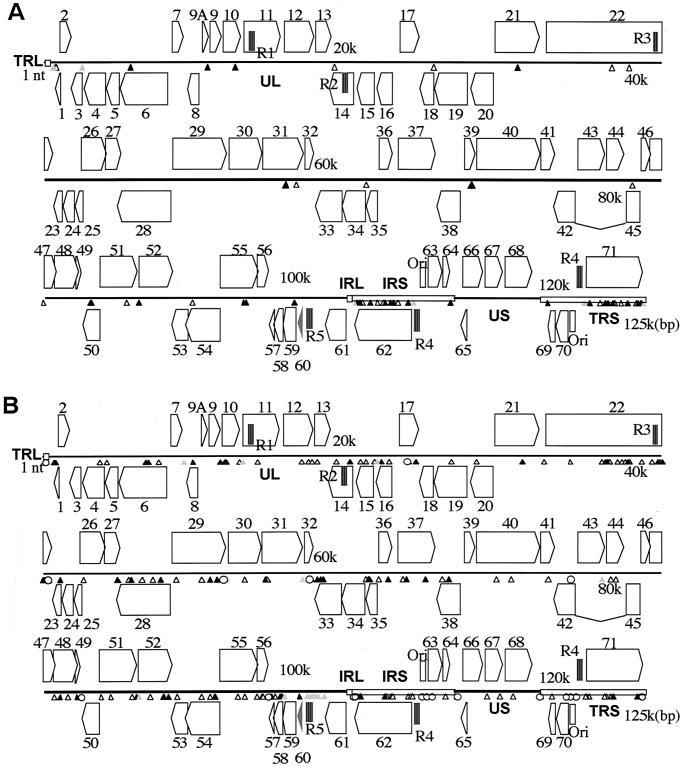
The entire VZV DNA sequences of V-Oka and P-Oka were determined mainly by the direct sequencing of PCR products. Thus, the determined sequences reflect the VZV DNA population in infected cells; that is, if mutated viruses also exist within a sample, more than two kinds of nucleotides would be recognized at one base position by this method. The genome sizes of both the Oka viruses were ca. 125 kbp, and >95% of the sequences were determined by this method on both the sense and antisense strands of the DNAs. When the complete DNA sequences of the Oka viruses were compared, only 42 bases were found to be different (Fig. 1A and Table 2), not including the substitutions in the TRS region, which are duplications of the IRS region. Two base substitutions were found in genes 10 and 14 for the first time in the present study, although we had missed them in our earlier study (15, 16). Base substitutions at 20 nucleotide positions in V-Oka caused amino acid conversions, and the base substitutions at 17 nucleotide positions were silent mutations. The remaining five base substitutions were found to be in noncoding regions. Thirty-one base substitutions in V-Oka were mixtures of two kinds of base at each of these positions (Table 2). Every base substitution was a G-versus-A or a C-versus-T conversion except for a G-versus-T conversion only at position 39227 in gene 22. Fifteen amino acid substitutions in V-Oka were partial, resulting in mixtures of two kinds of amino acids at each of these positions. Surprisingly, more than one-third (15 of 42) of the substitutions were found in gene 62 (or gene 71) and its flanking regions, and about half of the amino-acid substitutions (8 of 20) were within ORF 62. Thus, the gene 62 region was obviously the most variable region in the V-Oka virus. The major transactivating domains of IE62 are divided into five regions (regions 1 to 5) based on sequence homology in comparison with HSV-1 ICP4 (9, 35). Regions 4 and 5 contained one and three amino acid substitutions each, respectively.
FIG. 1.
Comparisons of complete DNA sequences. The genome is presented in sections; horizontal lines indicate unique regions, and open boxes indicate inverted repeat regions. ORFs 1 to 71 are illustrated as pentagons. ORFs 62 through 64 are duplications of ORFs 69 through 71. Solid, shaded, and open symbols show the base substitutions causing amino acid conversions, base substitutions in noncoding regions, and silent mutations, respectively. (A) Forty-two base substitutions between P-Oka and V-Oka. (B) One hundred seventy-six base substitutions between the Dumas virus and P-Oka. All 176 substitutions between the Dumas virus and P-Oka are contained within 218 substitutions between the Dumas virus and V-Oka; the 218 substitutions between the Dumas virus and V-Oka were the total of 176 plus 42 substitutions.
TABLE 2.
Base and amino acid substitutions found by comparing the complete DNA sequences of V-Oka and P-Okaa
| Positionb | Gene | Amino acid substitution(s)
|
|
|---|---|---|---|
| V-Oka | P-Oka | ||
| 560 | 5′ noncoding | C (NCR) | T (NCR) |
| 703 | 1 | T/C (Gln) | T (Gln) |
| 763 | 1 | T/C (Pro) | T (Pro) |
| 2515 | 3/4 | T/C (NCR) | T (NCR) |
| 5745 (945) | 6 | G (Pro) | A (Ser) |
| 10900 (91) | 9A | T/C (Trp/Arg) | T (Trp) |
| 12779 (207) | 10 | T/C (Ala/Val) | T (Ala) |
| 19431 | 14 | T/C (OCH/AMB) | T (OCH) |
| 26125 | 18 | G (Asn) | A (Asn) |
| 31732 (325) | 21 | T/C (Thr/Ile) | C (Thr) |
| 38036 | 22 | T/C (Thr) | T (Thr) |
| 39227 | 22 | T/G (Pro) | T (Pro) |
| 58595 (530) | 31 | A/G (Ile/Val) | A (Ile) |
| 59287 | 31 | A/G (Pro) | A (Pro) |
| 64067 | 35 | A/G (Ala) | A (Ala) |
| 71252 (207) | 39 | T/C (Met/Thr) | T (Met) |
| 82225 | 45 | A/G (Pro) | A (Pro) |
| 84091 | 47 | A/G (Glu) | G (Glu) |
| 87280 | 50 | A/G (Cys) | A (Cys) |
| 87306 (193) | 50 | T/C (Ser/Gly) | T (Ser) |
| 89734 | 51 | A/G (Thr) | A (Thr) |
| 90535 (15) | 52 | A/G (Ile/Val) | A (Ile) |
| 94167 | 54 | C (Leu) | T (Leu) |
| 97748 (585) | 55 | G/A (Ala/Thr) | G (Ala) |
| 97796 (601) | 55 | T/C (Cys/Arg) | T (Cys) |
| 101089 (44) | 59 | A/G (Leu/Pro) | A (Leu) |
| 105169 | 61/62 | A/G (NCR) | A (NCR) |
| 105310 (1275) | 62 | A/G (Leu/Ser) | A (Leu) |
| 105356 (1260) | 62 | C (Val) | T (Ile) |
| 105544 (1197) | 62 | G (Ala) | A (Val) |
| 105705 | 62 | C (Ala) | T (Ala) |
| 106262 (958) | 62 | C (Gly) | T (Arg) |
| 106710 | 62 | A/G (Ala) | A (Ala) |
| 107136 | 62 | C (Ala) | T (Ala) |
| 107252 (628) | 62 | C (Gly) | T (Ser) |
| 107599 (512) | 62 | A/G (Val/Ala) | A (Val) |
| 107797 (446) | 62 | A/G (Leu/Pro) | A (Leu) |
| 108111 | 62 | C (Pro) | T (Pro) |
| 108838 (99) | 62 | A/G (Met/Thr) | A (Met) |
| 109137 | 62/63 | A/G (NCR) | A (NCR) |
| 109200 | 62/63 | A/G (NCR) | A (NCR) |
| 111650 (29) | 64 | A/G (Gln/Arg) | A (Gln) |
Mutations in the repeat region are not shown here (see Fig. 2). The number at the base position refers to the sequence analysis of the Dumas virus. Base substitutions leading to amino acid conversions are indicated by filled cells. P-Oka has a base identical to that in the Dumas virus at every position. NCR, noncoding region; OCH, ocher; AMB, amber.
Numbers in parentheses refer to the amino acid number within a particular ORF.
There were 12 amino acid substitutions between the Oka viruses in the entire genome outside of gene 62. These existed in genes 6, 9A, 10, 21, 31, 39, 50, 52, 55, 59, and 64 (Fig. 1A and Table 2). The HSV-1 helicase-primase complex consists of three proteins encoded by UL52, UL8, and UL5 (8); their homologs in VZV are ORFs 6, 52, and 55, respectively. Interestingly, four amino acid substitutions were found in these proteins. One amino acid substitution was located near the carboxyl terminus of gene 6, which was Ser in P-Oka and Gly in V-Oka. Another substitution occurred in gene 52. In this case, the amino acid residue in P-Oka was Ile, which was incompletely altered to Val in V-Oka; V-Oka contained two kinds of base at a single position, resulting in two kinds of ORF 52 protein. The remaining two amino acid substitutions were located in ORF 55. An Ala at position 97748 and a Cys at position 97796 in P-Oka were partially substituted by Thr and Arg in V-Oka, respectively.
There was only one amino acid substitution among the seven known VZV surface glycoproteins gK, gC, gB, gH, gL, gI, and gE, encoded in genes 5, 14, 31, 37, 60, 67, and 68, respectively. gB contained an amino acid substitution in which an Ile in P-Oka was substituted by Val in about half of the virus population of V-Oka. Another substitution was found in ORF 50, which is homologous to the gene encoding the HSV-1 glycoprotein gM, but it has not been established whether gM exists and is functional in VZV. Gene 9A is homologous to HSV-1 UL49.5, which encodes a hydrophobic protein with only 91 amino acids (4). The gene product in VZV, the ORF 9A protein, is present in the membrane fraction of infected MeWo cells, although it is absent in the cytosolic fraction, and cells infected with a recombinant virus that is unable to express the ORF 9A protein produce smaller plaques than cells infected with the parental virus (50). A partial amino acid substitution was located at the end of the carboxyl terminus of ORF 9A protein. It is possible that this substitution affects the stability of the protein. ORF 10 protein is a tegument protein (24) that regulates positively to the IE62 gene promoter, and V-Oka contained both Ala and Val at a single position (i.e., 12779) in the middle of the protein. In the gene 21 protein, whose function is unknown, Thr in P-Oka was incompletely substituted by Ile in V-Oka as a result of a base substitution at position 31732. Amino acid substitutions also existed in genes 39 and 64. The HSV-1 homologue to the gene 39 product is the UL20 product, which is known to be a membrane protein, and the homologue to gene 64 is the US10 product, which is known to be a tegument protein, but the function of either of these products in VZV is not clear. Finally, an amino acid substitution was found in the gene 59-encoded uracil-DNA glycosylase, which is a DNA repair enzyme responsible for the removal of uracil residues misincorporated into DNA.
The DNA sequence differences of the Oka viruses were then compared with the Dumas strain. The G+C content was 46.1% for both V-Oka and P-Oka; this value is very close to that of the Dumas strain (46.0%). When the complete DNA sequences of V-Oka and P-Oka were compared with the Dumas strain, 213 and 171 base substitutions leading to 75 and 55 amino acid conversions, respectively (Fig. 1B), were found. All 176 substitutions between the Dumas virus and P-Oka are contained within 218 substitutions between the Dumas virus and V-Oka; 218 substitutions between the Dumas virus and V-Oka were the total of 176 plus 42 substitutions. In the entire DNA sequences, excluding the repeat regions, the mutation rates of P-Oka and V-Oka were 0.16 and 0.19% compared with the Dumas strain, respectively. Thus, P-Oka is genetically slightly closer to the Dumas strain than is V-Oka. Several regions in gene 22 and genes 55 through 61 were variable between the Dumas strain and P-Oka (Fig. 1B). The gene 62 region was an additional variable region when the Dumas strain was compared with V-Oka; 24 base substitutions existed in gene 62 and its flanking regions between V-Oka and the Dumas strain. At the amino acid level, the mutation rate in all of the ORFs between the Dumas strain and P-Oka was 0.16%, whereas that between the Dumas strain and V-Oka was 0.21%. V-Oka ORF62 possessed the greatest mutation rate (0.84%) among all of the ORFs compared with the Dumas strain.
(ii) Deletions and insertions in V-Oka.
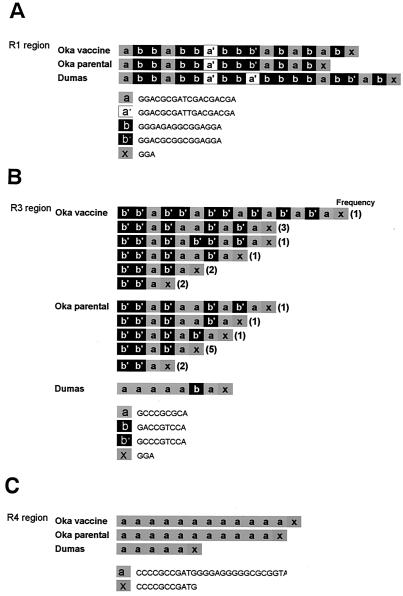
V-Oka had several deletions and insertions compared to P-Oka in certain regions, including the tandem repeat regions and an origin of DNA replication. Five repeat regions—R1, R2, R3, R4, and R5—lie in genes 11, 14, and 22, and in the flanking regions between genes 62 and 63 (and genes 70 and 71), and genes 60 and 61, respectively. There were length differences in regions R1, R3, and R4. In particular, differences in the length of the R1 region have been used to distinguish different VZV strains (18, 62). R1 is composed of four kinds of allele, i.e., a, a′, b, and b′, which are 15 or 18 bp in length and partial copies of them, x, which are 3 bp in length. V-Oka had 17 reiteration elements in the R1 region, whereas P-Oka had 15 elements (Fig. 2A). The R3 region consists of three kinds of allele, a, b, and b′, which are 9 bp in length, and partial copies of them, x, which are 3 bp. The DNA sequences of the R3 regions from both Oka viruses could not be completely determined by the direct sequencing of the PCR products. The sequences could be determined to just before the fourth element, b′b′a, with sense primers, and the last three elements, b′ax, with antisense primers. These results indicated that the R3 regions contained mixtures of multiple sequences from the fourth element to just before the last three elements. Thus, the R3 region appeared to be unstable, and each virus was represented by more than two clones containing different numbers and combination patterns in this region. Sequencing analysis of fragments containing the R3 region of each virus revealed that the numbers and types of combination patterns varied (Fig. 2B). All of the clones from the viruses had more than four elements. The maximum number of reiteration elements that we detected from V-Oka was 16, whereas that from P-Oka was 11. However, Davison and Scott reported that the R3 region might contain over 100 copies of the 9-bp element (9). Although we amplified the fragment containing the R3 region under various PCR conditions, we did not obtain any fragment containing this many elements. No frameshifts occurred in spite of the differences in the R1 and R3 regions located within the ORFs because the base numbers of elements were multiples of 3. R4 is located in both the IRS and TRS regions and consists of a single element, a, which is 27 bp in length, and a partial copy of it, x, which is 11 bp in length. V-Oka had 9 repeat elements in the R4 region, whereas P-Oka had 10 elements (Fig. 2c).
FIG. 2.
Comparisons of repeat regions. V-Oka has different structures in R1 (A), R4 (B), and R3 (C) compared to P-Oka. Both V-Oka and P-Oka contain multiple clones that have different patterns and combinations of repeat elements in R3.
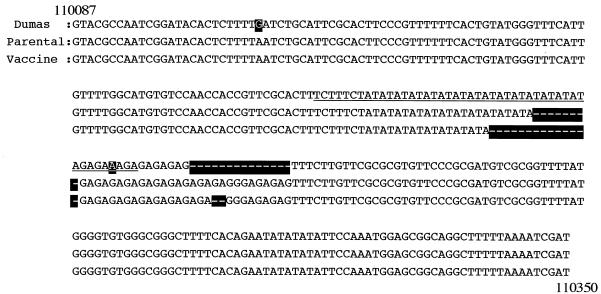
Finally, the VZV genome contains origin regions of replication in both the IRS and the TRS regions, and each origin region is ca. 270 bp in length (Fig. 3). P-Oka contained 269 bp in this region and was 8 bp longer than V-Oka. The Dumas strain has an almost perfect palindrome of 45 bp in the middle of the origin region (Fig. 3, underline) (9, 54), and both Oka viruses had additional sequence following the palindrome. P-Oka had part of the palindromic sequence (TA)12 and just following (GA)10, whereas V-Oka had a different sequence (TA)9 and (GA)9. The genome size of V-Oka was slightly longer than that of P-Oka due to these insertions and deletions. The whole genomes of V-Oka and P-Oka consisted of 125,076 and 125,069 bp, respectively.
FIG. 3.
Comparisons of DNA sequences in the origin region. An almost palindromic sequence (underline) is located in the middle of this region. P-Oka and V-Oka have deletions, (TA)4 and (TA)7, in the palindromic sequence, respectively. The other deletion site is in the (GA) repeat just after the palindromic sequence.
Transactivational activity of ORF 10 protein containing an amino acid substitution.
ORF 10 protein, which is a homolog of HSV-1 VP16, is a virion-associated transactivator (24). It is reported that the ORF 10 protein can stimulate the IE62 gene promoter but has no regulatory effect on the gene 4 promoter (43). A base substitution that existed within the HindIII site in ORF 10 caused an amino acid conversion in which an Ala in the middle of the ORF 10 protein in P-Oka was altered to a mixture of Ala and Val in V-Oka. When the PCR product containing the site from V-Oka was digested by HindIII, approximately half of the product was cleaved similar to P-Oka. To compare the transcriptional activity of the ORF 10 protein from clones with the amino-acid substitution, a transient luciferase assay was performed. The effector plasmids pCG10(+) and pCG10(−) had Ala and Val at the substituted positions, respectively. ORF 10 proteins from the V-Oka and P-Oka viruses could both transactivate the gene 62 promoter in CV1 cells in a dose-dependent fashion. The transactivational activity of pCG10(−) on the gene 62 promoter was slightly higher than that of pCG10(+), but this difference was not significant.
Plaque sizes and infectious center assay of cloned viruses.
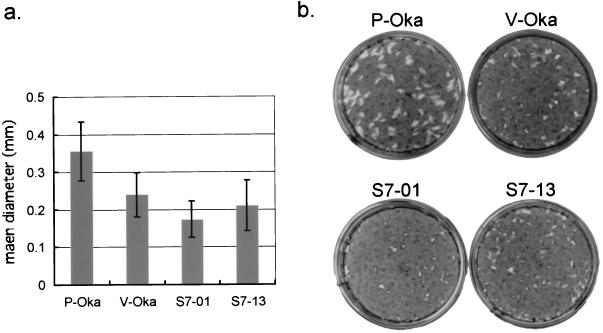
V-Oka was a mixture of different clones (15, 16). To obtain clones of V-Oka, plaque purification was carried out and 13 clones were obtained. Because we found 20 base replacements that led to amino acid differences between V-Oka and P-Oka as described above, these clones were sequenced. In gene 62, one of the clones, S7-01, had all eight amino acid substitutions compared to P-Oka. In contrast, another clone, S7-13, had the minimum number of substitutions compared to P-Oka, with only five amino acid differences, and the remaining three amino acids conserved with P-Oka (Table 3). Thus, these two clones were chosen to determine whether the mutations in gene 62 were correlated with viral growth. A comparison of the plaque sizes of S7-01, S7-13, V-Oka, and P-Oka showed that P-Oka gave rise to the largest plaques (Fig. 4). V-Oka and S7-13 showed relatively larger plaques than S7-01. In particular, S7-01 showed significantly smaller plaques compared with P-Oka (P < 0.01); the mean diameter of the plaques resulting from S7-01 infection (1.73 mm) was about half the size of those resulting from P-Oka (3.55 mm). V-Oka and S7-13 had smaller plaque diameters (2.40 and 2.10 mm, respectively) than did P-Oka. In addition, the cytopathic effect (CPE) of P-Oka appeared 2 days after infection, whereas the CPE of the other viruses appeared in 3 to 4 days. Thus, an infectious center assay was performed to examine the difference in cell-to-cell spreading of progeny viruses. HEL cells were infected with P-Oka, V-Oka, S7-01, or S7-13, and the numbers of infected cells from 0 to 5 days were titrated. In this assay, P-Oka exhibited an obviously faster spread than the other viruses in HEL cells. After 5 days, the numbers of cells infected by P-Oka were 2.1-, 4.2-, and 2.6-fold higher than the numbers infected by V-Oka, S7-01, and S7-13, respectively (Fig. 5a). These results suggested that the virus growth was slightly different among the V-Oka clones and that P-Oka grew best among these Oka viruses in HEL cells. On the other hand, in GPE cells, the plaque sizes of the viruses derived from Oka vaccine were not significantly different from those of its parental viruses (data not shown). Moreover, both Oka viruses showed similar propagation in cell-to-cell spreading by infectious center assay in GPE cells (Fig. 5b).
TABLE 3.
Amino acid substitutions in clones S7-01 and S7-13 relative to V-Oka and P-Oka
| Position | Gene | Amino acid substitution(s)
|
|||
|---|---|---|---|---|---|
| Oka parental | Oka vaccine | S7-01 | S7-13 | ||
| 5745 | 6 | A (Ser) | G (Pro) | G (Pro) | G (Pro) |
| 10900 | 9A | T (Trp) | T/C (Trp/Arg) | T (Trp) | C (Arg) |
| 12779 | 10 | T (Ala) | T/C (Ala/Val) | C (Val) | C (Val) |
| 31732 | 21 | C (Thr) | C/T (Thr/Ile) | T (Ile) | C (Thr) |
| 58595 | 31 | A (Ile) | A/G (Ile/Val) | G (Val) | A (Ile) |
| 71252 | 39 | T (Met) | T/C (Met/Thr) | C (Thr) | C (Thr) |
| 87306 | 50 | T (Ser) | T/C (Ser/Gly) | T (Ser) | T (Ser) |
| 90535 | 52 | A (Ile) | A/G (Ile/Val) | A (Ile) | G (Val) |
| 97748 | 55 | G (Ala) | G/A (Ala/Thr) | G (Ala) | A (Thr) |
| 97796 | 55 | T (Cys) | T/C (Cys/Arg) | C (Arg) | T (Cys) |
| 101089 | 59 | A (Leu) | A/G (Leu/Pro) | A (Leu) | A (Leu) |
| 105310 | 62 | A (Leu) | A/G (Leu/Ser) | G (Ser) | G (Ser) |
| 105356 | 62 | T (Ile) | C (Val) | C (Val) | C (Val) |
| 105544 | 62 | A (Val) | G (Ala) | G (Ala) | G (Ala) |
| 106262 | 62 | A (Arg) | C (Gly) | C (Gly) | C (Gly) |
| 107252 | 62 | T (Ser) | C (Gly) | C (Gly) | C (Gly) |
| 107599 | 62 | A (Val) | A/G (Val/Ala) | G (Ala) | A (Val) |
| 107797 | 62 | A (Leu) | A/G (Leu/Pro) | G (Pro) | A (Leu) |
| 108838 | 62 | A (Met) | A/G (Met/Thr) | G (Thr) | A (Met) |
| 111650 | 64 | A (Gln) | A/G (Gln/Arg) | G (Arg) | A (Gln) |
FIG. 4.
(a) Plaque sizes after infection with cell-free P-Oka, V-Oka, S7-01, and S7-13. (b) HEL cells were infected with each cell-free virus and cultured for 10 days. The cells were stained with methylene blue.
FIG. 5.
Infectious center assay of P-Oka, V-Oka, S7-01, and S7-13. HEL cells (a) and GPE cells (b) in 35-mm dishes were infected independently with similar titers of these four kinds of cell-free viruses, and then the cells were washed and treated with trypsin from 1 to 5 days postinfection. The trypsin-treated cells were diluted and transferred onto monolayers of uninfected HEL cells in 35-mm dishes, and the numbers of infected cells were assessed by counting the number of VZV plaques appearing after 7 days. The number of infected cells was normalized to the initial viral titer per dish; the fold increase indicates the number of infected cells spread from one initial infected cell at day 0.
Transactivational activities of mutant IE62s.
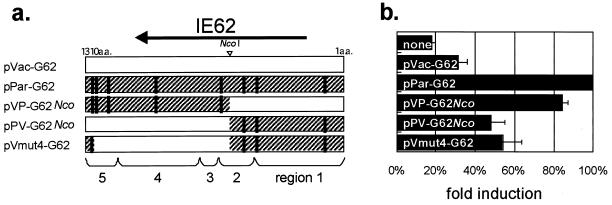
We next analyzed whether the transactivational activities of the IE62s of these clones differed from the parental activity. pLuc-Pol, containing the cloned VZV DNA polymerase gene promoter upstream of the luciferase ORF, was used as the reporter plasmid for transient luciferase assays, because the transactivation of this promoter by IE62 is well known. Five kinds of effector plasmids expressing IE62s were constructed (Fig. 6a). Two of them, pPar-G62 derived from P-Oka and pVac-G62 derived from V-Oka, were used in our previous study (15, 16). One clone containing an IE62 gene was selected from V-Oka and used as pVac-G62 because this clone had all eight amino acid mutations compared to pPar-G62. In addition, another clone was selected from V-Oka, called pVmut4-G62, which had the minimum four mutations; all of the other clones contained in V-Oka had these four mutations and one to four of the other mutations. pPV-G62Nco and pVP-G62Nco were chimeric IE62-expressing plasmids, which were bounded by an NcoI site. pPV-G62Nco expressed the amino terminus of parental IE62 and the carboxyl terminus of pVac-G62. pVP-G62Nco was constructed to contrast with pPV-G62Nco and expressed the amino terminus of pVac-G62 and the carboxyl terminus of pPar-G62. The IE62 genes of P-Oka, S7-01, and S7-13 corresponded with those of pPar-G62, pVac-G62, and pPV-G62Nco, respectively. V-Oka contained from four (like pVmut4-G62) to eight (like pVac-G62) amino acid mutations. The relative luciferase activity was set equal to 100% in cells, where the reporter plasmid pLuc-Pol was cotransfected with pPar-G62. The basal activity of the DNA polymerase promoter was 18% of the activity of parental IE62 (Fig. 6b). pVac-G62 and pVmut4-G62 showed 32 and 54% activities. Therefore, the mutant IE62s contained in the clones of V-Oka were likely to have lower activity than the parental IE62. The chimeric IE62 expressed by pVP-G62Nco transfection possessed an activity that was still high (84%), whereas the other chimera expressed by pPV-G62Nco showed 48% activity, suggesting that the first five amino acid mutations from the carboxyl terminus, rather than the other three mutations, were more important for transactivating the promoter. The mutation closest to the carboxyl terminus changed IE62's activity moderately but significantly. pPar-G62 and pVac-G62 exhibited the highest and lowest activities among the forms we analyzed, respectively, and pPV-G62Nco led to more effective luciferase expression than pVac-G62 but was not as effective as pPar-G62. These transient assay results roughly showed trends that were consistent with both the mean plaque sizes and the square roots of the number of infected cells determined in the infectious center assay. These findings support the possibility that viral growth and spreading may be regulated by the transactivational activity of IE62.
FIG. 6.
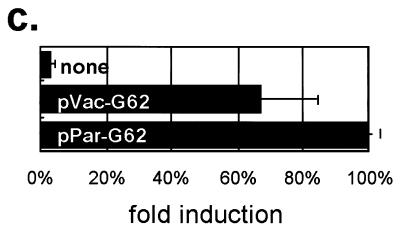
(a) Structure of IE62s. Amino acid residues are numbered from 1 to 1310 from the N to the C terminus. Vertical lines indicate the positions of amino acid changes compared with the IE62 from P-Oka. (b and c) Transactivational activity of parental and mutant IE62s on the DNA polymerase promoter in 293 cells (b) and GPE cells (c). Each of the five effector plasmids was transfected together with pLuc-Pol, and each luciferase activity was determined after 24 h. The basal luciferase activity, induced by no effector, was set equal to 1.0, and the other activities relative to that were given.
Transient luciferase assay with GPE cells were also performed to analyze the transactivational activities of the Oka vaccine IE62 and its parental IE62 in these cells. The assay showed that pVac-G62, the most heavily substituted, had 67% of the activity of pPar-G62 (Fig. 6c), suggesting that the transactivational activity of the IE62 of V-Oka was relatively higher in GEF cells compared to that found in other cells as described above.
Differentiation of the DNA sequence in gene 62 among clinical isolates.
Comparison of the DNA sequences of the Oka viruses showed that gene 62 was the most variable region in the VZV genes, as described above. Furthermore, V-Oka consisted of at least eight different virus clones (15). Thus, we determined the nucleotide sequences of gene 62 from clinical isolates and compared the sequences among different VZV strains (Fig. 7b). Four clinical isolates (RT-283, OZ-4, Inoue, and Kawaguchi) were derived from patients with varicella. The first two strains were recently isolated but the latter two were isolated more than 30 years ago. Three isolates (Z-4, Z-5, and Z-8) were obtained from patients with zoster. Only the RT-342 and RT-345 strains were obtained from vaccinated patients, as described in Materials and Methods. Seven bases (at positions 105356, 105544, 105705, 106262, 107136, 107252, and 108111) were conserved in the clinical isolates and were only different in V-Oka, and four of these led to amino acid substitutions that were specific for V-Oka. Some of the clones contained in V-Oka showed different bases at eight positions (i.e., 105169, 105310, 106710, 107599, 107797, 108838, 109137, and 109200) that were not seen in the clinical isolates, and four of them led to partial amino acid substitutions. In summary, all of the clinical isolates differed from V-Oka in 15 bases that were found to be different between V-Oka and P-Oka. Only one base substitution (at position 105451) causing an amino acid conversion was specific for the Oka viruses. Interestingly, an identical sequence was seen in RT-283, RT-342, and RT-345. These strains were isolated from patients in the same clinic; thus, it was likely that the same virus strain was circulating there.
FIG. 7.
(a) Structure of gene 62. Amino acid residues are numbered from 1 to 1310 from the N to the C terminus. Fifteen vertical lines indicate the positions of base changes between Oka vaccine and its parental virus. Bold or broken lines show base mutations with or without amino acid conversions, respectively. (b) Sequence analysis of gene 62 from the V-Oka vaccine, Oka parental, Dumas strains, and nine clonal isolates. Filled cells indicate base differences compared with V-Oka. All nine viruses have identical bases at these 15 positions, but V-Oka has different bases. R, mixture of G and A; M, mixture of A and C
Genetic differentiation of the Oka vaccine virus from other viruses by RFLP analysis.
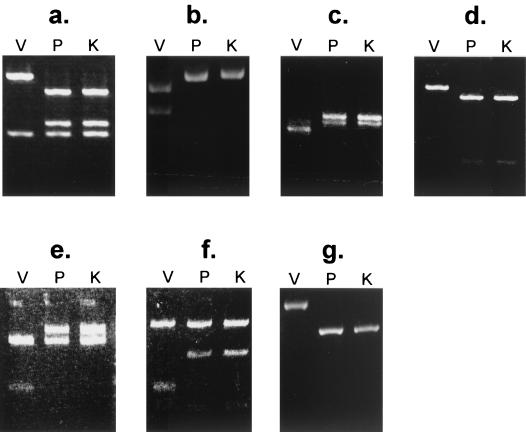
Comparison of the complete DNA sequences of the Oka viruses revealed 11 bases that were completely substituted by other bases. These bases (at positions 560, 5745, 26125, 94167, 105356, 105544, 105705, 106262, 107136, 107252, and 108111) were located within sites recognized by the restriction enzymes NlaIII, AluI, MaeII, BstXI, SfaNI, AccII, SacII, SmaI, BssHII, NaeI, and BsrI, respectively. A method for distinguishing V-Oka from other viruses, including P-Oka, was established in our previous study, which demonstrated that the cleavage sites for BssHII and NaeI were useful for identifying V-Oka (15). Here we sought to establish new methods for distinguishing the viruses by using the eight other restriction enzyme sites, and V-Oka was successfully identified by using all of these enzymes except for NlaIII and MaeII. First, the base substitution at position 5745 in gene 6, which encodes a putative subunit of the helicase-primase complex of V-Oka, resulted in the loss of a restriction site that is recognized by AluI (Fig. 8a). Immediately adjacent to this position, another site recognized by this enzyme was present in every virus and was predicted to be cleaved. The PCR product containing these sites in gene 6 of V-Oka was cleaved at only one site with the enzyme, resulting in two fragments, whereas that of P-Oka was cleaved at two sites by the enzyme, resulting in three fragments. The RFLP study was further performed with 54 clinical isolates. The results of this PCR-RFLP analysis demonstrated that the clinical isolates tested did not show a pattern identical to that of V-Oka. Similar results were obtained in six other experiments. P-Oka and all 54 clinical isolates contained cleavage sites for SfaNI and BsrI, whereas only V-Oka lost those sites. In contrast, four other sites, which were recognized by BstXI, AccII, SacII, and SmaI, were specific only for V-Oka (Fig. 8b to g) but not for the clinical isolates. Moreover, V-Oka could be distinguished from five clinical isolates such as RT-342 and RT-348, which were from vaccinated patients with clinical symptoms, by using some of these restriction enzyme sites in gene 62 (data not shown). Therefore, these findings suggested that these clinical symptoms were caused by wild-type viruses and not by V-Oka.
FIG. 8.
RFLP analysis of the PCR products from V-Oka, P-Oka, and 54 wild-type viruses. V-Oka (V) can be distinguished not only from P-Oka (P) but also from all wild-type viruses. The Kawaguchi strain (K) was used to represent the wild-type viruses by using AluI (a), BstXI (b), AccII (c), SfaNI (d), SacII (e), SmaI (f), and BsrI (g).
DISCUSSION
We found here that the VZV DNA sequences were quite highly conserved (>99.8%) among the Dumas strain, P-Oka, and V-Oka. In particular, there was only a 0.034% sequence difference between the two Oka viruses. However, clinical reactions after vaccination with the Oka varicella vaccine are very rare in healthy children. It was reported that V-Oka grows better than wild-type viruses, including the Oka parental virus, in GPE cells (34) and that the immune response in guinea pigs could be induced by V-Oka but not by wild-type strains (61). These results indicate that a sequence difference of ca. 0.034% of can make a biological difference in such properties as the pathogenicity and growth of VZV. In this and previous studies, gene 62 has emerged as having the potential to be involved in the attenuation of the Oka virus, based on the following remarkable features of V-Oka. First, gene 62 shows the highest divergence in both base and amino acid sequences when V-Oka is compared to P-Oka. In particular, comparison of the Oka viruses revealed that gene 62 and its duplicate gene (gene 71), which cover only 7% of the whole genome, contained more than one-third of the base substitutions, although the other three IE genes (genes 4, 61, and 63) had no substitutions. Second, comparisons of gene 62 from nine clinical isolates showed that all 15 base substitutions leading to eight amino acid conversions were specific for V-Oka. Third, the sequence analysis of a fragment including the gene 62 region from nine clones revealed that V-Oka is a mixture of at least eight different viruses that had a variety of mutations (15). Fourth, our previous studies had demonstrated that the vaccine IE62, which contained all eight amino acid substitutions, had less transactivational activity than parental IE62 in activating all three kinetic classes of VZV promoters in CV1 cells (15, 16). Fifth, S7-01 virus, which had mutations in all eight amino acids in IE62, showed slower virus spreading in HEL cells. Thus, the substitutions that have accumulated in gene 62 are most likely to be important for the differences in the replication and the attenuation of VZV. Because V-Oka had been passaged in guinea pig cells and also in human fibroblast cells at a low temperature (56), mutant viruses must have been selected and grown under selective pressure. The reason why so many amino acid substitutions were accumulated in gene 62 of V-Oka is still unclear, but it is possible that the mutant IE62s contained in V-Oka may have a competitive advantage over parental IE62 in interacting with some cellular transcription factors in guinea pig cells. The five amino acid substitutions closest to the carboxyl terminus in IE62 directly reduced its transactivational activity, and these locate in regions 4 and 5. Region 2 contains the DNA-binding domain (5), which is located between amino acids 472 and 646 (59, 60) and is essential for the protein's regulatory functions (58). A substitution of Lys residue 548, which lies within a highly conserved quartet of residues, WLQN, in region 2 drastically reduces the DNA-binding activity (58). Although the DNA-binding region of V-Oka had three amino acid mutations, no substitutions were found in the WLQN region. There were also no amino acid substitutions in other domains of IE62 that are important for its transcriptional activity, such as the proline-rich domain between regions 4 and 5 (5), the nuclear localization signal (5, 26), the potent activation domain in the N terminus (48), the kinase catalytic domain, and the ATP-binding domain (23). Thus, further studies are needed to determine the mechanism by which the expression of VZV genes is affected by the amino acid substitutions in IE62.
We detected insertions and a deletion in three repeat regions—R1, R3, and R4—that could potentially lead to different biological behaviors in V-Oka. R1 and R3 lie in ORF 11 and ORF 22, respectively, but both of these reiteration elements are multiples of 3 bp, so there were no frameshifts. The function of the ORF 11 protein is unclear, although it is homologous to HSV-1 UL47, a nucleotidylylated tegument phosphoprotein reported to modulate the activity of VP16 (6, 63, 64). It is difficult to differentiate V-Oka from wild-type viruses in Japan by analyzing R1 because a predominant R1 structure is contained in both the wild-type viruses and V-Oka (62). The R1 sequence in V-Oka determined by us is also identical to that in some viruses from patients with acute retinal necrosis syndrome (1). Thus, the deletion in the R1 region is not likely to be important for viral pathogenesis. R3 is unstable, as described above, and lies within the longest ORF in VZV, ORF 22. The ORF 22 product is homologous to the UL36 virion tegument phosphoprotein (36, 37). It is not clear whether the variable R3 exerts a function in ORF 22, because little is known about this gene. R4 lies in a noncoding region situated ca. 550 bp upstream from the transcription start site, ca. 620 bp upstream from the initiator codon, of gene 62. It is possible that the deletion of R4 in V-Oka affects the expression level of IE62 despite its far distance from the transcription start site. In contrast, Perera et al. (46) reported that IE62 could autoactivate a truncated IE62 promoter that contained only 45 bp upstream of the transcription start site and suggested that IE62 self-activation might be mediated by a TATA sequence-dependent mechanism. Thus, at least the autoactivating activity of IE62 is independent of R4. Finally, a comparison of the Oka viruses showed that V-Oka had a deletion of (TA)3 and GA in the origin regions in IRS and TRS. Plasmid replication experiments showed that the yields of deletion mutants that lost (TA)6, and (TA)10 in the corresponding region from the Dumas virus were 45 and 18%, respectively (54). Thus, the deletion in V-Oka may cause a reduced replication of VZV. Further studies are required to clarify the correlation between these candidates, including IE62, and pathogenicity. Recombinant virus could be prepared from viral DNA clones as previously reported (7, 32). Viral pathogenesis and proteins that are important for virus replication could be analyzed by using two systems: the SCID-hu mouse implanted with human fetal tissue (39, 40) and human mononuclear cells in umbilical cord blood infected with VZV (53). Studies are in progress to examine the pathogenesis of the IE62 mutant virus. The sequence differences presented here and the application of these techniques may provide new insights into mechanisms for the molecular pathogenesis of VZV infection.
Interestingly, differences in virus spreading to HEL cells were found among P-Oka, V-Oka, S7-01, and S7-13. P-Oka is known to be as virulent in vivo as other wild-type viruses. Naturally infected children who develop varicella have viremia that is detectable by virus isolation, whereas no viruses are isolated from healthy children immunized by the vaccine (3). Despite the different infection routes in these situations, the findings suggest that the vaccine virus cannot propagate in vivo to the same level as wild-type viruses. The behavior of V-Oka and its constituents both in vitro and in vivo differ from that of P-Oka and the wild-type viruses. Our data showed that viral pathogenicity was affected in association with 20 amino acid substitutions and slow spreading was associated with 15 of them, in ORFs 6, 10, 21, 31, 39, 55, 62, and 64, in the S7-01 virus. IE62 may be the most attractive candidate for causing the different biological properties of the vaccine. S7-01 had 14 amino acid substitutions, including all 8 amino acid substitutions in IE62. The IE62 of S7-01 is identical to a mutant IE62 that exhibited less transcriptional activity than the IE62 of P-Oka in CV1 cells in our previous study (15, 16), and similar results were obtained with the human 293 cell line (data not shown). It is possible that the lower activity of the mutant IE62 may be involved in the ineffective spreading or, moreover, the avirulence of the vaccine. On the other hand, S7-01 did not have mutations in genes 9A, 50, 52, 55, or 59. P-Oka not only is known to be virulent in vivo but also was more efficient in spreading in vitro than was S7-01 in the present study. Thus, the amino acid mutations in these gene products, which were the ORF 9A protein, two helicase-primase components, gM, and uracil-DNA glycosylase, did not seem to be important for efficient spreading. On the other hand, S7-13 exhibited moderately better growth than did S7-1, based on in the analysis of plaque sizes and spreading. Seven amino acids, in genes 21, 31, 55, 62, and 64, of S7-13 were found to be identical with P-Oka but not with S7-01. Three of these amino acids were in IE62, and when we expressed this mutant IE62 from pPV-G62Nco, it was shown to have <50% of the activity of parental IE62. These data indicate that the difference in viral spreading was likely to have been determined by the three mutations in IE62 rather than the four mutations in other gene products, ORF21 protein, gB glycoprotein, helicase-primase component, or ORF64 protein. Studies with recombinant VZVs will be needed to confirm the amino acid substitution(s) responsible for the differences in spreading activity.
Several candidates other than IE62 are potentially responsible for the biological differences in different forms of the virus. First, ORF 10 protein transactivates the IE62 gene promoter, although it does not transactivate the IE ORF 4 and ORF 61 promoters (43, 44). In the present study, both the parent and mutant ORF 10 proteins stimulated the IE62 promoter, and the amino acid substitution in ORF 10 failed to affect its transactivation activity. Thus, the mutant ORF 10 protein is unlikely to cause attenuation by repressing the expression level of IE62. Second, there were four amino acid substitutions in ORFs 6, 52, and 55, which are homologous to HSV-1 UL52, UL8, and UL5, respectively. ORFs 52 and 55 encode two kinds of protein each, as a result of partial amino acid substitutions, whereas ORF 6 is a single clone, as shown by RFLP analysis, indicating that eight (i.e., 23) isomeric complexes may exist by combining these three proteins. It is possible that different helicase-primase activities might exist among different isomeric complexes. Third, a base substitution led to an amino acid conversion in the uracil-DNA glycosylase. If this amino acid substitution were responsible for making the base substitutions in gene 62 in V-Oka, then uracil residues should misincorporate randomly in all of the genes, and substitutions should accumulate in dispensable genes rather than in essential genes. Nonetheless, more than one-third of the substitutions accumulated in gene 62, the homologue of HSV-1 ICP-4, which is essential in vitro (31). Therefore, the amino acid substitution in uracil-DNA glycosylase is not involved in promoting mutations during replication.
VZV glycoproteins are known to induce a strong humoral response both in natural infection and in vaccination with the Oka varicella vaccine. VZV produces at least seven glycoproteins, as described above. The expression level of gC, the gene 14 product, was dependent on the strain of VZV, and the gC level of V-Oka was much lower than that of wild-type viruses (25, 30). SCID-hu mouse experiments showed that a decrease in gC played a critical role in attenuation (39). However, we found no differences in its ORF or flanking regions except for a silent substitution in a termination codon of the gC gene: an ocher (TAA) incompletely altered to an amber (TAG). In our earlier study we found no evidence that the decrease in gC in V-Oka was caused by amino acid substitutions in IE62 because the gC promoter was unresponsive to IE62. The transactivational activity of IE62 is negatively regulated at later stages of the infectious cycle by its phosphorylation by the ORF66 protein kinase, which inhibits its nuclear import (23, 26). Therefore, vaccine IE62 is not directly involved with the decrease in gC by transactivating the gene to a lesser extent. We found amino acid substitution in only gB of V-Oka; i.e., an Ile in the gB of P-Oka was Ile plus Val in V-Oka. gB is the second most abundant and immunogenic glycoprotein in VZV. It appears to play a role in the attachment and penetration of particles, along with gH and gC, and can elicit a monoclonal antibody that neutralizes viral infectivity in vitro. VZV gB is associated with cell-to-cell infection, as is HSV-1 gB (41, 45). gB was also shown to have great fusogenic properties in the presence of gE (33). Nonetheless, it is thought that the amino acid substitution in gB is not crucial for the antigenicity of the vaccine because about half of the clones in V-Oka contain identical sequences to P-Oka in this gene; moreover, Ile and Val have similar properties, and this substitution would be unlikely to change the protein's properties in the remaining half of the clones. These findings indicate that the cell-mediated and humoral immunities led by the glycoproteins of V-Oka are identical to those of P-Oka without adapting to the heterogeneous host, guinea pig cells. V-Oka is likely to have few functional differences in viral attachment and penetration from P-Oka. Although it was reported that V-Oka shows a higher adsorption rate than P-Oka in GPE cells as determined by the infectious center assay (34), the higher adsorption of V-Oka appeared to be caused by the virus's capacity to replicate in guinea pig cells but not by the substitution in gB.
Some cases of chickenpox or shingles in immunocompromised individuals after vaccination have been reported. In rare cases, even healthy vaccine recipients develop rashes within a few weeks after vaccination. However, it has not been clear whether these cases were caused by the vaccine virus or by a wild-type virus. In addition, some reports concerning transmission of the vaccine virus have been reported (21, 29, 51). Therefore, it is epidemiologically important to differentiate the Oka vaccine virus from circulating wild-type viruses. At present, the Oka viruses, which include V-Oka and P-Oka, are identified by using restriction endonuclease digestion of extracted purified viral DNA (14, 19), analysis of repeat regions (17, 55), and RFLP assays after PCR (17, 20, 27, 28, 55, 57). We also have reported an identifying method that combines the single-strand conformational polymorphism of repeat region 2 (R2) with PstI cleavage of the PstI-site-less region (42). Although the Oka viruses can be distinguished from other clinical isolates by these methods, it is difficult to distinguish V-Oka from P-Oka. Therefore, we attempted to identify only V-Oka by another method that uses the simplified RFLP analysis by NaeI and BssHII (15, 16). This method was established based on the first known sequence differences we found in gene 62, and V-Oka could be distinguished not only from clinical isolates but also from P-Oka. Therefore, comparisons of the gene 62 sequence may enable the epidemiological typing of VZV strains. Thus, the comparison of the DNAs is important for quality control of varicella vaccine, molecular epidemiology and, in particular, the development and improvement of new vaccines.
The mechanism of attenuation is still unclear because the mutations were found not only in gene 62 but also in other genes. Therefore, further study to analyze more in detail by using recombinant viruses and suitable animals are needed to resolve the mechanism of attenuation of VZV.
REFERENCES
- 1.Abe, T., M. Sato, and M. Tamai. 2000. Variable R1 region in varicella zoster virus in fulminant type of acute retinal necrosis syndrome. Br. J. Ophthalmol. 84:193-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argaw, T., J. I. Cohen, M. Klutch, K. Lekstrom, T. Yoshikawa, Y. Asano, and P. R. Krause. 2000. Nucleotide sequences that distinguish Oka vaccine from parental Oka and other varicella-zoster virus isolates. J. Infect. Dis. 181:1153-1157. [DOI] [PubMed] [Google Scholar]
- 3.Asano, Y., N. Itakura, Y. Hiroishi, S. Hirose, T. Ozaki, K. Kuno, T. Nagai, T. Yazaki, K. Yamanishi, and M. Takahashi. 1985. Viral replication and immunologic responses in children naturally infected with varicella-zoster virus and in varicella vaccine recipients. J. Infect. Dis. 152:863-868. [DOI] [PubMed] [Google Scholar]
- 4.Barker, D. E., and B. Roizman. 1992. The unique sequence of the herpes simplex virus 1 L component contains an additional translated open reading frame designated UL49.5. J. Virol. 66:562-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baudoux, L., P. Defechereux, S. Schoonbroodt, M. P. Merville, B. Rentier, and J. Piette. 1995. Mutational analysis of varicella-zoster virus major immediate-early protein IE62. Nucleic Acids Res. 23:1341-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaho, J. A., C. Mitchell, and B. Roizman. 1994. An amino acid sequence shared by the herpes simplex virus 1 alpha regulatory proteins 0, 4, 22, and 27 predicts the nucleotidylylation of the UL21, UL31, UL47, and UL49 gene products. J. Biol. Chem. 269:17401-17410. [PubMed] [Google Scholar]
- 7.Cohen, J. I., and K. E. Seidel. 1993. Generation of varicella-zoster virus (VZV) and viral mutants from cosmid DNAs: VZV thymidylate synthetase is not essential for replication in vitro. Proc. Natl. Acad. Sci. USA 90:7376-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crute, J. J., T. Tsurumi, L. A. Zhu, S. K. Weller, P. D. Olivo, M. D. Challberg, E. S. Mocarski, and I. R. Lehman. 1989. Herpes simplex virus 1 helicase-primase: a complex of three herpes-encoded gene products. Proc. Natl. Acad. Sci. USA 86:2186-2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 10.Defechereux, P., L. Melen, L. Baudoux, M. P. Merville-Louis, B. Rentier, and J. Piette. 1993. Characterization of the regulatory functions of varicella-zoster virus open reading frame 4 gene product. J. Infect. Dis. 168:1330-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Disney, G. H., and R. D. Everett. 1990. A herpes simplex virus type 1 recombinant with both copies of the Vmw175 coding sequences replaced by the homologous varicella-zoster virus open reading frame. J. Gen. Virol. 71:2681-2689. [DOI] [PubMed] [Google Scholar]
- 12.Disney, G. H., T. A. McKee, C. M. Preston, and R. D. Everett. 1990. The product of varicella-zoster virus gene 62 autoregulates its own promoter. J. Gen. Virol. 71:2999-3003. [DOI] [PubMed] [Google Scholar]
- 13.Felser, J. M., P. R. Kinchington, G. Inchauspe, S. E. Straus, and J. M. Ostrove. 1988. Cell lines containing varicella-zoster virus open reading frame 62 and expressing the “IE” 175 protein complement ICP4 mutants of herpes simplex virus type 1. J. Gen. Virol. 69:2585-2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gelb, L. D., D. E. Dohner, A. A. Gershon, S. P. Steinberg, J. L. Waner, M. Takahashi, P. H. Dennehy, and A. E. Brown. 1987. Molecular epidemiology of live, attenuated varicella virus vaccine in children with leukemia and in normal adults. J. Infect. Dis. 155:633-640. [DOI] [PubMed] [Google Scholar]
- 15.Gomi, Y., T. Imagawa, M. Takahashi, and K. Yamanishi. 2000. Oka varicella vaccine is distinguishable from its parental virus in DNA sequence of open reading frame 62 and its transactivation activity. J. Med. Virol. 61:497-503. [DOI] [PubMed] [Google Scholar]
- 16.Gomi, Y., T. Imagawa, M. Takahashi, and K. Yamanishi. 2001. Comparison of DNA sequence and transactivation activity of open reading frame 62 of Oka varicella vaccine and its parental viruses. Arch. Virol. Suppl. 17:49-56. [DOI] [PubMed] [Google Scholar]
- 17.Hawrami, K., and J. Breuer. 1997. Analysis of United Kingdom wild-type strains of varicella-zoster virus: differentiation from the Oka vaccine strain. J. Med. Virol. 53:60-62. [DOI] [PubMed] [Google Scholar]
- 18.Hawrami, K., D. Harper, and J. Breuer. 1996. Typing of varicella-zoster virus by amplification of DNA polymorphisms. J. Virol. Methods 57:169-174. [DOI] [PubMed] [Google Scholar]
- 19.Hayakawa, Y., T. Yamamoto, K. Yamanishi, and M. Takahashi. 1986. Analysis of varicella-zoster virus DNAs of clinical isolates by endonuclease HpaI. J. Gen. Virol. 67:1817-1829. [DOI] [PubMed] [Google Scholar]
- 20.Hondo, R., and Y. Yogo. 1989. Strain variation of R5 direct repeats in the right-hand portion of the long unique segment of varicella-zoster virus DNA. Jpn. J. Exp. Med. 59:233-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes, P., P. LaRussa, J. M. Pearce, M. Lepow, S. Steinberg, and A. Gershon. 1994. Transmission of varicella-zoster virus from a vaccinee with leukemia, demonstrated by polymerase chain reaction. J. Pediatr. 124:932-935. [DOI] [PubMed] [Google Scholar]
- 22.Inchauspe, G., and J. M. Ostrove. 1990. Differential regulation by varicella-zoster virus (VZV) and herpes simplex virus type-1 trans-activating genes. Adv. Virus Res. 38:45-98. [DOI] [PubMed] [Google Scholar]
- 23.Kinchington, P. R., K. Fite, and S. E. Turse. 2000. Nuclear accumulation of IE62, the varicella-zoster virus (VZV) major transcriptional regulatory protein, is inhibited by phosphorylation mediated by the VZV open reading frame 66 protein kinase. J. Virol. 74:2265-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinchington, P. R., J. K. Hougland, A. M. Arvin, W. T. Ruyechan, and J. Hay. 1992. The varicella-zoster virus immediate-early protein IE62 is a major component of virus particles. J. Virol. 66:359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinchington, P. R., P. Ling, M. Pensiero, A. Gershon, J. Hay, and W. T. Ruyechan. 1990. A possible role for glycoprotein gpV in the pathogenesis of varicella-zoster virus. Adv. Exp. Med. Biol. 278:83-91. [DOI] [PubMed] [Google Scholar]
- 26.Kinchington, P. R., and S. E. Turse. 1998. Regulated nuclear localization of the varicella-zoster virus major regulatory protein, IE62. J. Infect. Dis. 178(Suppl. 1):S16-S21. [DOI] [PubMed] [Google Scholar]
- 27.LaRussa, P., O. Lungu, I. Hardy, A. Gershon, S. P. Steinberg, and S. Silverstein. 1992. Restriction fragment length polymorphism of polymerase chain reaction products from vaccine and wild-type varicella-zoster virus isolates. J. Virol. 66:1016-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.LaRussa, P., S. Steinberg, A. Arvin, D. Dwyer, M. Burgess, M. Menegus, K. Rekrut, K. Yamanishi, and A. Gershon. 1999. Polymerase chain reaction and restriction fragment length polymorphism analysis of varicella-zoster virus isolates from the United States and other parts of the world. J. Infect. Dis. 178(Suppl. 1):S64-S66. [DOI] [PubMed] [Google Scholar]
- 29.LaRussa, P., S. Steinberg, F. Meurice, and A. Gershon. 1997. Transmission of vaccine strain varicella-zoster virus from a healthy adult with vaccine-associated rash to susceptible household contacts. J. Infect. Dis. 176:1072-1075. [DOI] [PubMed] [Google Scholar]
- 30.Ling, P., P. R. Kinchington, W. T. Ruyechan, and J. Hay. 1991. A detailed analysis of transcripts mapping to varicella-zoster virus gene 14 (glycoprotein V). Virol. 184:625-635. [DOI] [PubMed] [Google Scholar]
- 31.Longnecker, R., and B. Roizman. 1986. Generation of an inverting herpes simplex virus 1 mutant lacking the L-S junction a sequences, an origin of DNA synthesis, and several genes, including those specifying glycoprotein E and the alpha 47 gene. J. Virol. 58:583-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mallory, S., M. Sommer, and A. M. Arvin. 1997. Mutational analysis of the role of glycoprotein I in varicella-zoster virus replication and its effects on glycoprotein E conformation and trafficking. J. Virol. 71:8279-8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maresova, L., T. J. Pasieka, and C. Grose. 2001. Varicella-zoster virus gB and gE coexpression, but not gB or gE alone, leads to abundant fusion and syncytium formation equivalent to those from gH and gL coexpression. J. Virol. 75:9483-9492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matsunaga, Y., K. Yamanishi, and M. Takahashi. 1982. Experimental infection and immune response of guinea pigs with varicella-zoster virus. Infect. Immun. 37:407-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGeoch, D. J., A. Dolan, S. Donald, and D. H. Brauer. 1986. Complete DNA sequence of the short repeat region in the genome of herpes simplex virus type 1. Nucleic Acids Res. 14:1727-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McNabb, D. S., and R. J. Courtney. 1992. Analysis of the UL36 open reading frame encoding the large tegument protein (ICP1/2) of herpes simplex virus type 1. J. Virol. 66:7581-7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McNabb, D. S., and R. J. Courtney. 1992. Characterization of the large tegument protein (ICP1/2) of herpes simplex virus type 1. Virology 190:221-232. [DOI] [PubMed] [Google Scholar]
- 38.Meier, J. L., X. Luo, M. Sawadogo, and S. E. Straus. 1994. The cellular transcription factor USF cooperates with varicella-zoster virus immediate-early protein 62 to symmetrically activate a bidirectional viral promoter. Mol. Cell. Biol. 14:6896-6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moffat, J. F., L. Zerboni, P. R. Kinchington, C. Grose, H. Kaneshima, and A. M. Arvin. 1998. Attenuation of the vaccine Oka strain of varicella-zoster virus and role of glycoprotein C in alphaherpesvirus virulence demonstrated in the SCID-hu mouse. J. Virol. 72:1542-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moffat, J. F., L. Zerboni, M. H. Sommer, T. C. Heineman, J. I. Cohen, H. Kaneshima, and A. M. Arvin. 1998. The ORF47 and ORF66 putative protein kinases of varicella-zoster virus determine tropism for human T cells and skin in the SCID-hu mouse. Proc. Natl. Acad. Sci. USA 95:11969-11974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Montalvo, E., and C. Grose. 1987. Assembly and processing of the disulfide-linked varicella-zoster virus glycoprotein gpII(140). J. Virol. 61:2877-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mori, C., R. Takahara, T. Toriyama, T. Nagai, M. Takahashi, and K. Yamanishi. 1998. Identification of the Oka strain of the live attenuated varicella vaccine from other clinical isolates by molecular epidemiologic analysis. J. Infect. Dis. 178:35-38. [DOI] [PubMed] [Google Scholar]
- 43.Moriuchi, H., M. Moriuchi, S. E. Straus, and J. I. Cohen. 1993. Varicella-zoster virus open reading frame 10 protein, the herpes simplex virus VP16 homolog, transactivates herpesvirus immediate-early gene promoters. J. Virol. 67:2739-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moriuchi, M., H. Moriuchi, S. E. Straus, and J. I. Cohen. 1994. Varicella-zoster virus (VZV) virion-associated transactivator open reading frame 62 protein enhances the infectivity of VZV DNA. Virology 200:297-300. [DOI] [PubMed] [Google Scholar]
- 45.Navarro, D., P. Paz, and L. Pereira. 1992. Domains of herpes simplex virus I glycoprotein B that function in virus penetration, cell-to-cell spread, and cell fusion. Virology 186:99-112. [DOI] [PubMed] [Google Scholar]
- 46.Perera, L. P. 2000. The TATA motif specifies the differential activation of minimal promoters by varicella-zoster virus immediate-early regulatory protein IE62. J. Biol. Chem. 275:487-496. [DOI] [PubMed] [Google Scholar]
- 47.Perera, L. P., J. D. Mosca, W. T. Ruyechan, and J. Hay. 1992. Regulation of varicella-zoster virus gene expression in human T lymphocytes. J. Virol. 66:5298-5304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Perera, L. P., J. D. Mosca, W. T. Ruyechan, G. S. Hayward, S. E. Straus, and J. Hay. 1993. A major transactivator of varicella-zoster virus, the immediate-early protein IE62, contains a potent N-terminal activation domain. J. Virol. 67:4474-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perera, L. P., J. D. Mosca, M. Sadeghi-Zadeh, W. T. Ruyechan, and J. Hay. 1992. The varicella-zoster virus immediate early protein, IE62, can positively regulate its cognate promoter. Virology 191:346-354. [DOI] [PubMed] [Google Scholar]
- 50.Ross, J., M. Williams, and J. I. Cohen. 1997. Disruption of the varicella-zoster virus dUTPase and the adjacent ORF9A gene results in impaired growth and reduced syncytia formation in vitro. Virology 234:186-195. [DOI] [PubMed] [Google Scholar]
- 51.Salzman, M. B., R. G. Sharrar, S. Steinberg, and P. LaRussa. 1997. Transmission of varicella-vaccine virus from a healthy 12-month-old child to his pregnant mother. J. Pediatr. 131:151-154. [DOI] [PubMed] [Google Scholar]
- 52.Schoonbroodt, S., J. Piette, L. Baudoux, P. Defechereux, B. Rentier, and M. P. Merville. 1996. Enhancement of varicella-zoster virus infection in cell lines expressing ORF4- or ORF62-encoded proteins. J. Med. Virol. 49:264-273. [DOI] [PubMed] [Google Scholar]
- 53.Soong, W., J. C. Schultz, A. C. Patera, M. H. Sommer, and J. I. Cohen. 2000. Infection of human T lymphocytes with varicella-zoster virus: an analysis with viral mutants and clinical isolates. J. Virol. 74:1864-1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stow, N. D., and A. J. Davison. 1986. Identification of a varicella-zoster virus origin of DNA replication and its activation by herpes simplex virus type 1 gene products. J. Gen. Virol. 67:1613-1623. [DOI] [PubMed] [Google Scholar]
- 55.Takada, M., T. Suzutani, I. Yoshida, M. Matoba, and M. Azuma. 1995. Identification of varicella-zoster virus strains by PCR analysis of three repeat elements and a PstI-site-less region. J. Clin. Microbiol. 33:658-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takahashi, M., T. Otsuka, Y. Okuno, Y. Asano, and T. Yazaki. 1974. Live vaccine used to prevent the spread of varicella in children in hospital. Lancet ii:1288-1290. [DOI] [PubMed] [Google Scholar]
- 57.Takayama, M., N. Takayama, N. Inoue, and Y. Kameoka. 1997. Application of long PCR method of identification of variations in nucleotide sequences among varicella-zoster virus isolates. J. Clin. Microbiol. 34:2869-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tyler, J. K., K. E. Allen, and R. D. Everett. 1994. Mutation of a single lysine residue severely impairs the DNA recognition and regulatory functions of the VZV gene 62 transactivator protein. Nucleic Acids Res. 22:270-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tyler, J. K., and R. D. Everett. 1994. The DNA binding domain of the varicella-zoster virus gene 62 protein interacts with multiple sequences which are similar to the binding site of the related protein of herpes simplex virus type 1. Nucleic Acids Res. 22:711-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu, C. L., and K. W. Wilcox. 1991. The conserved DNA-binding domains encoded by the herpes simplex virus type 1 ICP4, pseudorabies virus IE180, and varicella-zoster virus ORF62 genes recognize similar sites in the corresponding promoters. J. Virol. 65:1149-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yamanishi, K., Y. Matsunaga, T. Otsuka, and M. Takahashi. 1980. Immune response of guinea pigs to varicella vaccine strain (OKA) and wild strains. Biken J. 23:53-55. [PubMed] [Google Scholar]
- 62.Yoshida, M., T. Tamura, and M. Hiruma. 2000. Analysis of strain variation of R1 repeated structure in varicella-zoster virus DNA by polymerase chain reaction. J. Med. Virol. 58:76-78. [PubMed] [Google Scholar]
- 63.Zhang, Y., and J. L. McKnight. 1993. Herpes simplex virus type 1 UL46 and UL47 deletion mutants lack VP11 and VP12 or VP13 and VP14, respectively, and exhibit altered viral thymidine kinase expression. J. Virol. 67:1482-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang, Y., D. A. Sirko, and J. L. McKnight. 1991. Role of herpes simplex virus type 1 UL46 and UL47 in αTIF-mediated transcriptional induction: characterization of three viral deletion mutants. J. Virol. 65:829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]