Abstract
A single retroviral protein, termed Gag, is sufficient for assembly of retrovirus-like particles in mammalian cells. Gag normally selects the genomic RNA of the virus with high specificity; the nucleocapsid (NC) domain of Gag plays a crucial role in this selection process. However, encapsidation of the viral RNA is completely unnecessary for particle assembly. We previously showed that mutant murine leukemia virus (MuLV) particles that lack viral RNA because of a deletion in the cis-acting packaging signal (“Ψ”) in the genomic RNA compensate for the loss of the viral RNA by incorporating cellular mRNA. The RNA in wild-type and Ψ− particles was also found to be necessary for virion core structure. In the present work, we explored the role of RNA in MuLV particles that lack genomic RNA because of mutations in the NC domain of Gag. Using a fluorescent dye assay, we observed that NC mutant particles contain the same amount of RNA that wild-type virions do. Surprisingly enough, these particles contained large amounts of rRNAs. Furthermore, ribosomal proteins were detected by immunoblotting, and ribosomes were observed inside the particles by electron microscopy. The biological significance of the presence of ribosomes in NC mutant particles lacking genomic RNA is discussed.
Expression of a single retrovirus-coded protein, termed Gag, is sufficient for efficient assembly of virus-like particles (VLPs) in mammalian cells. During normal assembly, the Gag protein selects the viral genomic RNA for encapsidation into the nascent particle. Mutational studies have shown that this selection is due to recognition of a packaging signal, “Ψ,” in the 5′ end of the genomic viral RNA. Deletion of this signal severely impairs viral RNA packaging in the particle but does not interfere with particle assembly (reviewed in references 2 and 25).
Upon the release of the retroviral particle from the cell, the Gag protein is cleaved by the viral protease (PR) into a series of cleavage products. These products always include at least three proteins, i.e., matrix, capsid (CA), and nucleocapsid (NC). Retroviral NC proteins are highly basic and usually contain one or two zinc fingers, a motif found in nucleic acid-binding proteins. Packaging of the viral RNA depends upon an interaction between the NC domain of Gag and the Ψ signal. Thus, either deletion of Ψ from the RNA or mutation of the NC domain results in the assembly of VLPs that do not contain viral genomic RNA (reviewed in reference 2).
We have recently characterized the RNA content of Ψ− particles of the gammaretrovirus murine leukemia virus (MuLV). These particles are formed by wild-type Gag in mammalian cells in the absence of any RNA containing Ψ. It was found (17) that they contain nearly normal amounts of RNA, with cellular mRNA molecules largely replacing the genomic RNA that would be present in wild-type particles. Typical wild-type particles also contain small cellular RNA such as tRNA: we found that the tRNA population was unchanged in Ψ− particles and still represents 15 to 20% of the total RNA content, just as in wild-type particles. Our experiments also showed that RNA in both wild-type and Ψ− particles acts as scaffolding, so that digestion of immature viral capsids with RNase results in the solubilization of the Gag proteins.
In the present report, we present an analysis of the RNA content of MuLV particles that lack viral RNA because of mutations in the NC domain of Gag. Surprisingly, the RNA in these particles is predominantly rRNA. The experiments indicate that ribosomes are present in these particles. Possible explanations for the incorporation of ribosomes into the particles are discussed.
MATERIALS AND METHODS
Cell culture.
293T cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal calf serum, 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml). BHK-21 cells were cultured in Glasgow modified Eagle's medium (Gibco BRL), with 5% fetal calf serum, 10% tryptose phosphate broth, 10 mM HEPES (pH 7.4), 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml).
Plasmid constructs.
Wild-type Moloney MuLV pRR88 and the PR- active-site mutant D32L proviral clones used in this study are in a plasmid vector, pGCcos3neo, derived from pSV2Neo (24) and have been described previously (11). C26S, C29S, C26S/C29S, Y28S, C39S, C17− (previously designated R44ter), NC(Δ8-11), and NC(Δ16-23) NC mutant clones have all been described previously (10, 12, 21). C37− contains a termination codon at position 24 of NC. All of these mutants were in the full-length pRR88 plasmid.
The Semliki Forest virus (SFV) vector constructs pSFVC-Pr65gag and pSFV1 have been described previously (16) and were a kind gift of Henrik Garoff. The mutations in NC listed above were placed into pSFVC-Pr65gag, creating pSFVGagΔNC(16-23), pSFVGagC39S, pSFVGagC17−, and pSFVGagC37− plasmids. When these vectors are expressed in BHK cells, they produce immature Gag particles to which we will refer as VLPs.
Mammalian cell transfection and virus production.
Culture fluids containing virus particles were harvested at 24, 48, and 72 h after transfection of 293T cells (14) and were clarified by filtration (0.45-μm pore size; Nalge). Ten micrograms of DNA plasmid was used to transfect 106 cells per 10-cm culture dish.
SFV-derived plasmids were linearized with SpeI and were transcribed with SP6 polymerase (Promega). The transcripts were introduced into BHK cells by electroporation (8). The cells were then plated on 10-cm tissue culture dishes and incubated at 37°C. The supernatant containing the VLPs was harvested at 24 h after electroporation and clarified by centrifugation at low speed (1,000 × g).
Immunoblotting.
Relative amounts of virus were measured by immunoblotting with rabbit antiserum against MuLV p30CA, using the enhanced chemiluminescence reagent (Amersham Life Science). The quantification of the enhanced chemiluminescence signal on the membrane was performed either by scanning the X-ray film or by measuring the chemiluminescence with a phosphorimager (Bio-Rad) with the quantification program Quantity One (Bio-Rad). Wild-type and NC mutant virions that undergo maturation were compared using p30CA, while PR- and SFV-derived immature VLPs were compared with respect to Pr65Gag levels. In all cases, several dilutions of the samples were compared on the blot, in order to find the dilution of the mutant virus preparation matching the amount of virus in the wild-type preparation. Comparisons of RNAs and proteins in virus preparations were always made after the relative amounts of viral proteins (Gag or CA) in the preparations were determined by immunoblotting analysis; thus, comparisons were always performed on equal amounts of VLPs.
Human ribosomal P proteins were detected with a human antiribosomal P protein autoantibody (Immunovision) (9).
Virus preparation and RNA isolation.
Virions and VLPs were purified from filtered culture supernatants by pelleting through a cushion of 20% sucrose-TNE (100 mM NaCl, 10 mM Tris hydrochloride [pH 7.4], and 1 mM EDTA) at 25,000 rpm for 50 min at 4°C in a Beckman SW28 rotor. The virus pellets were resuspended for 1 h at 4°C in TNE and were either stored at −20°C for protein quantification or disrupted by addition of 1 volume of a 2× concentrated lysis buffer containing 100 mM Tris hydrochloride (pH 7.4), 20 mM EDTA, 2% sodium dodecyl sulfate, 200 mM NaCl, and 200 μg of proteinase K per ml, followed by an incubation of 30 min at 37°C. The RNAs were then extracted by phenol-chloroform and were precipitated with 3 volumes of 100% ethanol, 0.3 M sodium acetate (pH 5.2), and 0.02% linear acrylamide.
Mock preparations were obtained from 293T cells transfected with the empty pGCcos3neo vector or from BHK cells electroporated with pSFV-LacZ vector and were treated exactly as in the viral preparations.
Poly(A)+ RNA purification.
Poly(A)+ RNAs were purified from total viral RNAs by using a poly(A)+ RNA isolation kit (Roche). Mock-transfected cultures always gave values that were ≤ 2% of the experimental values.
Ribosomes and rRNA purification.
293T cells, in exponential-phase growth, were lysed with TNT buffer (0.02 M Tris, pH 7.5-0.2 M NaCl -1% Triton X-100) and were centrifuged at 4°C for 10 min at 18,000 × g to remove nuclei and other cellular organelles. The supernatant was then ultracentrifuged at 4°C for 1 h at 100,000 × g to pellet ribosomes from the clarified cell cytoplasm. Half of the pellet containing ribosomes was resuspended in TNT buffer to use as the ribosome solution. The protein concentration was determined by using the Coomassie plus protein assay (Pierce). The other half of the pellet was resuspended in lysis buffer, and the rRNAs were extracted by phenol-chloroform and precipitated, as described above. The rRNA concentration was determined by measuring optical density at 260 nm.
RNA quantification.
Amounts of total RNA extracted from viral particles and poly(A)+ RNA were measured using the Ribogreen quantification kit (Molecular Probes), as described elsewhere (17). Small RNAs were quantified by measuring the intensity of the spots on the gels, after RNA end labeling, by the Molecular Imager FX phosphorimager (Bio-Rad).
RNA analysis.
RNAs were analyzed by denaturing Northern blots as described earlier (22). RNA samples were heated for 10 min at 65°C in denaturing RNA loading dye before electrophoresis on a 0.9% agarose denaturing gel. The 32P-labeled MuLV cDNA probe was generated from the entire Moloney MuLV proviral clone pRR88, which had been digested by XbaI, by using a random primer cDNA kit (Roche).
The 32P-labeled Xenopus laevis ribosomal cDNA probe was generated from the entire clone, pXlr101a (a kind gift of Barbara Sollner-Webb, John Hopkins University), which was digested by HindIII prior to labeling, using the random primer cDNA kit.
The RNA end-labeling technique was previously described (17). RNA samples were labeled by 32P-pCp with T4 RNA ligase and were analyzed on a denaturing 1% agarose gel. This procedure adds a single 32P-pCp group to the 3′ end of the RNA molecule.
Retrovirus core isolation and RNase A treatment.
VLPs, isolated as described above, were resuspended in 50 mM Tris, pH 7.4, and 100 mM NaCl (TN buffer) with 1% NP-40 + 1% Triton X-100 or in TN buffer with no detergent and were incubated at 37°C for 40 min in a final volume of 20 μl. Afterwards, the samples were incubated at 37°C for 2 h in the presence or in the absence of RNase A (50 μg from Roche) in TN buffer in a final volume of 30 μl. The samples were then layered onto 10 μl of 20% sucrose-TNE cushions and were centrifuged in a microcentrifuge tube at 18°C for 1 h at 18,000 × g. The Gag proteins present in the supernatant (S) and pellet (P) were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and were analyzed by immunoblotting with a rabbit anti-MuLV CA antiserum, as described above.
TEM.
Detailed procedures for transmission electron microscopy (TEM) were previously described (26). The procedures were modified as follows: 293T cells were trypsinized, washed once with phosphate-buffered saline, and pelleted by centrifugation for 5 min at 4°C, 185 × g, in a J-75 Beckman rotor. Cells were fixed in 2% glutaraldehyde in cacodylate buffer (0.1 M, pH 7.4). The cell pellet was rinsed in cacodylate buffer, postfixed in 1% osmium in the same buffer, and dehydrated in graded ethanol and propylene oxide. The pellet was infiltrated overnight in a 1:1 mixture of 100% propylene oxide and epoxy resin (Electron Microscope Science). The pellet was then embedded in pure resin and cured at 55°C. The cured block was thin-sectioned and stained with uranyl acetate and lead citrate. The digital images were obtained with an electron microscope (Hitachi H7000) equipped with a digital camera system (Gatan).
RESULTS
(i) Total RNA content of MuLV NC mutants.
We analyzed virions from a series of NC mutants with respect to their RNA content. The mutants are depicted in Fig. 1. As indicated, C39S is a mutant in which the zinc finger in NC has been effectively destroyed by the replacement of one of the cysteine residues with serine (12). NC (Δ16-23) is a deletion of eight residues in a highly basic region on the N-terminal side of the zinc finger (21). The mutations in C39S and Δ16-23 do not prevent the maturation of Gag by the viral PR after virus release. In addition, we analyzed two C-terminal truncations of MuLV Gag. C37− lacks the 37 C-terminal amino acids of Gag, and C17− (also named R44ter [10]) lacks the last 17 residues of Gag. These two mutations prevent the translation of the pol gene and thus lack PR; therefore, the Gag protein in these particles does not undergo cleavage after the particles are released. None of these mutants, except C17−, packages significant amounts of viral RNA, and all, of course, are noninfectious.
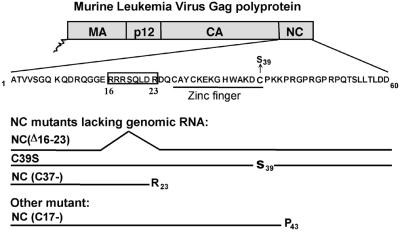
FIG. 1.
MuLV NC mutants used in this study. The NC domain is located at the C terminus of the Gag polyprotein of MuLV. The MuLV NC protein, also called NCp10, is a 60-amino-acid protein and contains one zinc finger. The NC mutants used most in this study are the “C39S” mutant carrying a point mutation in the zinc finger, the NC(Δ16-23) mutant with a deletion of residues R16 to R23, and the C37− mutant, which is truncated after R23. The NC domain of C37− Gag protein is missing 37 amino acids of NC. These mutants lack viral genomic RNA. Another mutant, C17−, is missing the 17 C-terminal amino acids of the NC domain of Gag but still packages some viral RNA. MA, matrix.
Total RNA in the NC mutant particles lacking genomic RNA was measured by the Ribogreen assay in comparison to wild-type or PR−. The results showed that all NC mutant particles contain roughly the same amount of total RNA as do wild-type particles (Table 1). Since the viral RNA is not packaged, some other, cell-derived, RNAs must be present in these mutant particles. We tested the particles for the presence of poly(A)+ mRNA; it was previously found that Ψ− MuLV particles contain cellular mRNA in place of the viral RNA (17). However, these assays showed that the mutant particles contain very small amounts of poly(A)+ RNA (Table 1) (except C17−, which contains a substantial amount of viral RNA). Thus, unlike Ψ− particles, the NC mutant particles contain very little mRNA.
TABLE 1.
RNA content of NC mutant virions, produced by 293T cells, deficient in viral RNA packaginga
| MuLV virion | Packaging of genomic viral RNA | Total RNA content (%) (by Ribogreen) | Poly(A)+ mRNA content (%) (by Ribogreen) | rRNA content in particles (by Northern blot) | Small RNA content (%) (by phosphorimager) |
|---|---|---|---|---|---|
| Wild type | ++ | 100 | 100 | − | 100 |
| ψ−b | − | 60 ± 8 | 35 ± 6 | − | 86 ± 14 |
| C39S | −c | 101 ± 23 | 6 ± 0.8 | + | 104 ± 21 |
| NC(Δ16-23) | −d | 118 ± 9 | 8 ± 0.01 | ++ | 107 ± 46 |
| PR− | ++e | 111 ± 13 | 100 | − | 100 |
| C17− | +e | 91 ± 23 | 65 ± 20 | − | 55 ± 15 |
| C37− | −e | 83 ± 9 | 2 ± 2 | ++ | 142 ± 55 |
Results are expressed relative to RNA content of wild-type or PR− virions, determined by Ribogreen assay (or by phosphorimager analysis of the bands of small RNA on the gels). Each sample contains the same amount of p30CA (wild type, C39S, and NC[Δ 16-23]) or Pr65Gag (PR−, C17−, and C37−). Each value is expressed as a percentage of wild-type or PR− RNA content and represents the average (± standard deviation) of values obtained from at least two independent experiments. Each experimental value shown has been reduced by subtraction of the RNA content of the mock preparation, i.e., supematant harvested from 293T cells transfected with pGCcos3neo vector (equal to 6% ± 1% for total RNA, 1% for poly[A]+ mRNA, and 21% ± 3% for small RNA quantifications). For comparison, the presence (++, 100, or +, ∼50%) or the absence (−, less than 1%) of viral genomic RNA and rRNA in virions is indicated.
ψ− RNA content was added in the table for comparison (17).
As previously determined (12).
As previously determined (21).
Viral RNA content determined by Northern bloting (data not shown) or RNA end labeling (Fig.2A).
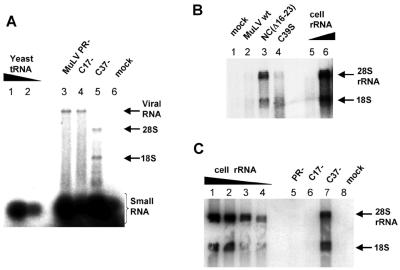
We attempted to identify the RNA species in the mutant particles by labeling the RNA extracted from the particles with 32P-pCp and T4 RNA ligase, as previously described (17). Figure 2A shows results for one mutant, C37−, compared with those for PR−. We found that the mutant particles contained roughly the same amount of small RNAs as did PR− particles (see Table 1 for quantitation). However, there were prominent bands of labeled RNA migrating at positions of ∼18S and ∼28S RNA in denaturing gels, as shown for the C37− NC mutant (Fig. 2A, lane 5). (The end-labeling profile of RNA extracted from NC mutants was frequently less clear than in Fig. 2A; there was often a smear down the lane in addition to the 28S and 18S bands [data not shown]). When the radioactivity in the 18S and 28S bands in C37− particles (lane 5) was counted and combined, it was found to be 1.9 times that in the viral RNA band in the PR control (lane 3). When the sizes of these end-labeled RNAs are taken into account, the mass of the 18S and 28S RNA in the mutant particles was found to represent ∼60% that of the viral genomic RNA in PR− particles. This value seemed to vary from 40 to 75% from one NC mutant to another (C39S and NC[Δ16-23] mutants, respectively). These results showed that viral RNA is principally replaced by 18S and 28S RNA in these NC mutant particles. All NC mutant particles contain roughly the same amount of small RNA as the control, except for C17−, which contains somewhat less (Table 1).
FIG. 2.
RNA analysis of MuLV NC mutants. (A) RNA end labeling. RNA samples extracted from MuLV particles were labeled at their 3′end with [32P]pCp by T4 RNA ligase and were analyzed on a denaturing agarose gel. RNA from PR−, C17−, and C37− virions are shown in lanes 3 to 5, respectively. Yeast RNA was used as control for small RNA labeling (lanes 1 and 2, 0.1 μg and 0.05 μg). A mock preparation corresponding to the same volume of cell supernatant as the virion preparations is shown in lane 6. The rRNAs, genomic viral RNA, and small cellular RNA are indicated. (B and C) Identification of cellular rRNA by Northern blotting using a ribosomal cDNA probe. rRNAs were purified from 293T cells and serve as controls (B, lanes 5 and 6; respectively, 20 and 200 ng; C, lanes 1 through 4; respectively, 500, 250, 125, and 60 ng). 28S (∼5 kb) and 18S (∼1.5 kb) rRNAs are indicated by arrows. RNAs extracted from MuLV wild type (wt) (B, lane 2), NC(Δ16-23) mutant (B, lane 3), and the C39S mutant (B, lane 4) and from PR− (C, lane 5), C17− (C, lane 6), and C37− (C, lane 7) virions were analyzed. Supernatant from 293T cells transfected with the empty vector pCGcos3neo is the “mock” and corresponds to 10 ml (B, lane 1) and 6 ml (C, lane 8) of cell media.
(ii) NC mutants lacking genomic RNA package rRNA.
Since end-labeling analysis showed that the NC mutant particles contain significant amounts of ∼28S and ∼18S RNAs (Fig. 2A), we tested them for the presence of rRNA by Northern blotting. As shown in Fig. 2B and C, we found abundant 18S and 28S rRNA in NC(Δ16-23), C39S, and C37− particles (Fig. 2B, lanes 3 and 4, and C, lane 7, respectively). In contrast, very little rRNA was detected in the wild-type (Fig. 2B, lane 2) or PR− (Fig. 2C, lane 5) control particles, in the C17− mutant particles (Fig. 2C, lane 6), or in the mock cell supernatants (Fig. 2B, lane 1, and C, lane 8). It seems likely that 18S and 28S rRNAs are present in equal proportions in the virions, although the 18S band is less prominent in the Northern blot than in the 28S band (e.g., Fig. 2B, lane 3), as it is for the cellular rRNA (Fig. 2B, lane 6). This may reflect the composition of the rRNA probe, the relative efficiency with which it recognizes 18S and 28S rRNA, or a difference in transfer efficiency of the two RNAs. These results showed that NC mutants lacking viral genomic RNA encapsidate high levels of rRNA.
(iii) NC mutants contain ribosomal proteins.
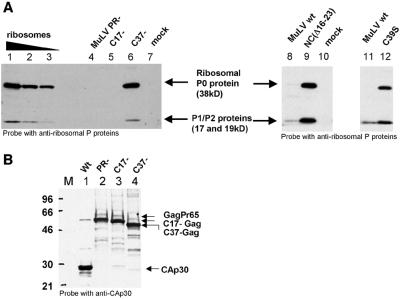
Since the NC mutants encapsidated both rRNAs, we explored the possibility that these particles contained ribosomes. We therefore examined the NC mutant particles lacking viral RNA for the presence of ribosomal proteins. As shown by immunoblotting (Fig. 3A), particles packaging viral RNA, i.e., wild-type, PR−, and C17−, did not contain detectable amounts of ribosomal P0 protein (Fig. 3A, lanes 8 and 11, and Fig. 4 and 5, respectively) or contained only traces of P1 and P2 proteins, like the wild type (Fig. 3A, bottom of lanes 8 and 11), while all the NC mutant particles lacking viral RNA contained high levels of these ribosomal proteins (Fig. 3A, lanes 6, 9, and 12). These comparisons were, of course, performed on equal amounts of virions, as illustrated by an immunoblot of some of the samples shown in Fig. 3A using antiserum against p30CA (Fig. 3B).
FIG. 3.
Immunoblotting with antiribosomal P proteins. (A) Identification of the cellular ribosomal P proteins (proteins of the 60S subunit of the ribosome) by Western blot using human antiribosomal P protein antiserum. P0 is 38 kDa, P1 is 19 kDa, and P2 is 17 kDa. Ribosomes purified from 293T cells were used as positive control (lanes 1 to 3, i.e., 250, 100, and 50 ng of proteins, respectively). An equal amount of virions is loaded on each lane, as determined by MuLV Gag or CA quantification by immunoblot using a p30CA antiserum (example in panel B). Lane 4, PR− MuLV; lane 5, C17−; and lane 6, C37−. Lanes 8 and 11, MuLV wild type; lane 9, NC(Δ16-23); and lane 12, C39S. The mock preparations correspond to the same volume of culture media as for C37− (lane 7) or NC(Δ16-23) virions (lane 10). wt, wild type. (B) Western blot of several samples used in panel A, probed with a p30CA antiserum. Lane M, protein molecular weight marker RPN756 (Amersham Pharmacia) (in kilodaltons). Lane 1, wild-type MuLV; lanes 2, 3, and 4 are PR−, C17−, and C37− mutants, respectively.
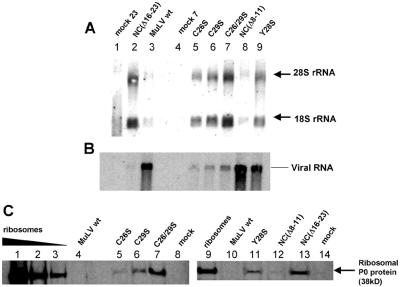
FIG. 4.
Other NC mutant virions contain ribosomes. (A) Identification of cellular rRNA by Northern blotting. 28S (∼5 kb) and 18S (∼1.5 kb) rRNAs are indicated. RNAs extracted from MuLV wild-type (wt) (lane 3), the C26S mutant (lane 5), and C29S (lane 6) and from the C26/29S double mutant (lane 7), NC(Δ 8-11) mutant (lane 8), and Y28S mutant (lane 9) virions were analyzed. NC(Δ16-23) mutant (A, lane 2) serves as a positive control. The mock lanes correspond to 23 ml (A, lane 1) and 7 ml (A, lane 4) of cell media, the same volumes that were used for NC(Δ16-23) and C26S virions, respectively. (B) Genomic viral RNA contained in NC mutant virions analyzed by Nothern blotting. The same membrane as shown in panel A was reprobed with a full-length MuLV proviral DNA probe. (C) Identification of the cellular ribosomal P0 protein (38kD) by Western blot using an human antiribosomal P protein antiserum. Ribosomes purified from 293T cells were used as positive control (lanes 1 to 3, respectively; 2, 0.5, and 0.2 μg of proteins). An equal amount of virions is loaded on each lane, based on p30CA quantification. Lanes 4 and 10, MuLV wild type; lane 5, C26S; lane 6, C29S; lane 7, C26/29S; lane 11, Y28S; lane 12, NC(Δ 8-11); and lane 13, NC(Δ16-23). The mock lane (lane 14) corresponds to the same volume of culture media that the NC(Δ16-23) virions do.
FIG. 5.
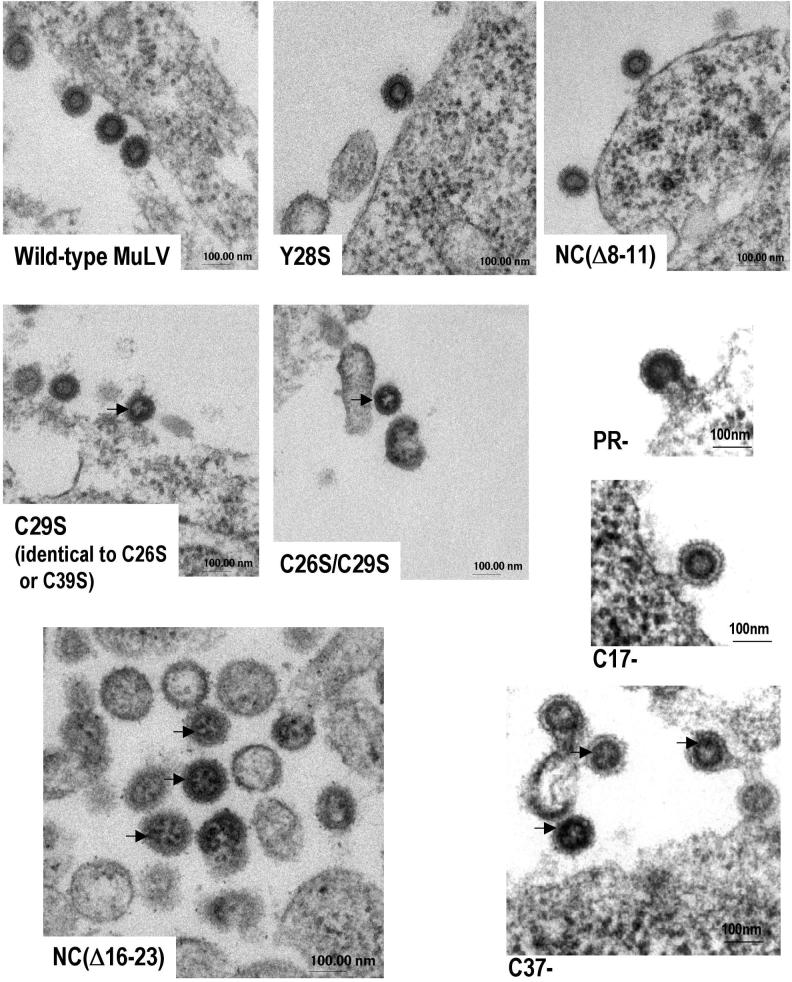
TEM thin sections of 293T cells expressing NC mutant virions. Arrows show round dark spots under the dense ring, indicating ribosomes inside the particles. The ring is the layer of immature Gag proteins of the virion core (18).
Since these proteins belong to the 60S subunit of the ribosome, these results show that ribosomal proteins, as well as rRNA, were incorporated in the particles. We also noticed that those mutants with the most rRNA also contained the most ribosomal proteins (C39S < NC[Δ16-23]). This correlation strongly suggests that the protein and RNA are packaged in the virions together, presumably in intact ribosomes or ribosomal subunits.
In order to determine whether other NC mutants exhibited this phenotype, we also analyzed the rRNA and ribosomal protein content of a number of other NC mutant particles. First, we tested the point mutants C26S, C29S, and C26/29S and Y28S. These are all within the zinc finger in NC, and all of them profoundly reduce the packaging of viral RNA (12, 13) (Fig. 4B, lanes 5, 6, 7, and 9, respectively). We found that these particles contained rRNA (Fig. 4A, lanes 5, 6, 7, and 9). In contrast, the NC(Δ 8-11) deletion mutant, which packages viral RNA and is infectious (21) (Fig. 4B, lane 8), contained no more rRNA than the traces found in wild-type particles (Fig. 4A, compare lane 3 with lane 8). No rRNA could be detected in 23 ml of mock virus preparation, suggesting that the RNA in virus pellets is not derived from cell debris (Fig. 4A, lane 1).
We tested the same panel of mutants for the ribosomal P0 protein. As shown in Fig. 4C, the protein could be detected in all of the mutants (lanes 5, 6, 7, 11, and 13), except NC(Δ 8-11) (lane 12), but not in the wild-type control (lane 4 or 10) or in the mock preparation (lane 14). We conclude that all of the mutants in our panel that fail to package viral RNA because of a defect in NC package both rRNA and ribosomal proteins, probably in the form of ribosomes.
(iv) Ribosomes inside NC mutant particles.
To determine whether ribosomes were located inside the particles, we examined thin sections of 293T cells expressing NC mutants and wild-type particles by electron microscopy. We found that the majority of the budding particles of those NC mutants that lack viral RNA (C29S, C26S, C26S/C29S, C39S, NC[Δ16-23], and C37−) contained a grainy interior, with “dark spots” within them. These dark spots resembled the ribosomes visible in the cytoplasm of the virus-producing cells and seemed to be located just under the bilayer of the immature capsid (dark ring representing the layer of Gag proteins), as indicated by the arrows (Fig. 5). The spots were not observed inside wild-type, PR−, C17−, or NC(Δ 8-11) particles and were hard to detect inside Y28S. These data lend further strong support for the conclusion that NC mutant particles that do not package the viral genomic RNA encapsidate ribosomes.
(v) Analysis of cellular RNA in SFV-derived MuLV NC mutant Gag VLPs.
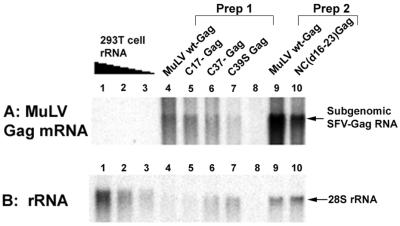
We previously analyzed the RNA content of VLPs produced in cells by the expression of MuLV Gag protein from an SFV-derived vector. This vector produces two mRNA species at extraordinary levels (the SFV genomic RNA and the SFV subgenomic RNA); indeed, the entire mRNA pool in these cells is principally composed of these two species. We found that the particles formed by wild-type MuLV Gag under these conditions encapsidated these two species, despite the absence of any retroviral packaging signals in these RNAs (17), with a preference for the 2.4-kb subgenomic mRNA coding for Gag. A unique advantage of this system is that, because these mRNAs are so abundant in the cytoplasm, we can easily detect their incorporation in retrovirus particles. We analyzed the RNAs packaged by MuLV NC mutants under these conditions. The RNAs, extracted from Gag VLPs, were separated by electrophoresis on denaturing gels and identified by Northern analysis. As shown in Fig. 6A, some of the SFV-derived, 2.4-kb mRNA was encapsidated by the mutant particles, but in each case (except perhaps C17−, lane 5) significantly less than in the particles formed by expression of wild-type Gag (compare lane 4 in Prep1 with lanes 5, 6, and 7, and compare lane 9 in Prep 2 with lane 10). Conversely, while there was some 28S rRNA detectable in the wild-type particles (Fig. 6B, lanes 4 and 9), far more was present in the particles formed from C37− Gag (lane 6), C39S Gag (lane 7), and NC(Δ16-23) Gag (lane 10). The data in Fig. 6A and B show that the less that the NC mutant VLPs packaged mRNA, the more they packaged 28S rRNA. This observation is consistent with the measurement of total RNA by the Ribogreen assay, showing that wild-type and mutant particles contained similar amounts of total RNA (Table 2). These results suggest that rRNA replaced mRNA and show clearly that the nature of the RNA packaged in the particles is determined by the NC domain of the Gag protein; these properties are seen in particles formed by expression of an SFV vector as well as in particles obtained by transfection of proviral clones.
FIG. 6.
Northern blots on SFV-derived Gag VLPs. RNA from wild-type Gag VLPs (lane 4), C17− Gag (lane 5), C37− Gag (lane 6), and C39S Gag (lane 7) VLPs of Prep1 and wild-type (wt)-Gag (lane 9) and NC(Δ16-23)-Gag (lane 10) VLPs of Prep2 were analyzed. VLP preparations from BHK-21 cells electroporated with pSFV-LacZ RNA are “mock” (lane 8). Lane 8 corresponds to 5 ml of cell media. (Lanes 4, 5, 6, and 7 of Prep1 contain equal amounts of particles; lanes 9 and 10 of Prep2 also contain equal amounts. However, levels of virus were different between Prep1 and Prep2). (A) Identification of SFV-Gag mRNA by Northern blot using a full-length proviral MuLV probe. (B) Identification of rRNA by Northern blot: the same membrane as in panel A was reprobed with a ribosomal DNA probe. SFV subgenomic Gag mRNA (2.4 kb) and 28S (∼5 kb) rRNA are indicated. rRNA extracted from 293T cells served as control (lanes 1 to 3, respectively, 1.2, 0.6 and 0.3 μg).
TABLE 2.
RNA content of MuLV wild-type Gag and NC mutant Gag SFV-derived VLPsa
| Gag protein | Total RNA content (%) (by Ribogreen assay) | SFV subgenomic mRNA content (by Northern blot) | rRNA content in VLPs (by Northern blot) |
|---|---|---|---|
| MuLV wild type | 100 | ++ | +/− |
| C17− | 102 ± 1 | ++ | +/− |
| C37− | 92 ± 2 | + | + |
| C39S | 107 ± 1 | + | + |
| NC(Δ 16-23) | 118 ± 25 | + | + |
| Mock BHK cells | 25 ± 5 | − | − |
Total RNA content results are expressed relative to RNA content of wild-type Gag SFV-derived VLPs. ++, VLPs contain the RNA species shown by Northern bloting; +, contain fever RNA species than do ++ VLPs, +/+, contain traces of the RNA species.
(vi) Stability of MuLV NC mutant cores to detergent and their sensitivity to RNase.
A previous analysis of Ψ− MuLV particles (17) showed that they contain mRNA in place of genomic RNA. In addition, it was found that the mRNA, like genomic RNA in wild-type virions, is a structural element in the particles: the treatment of detergent-disrupted immature particles with RNase rendered the Gag protein largely soluble. Thus, it was of interest to determine whether the RNA in NC mutant particles, which is predominantly rRNA, is also a structural element in the particles. We used the SFV-derived expression system (Fig. 6) for these experiments, since it produces relatively high levels of particles and since these particles, lacking PR, do not undergo proteolytic maturation.
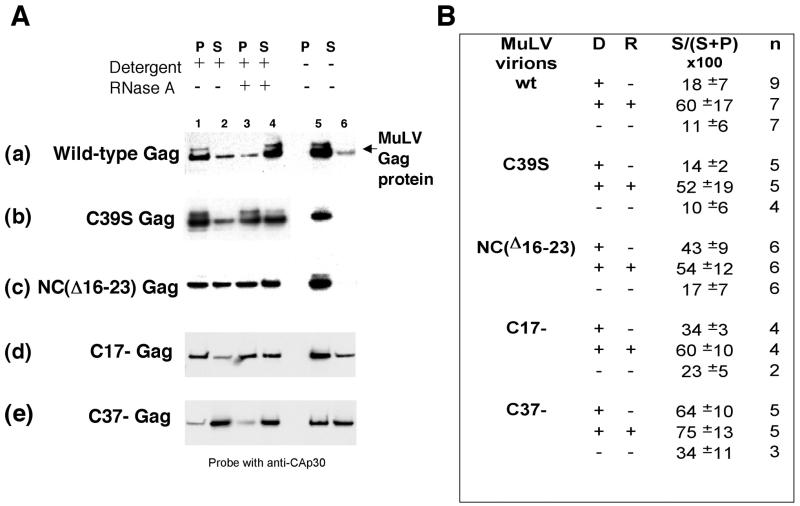
In our initial experiments, we attempted to reproduce the RNase results originally obtained using virions produced by transient transfection of PR-MuLV genomes in 293T cells (17). We found that the immature cores obtained from VLPs produced by expression of SFV-derived wild-type Gag in BHK cells were more difficult to disrupt than were the cores from 293T cells. Therefore, the virion membrane was stripped with a detergent cocktail made of 1% NP-40 and 1% Triton X-100 rather than of 1% NP-40 alone. Under these conditions, wild-type Gag cores were sensitive to RNase A, as shown by the appearance of Gag proteins in the soluble fraction (Fig. 7A, gel a, lane 4). Under the same conditions, C39S Gag cores (Fig. 7A, gel b) and C17− cores (Fig. 7A, gel d) behaved like the wild type (in some experiments, C17− Gag cores appeared more detergent sensitive than did wild-type Gag VLPs). Quantitation of results obtained in multiple experiments is shown in Fig. 7B.
FIG. 7.
Effects of detergent and RNase A on NC mutant core stability. Gag VLPs were incubated with or without detergent (1% NP-40 + 1% Triton X-100) and were then incubated in the presence or the absence of RNase A before fractionation by centrifugation. The Gag proteins present in the supernatant (S) and pellet (P) were fractionated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and were analyzed by immunoblotting with rabbit anti-MuLV CA antiserum (A). Gels in panel A: a, wild-type Gag VLPs; b, C39S Gag VLPs; c, NC(Δ16-23)-Gag VLPs; d, C17− Gag VLPs; and e, C37− Gag VLPs. Lanes 1 to 4, virion cores in detergent; lanes 5 to 6, no detergent; and lanes 3 and 4, + RNase A. Panel B represents the quantification of gels from multiple experiments like that shown in panel A. The bands representative of Gag in supernatants (S) and pellets (P) were scanned by densitometry and were quantified by the Bio-Rad Quantity One software. The percentage of Gag released in supernatant can be evaluated for each mutant in the presence (+) or in the absence (−) of detergent (D) and RNase (R). n is the number of experiments.
In contrast, NC(Δ16-23) Gag VLPs were highly detergent sensitive: NC(Δ16-23) Gag proteins were partially soluble even without digestion with RNase A (Fig. 7A, gel c, lanes 1 and 2). RNase treatment caused only a slight increase in the amount of Gag found in the soluble fraction (Fig. 7A, gel c, lane 4). These results suggest that NC(Δ16-23) Gag-Gag or Gag-RNA interactions are weaker than in wild-type Gag VLPs. Similarly, C37− Gag VLPs seemed quite fragile in our experiments: on average, more than 50% of C37− Gag proteins were soluble after detergent treatment (Fig. 7A, gel e, lanes 1 and 2). The RNase treatment had little or no additional effect (Fig. 7A, gel e, lanes 3 and 4). In fact, C37− Gag VLPs were not even very stable after centrifugation and resuspension in TNE buffer (7A, gel e, lane 6). Overall, these results showed that some mutations in the NC domain of Gag affected virion core stability and weakened Gag-RNA and/or Gag-Gag interactions but that, for some mutants, RNA seemed to still play a structural role in Gag VLPs. Correct interactions between RNA and NC are necessary for maintaining virion core stability.
DISCUSSION
The results of this work can be summarized as follows: MuLV NC mutants lacking viral genomic RNA contain the same amount of total RNA as do wild-type MuLV particles (Table 1). While wild-type particles package viral RNA, these NC mutant particles mainly encapsidate rRNA and very little cellular poly(A)+ mRNA (Fig. 2 and 4; Table 1). Furthermore, these mutants also contain ribosomal proteins (Fig. 3 and 4), and electron-dense ribosomes are visible by electron microscopy in thin sections of virus particles from transiently transfected 293T cells (Fig. 5). rRNAs were also observed in Gag VLPs (obtained by expression of Gag in an SFV vector) containing mutations in the NC domain of Gag (Fig. 6). Finally, the treatment of VLP cores with RNase revealed that, at least in some of the mutants, RNA seems to be a structural element in Gag VLP cores (Fig. 7).
We have compared the mutant particles with wild-type virus by several quantitative and semiquantitative approaches. Analysis of end-labeled RNA profiles (Fig. 2A) indicates that there is ∼60% as much rRNA (in mass) in C37− mutant virions as genomic RNA in wild-type particles. The end-labeling experiments also showed that the mutant particles contain virtually the same amount of small cellular RNAs as do wild-type or PR− particles, except for C17− (Table 1); these RNAs constitute ∼20% of the mass of RNA in wild-type particles (2). Ribogreen assay measurements showed that the mutant particles contain the same total amount of RNA (per amount of Gag protein) as do wild-type particles (Table 1). Therefore, rRNA is evidently the predominant RNA species in mutant particles. Since a ribosome contains ∼6.7 kb of high-molecular-weight RNA and since a wild-type particle should contain an ∼16-kb dimer of genomic RNA molecules, the data suggest that mutant particles contain, on the average, two or three ribosomes. This average seems reasonably consistent with the electron microscopy observations (Fig. 5).
Our assays also indicated that the mutant particles contain, in addition to rRNA, a low level of poly(A)+ mRNA (<10% of the amount of poly[A]+ mRNA found in wild-type particles) (Table 1). It seems possible that these particles contain polysomes; for example, a polysome composed of three ribosomes translating an mRNA for a 300-residue protein would be <5% poly(A)+ mRNA.
The present results are in striking contrast with those obtained with Ψ− MuLV particles, which are assembled from wild-type Gag protein in cells lacking viral RNA. In the latter case, the RNA in the particles is principally cellular mRNA (17). The mRNA in these particles (and the genomic RNA in wild-type particles) was shown to be a structural element in the particles, since digestion of detergent-treated immature capsids with RNase led to the solubilization of the viral Gag protein (17). It was thus of great interest to know whether the RNA in NC mutant particles played a similar structural role. It is conceivable that the RNA in a ribosome could interact with other nonribosomal proteins, such as Gag, since a large fraction of the outer surface of a ribosome consists of RNA rather than protein (20). We found that C39S Gag VLP capsids, like those composed of wild-type Gag proteins, are sensitive to RNase (Fig. 7). These results are consistent with the idea that these VLPs are assembled on a scaffold of rRNA, despite the absence of an intact zinc finger in the NC domain of C39S Gag protein. C17− mutant particles also behaved like wild-type particles in these tests (Fig. 7D); thus, the 17 C-terminal amino acids of NC (including the basic residues R44, R47, and R50) are not essential for core stability.
A different result was obtained with NC(Δ16-23) and C37− Gag VLP cores. We found that these particles were disrupted to a significant extent simply by the detergent treatment that was used to expose the capsid to RNase. These findings suggest that the basic region on the N-terminal side of the zinc finger (i.e., from R16 to R23 of NC) and the region of NC on the C-terminal side of residue 23 are both critical for core stability; these residues might participate in Gag-Gag or Gag-RNA interactions that maintain the structure of the wild-type Gag particle. It has been previously reported that basic residues of NC proteins, flanking the zinc finger, are crucial in RNA packaging, in the initiation of reverse transcription (15, 21), and in particle assembly (4, 7). Our results are consistent with these earlier reports. The fact that the NC(Δ16-23) and C37− Gag VLP cores are more sensitive than wild-type controls to mild detergent treatment further highlights the significance of the NC domain in particle structure, suggesting that the protein-protein interactions involving the matrix, p12, and CA domains are all relatively weak interactions. It is remarkable to note the correlation between the number of basic residues lost in the mutants and the fragility of the particles: C37− is missing five basic residues, NC(Δ16-23) is missing four, and C17− is missing three.
How can we explain the presence of ribosomes in the mutant particles? Several hypotheses can be entertained. One possibility is that the ribosomes enter the particles passively, as constituents of the cytoplasm. One other class of virus, i.e., arenavirus, contains ribosomes (5). Arenavirus particles acquire ribosomes during budding from the plasma membrane of infected BHK-21 cells, and the ribosomes are apparently not required for infectivity.
A second hypothesis, alluded to above, is that rRNA in ribosomes interacts directly with the mutated NC domain of Gag and acts as scaffolding for the mutant particles, just as viral RNA and cellular mRNA do for particles assembled from wild-type Gag (17). Perhaps the wild-type NC domain is responsible for the ability of Gag to discriminate between mRNA and rRNA; in the absence of this function, rRNA (by far the most abundant RNA in the cell) is the “default” packageable RNA and scaffold. Careful examination of electron micrographs of the mutant particles lends some support to this possibility: the small electron-dense spots that we believe to be ribosomes are located just under the dark ring of Gag protein in the budding particle (Fig. 5, particularly the NC[Δ16-23], C37−, and C26S images). We found it difficult to test this possibility experimentally by RNase digestion, as the mutant particles proved to be sensitive to the mild detergent used to expose the immature capsid to RNase. It is also possible that the mutant Gag proteins interact with ribosomal proteins rather than rRNA. The Z protein of the arenavirus lymphocytic choriomeningitis virus interacts with the P0 protein in the nucleus of infected cells and is also found within the virions (3). Indeed, nucleolin, a cellular protein involved in ribosome assembly, has been shown to interact with MuLV NC and is incorporated into virions via this interaction (1). It is also interesting that alphaviruses use ribosomes for the uncoating process of their viral mRNA, an event that occurs in the cytoplasm of infected cells after virus entry (23).
The third hypothesis recognizes the possibility that the mutant particles might contain some mRNA as well as ribosomes, i.e., that the particles package polysomes. According to this model, wild-type Gag normally binds to viral RNA that is being translated (6, 19) and, in incorporating it into the particle, removes the ribosomes from it. In other words, the affinity of wild-type Gag for viral RNA and for mRNA in the absence of viral RNA is higher than the affinity of a ribosome for these RNAs, so that Gag is able to successfully compete with ribosomes for these RNAs: the binding of a wild-type NC domain displaces ribosomes from the viral RNA or mRNA. In contrast, Gag molecules with defective NC domains have not only lost the ability to recognize Ψ but are weaker competitors for these RNAs; they form particles with mRNA but fail to displace ribosomes from the mRNA. The presence of some mRNA, in addition to rRNA, in the mutant particles is evident in Gag VLPs produced by the SFV gene expression system (Fig. 6A). This hypothesis is consistent with the idea that mRNA molecules act as scaffolding in mutant particles, even though these particles contain far more rRNA than mRNA.
In conclusion, we have found that MuLV particles with defects in the NC domain of Gag contain rRNA, evidently as a component of ribosomes. This result suggests that one function of the wild-type NC domain is to exclude ribosomes from particles during assembly. We are presently trying to determine whether Gag interacts with ribosomes and also whether the ribosomes in the particles are engaged in translation. We hope that these experiments will clarify the significance of their presence in NC mutant particles.
Acknowledgments
We thank Henrik Garoff for the gift of SFV-derived vectors, Barbara Sollner-Webb for the plasmid-containing ribosomal cDNA sequence, Sandra Ernst for the cloning of C17− and C37− MuLV proviral clones, and Dave Ott and Cathy Hibbert for insightful comments on the manuscript.
Electron microscopy was supported by contract no. N01-CO-12400.
REFERENCES
- 1.Bacharach, E., J. Gonsky, K. Alin, M. Orlova, and S. P. Goff. 2000. The carboxy-terminal fragment of nucleolin interacts with the nucleocapsid domain of retroviral gag proteins and inhibits virion assembly. J. Virol. 74:11027-11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. [DOI] [PubMed] [Google Scholar]
- 3.Borden, K. L. B., E. J. Campbelldwyer, G. W. Carlile, M. Djavani, and M. S. Salvato. 1998. Two RING finger proteins, the oncoprotein PML and the arenavirus Z protein, colocalize with the nuclear fraction of the ribosomal P proteins. J. Virol. 72:3819-3826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowzard, J. B., R. P. Bennett, N. K. Krishna, S. M. Ernst, A. Rein, and J. W. Wills. 1998. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J. Virol. 72:9034-9044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchmeier, M., M. Bowen, and C. Peters. 2001. Arenaviridae: the viruses and their replication, p. 1635-1668. In D. M. Knipe and P. M. Howley (ed.), Fields virology, vol. 2. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 6.Butsch, M., and K. Boris-Lawrie. 2002. Destiny of unspliced retroviral RNA: ribosome and/or virion? J. Virol. 76:3089-3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74:3046-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ekstrom, M., H. Garoff, and H. Andersson. 1998. Semliki Forest virus expression system, p. 218-224. In J. E. (ed.), Cell biology: a laboratory handbook, 2nd ed., vol. 4. Academic Press, New York, N.Y.
- 9.Elkon, K. B., A. P. Parnassa, and C. L. Foster. 1985. Lupus autoantibodies target ribosomal P proteins. J. Exp. Med. 162:459-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu, W., B. A. Ortiz-Conde, R. J. Gorelick, S. H. Hughes, and A. Rein. 1997. Placement of tRNA primer on the primer-binding site requires pol gene expression in avian but not murine retroviruses. J. Virol. 71:6940-6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu, W., and A. Rein. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 67:5443-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorelick, R. J., L. E. Henderson, J. P. Hanser, and A. Rein. 1988. Point mutants of Moloney murine leukemia virus that fail to package viral RNA: evidence for specific RNA recognition by a “zinc finger-like” protein sequence. Proc. Natl. Acad. Sci. USA 85:8420-8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorelick, R. J., S. M. Nigida, Jr., L. O. Arthur, L. E. Henderson, and A. Rein. 1991. Roles of nucleocapsid cysteine arrays in retroviral assembly and replication: possible mechanisms in RNA encapsidation, p. 257-272. In A. Kumar (ed.), Advances in molecular biology and targeted treatment for AIDS. Plenum Press, New York, N.Y.
- 14.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 15.Housset, V., H. De Rocquigny, B. P. Roques, and J. L. Darlix. 1993. Basic amino acids flanking the zinc finger of Moloney murine leukemia virus nucleocapsid protein NCp10 are critical for virus infectivity. J. Virol. 67:2537-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, K. J., and H. Garoff. 1996. Production of infectious recombinant Moloney murine leukemia virus particles in BHK cells using Semliki Forest virus-derived RNA expression vectors. Proc. Natl. Acad. Sci. USA 93:11658-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muriaux, D., J. Mirro, D. Harvin, and A. Rein. 2001. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 98:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nermut, M. V., and D. J. Hockley. 1996. Comparative morphology and structural classification of retroviruses. Curr. Top. Microbiol. Immunol. 214:1-24. [DOI] [PubMed] [Google Scholar]
- 19.Poon, D. T., E. N. Chertova, and D. E. Ott. 2002. Human immunodeficiency virus type 1 preferentially encapsidates genomic RNAs that encode Pr55(Gag): functional linkage between translation and RNA packaging. Virology 293:368-378. [DOI] [PubMed] [Google Scholar]
- 20.Ramakrishnan, V., and P. B. Moore. 2001. Atomic structures at last: the ribosome in 2000. Curr. Opin. Struct. Biol. 11:144-154. [DOI] [PubMed] [Google Scholar]
- 21.Rein, A., D. P. Harvin, J. Mirro, S. M. Ernst, and R. J. Gorelick. 1994. Evidence that a central domain of nucleocapsid protein is required for RNA packaging in murine leukemia virus. J. Virol. 68:6124-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Press, Cold Spring Harbor, N.Y.
- 23.Singh, I., and A. Helenius. 1992. Role of ribosomes in Semliki Forest virus nucleocapsid uncoating. J. Virol. 66:7049-7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Southern, P. J., and P. Berg. 1982. Transformation of mammalian cells to antibiotic resistance with a bacterial gene under control of the SV40 early region promoter. J. Mol. Appl. Genet. 1:327-341. [PubMed] [Google Scholar]
- 25.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In J. M. Coffin, S. H. Hughes, and H. E. Varmus (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, N.Y. [PubMed]
- 26.Tobin, G. J., K. Nagashima, and M. A. Gonda. 1996. Immunologic and ultrastructural characterization of HIV pseudovirions containing Gag and Env precursor proteins engineered in insect cells. Methods 10:208-218. [DOI] [PubMed] [Google Scholar]