Abstract
The orthopoxvirus gene p4c has been identified in the genome of the vaccinia virus strain Western Reserve. This gene encodes the 58-kDa structural protein P4c present on the surfaces of the intracellular mature virus (IMV) particles. The gene is disrupted in the genome of cowpox virus Brighton Red (BR), demonstrating that although the P4c protein may be advantageous for virus replication in vivo, it is not essential for virus replication in vitro. Complementation and recombination analyses with the p4c gene have shown that the P4c protein is required to direct the IMV into the A-type inclusions (ATIs) produced by cowpox virus BR. The p4c gene is highly conserved among most members of the orthopoxvirus genus, including viruses that produce ATIs, such as cowpox, ectromelia, and raccoonpox viruses, as well as those such as variola, monkeypox, vaccinia, and camelpox viruses, which do not. The conservation of the p4c gene among the orthopoxviruses, irrespective of their capacities to produce ATIs, suggests that the P4c protein provides functions in addition to that of directing IMV into ATIs. These findings, and the presence of the P4c protein in IMV but not extracellular enveloped virus (D. Ulaeto, D. Grosenbach, and D. E. Hruby, J. Virol. 70:3372-3377, 1996), suggest a model in which the P4c protein may play a role in the retrograde movement of IMV particles, thereby contributing to the retention of IMV particles within the cytoplasm and within ATIs when they are present. In this way, the P4c protein may affect both viral morphogenesis and processes of virus dissemination.
One of the unusual features of the poxviruses is their ability to produce virus particles of more than one type. The majority of the orthopoxviruses can produce infectious virus particles of four major types differing in either surface structure or site of accumulation. Most abundant, often representing >90% of virus progeny, are the intracellular mature virus (IMV) particles, which are generally thought to possess a single membrane (6, 18, 22), though an alternative view is that this membrane consists of two membranes in tight apposition (16, 17, 70). A small proportion of the IMVs are converted to intracellular enveloped viruses (IEVs), which are IMVs wrapped in double membranes derived from the trans-Golgi network or tubular endosomes (20, 30, 63, 75). The IEVs are transported to the cell surface, where their outer membranes fuse with the plasma membrane to produce either the released extracellular enveloped viruses (EEVs) or the cell-associated enveloped viruses (CEVs), which remain attached to the cell surface (4). Those IMVs that are not converted into IEVs, CEVs, or EEVs remain in the cytoplasm of the cell, either as free particles or as particles embedded within A-type inclusions (ATIs). The ATIs are large, well-defined proteinaceous bodies produced in cells infected with certain strains of cowpox, ectromelia, raccoonpox, fowlpox, or canarypox viruses (9, 28, 37, 45, 53). The four different kinds of virus particles have distinct physical, immunological, and biological properties (reviewed in reference 48), suggesting that particles of each type provide some advantages for virus replication; however, the exact in vivo roles of particles of each of these types are not fully understood.
The least complex forms of infectious particles are the IMVs, which also constitutes the kernels of the other, more elaborate particles. To provide these two different functions, the proteins on the surfaces of the IMVs are involved in a variety of processes, including the formation of the IMV, directing the incorporation of the IMV within IEV or the ATI, and facilitating viral infection of cells. In keeping with the multiplicity of roles, >12 proteins that participate in these processes have been identified either at or near the surface of the IMV (reviewed in references 48 and 68).
One of the first IMV surface proteins to be identified was the P4c protein, one of a triplet of major structural proteins (designated 4a, 4b, and P4c) with apparent molecular masses of about 60 kDa. These proteins were first resolved in analyses of the proteins of the IMV particles of the Western Reserve strain of vaccinia virus (VV-WR) by Katz and Moss (40, 41). Subsequently, the P4c protein was shown to be among the last proteins incorporated into the outer surface membranes of these particles (39, 61, 62). The 4a and 4b proteins, which are generated by proteolytic cleavage of the precursor proteins P4a and P4b, function as the two major proteins of the core of the virus particle (49, 61).
Various functions have been attributed to the P4c protein. First, it was proposed to be a component of the surface tubules visualized on the surfaces of virus particles by conventional electron microscopy (72, 73, 82), though surface tubules have been visualized on virions lacking intact P4c (31, 32) and the existence of the tubules has not been confirmed by cryoelectron microscopy studies (10). Second, the P4c protein has been implicated as a factor directing the inclusion of virus particles into the ATIs. ATIs have two major phenotypes, those that contain virus particles, the V+ ATIs, and those that lack virus particles, the V− ATIs (28, 29). The V phenotype is a strain-specific property (28, 29). Usually only mature virions are found in ATIs, but immature virions whose maturation has been arrested by treatment with rifampin can associate with ATIs (30). At least two genes are required for the formation of V+ ATIs: one, the ati gene (52, 53), encoding the ATI matrix protein and one or more encoding the protein(s) required for the inclusion of virus particles, previously designated either the “In” or the “VO” factor. Cells coinfected with V− and V+ virus strains produce almost entirely V+ ATIs, in which both V+ and V− viruses are present (23, 24, 29). Fusions between cells infected with V+ or V− viruses were also used to study viral association with ATIs (67). These studies showed that the VO factor is synthesized late in infection and has rapid attachment to V+ viruses and that the residual portion in the cytoplasm attaches to V− viruses after fusion. In comparing the 16 most abundant proteins of V+ and V− particles of cowpox virus and vaccinia virus, Shida et al. (67) noted a correlation between the amount of the P4c protein present in either the particle or virus-free cell extracts and the VO activity, suggesting that the p4c protein might constitute the VO factor. Third, Ulaeto et al. (76) have provided evidence that the P4c protein is an IMV-specific protein, suggesting that IMVs that acquire the P4c protein may not differentiate into IEVs, CEVs, or EEVs.
In this study, we have identified the orthopoxvirus gene encoding the P4c protein. This gene is highly conserved among members of the orthopoxvirus genus. The identification of this gene has allowed us to confirm that one of the roles of this protein is to direct the insertion of virus particles into the ATI bodies produced in cells infected with orthopoxviruses encoding the Ati protein.
MATERIALS AND METHODS
Viruses and cells.
VV-WR (provided by W. K. Joklik), the Copenhagen strain of vaccinia virus (provided by R. Drillien), and the Brighton Red strain of cowpox virus (CPV-BR) were used in this study. Strain 143 human osteosarcoma cells or STO cells (mouse embryonic fibroblasts; ATCC strain CRL 1503) were used for virus culture and the selection of recombinant viruses. For protein analyses, vaccinia viruses were grown in spinner cultures of HeLa S3 cells cultured in Joklik's modification of minimal essential medium (Gibco-Invitrogen) containing 7.5% calf serum and purified as previously described (36).
Plasmid and recombinant virus constructions.
Escherichia coli strains TG1 and JM103, originally provided by G. Winter (MRC Laboratory of Molecular Biology, Cambridge, United Kingdom), were cultured in Luria broth or tryptone-yeast extract broth. DNA fragments were inserted into plasmids derived from pGEM7zf+ (Promega Biotec, Madison, Wis.), pUC4K, or pUC19 (79) using standard techniques (57).
To subclone the VV-WR p4c gene, the 6.6-kbp XhoI-SalI fragment (containing the p4c gene) from the XhoI E fragment of VV-WR was inserted into the XhoI site in pGEM7zf+, generating plasmid p1540. The 1.8-kbp EcoRV fragment from p1540 containing the entire p4c gene under the transcriptional control of its native promoter was inserted (using HindIII linkers) into the HindIII site of pGEM7zf+, generating plasmid p1542.
To generate a VV-WR containing an inactivated p4c gene, the following procedure was used. The 1.8-kbp HindIII fragment containing the p4c gene from the DNA of plasmid p1542, was inserted into the HindIII site of a pUC19 derivative that lacks the rest of the pUC19 polylinker. The 2.0-kbp EcoRI fragment from plasmid pTK61-gpt (11), containing the E. coli xanthine-guanine phosphoribosyltransferase (gpt) gene under the control of the promoter of the vaccinia virus 7.5K gene, was inserted into the EcoRI site in the p4c gene to create plasmid p1546. This plasmid was used to construct a recombinant vaccinia virus containing a p4c gene disrupted by a functional copy of the gpt gene. The resulting vaccinia virus recombinant, A504, was isolated by positive gpt selection according to the method of Falkner and Moss (11). The presence of the insert of the gpt gene was verified by DNA hybridization analyses (71) of DNA extracted from cells infected with the recombinant virus. A wild-type p4c revertant of vaccinia virus A504 was generated by recombination of plasmid p1542 DNA with the DNA of virus A504. The resulting recombinant vaccinia virus, A507, was isolated using negative gpt selection as described previously (34).
To create a CPV-BR recombinant expressing the p4c gene, the 1.8-kbp HindIII fragment containing the p4c gene from p1542 was inserted (using ClaI linkers) into the ClaI site of plasmid p1249 (33), which contains the vaccinia virus thymidine kinase gene in pUC4K. This plasmid, p1544, was used to insert a functional copy of the p4c gene into the thymidine kinase gene of CPV-BR. The resulting recombinant cowpox virus, A505, was isolated by selection for viruses lacking a functional thymidine kinase as described previously (44). The presence of the insert in the tk gene was verified by DNA hybridization analyses (71) of DNA extracted from cells infected with the tk mutant virus isolates.
Additional plasmids containing partial deletions of the p4c gene (from plasmid p1542) were generated for use as templates in nucleotide sequence determinations. Plasmid p1548 was created by the deletion of p1542 from the EcoRI site in the p4c gene through the end of the gene to the EcoRI site in the pGEM7zf+ vector. A deletion from the BamHI site in the pGEM7zf+ vector through the start of the p4c gene to the AccI site in the p4c gene generated p1550. Plasmid p1552 was created by deleting the sequences in p1542 from the AccI site in the p4c gene through to the end of the gene to the ClaI site in the pGEM7zf+ vector.
Standard methods (3, 60, 74, 79) were used to determine the nucleotide sequences of both strands of the EcoRV fragments containing the p4c genes of CPV-BR and VV-WR.
Metabolic labeling of proteins in infected cells.
Human 143 cells (106) in monolayer cultures were infected with a virus at a multiplicity of 5 PFU/cell. Eighteen hours after infection, the medium was removed and nascent proteins were labeled by the addition of 20 μCi of [35S]methionine in 200 μl of Eagle's minimal essential medium at 37°C for 30 min. The medium was removed, the cells were washed twice with cold phosphate-buffered saline, and the cells were lysed with 0.3 ml of buffer containing 50 mM Tris (pH 7.0), 30 mM dithiothreitol, 5 mM EDTA, 1% sodium dodecyl sulfate (SDS), and 10% glycerol. Each lysate was passed through a 25-gauge needle five times to shear the DNA in the sample.
Electron microscopy of infected cells.
Electron microscopy of cells infected with poxvirus was performed as described previously (47). For each infection, human 143 cells at 80% confluence in two 30-cm2 dishes were infected either with a multiplicity of 10 PFU/cell for single-virus infections or with a multiplicity of 5 PFU of each virus/cell for coinfections of two different viruses. The cells were fixed 18 h after infection by washing them twice with 3 ml of cold phosphate-buffered saline and adding 3 ml of CS buffer, a buffer of 0.1 M sodium cacodylate containing 3.4% sucrose (pH 7.4; 300 mosM), containing 2% glutaraldehyde. After glutaraldehyde fixation for 5 min, the cells were scraped from the dishes with a rubber scraper and centrifuged at 10,000 × g for 5 min in an Eppendorf microcentrifuge. The cells were allowed to sit undisturbed as a pellet in the tube for an additional hour. The supernatant was removed with a pipette, and the cells were dried with a pointed piece of filter paper. The tip of the tube was cut off with a razor blade, and the pellet was placed onto Parafilm, where it was further dried to a thick paste. It was then divided into clumps (0.5 mm3), and each clump was encased in molten agar cooled to 40°C. After the agar solidified (10 min), the cell pellets were washed three times, each time for 3 min, with CS buffer. The cell pellets were postfixed with 1% osmium tetroxide-0.1 M sodium cacodylate buffer for 1 h. They were washed twice, each time for 10 min, in CS buffer. Two additional 10-min washes were performed with 0.11 M sodium veronal acetate buffer, pH 7.4. Following en bloc staining with 1% uranyl acetate-0.11 M sodium veronal acetate buffer for 1 h, the pellets were washed with 0.11 M sodium veronal acetate buffer, pH 7.4, for 5 min and then briefly with distilled water. The pellets were dehydrated in a graded series of ethanol (10 min per wash), 30, 60, 80, and 95%, concluding with three final washes in 100% ethanol for 10 min. The pellets were infiltrated with a mixture of 50% Spurr's resin in absolute ethanol for 1 h followed by five changes of 100% resin for 10 min each. The resin was polymerized by baking it at 70°C for 8 h. Ultrathin sections were cut on a Reichert-Jung ultracut E microtome using a diamond knife. They were poststained in aqueous saturated uranyl acetate for 5 min, washed in water, and further stained in lead citrate for 2 min. After a final wash in water, the sections were viewed and photographed in a Philips EM300 electron microscope.
Nucleotide sequence accession numbers.
The nucleotide sequences of the EcoRV fragments of the PstI N fragment of VV-WR DNA and the corresponding region of CPV-BR DNA reported in this article have been assigned GenBank accession numbers AF464893 and AF464894.
RESULTS
Identification of vaccinia and cowpox virus p4c genes.
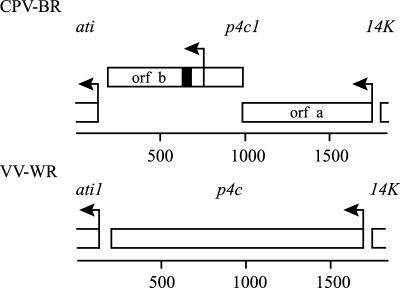
Previous analyses of the nucleotide sequence of the gene immediately upstream of the ati gene in CPV-BR identified part of an open reading frame (Fig. 1, orf b) with the potential to encode a protein containing 28 consecutive aspartic acids (52). Further sequence analysis revealed that this open reading frame lacks an obvious transcriptional promoter region upstream of the predicted initiation codon. In addition, it narrowly overlaps an upstream open reading frame (Fig. 1, orf a) which begins with the sequence TAAATG, the consensus nucleotide sequence for the transcriptional start site of late promoters of poxviruses (7, 56, 81). An insertion of a single base in the overlapping region can put these two coding regions in frame. These features suggested that the two open reading frames might be remnants of a single late gene predicted to encode a 58-kDa protein, possibly a virion protein, because many late proteins are structural components of the virus particles. These properties suggested that the late gene might encode the P4c protein, because this is a structural protein of about 60 kDa known to be present in some, but not all, strains of cowpox virus (31, 67). Furthermore, although this protein had been identified in the virions of orthopoxviruses of several types, its gene had not been identified. In the genome of CPV-BR, as recently described (GenBank accession number AF482758), open reading frame a corresponds to open reading frame CPXV161, and the part of open reading frame b downstream of the first potential initiation codon corresponds to open reading frame CPXV159.
FIG. 1.
The p4c gene is intact in the genome of VV-WR but disrupted in the genome of CPV-BR. The rectangles correspond to the open reading frames of the ati, p4c, and 14K genes, where mutant alleles are designated ati1 and p4c1. The arrows indicate the position of the first potential initiation codon within each open reading frame. Each of these initiation codons, except for that in orf b of p4c1, is contained within a predicted late promoter element. The solid rectangle corresponds to the position of the 28 GAT repeat sequence predicted to encode a 28-aspartic-acid repeat (52). The scale is in base pairs.
The P4c protein was first described in VV-WR (40, 61). To determine if the VV-WR gene encoding the P4c protein is located in the corresponding position (upstream of the ati locus) in the vaccinia virus genome, we first analyzed the DNA sequence of this region. This analysis identified an open reading frame predicted to encode a 58-kDa protein corresponding to the hypothetical product of an in-frame linkage of the two overlapping open reading frames in the cowpox virus DNA (Fig. 1). A specific genomic number has not been assigned to this open reading frame, because its numerical position among the open reading frames in the VV-WR genome is not known.
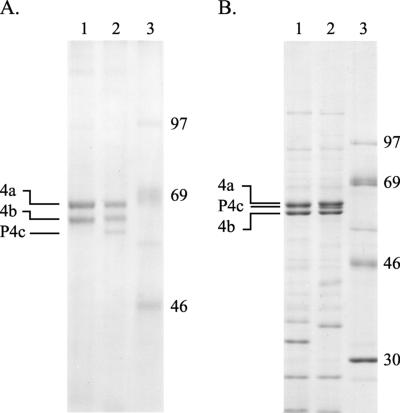
Analyses of the protein components of purified virions of VV-WR and CPV-BR confirmed both the presence of a protein with the expected properties (relative abundance and electrophoretic mobilities) of the P4c protein in the vaccinia virions and its absence from the cowpox virions. A continuous phosphate-buffered electrophoresis system similar to those used originally to identify the P4c protein resolves the P4c protein (apparent molecular mass, 61 kDa) from the 4a and 4b proteins in the vaccinia virus sample (Fig. 2A, lane 2). The P4c protein is not present among proteins of cowpox virus (Fig. 2A, lane 1). This difference is also apparent if the proteins are resolved by electrophoresis using the discontinuous buffer system described by Laemmli (43). However, in that system, the relative electrophoretic mobilities of the 4a, 4b, and P4c proteins are altered such that, for example, the 4a protein migrates with an apparent molecular mass of 61 kDa versus 59 kDa for the P4c protein (Fig. 2B, lane 2). These alterations in the relative electrophoretic mobilities of the 4a, 4b, and P4c proteins in different electrophoresis buffer systems are distinguishing characteristics of these proteins (51).
FIG. 2.
The P4c protein is present in VV-WR particles but absent from particles of CPV-BR. Proteins present in virus particles (16 μg per sample), purified as described previously(36), were characterized as follows. (A) Virus particles were solubilized by boiling them for 3 min in a buffer containing 0.1 M sodium phosphate (pH 7.2), 6 M urea, 1 mM EDTA, 2% SDS, 5% β-mercaptoethanol, and 10% glycerol. The solubilized proteins were resolved by electrophoresis in a 10% polyacrylamide gel containing 0.1 M sodium phosphate, 6 M urea, 1 mM EDTA, and 0.1% SDS (61) and visualized with Coomassie brilliant blue R-250 stain. (B) Virus particles were solubilized by boiling them for 3 min in a buffer containing 50 mM Tris (pH 7.0), 1% SDS, 30 mM dithiothreitol, 5 mM EDTA, and 10% glycerol. The solubilized proteins were resolved by electrophoresis in a 10% polyacrylamide gel using a discontinuous buffer system as described previously (43) and visualized with Coomassie brilliant blue R-250 stain. Lanes 1, proteins of CPV-BR; lanes 2, proteins of VV-WR; lanes 3, protein markers (in kilodaltons).
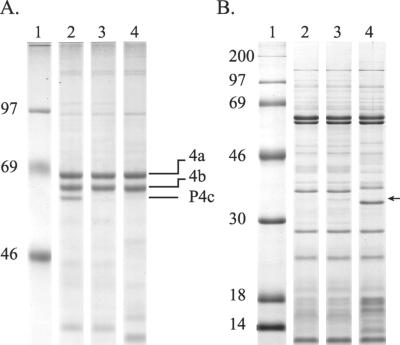
To confirm that the gene identified in the vaccinia virus genome encodes the P4c protein, a recombinant vaccinia virus (A504) was constructed in which this gene was disrupted by the insertion of a DNA fragment containing the gpt gene under the transcriptional control of an early poxvirus promoter (11). Analysis of the proteins of purified virus particles showed that the insertional inactivation of the predicted p4c gene in vaccinia virus A504 led to the production of virus particles lacking the P4c protein (Fig. 3A, lane 3). The A504 virus particles also contained an ∼34-kDa protein (Fig. 3B, lane 3) that may be the product of the disrupted p4c gene, corresponding to the N-terminal portion of the P4c protein encoded by the region upstream of the insertion point of the gpt gene.
FIG. 3.
Insertional inactivation of the p4c gene leads to loss of intact P4c protein in the purified virus. Proteins present in virus particles (16 μg per sample), purified as described previously(36), were characterized as described in the legend to Fig. 2. (A) Solubilized proteins were resolved by electrophoresis in an 8% polyacrylamide gel containing a continuous phosphate buffer system (61). (B) Solubilized proteins were resolved by electrophoresis in a 12.5% polyacrylamide gel using a discontinuous buffer system as described previously (43). Lanes 1, protein markers (sizes in kilodaltons); lanes 2, proteins of VV-WR; lanes 3, proteins of vaccinia virus A504; lanes 4, proteins of CPV-BR. The arrow indicates the position of an ∼34-kDa protein present only in vaccinia virus A504 (panel B, lane 3).
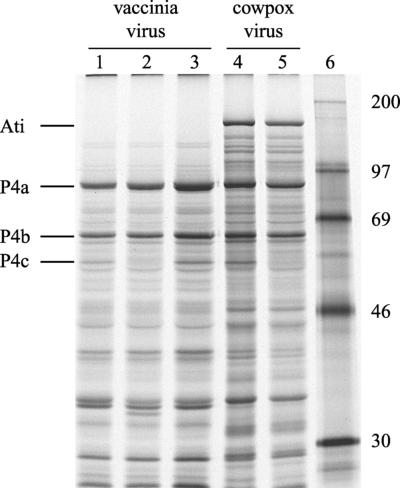
The expression of the p4c gene can also be detected in virus-infected cells pulse-labeled with [35S]methionine during the late phase of virus replication, when most newly synthesized proteins are virus encoded. In the absence of a chase period, the nascent P4a and P4b proteins are not proteolytically processed to the forms 4a and 4b, which can obscure the P4c protein in one-dimensional gel electrophoretic analyses. The expression of viral proteins in cells infected with different orthopoxviruses that contain or lack the predicted p4c gene is shown in Fig. 4. A late protein with electrophoretic mobilities identical to those of the virion P4c protein is synthesized in cells infected with VV-WR (lane 1), absent in cells infected with vaccinia virus A504 (lane 2), and present in cells infected with vaccinia virus A507, a revertant of virus A504 (lane 3). An ∼34-kDa protein that may be the product of the disrupted p4c gene is also evident among proteins synthesized in cells infected with vaccinia virus A504 (lane 2) but not the revertant virus A507 (lane 3). As expected, CPV-BR does not direct the synthesis of a 59-kDa P4c protein (lane 5), but cowpox virus A505, containing a copy of the vaccinia virus p4c gene including its predicted promoter region, does (lane 4).
FIG. 4.
Expression of the P4c protein in cells infected with recombinant vaccinia and cowpox viruses. Human 143B cells were infected and metabolically labeled with [35S]methionine for 30 min 18 h after infection, as described in Materials and Methods. The solubilized labeled proteins were resolved by electrophoresis in a 12.5% polyacrylamide gel using a discontinuous buffer system as described previously (43) and visualized by autoradiography of the dried gel. The lanes contain proteins from cells infected with VV-WR (lane 1), vaccinia virus A504 (lane 2), vaccinia virus A507 (lane 3), cowpox virus A505 (lane 4), and CPV-BR (lane 5). Lane 6 contains protein markers (in kilodaltons).
Collectively, these results show that the open reading frames immediately upstream of the ati genes in vaccinia and cowpox viruses are the coding regions of the p4c genes in these viruses. In CPV-BR, we designated this gene p4c1 to indicate that this is a variant of the wild-type p4c gene.
The P4c protein facilitates the formation of V+ ATI bodies.
The identification of the p4c gene allowed us to test the hypothesis that the P4c protein is required for V+ ATI formation. The hypothesis predicts that orthopoxviruses lacking a functional p4c gene will have a V− phenotype, whereas orthopoxviruses containing a functional p4c gene will have a V+ phenotype.
First, we used complementation assays, as described by Ichihashi and Matsumoto (29), to examine these predictions. Cells were infected with one or two strains of poxviruses. Eighteen hours later, the cells were fixed, stained, and examined by electron microscopy to identify the phenotype of the ATIs present in the cytoplasm. The ATIs of at least 30 cells were examined in each sample. If all of the ATIs were V−, the phenotype of the virus was scored as V−. The phenotype of the virus was scored as V+ if any of the ATIs were V+, though in practice, infection of a cell with a V+ virus resulted in the production of ATIs of which >90% contained virus particles. Ichihashi and Matsumoto (29) similarly noted the dominance of the inclusion of particles over the exclusion of particles in complementation assays.
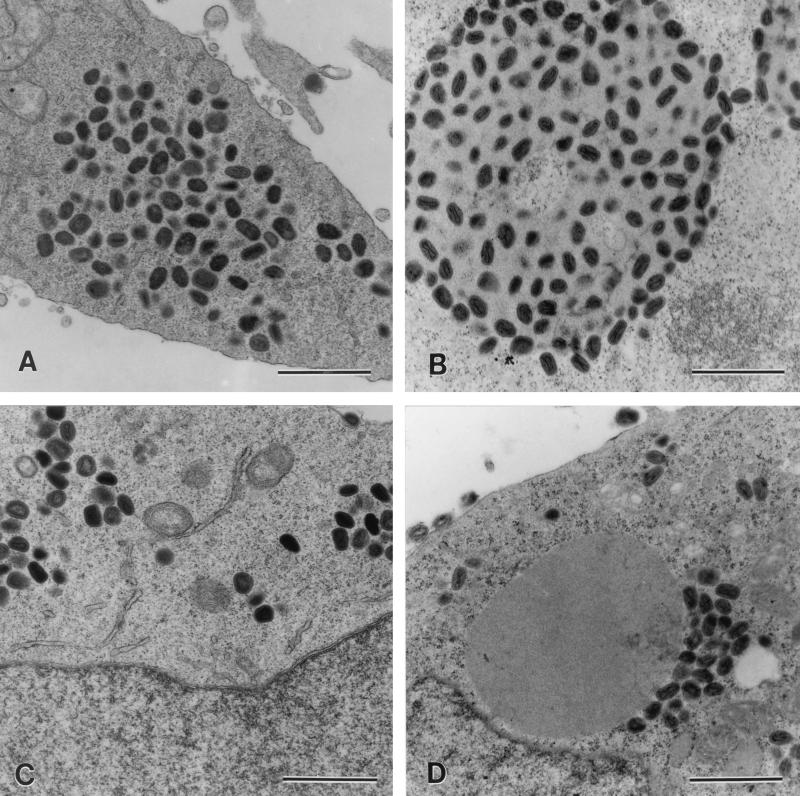
Complementation assays were initially used to determine the V phenotypes of two strains of vaccinia virus whose genotypes with respect to both the p4c gene and the ati gene are known. VV-WR contains the ati1 gene and the intact p4c gene. The vaccinia virus ati1 gene produces a variant of the cowpox virus ati gene containing a stop codon that results in the synthesis of a truncated 94-kDa version of the ATI protein that does not form ATIs (1, 8, 53). Vaccinia virus strain Copenhagen contains partially deleted variants of the ati and p4c genes. Major portions of both genes are missing so that the 5′ end of the residual portion of the p4c gene is fused to the 3′ end of the ati gene, generating a fusion gene, Φ(p4c′-Δati), containing the A26L open reading frame (14). CPV-BR contains the ati and p4c1 genes, where the ati gene is functional, producing a 160-kDa protein that forms ATIs (52, 53), and the p4c1 gene is the disrupted form of the p4c gene shown in Fig. 1. Human 143B cells were infected with VV-WR or vaccinia virus Copenhagen, either alone or together with CPV-BR. As expected, neither VV-WR nor vaccinia virus Copenhagen produced ATIs (Fig. 5A and C). Coinfection of VV-WR (ati1 p4c) with CPV-BR (ati p4c1) resulted in the formation of V+ ATIs (Fig. 5B), whereas the coinfection of vaccinia virus Copenhagen [Φ(p4c′-Δati)] with CPV-BR (ati p4c1) resulted in the formation of V− ATIs (Fig. 5D).
FIG. 5.
VV-WR expressing the p4c gene can provide a factor needed to direct the inclusion of IMV within ATIs. Human 143B cells were infected with 10 PFU of one strain of poxvirus/cell (A and C) or with 5 PFU each of two strains of poxvirus/cell (B and D). Eighteen hours after the cells were infected, they were fixed, stained, sectioned, and examined by electron microscopy to identify the phenotype of the ATIs present in the infected cells. Shown are sections of cells infected with VV-WR (p4c ati1) (A); VV-WR and CPV-BR (p4c1 ati), where the large mass is a V+ ATI (B); vaccinia virus Copenhagen [Φ(p4c′-Δati)] (C); and vaccinia virus Copenhagen and CPV-BR, where the large mass is a V− ATI (D). Bars, 1,000 nm.
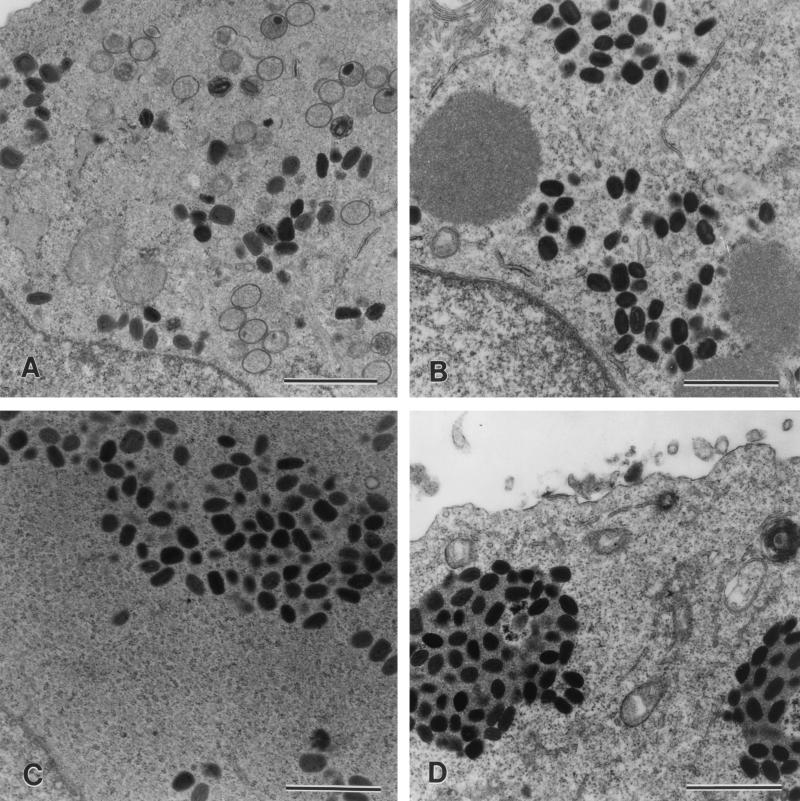
Insertional inactivation of the p4c gene in VV-WR, producing vaccinia virus A504, abrogated the ability of this virus to induce the formation of V+ ATIs in coinfections with CPV-BR (Fig. 6B). A revertant of this virus, vaccinia virus A507, which was generated by repairing the disrupted p4c gene of vaccinia virus A504 by recombination with the wild-type p4c gene in plasmid p1542, regained the ability both to express the P4c protein (Fig. 4, lane 3) and to induce the formation of V+ ATIs in cells coinfected with CPV-BR (Fig. 6D).
FIG. 6.
The p4c gene is required for the formation of V+ ATIs in cells coinfected with VV-WR and CPV-BR. Human 143B cells were infected with either 10 PFU of one strain of poxvirus/cell (A and C) or 5 PFU each of two strains of poxvirus/cell (B and D). Eighteen hours after the cells were infected, they were fixed, stained, sectioned, and examined by electron microscopy. Shown are sections of cells infected with vaccinia virus A504 (p4c::gpt ati1) containing a p4c gene disrupted by the insertion of a selectable marker gene, the gpt gene (A); vaccinia virus A504 and CPV-BR (p4c1 ati), where the large masses are V− ATIs (B); vaccinia virus A507 (p4c ati1), which is a revertant of vaccinia virus A504 containing an intact p4c gene (C); and vaccinia virus A507 and CPV-BR, where the large masses are V+ ATIs (D). Bars, 1,000 nm.
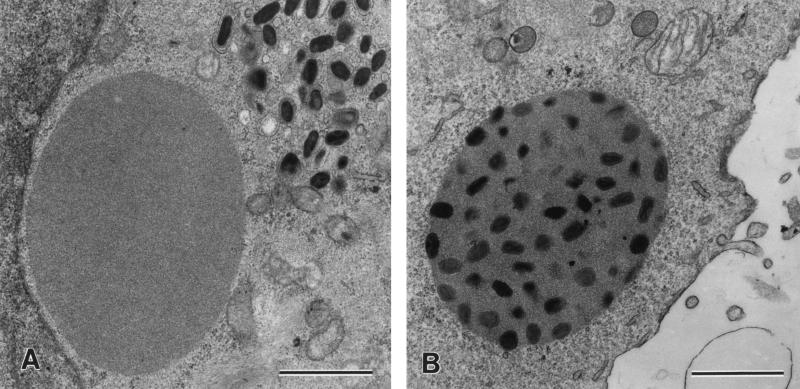
These results predicted that a recombinant CPV-BR expressing the VV-WR p4c gene would have a V+ phenotype. To test this hypothesis, we examined the phenotype of recombinant cowpox virus A505, which contains a functional copy of the VV-WR p4c gene under the transcriptional control of its endogenous promoter. This p4c gene was recombined into the thymidine kinase gene of the cowpox virus as described in Materials and Methods, generating cowpox virus A505 (ati p4c1 p4c), which expresses the vaccinia virus P4c protein (Fig. 4, lane 4). As shown in Fig. 7, the insertion of the functional p4c gene into the genome of CPV-BR resulted in the embedding of virus particles within the ATIs.
FIG. 7.
Insertion of the vaccinia virus p4c gene into the genome of CPV-BR is sufficient to convert the phenotype of the ATIs from V− to V+. Human 143B cells were infected with 10 PFU of cowpox virus/cell. Eighteen hours after infection, the cells were fixed, stained, sectioned, and examined by electron microscopy. Shown are sections of cells infected with CPV-BR (p4c1 ati), which produces V− ATIs (A), and cowpox virus A505 (p4c1 p4c ati), which is a recombinant cowpox virus containing the VV-WR p4c gene under the control of its own promoter (B). ATIs in cells infected with cowpox virus A505 have a V+ phenotype. Bars, 1,000 nm.
DISCUSSION
This study has identified the orthopoxvirus p4c gene and confirmed that one of the roles of this gene is to direct the inclusion of IMV particles within the ATIs.
The p4c gene is highly conserved among most if not all species of orthopoxvirus, including those that do not produce ATIs. For example, the p4c gene is present in the genomes of strains of variola virus major and variola virus minor that have been characterized by nucleotide sequence analysis (46, 64, 66), where the protein (encoded by open reading frame A29L or A30L, depending on the virus strain) shows ∼94% (473 of 500 amino acids) identity with the vaccinia virus P4c protein. The p4c gene is present as open reading frame A28L in monkeypox virus strain Zaire-96-I-16, where it is predicted to encode a protein with ∼91% (476 of 520 amino acids) identity to the vaccinia virus P4c protein (65). It is present in camelpox virus M-96 (19), where it is predicted to show 95% identity (476 of 500 amino acids) to the vaccinia virus P4c protein. It is also present in a variety of characterized VV-WR isolates, as well as the IHD-J strain of vaccinia virus (72). These data are consistent with those of early phenotypic analyses showing that 18 different strains of cowpox, vaccinia, ectromelia, variola, and monkeypox viruses each expressed an inclusion factor capable of directing the formation of V+ ATIs during coinfection with a V− cowpox virus (27). The high degree of sequence conservation of the p4c gene among the various types of orthopoxvirus, irrespective of whether the virus produces ATIs, suggests that the P4c protein is advantageous for viral replication in vivo, even in the absence of ATIs. The disruption of the p4c gene in many of the well-characterized strains and isolates of vaccinia virus, such as the Copenhagen strain (14), the MVA strain (2), the Tian Tan strain (GenBank accession number AF095689), and at least one isolate of the WR strain (1), may reflect the passage histories of these viruses. Whereas the expression of the p4c gene during replication in vivo may be advantageous to the virus, this may not be the case for virus replication in tissue culture.
The predicted sequences of the P4c proteins yield few insights into the structures and functions of these proteins. The PSORT II algorithm (50) predicts one potential transmembrane portion between residues 112 and 130, with the N-terminal portion of the protein predicted to be within the inner side of the IMV membrane. Consistent with this model, a novel protein of about 34 kDa, which may correspond to the predicted 37-kDa product of the insertionally inactivated p4c gene (equivalent to the N-terminal portion of the P4c protein), is present both in the purified particles of A504 virus (Fig. 3B, lane 3) and among proteins synthesized during the late phase of replication of A504 (Fig. 4, lane 2). This model suggests that the COOH-terminal portion of the P4c protein is required to facilitate the embedding of IMV particles within the ATIs. In this context, two features of the COOH-terminal regions of the P4c proteins are noteworthy. First, this portion of the cowpox virus P4c protein contains the region predicted to contain 28 consecutive aspartic acid residues. A similarly positioned tract of 23 aspartic acids is present in the predicted sequence of the monkeypox virus P4c protein (65). However, this region is absent from the P4c proteins of variola virus and vaccinia virus, showing that this polyaspartate tract is not required for the V+ phenotype. The presence or absence of the polyaspartate tract will affect the molecular masses of the P4c proteins, giving rise to P4c proteins that are slightly larger in cowpox and monkeypox viruses than in some of the other orthopoxviruses. Second, the amino acid sequence of a region near the COOH terminus of the VV-WR P4c protein (441-CCDTAAVDRLEHHIETLGQYAVILARKINMQT-472) shows 44% identity (underlined amino acids) with the segment of the 14K fusion protein that binds to the 21-kDa protein (the A17L protein in VV-WR) in the membrane of the IMV (78). Whether this region, or another region of the P4c protein, mediates an association between the P4c protein and other proteins in the IMV membrane remains to be determined. Ichihashi (25) previously reported that the P4c protein associates via a disulfide linkage with a 16-kDa viral membrane protein.
The presence of the P4c protein on the surface of the IMV is consistent with its role in the process that places virus particles into the ATIs. However, the mechanism involved in this process has yet to be determined. Ulaeto et al. (76) have shown that a 59-kDa protein is a specific marker for IMV of vaccinia virus (IHD-J strain). Antibodies against the 59-kDa IMV protein specifically precipitated the P4c protein encoded by cowpox virus A505, confirming the coidentity of the 59-kDa and P4c proteins in these two studies. The P4c protein was not detected in EEV, leading Ulaeto and colleagues to suggest that virion acquisition of the P4c protein marks this particle for maturation as an IMV, presumably either by the prevention of processes leading to the formation of IEV particles or by the facilitation of an alternative maturation pathway.
The IEV particles are produced after anterograde microtubule-mediated transport of IMVs to the sites where they become enveloped by membranes of the trans-Golgi network or tubular endosomes. This is a process that is mediated by direct or indirect interactions between the 14K protein and kinesin motors (59). Subsequently, the IEVs are propelled toward the plasma membrane by kinesin-dependent movement on microtubules, finally being propelled to and through the plasma membrane by actin bundles (5, 13, 21, 54, 55, 58, 77, 80). In contrast, the IMVs are either localized within the cytoplasm, often in the perinuclear region, or embedded within ATIs.
For the same reasons that active transport mechanisms are needed to move IMVs and IEVs to the plasma membrane, similar mechanisms may be necessary for the retrograde transport of both IMVs and the viral ATI proteins to the sites where the IMVs and ATIs colocalize. Several features of the cytoskeleton, the IMV, and ATIs are consistent with such a model. First, the use of microfilaments to move virus particles in one direction suggests that the virus may use the microfilaments to move in the opposite direction, because movement of cargoes (virus particles, organelles, mRNAs, protein, and nucleoprotein complexes) by kinesin superfamily proteins, myosins, and dyneins along a microfilament can occur in either the forward or reverse direction, depending on the nature of the motor protein (reviewed in references 15 and 38). Viruses of several types, including poxviruses, are now known to employ microfilament-mediated transport mechanisms to move subviral components in anterograde or retrograde directions (54), with the herpesviruses providing prime examples of viruses that employ both modes of transport to effect either entry or egress from the infected cell (reviewed in reference 69). Second, when P4c is absent in cells containing ATIs, there is a clear segregation of IMVs from the V− ATIs, with IMVss typically observed close to the ATIs but not within the ATIs themselves (Fig. 5 to 7). However, in cells containing both P4c and ATIs, most of the IMVs are observed within the ATIs in a cytoplasm containing immature virus particles but largely devoid of IMVs. Similar distributions are evident in micrographs of previous studies of V+ and V− ATIs (23, 26, 29, 30, 67). Indeed, in some previous studies, particularly those involving fusions between cells containing V− ATIs and cells containing virus expressing “inclusion factor,” it was common to find a special type of ATI, designated V+′ ATIs, in which the ATI lacks internal IMVs but is completely encrusted with IMVs on its surface within a field of cytoplasm devoid of IMVs (67). Furthermore, in these assays yielding V+ ATIs, all ATIs in the cell were V+ or V+′ (29, 67). These features are consistent with a mechanism involving the active transport of P4c-containing IMVs to the ATI. Such a mechanism could efficiently segregate IMVs containing P4c from IMVs lacking P4c and destined to become IEV, consistent with the results of Ulaeto et al. (76). Third, the ATIs themselves resemble aggresomes, which are microtubule-dependent cytoplasmic inclusion bodies that are metabolically stable, insoluble aggregates of proteins commonly found in cells within lesions in a variety of degenerative diseases (reviewed in references 35 and 42).
According to this model, one function of the P4c protein in orthopoxviruses, including those such as variola virus that encode a P4c protein but not an Ati protein, may be to direct the retention of most progeny viruses as IMVs rather than IEVs, CEVs, or EEVs. Retention of IMVs within ATIs takes this sequestration one step further. A mechanism to promote the intracellular retention of progeny virus as IMV particles may seem to be counterproductive, because during the early phase of an infection in vivo, EEVs may be the most efficient particles for dissemination of the virus in the host (reviewed in reference 68). However, IMV, as the more robust particle, may be more important than EEV for transmission of the virus from host to host (77). In addition, even for transmission within a host, as adaptive immune responses come into play, the dissemination of cells containing virus, rather than the transport of free virus, may become the more important mode of virus dissemination (reviewed in reference 12). This form of viral transport, which has important ramifications for viral pathogenesis, could be facilitated by the production of the P4c protein.
The identification of the p4c gene will allow these models of the potential roles of the P4c protein in viral morphogenesis and pathogenesis to be tested.
Acknowledgments
We are grateful to Susan Hester for her help in preparing the electron micrographs. The shared core facilities of the Duke University Comprehensive Cancer Center were used in this study.
This work was supported by grants AI32982, 2T32CA09111-16A10051, and 5T32GM07184-14 021 (T.A.M.) from the National Institutes of Health.
REFERENCES
- 1.Amegadzie, B. Y., J. R. Sisler, and B. Moss. 1992. Frame-shift mutations within the vaccinia virus A-type inclusion protein gene. Virology 186:777-782. [DOI] [PubMed] [Google Scholar]
- 2.Antoine, G., F. Scheiflinger, F. Dorner, and F. G. Falkner. 1998. The complete genomic sequence of the modified vaccinia Ankara strain: comparison with other orthopoxviruses. Virology 244:365-396. [DOI] [PubMed] [Google Scholar]
- 3.Biggin, M. D., T. J. Gibson, and G. F. Hong. 1983. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc. Natl. Acad. Sci. USA 80:3963-3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasco, R., and B. Moss. 1992. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J. Virol. 66:4170-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cudmore, S., P. Cossart, G. Griffiths, and M. Way. 1995. Actin-based motility of vaccinia virus. Nature 378:636-638. [DOI] [PubMed] [Google Scholar]
- 6.Dales, S., and E. H. Mosbach. 1968. Vaccinia as a model for membrane biogenesis. Virology 35:564-583. [DOI] [PubMed] [Google Scholar]
- 7.Davison, A. J., and B. Moss. 1989. Structure of vaccinia virus late promoters. J. Mol. Biol. 210:771-784. [DOI] [PubMed] [Google Scholar]
- 8.de Carlos, A., and E. Paez. 1991. Isolation and characterization of mutants of vaccinia virus with a modified 94-kDa inclusion protein. Virology 185:768-778. [DOI] [PubMed] [Google Scholar]
- 9.Downie, A. W. 1939. A study of the lesions produced experimentally by cowpox virus. J. Pathol. Bacteriol. 48:361-379. [Google Scholar]
- 10.Dubochet, J., M. Adrian, K. Richter, J. Garces, and R. Wittek. 1994. Structure of intracellular mature vaccinia virus observed by cryoelectron microscopy. J. Virol. 68:1935-1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Falkner, F. G., and B. Moss. 1988. Escherichia coli gpt gene provides dominant selection for vaccinia virus open reading frame expression vectors. J. Virol. 62:1849-1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fenner, F., D. A. Henderson, I. Arita, Z. Jezek, and I. D. Ladnyi. 1988. The clinical features of smallpox, p. 1-68. In F. Fenner, D. A. Henderson, I. Arita, Z. Jezek, and I. D. Ladnyi (ed.), Smallpox and its eradication. World Health Organization, Geneva, Switzerland.
- 13.Geada, M. M., I. Galindo, M. M. Lorenzo, B. Perdiguero, and R. Blasco. 2001. Movements of vaccinia virus intracellular enveloped virions with GFP tagged to the F13L envelope protein. J. Gen. Virol. 82:2747-2760. [DOI] [PubMed] [Google Scholar]
- 14.Goebel, S. J., G. P. Johnson, M. E. Perkus, S. W. Davis, J. P. Winslow, and E. Paoletti. 1990. The complete DNA sequence of vaccinia virus. Virology 179:247-266, 517-563. [DOI] [PubMed] [Google Scholar]
- 15.Goldstein, L. S. 2001. Molecular motors: from one motor many tails to one motor many tales. Trends Cell Biol. 11:477-482. [DOI] [PubMed] [Google Scholar]
- 16.Griffiths, G., N. Roos, S. Schleich, and J. K. Locker. 2001. Structure and assembly of intracellular mature vaccinia virus: thin-section analyses. J. Virol. 75:11056-11070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffiths, G., R. Wepf, T. Wendt, J. K. Locker, M. Cyrklaff, and N. Roos. 2001. Structure and assembly of intracellular mature vaccinia virus: isolated-particle analysis. J. Virol. 75:11034-11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grimley, P. M., E. N. Rosenblum, S. J. Mims, and B. Moss. 1970. Interruption by rifampin of an early stage in vaccinia virus morphogenesis: accumulation of membranes which are precursors of virus envelopes. J. Virol. 6:519-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gubser, C., and G. L. Smith. 2002. The sequence of camelpox virus shows it is most closely related to variola virus, the cause of smallpox. J. Gen. Virol. 83:855-872. [DOI] [PubMed] [Google Scholar]
- 20.Hiller, G., and K. Weber. 1985. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J. Virol. 55:651-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hollinshead, M., G. Rodger, H. Van Eijl, M. Law, R. Hollinshead, D. J. Vaux, and G. L. Smith. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154:389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollinshead, M., A.Vanderplasschen, G. L. Smith, and D. J. Vaux. 1999. Vaccinia virus intracellular mature virions contain only one lipid membrane. J. Virol. 73:1503-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ichihashi, Y. 1968. Genetic character of pox virus A-type inclusion. Virus (Kyoto) 18:237-247. [Google Scholar]
- 24.Ichihashi, Y. 1969. Genetic study of poxgroup viruses. Virus (Kyoto) 19:155-163. [Google Scholar]
- 25.Ichihashi, Y. 1981. Unit complex of vaccinia polypeptides linked by disulfide bridges. Virology 113:277-284. [DOI] [PubMed] [Google Scholar]
- 26.Ichihashi, Y., and S. Dales. 1973. Biogenesis of poxviruses: relationship between a translation complex and formation of A-type inclusions. Virology 51:297-319. [DOI] [PubMed] [Google Scholar]
- 27.Ichihashi, Y., and T. Kitamura. 1976. A-type inclusion markers of wild white and other variola-related poxviruses. Jpn. J. Med. Sci. Biol. 29:221-225. [DOI] [PubMed] [Google Scholar]
- 28.Ichihashi, Y., and S. Matsumoto. 1966. Studies on the nature of Marchal bodies (A-type inclusion) during ectromelia virus infection. Virology 29:264-275. [DOI] [PubMed] [Google Scholar]
- 29.Ichihashi, Y., and S. Matsumoto. 1968. The relationship between poxvirus and A-type inclusion body during double infection. Virology 36:262-270. [DOI] [PubMed] [Google Scholar]
- 30.Ichihashi, Y., S. Matsumoto, and S. Dales. 1971. Biogenesis of poxviruses: role of A-type inclusions and host cell membranes in virus dissemination. Virology 46:507-532. [DOI] [PubMed] [Google Scholar]
- 31.Ichihashi, Y., and M. Oie. 1980. Adsorption and penetration of the trypsinized vaccinia virion. Virology 101:50-60. [DOI] [PubMed] [Google Scholar]
- 32.Ichihashi, Y., T. Tsuruhara, and M. Oie. 1982. The effect of proteolytic enzymes on the infectivity of vaccinia virus. Virology 122:279-289. [DOI] [PubMed] [Google Scholar]
- 33.Ink, B. S., and D. J. Pickup. 1989. Transcription of a poxvirus early gene is regulated both by a short promoter element and by a transcriptional termination signal controlling transcriptional interference. J. Virol. 63:4632-4644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Isaacs, S. N., G. J. Kotwal, and B. Moss. 1990. Reverse guanine phosphoribosyltransferase selection of recombinant vaccinia viruses. Virology 178:626-630. [DOI] [PubMed] [Google Scholar]
- 35.Johnston, J. A., C. L. Ward, and R. R. Kopito. 1998. Aggresomes: a cellular response to misfolded proteins. J. Cell Biol. 143:1883-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joklik, W. K. 1962. The purification of four strains of poxvirus. Virology 18:9-18. [DOI] [PubMed] [Google Scholar]
- 37.Kamahora, J., Y. Sato, S. Kato, and K. Hagiwara. 1956. Inclusion bodies of the vaccinia virus. Proc. Soc. Exp. Biol. Med. 97:43-47. [DOI] [PubMed] [Google Scholar]
- 38.Kamal, A., and L. S. Goldstein. 2002. Principles of cargo attachment to cytoplasmic motor proteins. Curr. Opin. Cell Biol. 14:63-68. [DOI] [PubMed] [Google Scholar]
- 39.Katz, E., and E. Margalith. 1973. Location of vaccinia virus structural polypeptides on the surface of the virus particle. J. Gen. Virol. 18:381-384. [DOI] [PubMed] [Google Scholar]
- 40.Katz, E., and B. Moss. 1970. Vaccinia virus structural polypeptide derived from a high-molecular-weight precursor: formation and integration into virus particles. J. Virol. 6:717-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katz, E., and B. Moss. 1970. Formation of a vaccinia virus structural polypeptide from a higher molecular weight precursor: inhibition by rifampicin. Proc. Natl. Acad. Sci. USA 66:677-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kopito, R. R. 2000. Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol. 10:524-530. [DOI] [PubMed] [Google Scholar]
- 43.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 44.Mackett, M., G. L. Smith, and B. Moss. 1984. General method for production and selection of infectious vaccinia virus recombinants expressing foreign genes. J. Virol. 49:857-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Marchal, J. 1930. Infectious ectromelia. A hitherto undescribed virus disease in mice. J. Pathol. Bacteriol. 33:713-728. [Google Scholar]
- 46.Massung, R. F., J. J. Esposito, L. I. Liu, J. Qi, T. R. Utterback, J. C. Knight, L. Aubin, T. E. Yuran, J. M. Parsons, and V. N. Loparev. 1993. Potential virulence determinants in terminal regions of variola smallpox virus genome. Nature 366:748-751. [DOI] [PubMed] [Google Scholar]
- 47.Miller, S. E. 1986. Detection and identification of viruses by electron microscopy. J. Electron Microsc. Tech. 4:265-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In D. M. Knipe, P. M. Howley, D. E. Griffin, M. A. Martin, R. A. Lamb, B. Roizman, and S. E. Straus (ed.), Fields virology. Lipincott Williams and Wilkins, Philadelphia, Pa.
- 49.Moss, B., and E. N. Rosenblum. 1973. Protein cleavage and poxvirus morphogenesis: tryptic peptide analysis of core precursors accumulated by blocking assembly with rifampicin. J. Mol. Biol. 81:267-269. [DOI] [PubMed] [Google Scholar]
- 50.Nakai, K., and P. Horton. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24:34-36. [DOI] [PubMed] [Google Scholar]
- 51.Oie, M., and Y. Ichihashi. 1981. Characterization of vaccinia polypeptides. Virology 113:263-276. [DOI] [PubMed] [Google Scholar]
- 52.Patel, D. D., and D. J. Pickup. 1987. Messenger RNAs of a strongly-expressed late gene of cowpox virus contain 5′-terminal poly(A) sequences. EMBO J. 6:3787-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patel, D. D., D. J. Pickup, and W. K. Joklik. 1986. Isolation of cowpox virus A-type inclusions and characterization of their major protein component. Virology 149:174-189. [DOI] [PubMed] [Google Scholar]
- 54.Ploubidou, A., V. Moreau, K. Ashman, I. Reckmann, C. Gonzalez, and M. Way. 2000. Vaccinia virus infection disrupts microtubule organization and centrosome function. EMBO J. 19:3932-3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rietdorf, J., A. Ploubidou, I. Reckmann, A. Holmstrom, F. Frischknecht, M. Zettl, T. Zimmermann, and M. Way. 2001. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat. Cell Biol. 3:992-1000. [DOI] [PubMed] [Google Scholar]
- 56.Rosel, J. L., P. L. Earl, J. P. Weir, and B. Moss. 1986. Conserved TAAATG sequence at the transcriptional and translational initiation sites of vaccinia virus late genes deduced by structural and functional analysis of the HindIII H genome fragment. J. Virol. 60:436-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 58.Sanderson, C. M., F. Frischknecht, M. Way, M. Hollinshead, and G. L. Smith. 1998. Roles of vaccinia virus EEV-specific proteins in intracellular actin tail formation and low pH-induced cell-cell fusion. J. Gen. Virol. 79:1415-1425. [DOI] [PubMed] [Google Scholar]
- 59.Sanderson, C. M., M. Hollinshead, and G. L. Smith. 2000. The vaccinia virus A27L protein is needed for the microtubule-dependent transport of intracellular mature virus particles. J. Gen. Virol. 81:47-58. [DOI] [PubMed] [Google Scholar]
- 60.Sanger, F., A. R. Coulson, B. G. Barrell, A. J. Smith, and B. A. Roe. 1980. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J. Mol. Biol. 143:161-178. [DOI] [PubMed] [Google Scholar]
- 61.Sarov, I., and W. K. Joklik. 1972. Studies on the nature and location of the capsid polypeptides of vaccinia virions. Virology 50:579-592. [DOI] [PubMed] [Google Scholar]
- 62.Sarov, I., and W. K. Joklik. 1973. Isolation and characterization of intermediates in vaccinia virus morphogenesis. Virology 52:223-233. [DOI] [PubMed] [Google Scholar]
- 63.Schmelz, M., B. Sodeik, M. Ericsson, E. J. Wolffe, H. Shida, G. Hiller, and G. Griffiths. 1994. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J. Virol. 68:130-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shchelkunov, S. N., S. S. Marennikova, V. M. Blinov, S. M. Resenchuk, A. V. Tetmenin, V. E. Chizhikov, V. V. Gutorov, P. F. Safronov, R. K. Kurmanov, and L. S. Sandakhchiev. 1993. Entire coding sequence of the variola virus. Dokl. Akad. Nauk. 328:629-632. [PubMed] [Google Scholar]
- 65.Shchelkunov, S. N., A. V. Totmenin, I. V. Babkin, P. F. Safronov, O. I. Ryazankina, N. A. Petrov, V. V. Gutorov, E. A. Uvarova, M. V. Mikheev, J. R. Sisler, J. J. Esposito, P. B. Jahrling, B. Moss, and L. S. Sandakhchiev. 2001. Human monkeypox and smallpox viruses: genomic comparison. FEBS Lett. 509:66-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shchelkunov, S. N., A. V. Totmenin, V. N. Loparev, P. F. Safronov, V. V. Gutorov, V. E. Chizhikov, J. C. Knight, J. M. Parsons, R. F. Massung, and J. J. Esposito. 2000. Alastrim smallpox variola minor virus genome DNA sequences. Virology 266:361-386. [DOI] [PubMed] [Google Scholar]
- 67.Shida, H., K. Tanabe, and S. Matsumoto. 1977. Mechanism of virus occlusion into A-type inclusion during poxvirus infection. Virology 76:217-233. [DOI] [PubMed] [Google Scholar]
- 68.Smith, G. L., and A. Vanderplasschen. 1998. Extracellular enveloped vaccinia virus. Entry, egress, and evasion. Adv. Exp. Med. Biol. 440:395-414. [PubMed] [Google Scholar]
- 69.Sodeik, B. 2000. Mechanisms of viral transport in the cytoplasm. Trends Microbiol. 8:465-472. [DOI] [PubMed] [Google Scholar]
- 70.Sodeik, B., R. W. Doms, M. Ericsson, G. Hiller, C. E. Machamer, W. van ′t Hof, G. van Meer, B. Moss, and G. Griffiths. 1993. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J. Cell Biol. 121:521-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 72.Stern, W., and S. Dales. 1976. Biogenesis of vaccinia: isolation and characterization of a surface component that elicits antibody suppressing infectivity and cell-cell fusion. Virology 75:232-241. [DOI] [PubMed] [Google Scholar]
- 73.Stern, W., and S. Dales. 1976. Biogenesis of vaccinia: relationship of the envelope to virus assembly. Virology 75:242-255. [DOI] [PubMed] [Google Scholar]
- 74.Tabor, S., and C. C. Richardson. 1990. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Effect of pyrophosphorolysis and metal ions. J. Biol. Chem. 265:8322-8328. [PubMed] [Google Scholar]
- 75.Tooze, J., M. Hollinshead, B. Reis, K. Radsak, and H. Kern. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163-178. [PubMed] [Google Scholar]
- 76.Ulaeto, D., D. Grosenbach, and D. E. Hruby. 1996. The vaccinia virus 4c and A-type inclusion proteins are specific markers for the intracellular mature virus particle. J. Virol. 70:3372-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.van Eijl, H., M. Hollinshead, G. Rodger, W. H. Zhang, and G. L. Smith. 2002. The vaccinia virus F12L protein is associated with intracellular enveloped virus particles and is required for their egress to the cell surface. J. Gen. Virol. 83:195-207. [DOI] [PubMed] [Google Scholar]
- 78.Vazquez, M. I., G. Rivas, D. Cregut, L. Serrano, and M. Esteban. 1998. The vaccinia virus 14-kilodalton (A27L) fusion protein forms a triple coiled-coil structure and interacts with the 21-kilodalton (A17L) virus membrane protein through a C-terminal alpha-helix. J. Virol. 72:10126-10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vieira, J., and J. Messing. 1982. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19:259-268. [DOI] [PubMed] [Google Scholar]
- 80.Ward, B. M., and B. Moss. 2001. Visualization of intracellular movement of vaccinia virus virions containing a green fluorescent protein-B5R membrane protein chimera. J. Virol. 75:4802-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weir, J. P., and B. Moss. 1987. Determination of the transcriptional regulatory region of a vaccinia virus late gene. J. Virol. 61:75-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wilton, S., A. R. Mohandas, and S. Dales. 1995. Organization of the vaccinia envelope and relationship to the structure of intracellular mature virions. Virology 214:503-511. [DOI] [PubMed] [Google Scholar]