Abstract
A kinetic study of the alkaline transition of DNA, in clearly defined physico-chemical conditions, is presented, which allows us to identify, within the alkaline transition region, different pH ranges, corresponding to different ratelimiting factors. This analysis brings into consideration three distinct intervals of time which characterize the whole process, namely the time necessary for full hyperchromicity to be reached, the time required for strand separation in the case of a single DNA molecule, and the time for complete denaturation to be reached in the case of a DNA solution.
The results obtained from ultracentrifugal, and spectrophotometric measurements, involving rapid mixing experiments, seem to indicate the following conclusions: whereas, in the lower pH ranges considered within the transition region, the denaturation process is limited by the first time constant, this same constant becomes extremely short at higher pH. On the other hand the fact that, in the higher pH range, the second and third time constants do not coincide (the time to unwind a single T2 DNA molecule being at least one order of magnitude shorter than the time required for bulk denaturation to be reached) suggests that in this pH range the overall denaturation rate is limited by a statistical process governing the initiation of unwinding.
These observations are discussed in terms of a model in which the unwinding energy is given by the electrostatic repulsions which originate in the deprotonated DNA molecule. The model itself suggests some experiment which seem to confirm it.
Full text
PDF

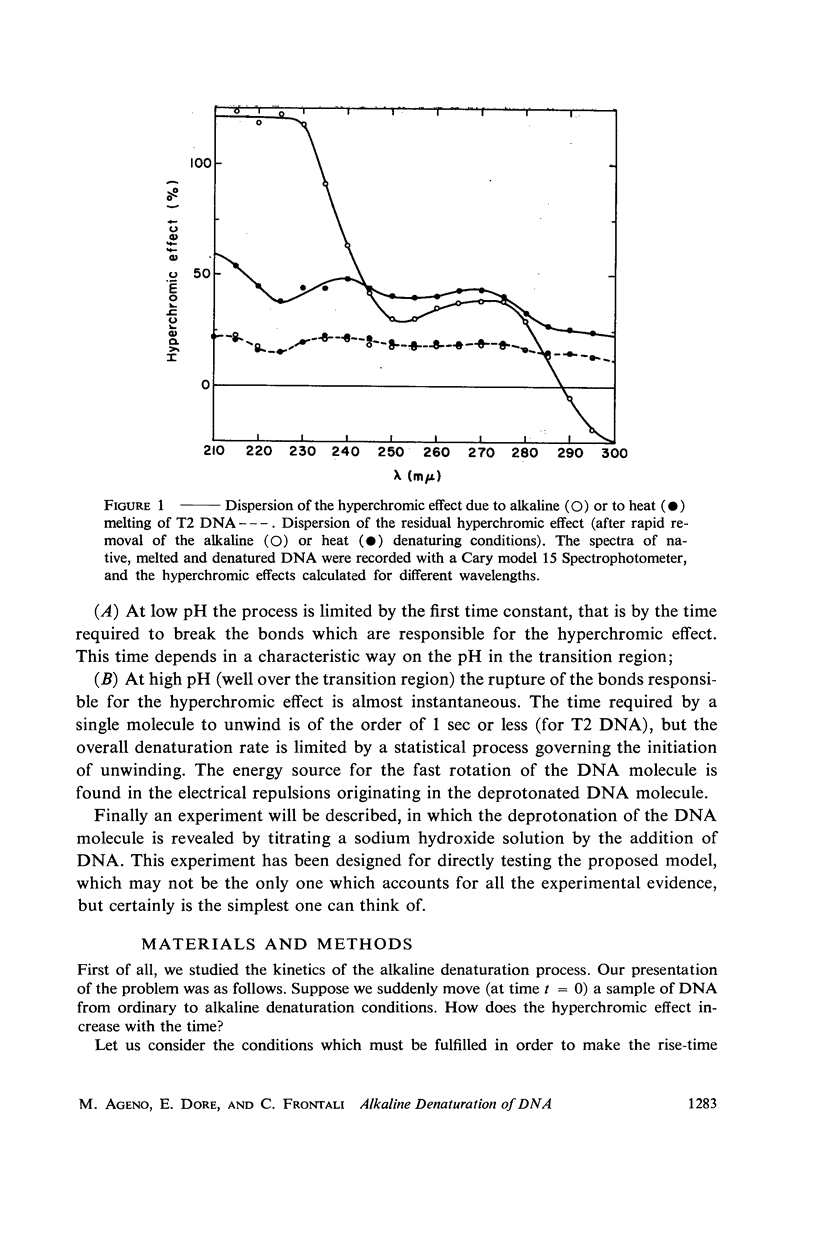

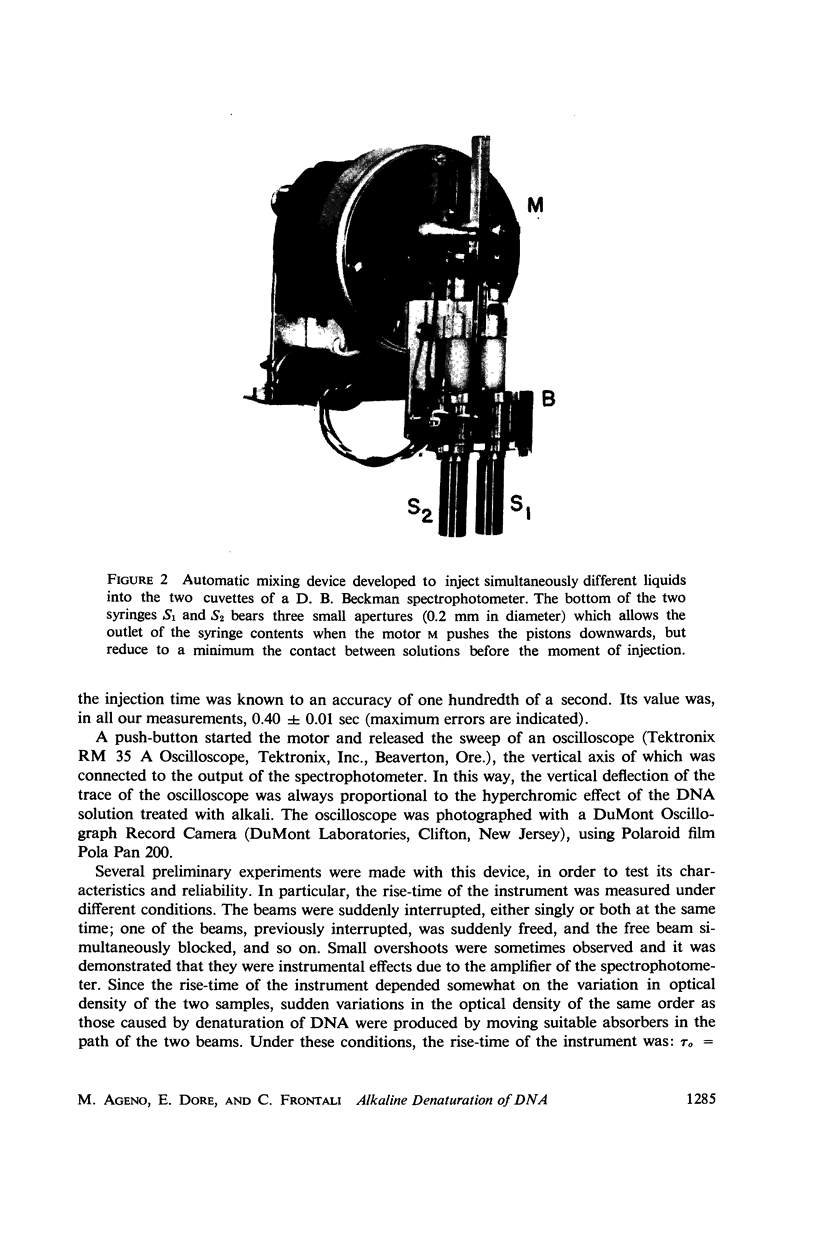
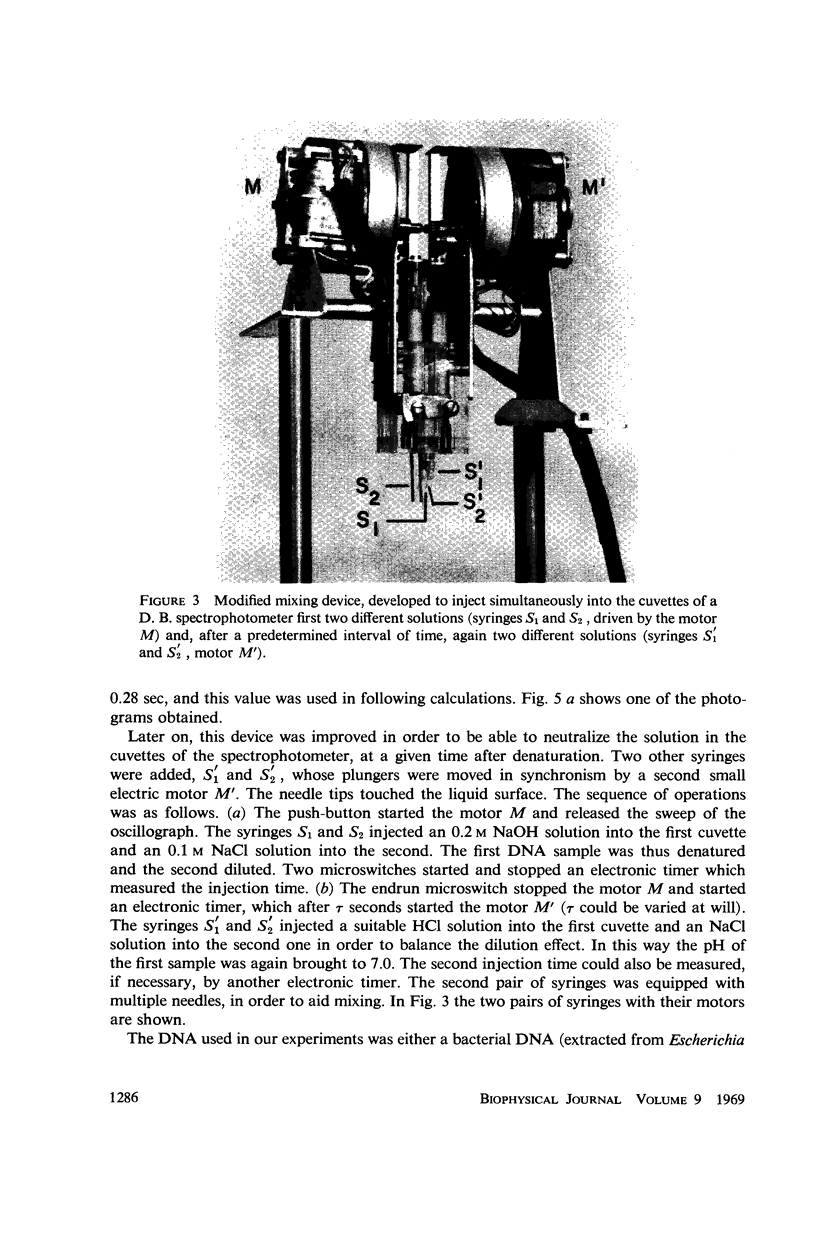

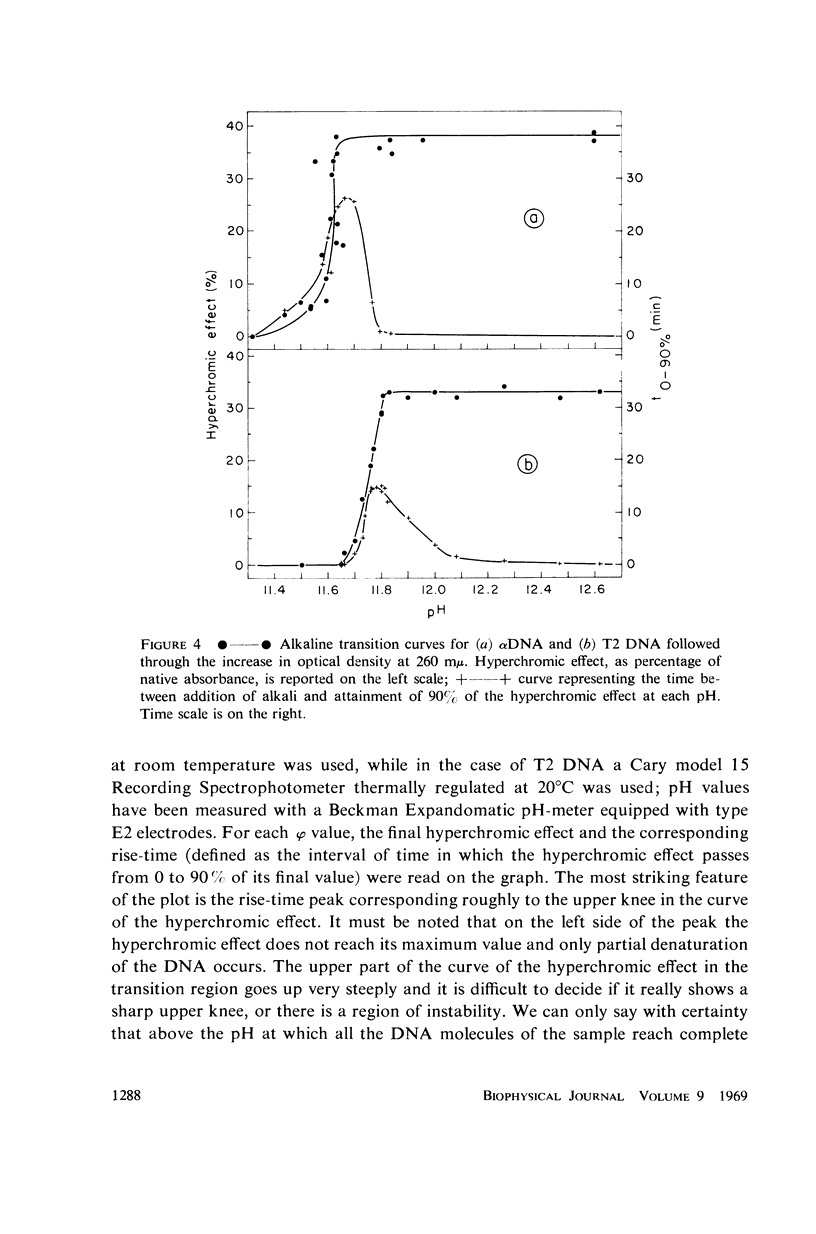


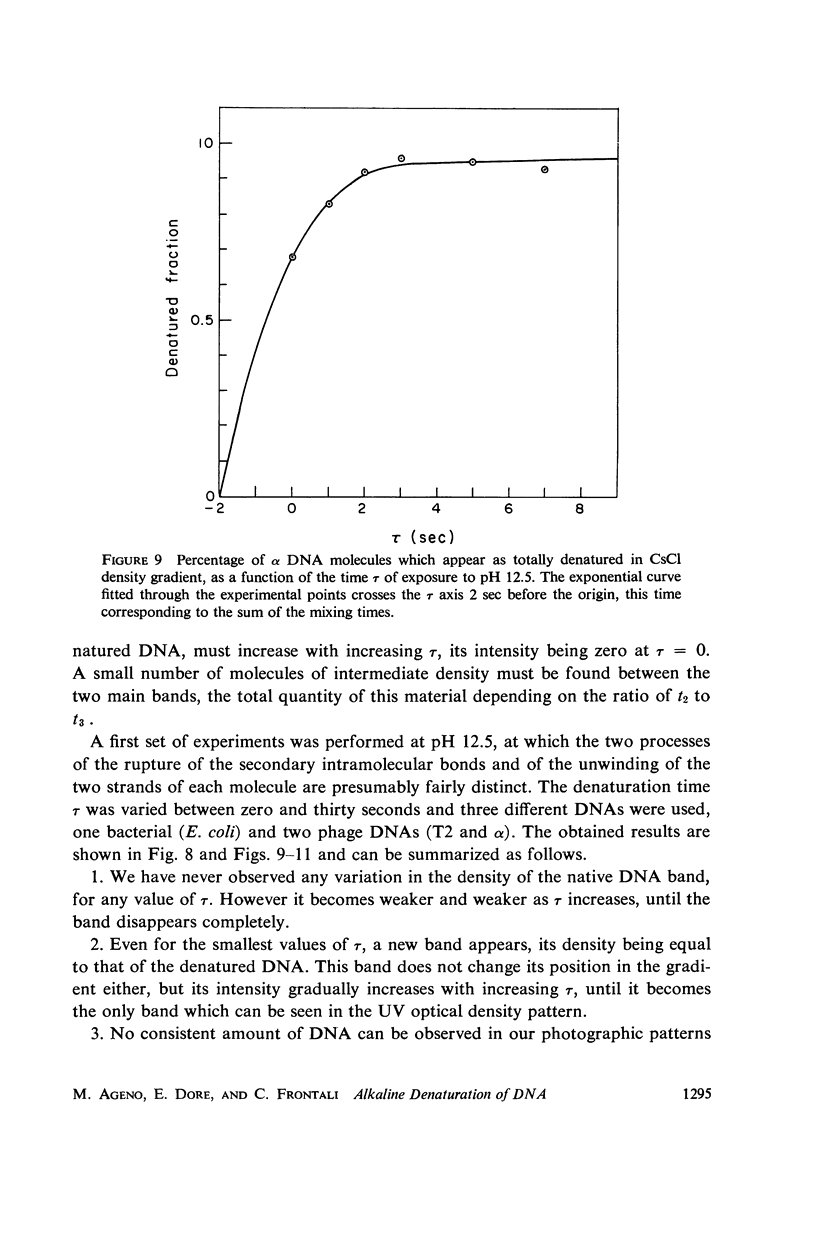
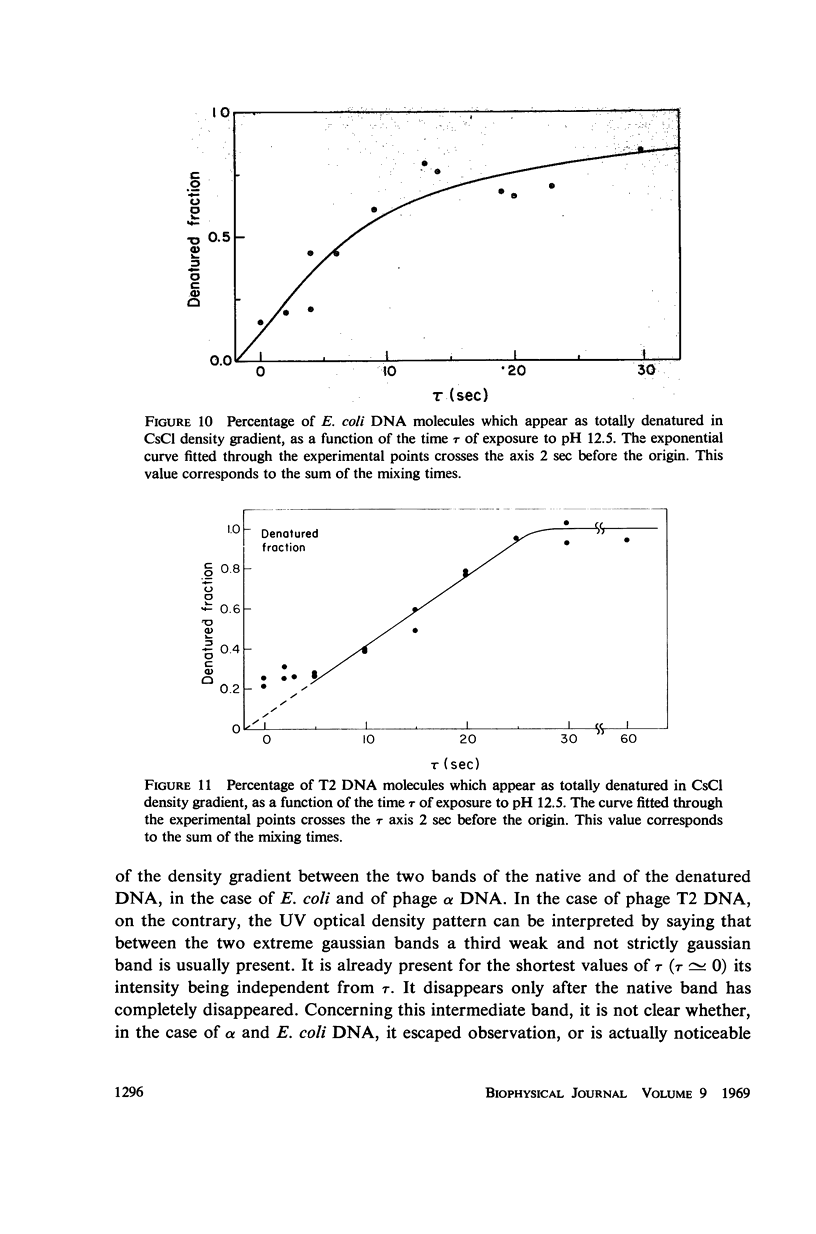

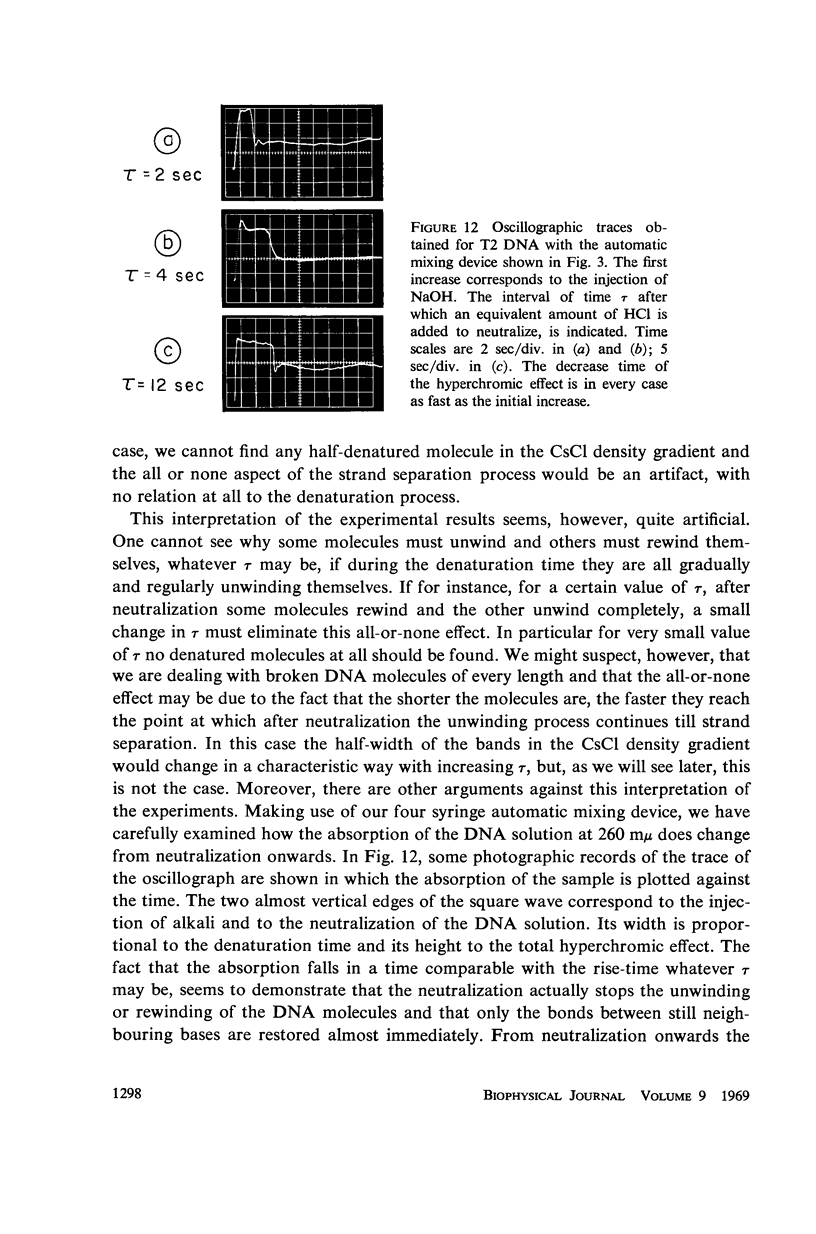


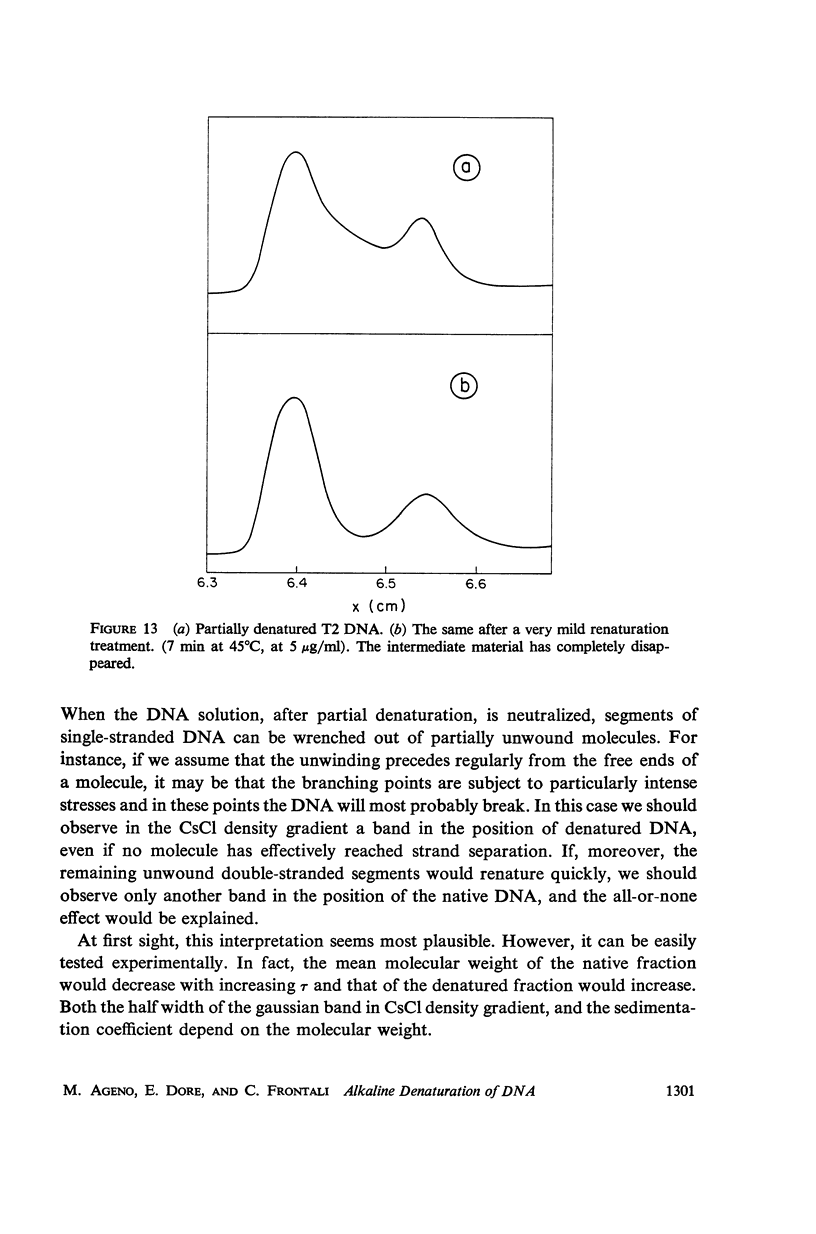
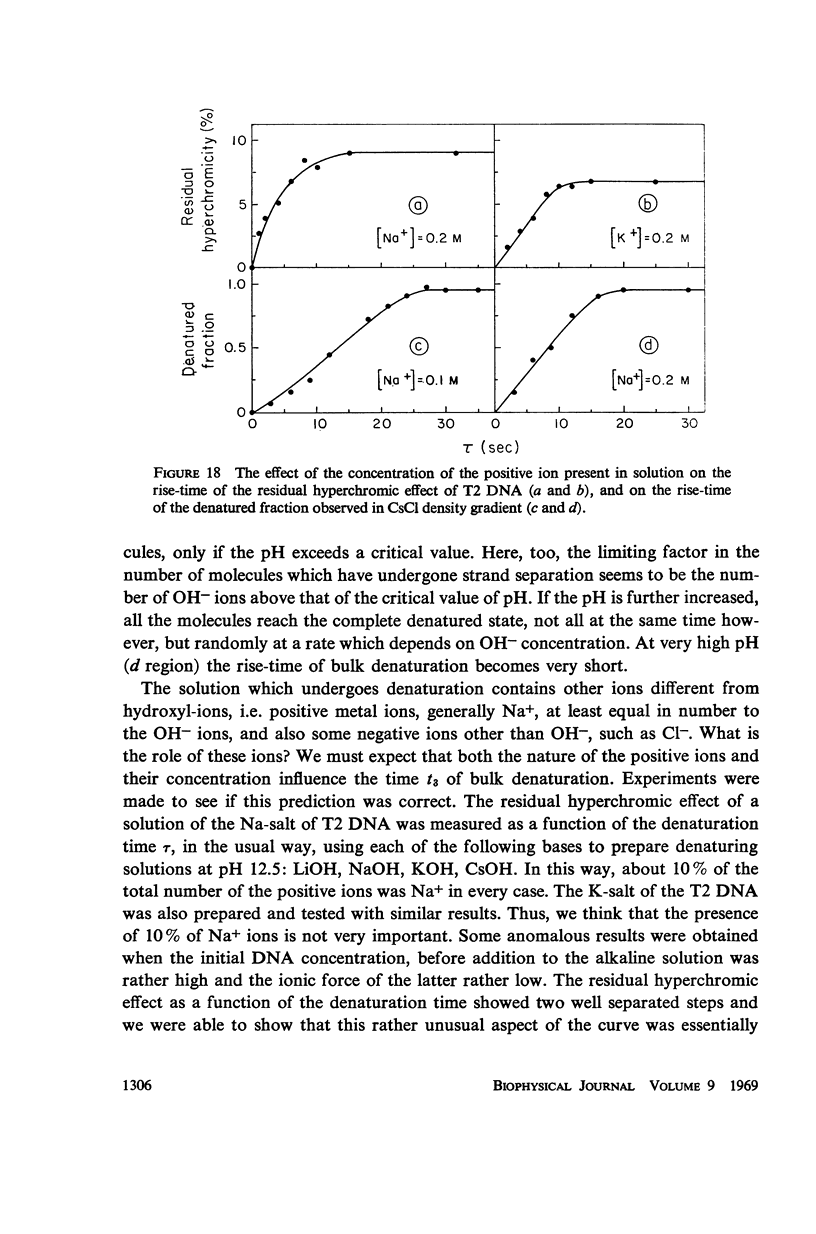



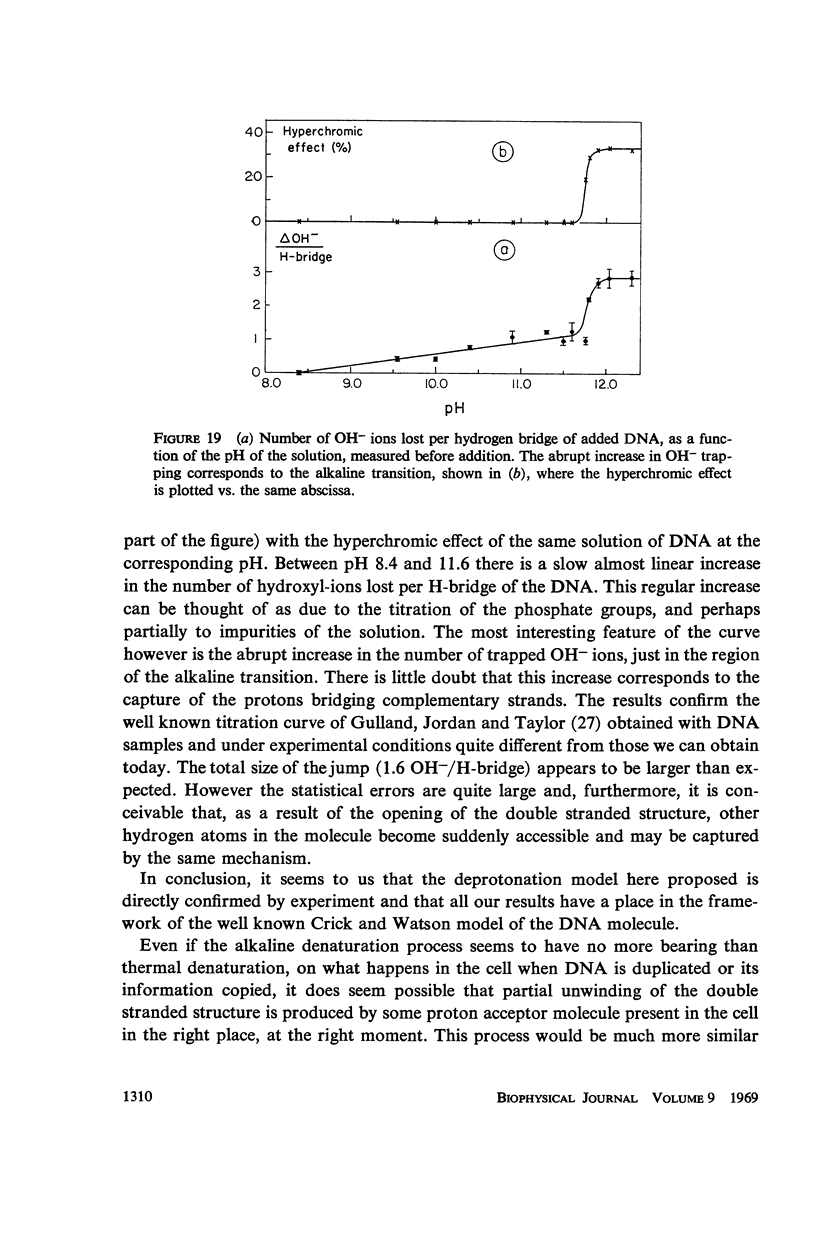

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- CAIRNS J. The bacterial chromosome and its manner of replication as seen by autoradiography. J Mol Biol. 1963 Mar;6:208–213. doi: 10.1016/s0022-2836(63)80070-4. [DOI] [PubMed] [Google Scholar]
- CROTHERS D. M., KALLENBACH N. R., ZIMM B. H. THE MELTING TRANSITION OF LOW-MOLECULAR-WEIGHT DNA: THEORY AND EXPERIMENT. J Mol Biol. 1965 Apr;11:802–820. doi: 10.1016/s0022-2836(65)80037-7. [DOI] [PubMed] [Google Scholar]
- CROTHERS D. M. THE KINETICS OF DNA DENATURATION. J Mol Biol. 1964 Sep;9:712–733. doi: 10.1016/s0022-2836(64)80177-7. [DOI] [PubMed] [Google Scholar]
- CROTHERS D. M., ZIMM B. H. THEORY OF THE MELTING TRANSITION OF SYNTHETIC POLYNUCLEOTIDES: EVALUATION OF THE STACKING FREE ENERGY. J Mol Biol. 1964 Jul;9:1–9. doi: 10.1016/s0022-2836(64)80086-3. [DOI] [PubMed] [Google Scholar]
- Eigner J., Doty P. The native, denatured and renatured states of deoxyribonucleic acid. J Mol Biol. 1965 Jul;12(3):549–580. doi: 10.1016/s0022-2836(65)80312-6. [DOI] [PubMed] [Google Scholar]
- Farchi G., Giucci D. Sui metodi di interpolazione di dati sperimentali. Ann Ist Super Sanita. 1966;2(4):722–730. [PubMed] [Google Scholar]
- Felsenfeld G., Hirschman S. Z. A neighbor-interaction analysis of the hypochromism and spectra of DNA. J Mol Biol. 1965 Sep;13(2):407–427. doi: 10.1016/s0022-2836(65)80106-1. [DOI] [PubMed] [Google Scholar]
- GEIDUSCHEK E. P. "Reversible" DNA. Proc Natl Acad Sci U S A. 1961 Jul 15;47:950–955. doi: 10.1073/pnas.47.7.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GEIDUSCHEK E. P. On the factors controlling the reversibility of DNA denaturation. J Mol Biol. 1962 Jun;4:467–487. doi: 10.1016/s0022-2836(62)80103-x. [DOI] [PubMed] [Google Scholar]
- Hayashi M. N., Hayashi M. Participation of a DNA-RNA hybrid complex in in vivo genetic transcription. Proc Natl Acad Sci U S A. 1966 Mar;55(3):635–641. doi: 10.1073/pnas.55.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidson C. Deoxyribonucleic acid secondary structure in the region of the replication point. J Mol Biol. 1966 May;17(1):1–9. doi: 10.1016/s0022-2836(66)80089-x. [DOI] [PubMed] [Google Scholar]
- Levinthal C., Crane H. R. ON THE UNWINDING OF DNA. Proc Natl Acad Sci U S A. 1956 Jul;42(7):436–438. doi: 10.1073/pnas.42.7.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MANDELL J. D., HERSHEY A. D. A fractionating column for analysis of nucleic acids. Anal Biochem. 1960 Jun;1:66–77. doi: 10.1016/0003-2697(60)90020-8. [DOI] [PubMed] [Google Scholar]
- STUDIER F. W. SEDIMENTATION STUDIES OF THE SIZE AND SHAPE OF DNA. J Mol Biol. 1965 Feb;11:373–390. doi: 10.1016/s0022-2836(65)80064-x. [DOI] [PubMed] [Google Scholar]
- Schildkraut C. Dependence of the melting temperature of DNA on salt concentration. Biopolymers. 1965;3(2):195–208. doi: 10.1002/bip.360030207. [DOI] [PubMed] [Google Scholar]
- Subirana J. A. Kinetics of renaturation of denatured DNA. II. Products of the reaction. Biopolymers. 1966;4(2):189–200. doi: 10.1002/bip.1966.360040205. [DOI] [PubMed] [Google Scholar]
- WATSON J. D., CRICK F. H. The structure of DNA. Cold Spring Harb Symp Quant Biol. 1953;18:123–131. doi: 10.1101/sqb.1953.018.01.020. [DOI] [PubMed] [Google Scholar]




