Abstract
Nipah virus (NiV) and Hendra virus (HeV) are novel paramyxoviruses from pigs and horses, respectively, that are responsible for fatal zoonotic infections of humans. The unique genetic and biological characteristics of these emerging agents has led to their classification as the prototypic members of a new genus within the Paramyxovirinae subfamily called Henipavirus. These viruses are most closely related to members of the genus Morbillivirus and infect cells through a pH-independent membrane fusion event mediated by the actions of their attachment (G) and fusion (F) glycoproteins. Understanding their cell biological features and exploring the functional characteristics of the NiV and HeV glycoproteins will help define important properties of these emerging viruses and may provide new insights into paramyxovirus membrane fusion mechanisms. Using a recombinant vaccinia virus system and a quantitative assay for fusion, we demonstrate NiV glycoprotein function and the same pattern of cellular tropism recently reported for HeV-mediated fusion, suggesting that NiV likely uses the same cellular receptor for infection. Fusion specificity was verified by inhibition with a specific antiserum or peptides derived from the α-helical heptads of NiV or HeV F. Like that of HeV, NiV-mediated fusion also requires both F and G. Finally, interactions between the glycoproteins of the paramyxoviruses have not been well defined, but here we show that the NiV and HeV glycoproteins are capable of highly efficient heterotypic functional activity with each other. However, no heterotypic activity was observed with envelope glycoproteins of the morbilliviruses Measles virus and Canine distemper virus.
The paramyxoviruses are enveloped, negative-stranded RNA-containing viruses and include a variety of important human and animal pathogens. These viruses contain two membrane-anchored envelope glycoproteins needed for efficient infection of a receptive host cell: an attachment glycoprotein which may be designated either the hemagglutinin-neuraminidase protein (HN), the hemagglutinin protein (H), or the G protein, depending on the particular paramyxovirus species, and the F glycoprotein, which facilitates the pH-independent membrane fusion event between the virion and host cell during virus infection, resulting in the entry of the nucleocapsid into the cytoplasm (reviewed in references 27 and 29). In a related process, cells expressing the fusion and attachment glycoproteins at their surfaces can mediate the formation of giant cells (syncytia). For most paramyxoviruses, efficient membrane fusion requires the presence of both the fusion and attachment glycoproteins, with the exception of the detectable F-mediated fusion in the absence of HN seen with the simian virus 5 (SV5) system (42). The details of how the attachment and fusion glycoproteins of the paramyxoviruses function in concert in mediating membrane fusion are not fully understood. For the most part, this interaction is type specific, and membrane fusion activity mediated by coexpression (mixing) of the fusion and attachment glycoproteins from different paramyxoviruses (heterotypic) is rarely seen. Although some examples have been noted, the potency of this fusion process is considerably lower than that mediated by the fusion and attachment glycoproteins from the same virus (homotypic) (2, 40).
To date, more is known about the important functional domains of the fusion glycoproteins that are involved in driving virion-host cell membrane fusion and their predicted fusogenic conformations than about the attachment glycoproteins. The paramyxovirus fusion proteins are type I membrane glycoproteins existing as trimeric oligomers with considerable hydrophobicity, and the attachment glycoproteins are type II proteins with a tetrameric oligomeric configuration (12, 36, 37, 47). Both proteins contain several potential N-linked glycosylation recognition sequences. Although it is generally presumed that the attachment protein must contact the fusion protein to induce conformational changes in F, evidence of a physical association between these glycoproteins has been observed with limited success and only with Newcastle disease virus (NDV) (49), Human parainfluenza virus (hPIV) (56), and, most recently, Measles virus (MeV) (44).
Recently, two newly emerging paramyxoviruses that were identified in cases of severe respiratory and encephalitic diseases in animals and humans have been described; they are now known as Hendra virus (HeV) and Nipah virus (NiV) (reviewed in reference 15). HeV emerged in 1994 and was transmitted to humans by close contact with horses; NiV emerged in 1999 and was passed from pigs to humans. Both are unusual among the paramyxoviruses in their abilities to infect and cause potentially fatal disease in a number of host species, including humans. Both viruses also have exceptionally large genomes and are genetically closely related yet distinct from all other paramyxovirus family members and distantly related to viruses in the genus Morbillivirus (52). The reclassification of HeV and NiV into the new Henipavirus genus was due to their unique biological and genetic features, and they are categorized as biological safety level-4 (BSL-4) pathogens, which severely limits the number of laboratory facilities capable of studying them.
HeV and NiV have a fusion (F) glycoprotein and an attachment (G) glycoprotein which lacks both hemagglutinin and neuraminidase activities. In initial studies we devised a quantitative assay for measuring viral glycoprotein-mediated membrane fusion with the F and G glycoproteins of HeV and demonstrated that both envelope glycoproteins were required to mediate fusion with host target cell membranes. Unlike other paramyxoviruses, HeV demonstrated a broad species tropism in vitro for both virus infection and membrane fusion. Protease treatment of target cells completely abolished HeV-mediated fusion, suggesting that the virus was employing a cell surface protein as its receptor (4).
Here we describe an examination of the NiV envelope glycoproteins and how these proteins compare to those of HeV. We demonstrate that NiV, like HeV, has a broad species tropism in vitro. HeV and NiV have the same receptor recognition pattern in the cell lines tested and also seem to have proportional fusion rates within each receptor-positive cell line, suggesting that HeV and NiV may use the same cellular receptor for virus entry. NiV fusion was also potently inhibited by peptides derived from either the HeV or NiV C-terminal heptad repeats of F, providing additional evidence for a conserved fusion mechanism for HeV and NiV compared to other paramyxoviruses. We have also examined the compatibility of the F and G glycoproteins of HeV and NiV, and we show that they are functionally closely related through the demonstration of highly efficient heterotypic membrane fusion activity. The efficiency of the heterotypic membrane fusion was correlated to the F envelope glycoprotein used. Together, our observations highlight some distinct differences and unique features of HeV and NiV in comparison to other members of the Paramyxoviridae and these observations may aid in our understanding of the mechanisms behind the emergence and cross-species transmission of these new infectious disease threats as well as afford new opportunities to dissect the underlying details of the paramyxovirus-mediated membrane fusion process.
MATERIALS AND METHODS
Cells and culture conditions.
The following cell lines were obtained from the American Type Culture Collection: HeLa (ATCC CCL 2), BSC-1 (ATCC CCL 26), HuTK−143B (TK−) (ATCC CRL 8303), RK-13 (rabbit) (ATCC CCL 37), Equus caballus (horse) (ATCC CCL-57), Sus scrofa (pig) (ATCC CL-101), and Tadarida brasilliensis (bat) (ATCC CCL-88). Primary chicken embryo fibroblasts (CEF) and baby hamster kidney (BHK) cells were provided by Norman Cooper, National Institutes of Health, Bethesda, Md. The 3T3, cat embryo, and duck embryo cell lines were provided by Jay A. Levy, University of California—San Francisco. The A3.01 and A3.02 cell lines were provided by Paul Kennedy, National Institutes of Health. The Hut 102, MT2, MT4, and CEM human T-cell lines were provided by Chou-Zen Giam, Uniformed Services University of the Health Sciences (USUHS), Bethesda, Md. The human osteosarcoma (HOS) and PM-1 cell lines were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program, and the human glioblastoma cell line U373-MG was provided by Adam P. Geballe, Fred Hutchinson Cancer Research Center, Seattle, Wash. (23).
Culture conditions.
HeLa, 3T3, BHK, HOS, and U373 cell monolayers were maintained in Dulbecco's modified Eagle's medium (Quality Biologicals, Gaithersburg, Md.) supplemented with 10% cosmic calf serum (CCS) (HyClone, Logan, Utah) and 2 mM l-glutamine. BS-C-1, TK−, and CEF cell monolayers were maintained in Eagle's minimal essential medium (EMEM) (Quality Biologicals) supplemented with 10% CCS and 2 mM l-glutamine (EMEM-10). Duck embryo monolayers, cat embryo cells, A3.01 cells, A2.01 cells, PM-1 cells, Hut 102 cells, MT2 cells, MT4cells, and CEM cells were maintained in RPMI 1640 (Quality Biologicals) supplemented with 10% CCS and 2 mM l-glutamine. Rabbit and horse cell monolayers were maintained in enriched EMEM (Quality Biologicals) supplemented with 10% CCS, 1 mM sodium pyruvate, and 2 mM l-glutamine. Bat cell monolayers were maintained in enriched EMEM containing 0.85 g of sodium bicarbonate/liter, 2 mM l-glutamine, and 10% CCS. Pig cell monolayers were maintained in Medium 199 (Quality Biologicals) containing 1.5 g of sodium bicarbonate/liter, 2 mM l-glutamine, and 3% CCS. All cell cultures were maintained at 37°C under a humidified 5% CO2 atmosphere.
Plasmids and recombinant vaccinia viruses.
For expression of recombinant NiV F and G glycoproteins, the F and G glycoprotein open reading frames (ORFs) were subcloned into the vaccinia virus promoter-driven expression vector pMC02 (8) from existing Escherichia coli expression plasmids. The NiV F ORF was initially PCR amplified from randomly primed NiV cDNA by using primers 5′-CGCGGATCCTCGACAATGGTAGTTATACTTG-3′ (NiV-F5-Bam) and 5′-GGTTGAAGCTTCAATCTGAATACACTATGTCC-3′ (NiV-F3-Hind), designed on the basis of the published NiV genome sequence (21). After gel purification, the PCR product was digested with BamHI and HindIII and cloned into the same sites of the E. coli expression vector pRSET-A (Invitrogen Corp., Carlsbad, Calif.). The NiV G ORF was cloned by using a similar strategy: PCR primers 5′-CGCGGATCCTTCAAGAAAATGCCGGCAGAA-3′ (NiV-G5-Bam) and 5′-GGTTGAAGCTTATGTACATTGCTCTGGTATC-3′ (NiV-G3-Hind) were used for initial amplification from random NiV cDNA, and the purified digested product was cloned into pRSET-A. The F and G gene coding regions were then transferred by PCR amplification into the vaccinia virus vector pMC02 by using primers 5′-GTCGACCCACATGGTAGTTATACTTGACAAGAGATGTTAT-3′ (NVFS) and 5′-GTCGACAGCCGGATCAAGCTTCAATCTGAATACACTATG-3′ (NVFAS) for NiV F and primers 5′-CTCGAGCCACATGCCGGCAGAAAACAAGAAAGTTAGATTCGAAAATACT-3′ (NVGS) and 5′-CTCGAGTAGCAGCCGGATCAAGCTTATGTACATTGCTCTGGTATC-3′(NVGAS) for NiV G with Accupol DNA polymerase (PGS Scientifics Corp., Gaithersburg, Md.). These primers generated a PCR product for the NiV F ORF flanked by SalI sites and a PCR product for the NiV G ORF flanked by XhoI sites, which were gel purified (Qiagen, Valencia, Calif.) and subcloned into the TOPO plasmid vector (Invitrogen). The TOPO NiV F construct was digested with SalI, and the TOPO NiV G construct was digested with XhoI; both fragments were gel purified (Qiagen) and subcloned into the SalI site of pMC02. All constructs were initially screened by restriction digestion and further verified by sequencing. The recombinant viruses were then obtained by standard techniques employing tk selection and Escherichia coli beta-glucuronidase (GUS) staining (7). Briefly, CV-1 cells were transfected with either pMC02 NiV F or pMC02 NiV G by using a calcium phosphate transfection kit (Promega Corp., Madison, Wis.). These monolayers were then infected with the Western Reserve (WR) wild-type strain of vaccinia virus at a multiplicity of infection (MOI) of 0.05 PFU/cell. After 2 days the cell pellets were collected as crude recombinant virus stocks. TK− cells were infected with the recombinant crude stocks in the presence of 25 μg of 5-bromo-2′-deoxyuridine (BrdU) (Calbiochem, La Jolla, Calif.)/ml. After 2 h the virus was replaced with an EMEM-10 overlay containing 1% low-melting-point agarose (Life Technologies, Gaithersburg, Md.) and 25 μg of BrdU/ml. After 2 days of incubation an additional EMEM-10 overlay containing 1% low-melting-point agarose, 25 μg of BrdU/ml, and 0.2 mg of 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (X-Gluc) (Clontech, Palo Alto, Calif.)/ml was added. Within 24 to 48 h blue plaques were evident; these were picked, and recombinant virus was subjected to two more rounds of double selection and plaque purification. The recombinant vaccinia viruses vKB7 (NiV F) and vKB6 (NiV G) were then amplified and purified. Recombinant vaccinia viruses vKB1 (HeV F) and vKB2 (HeV G) were produced in a manner similar to that previously described (4). The recombinant vaccinia viruses vT7-HMV, encoding H of MeV, vT7-FMV, encoding F of MeV, vT7-HCDV, encoding H of Canine distemper virus (CDV), and vT7-FCDV, encoding F of CDV, have been described previously (40). Bacteriophage T7 RNA polymerase was produced by infection with vTF7-3, which contains the T7 RNA polymerase gene linked to a vaccinia virus promoter (18). The E. coli lacZ gene linked to the T7 promoter was introduced into cells by infection with the recombinant vaccinia virus vCB21R-LacZ, which has been described previously (1).
Cell fusion assays.
Fusion between envelope glycoprotein-expressing and target cells was measured by a reporter gene assay in which the cytoplasm of one cell population contained vaccinia virus-encoded T7 RNA polymerase and the cytoplasm of the other contained the E. coli lacZ gene linked to the T7 promoter. β-Galactosidase (β-Gal) is synthesized only in fused cells (6, 39). Vaccinia virus-encoded proteins were produced by infecting cells (at an MOI of 10) and incubating infected cells at 31°C overnight (3). Cell fusion reactions were conducted with the various cell mixtures in 96-well plates at 37°C. Typically, the ratio of envelope glycoprotein-expressing cells to target cells was 1:1 (total cells per well, 2 × 105; total volume, 0.2 ml). Cytosine arabinoside (40 μg/ml) was added to the fusion reaction mixture to reduce nonspecific β-Gal production (3). For quantitative analyses, Nonidet P-40 was added (final concentration, 0.5%) at 2.5 h, and aliquots of the lysates were assayed for β-Gal at ambient temperature with the substrate chlorophenol red-d-galactopyranoside (Roche Diagnostics Corp., Indianapolis, Ind.). For NiV-mediated cell fusion assays, either we infected cells with the appropriate vaccinia virus encoding the NiV F or G or we transfected cell monolayers with the pMC02-based plasmid constructs containing these genes followed by a 2-h infection with WR vaccinia virus. Transfection of monolayers was performed with DOTAP (Roche Diagnostics Corp.). For inhibition by peptides, serial dilutions of peptides were made and added to envelope glycoprotein-expressing effector cells immediately prior to the addition of target cell populations. For inhibition by NiV-specific antisera, serial dilutions of the various rabbit sera were made and added to NiV glycoprotein-expressing effector cells just prior to the addition of target cells. All assays were performed in duplicate, and fusion results were calculated and expressed as rates of β-Gal activity (change in optical density at 570 nm per min × 1,000) (39).
Peptide synthesis.
The following hydrophilic peptide sequence was chosen for synthesis and immunization based on analysis of the hydrophobicity plot of the NiV G glycoprotein: CKSNGGGYNQHQLALRSIEKGRYDK (NiV G1; amino acids 324 to 347). The following peptide sequences, corresponding to the C-terminal α-helical heptad domains of the HeV F and NiV F glycoproteins, were chosen for synthesis: PPVYTDKVDISSQISSMNQSLQQSKDYIKEAQKILDTVNPSL (HeV FC2) and PPVFTDKVDISSQISSMNQSLQQSKDYIKEAQRLLDTVNPSL (NiV FC1). A scrambled version of the 42-amino-acid peptide HeV FC2 was also synthesized for use as a control (YVKTLKPDVSISQSMIQLQSKPYQIEQKSNDLTNSPVSDIDA) (ScHeV FC2). Each peptide was synthesized on an Applied Biosystems model 433 Peptide Synthesizer using 0.45 M 1H-benzotriazole tetramethyluronium hexafluorophosphate (HBTU)-0.5 M N-hydroxybenzotriazole • H2O (HOBt) in dimethyl formamide for activation on a hydroxymethylphenoxymethyl-copolystyrene-1% divinylbenzene resin. Upon completion of synthesis, the resin was washed twice with dichloromethane, followed by three washes with methanol, and allowed to dry. Cleavage of the peptide from the resin was obtained by using Reagent R (90% trifluoroacetic acid, 5% thioanisole, 3% 1,2-ethanedithiol, and 2% anisole) at room temperature for 3 h. The peptide was isolated from the mixture by vacuum filtration through a sintered glass funnel into cold ethyl ether, which permitted precipitation. The peptide and ether were transferred to a 50-ml centrifuge tube and centrifuged. The peptide pellet was resuspended in cold ether and centrifuged three separate times to remove residual scavengers and acid. After the third wash, the pellet was allowed to dry completely. Once dry, the peptide was resuspended in 95% water-5%CH3CN, the pH was adjusted to ∼7 by using dilute NH4OH, and the solution was frozen at −20°C and lyophilized.
Metabolic labeling and immunoprecipitation.
For labeling of NiV glycoproteins expressed by recombinant vaccinia viruses, HeLa cells were infected at an MOI of 10 PFU/cell. At 6 h postinfection, monolayers were washed, overlaid with methionine- and cysteine-free minimal essential medium (MEM) (Life Technologies) containing 2.5% dialyzed fetal calf serum (Life Technologies) and 100 μCi of [35S]ProMix (Amersham Pharmacia Biotech, Piscataway, N.J.)/ml, and incubated overnight. Cells were lysed in 100 mM Tris-HCl (pH 8.0)-100 mM NaCl-1% Triton X-100, and nuclei were removed by centrifugation. Typically, 0.5 to 1.0 μl of the antiserum or normal rabbit serum was utilized per immunoprecipitation. Incubations for at least 1 h at 4°C were followed by addition of protein G-Sepharose 4 (Amersham Pharmacia Biotech) for at least 30 min at room temperature. Complexes were washed twice with lysis buffer (100 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1% Triton X-100) and once with DOC buffer (100 mM Tris-HCl [pH 8.0], 100 mM NaCl, 0.1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]). Proteins were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) (10% acrylamide) and visualized by autoradiography. The HeV F2 peptide-specific antiserum was generated in previous studies (4). Sera from a rabbit immunized with gamma-irradiated NiV were used in radioimmunoprecipitation and cell fusion assays.
Western blot analysis.
HeLa cell monolayers were infected overnight at an MOI of 10 with a wild-type vaccinia virus or a recombinant vaccinia virus encoding NiV F or Ni G. Cells were extracted with 1% Triton X-100 in 100 mM Tris-HCl (pH 8.0)-100 mM NaCl, and nuclei were removed by centrifugation. Samples were prepared by boiling in sample buffer containing 2-mercaptoethanol. Extracts from 5 × 104 cells (total) were loaded per well onto an SDS-10% PAGE gel. Following transfer to nitrocellulose paper, the blot was probed with an HeV F2-specific rabbit antiserum. The blot was then incubated with horseradish peroxidase-conjugated rabbit anti-rabbit immunoglobulin G and developed with the SuperSignal chemiluminescence kit (Pierce).
Disclaimer.
The views expressed in this report are solely those of the authors, and they do not represent official views or opinions of the Department of Defense or the USUHS.
(K. N. Bossart performed this work as partial fulfillment of the requirements of the Ph.D. program in Microbiology and Immunology of the USUHS.)
RESULTS
Expression of henipavirus F and G glycoproteins.
To examine the functional and biochemical properties of the NiV and HeV envelope glycoproteins, the proteins responsible for host cell attachment and virus entry, we have employed the vaccinia virus-based recombinant expression system. The use of this system for study of the envelope glycoproteins of HeV was described recently (4). Here, for production of recombinant vaccinia virus-expressed NiV envelope glycoproteins, the putative glycoprotein ORFs for NiV F and G were subcloned into the vaccinia virus promoter-driven expression vector pMC02 (8) and recombinant vaccinia viruses were generated by standard techniques as detailed in Materials and Methods. NiV envelope glycoproteins F and G were produced in cell culture by infection with recombinant vaccinia viruses. Shown in Fig. 1A are immunoprecipitation results for recombinant vaccinia virus-expressed NiV F, NiV G, or both NiV F and NiV G, using NiV- or HeV-specific antiserum. Vaccinia virus-expressed NiV F appeared as the precursor protein, F0, and as the processed F1 subunit. The F2 subunit (∼19 kDa) was not readily detected under these conditions, most likely owing to a combination of the amount of protein and the specific activity of the metabolically labeled polypeptide. This profile of NiV F was quite similar to that of recombinant vaccinia virus-expressed HeV F (4), which is also shown for comparison (Fig. 1B), as well as to those of several other paramyxovirus F glycoproteins (2, 40, 56), with apparent molecular sizes of ∼61 kDa for F0 and ∼49 kDa for F1, and was also similar to that of the F polypeptides derived from purified HeV particles (33, 38, 53). Vaccinia virus-expressed NiV G had an apparent molecular size of ∼72 to 75 kDa, similar to, yet distinctly lower than, those of recombinant vaccinia virus-expressed HeV G (Fig. 1B) (4) and HeV G derived from purified HeV virions (38, 53), which have molecular sizes of ∼74 to 77 kDa. In general, these molecular sizes of both HeV and NiV G are similar to those of the attachment glycoproteins (H) from MeV and CDV (40). Shown in Fig. 1C are recombinant vaccinia virus-expressed NiV and HeV F glycoproteins detected by Western blotting using an HeV F2 peptide-specific antiserum. NiV F appears to be more processed than HeV F, as determined by a marked reduction of the NiV F0 species. This is consistent with the relative intensities of F0 and F1 observed in Fig. 1A and B. For NiV the intensities of F0 and F1 are similar, but for HeV there is more F0 than F1. F0 and F2 for both HeV and NiV migrate close to their predicted molecular sizes of ∼61 and ∼19 kDa, respectively.
FIG. 1.
Expression of recombinant NiV F and G glycoproteins. The NiV F and G glycoprotein ORFs were subcloned into the vaccinia virus promoter-driven expression vector pMC02 (8), and recombinant viruses were made (see Materials and Methods). HeLa cells were infected with NiV F- or G-encoding viruses and incubated 16 h at 37°C. Beginning at 6 h postinfection, cells were either labeled overnight with [35S]methionine-cysteine for immunoprecipitation or cultured in medium alone for Western blotting. Lysates were prepared in a buffer containing Triton X-100 and clarified by centrifugation. Immunoprecipitation was performed with a rabbit anti-NiV or a rabbit anti-HeV antiserum followed by protein G-Sepharose. Western blotting was performed with a rabbit polyclonal antiserum against a synthetic F2 peptide (see Materials and Methods). The metabolically labeled proteins were resolved by SDS-10% PAGE under reducing conditions and detected by fluorography; lysates for Western blotting were resolved by SDS-10% PAGE under reducing conditions and detected by chemiluminescence. (A and B) Immunoprecipitation; (C) Western blotting.
Membrane fusion tropism mediated by HeV and NiV F and G glycoproteins.
Adaptation of a previously developed reporter gene assay capable of quantitatively measuring cell fusion mediated by the viral envelope glycoproteins of either HeV or NiV has afforded several avenues of investigating the nature of these otherwise BSL-4-restricted agents. This system is based on gene expression using the recombinant vaccinia virus system (3, 39), where in addition to expression of the viral envelope glycoproteins and viral receptors on effector and target cell populations, respectively, one cell population also expresses bacteriophage T7 RNA polymerase and the other expresses a T7 promoter-driven E. coli lacZ cassette (see Materials and Methods). Thus, cell fusion results in the specific production of β-Gal, which can be quantified. This assay has proven especially useful in the study of envelope glycoproteins derived from viruses which employ a pH-independent mechanism of membrane fusion for virion entry (2, 11, 17, 26, 40, 41, 48, 50). For use in this assay, NiV glycoprotein-expressing effector cells were prepared and mixed with various target cell populations. Typically, the target and effector cell populations are assessed in duplicate or triplicate in 96-well-plate format and are incubated 2 to 4 h following mixing. Cell lysates are prepared and processed for β-Gal quantification. Initial experiments using HeLa cells for NiV F and G expression (effector cells) and BSC-1 and HeLa cells as putative receptor-positive cells (target cells) rapidly revealed that, as with HeV, HeLa cells were nonpermissive for NiV-mediated fusion and that NiV requires both the F and G envelope glycoproteins to mediate fusion with receptor-positive cell lines (data not shown). Since HeLa cells were not permissive for NiV-mediated fusion, they were selected for expression of NiV F and G in subsequent experiments (effector cells). The evaluation of host cell tropism by measuring cell fusion was then expanded to include a variety of target cells, including those that had previously been examined for their abilities to support HeV-mediated fusion. Figure 2A shows that both NiV F and G are needed to mediate cell fusion and that a wide panel of cell lines from a variety of animal species appear to have the NiV receptor on their cell surfaces. Shown in Fig. 2B are HeV- and NiV-mediated fusion results with additional human T-cell lines and U373, a human glioblastoma cell line, as target cells. The MT2 cell line is the first T-cell line examined that appears to express the HeV and NiV receptor. These data also demonstrate that the U373 cell line supported the highest level of NiV-mediated cell fusion, which may reflect the neural tropism of the virus and the subsequent pathology seen in NiV-infected humans and animals (9, 31, 34). For these reasons, U373 cells were included as an important target cell line in subsequent experiments. Although HeV fusion rates are not shown in Fig. 2A, NiV F and G were able to mediate fusion with the same target cell populations used by HeV, and for both NiV and HeV, BSC-1 (monkey kidney), U373, BHK 21, and cat embryo cells supported the highest levels of fusion. Together these findings suggest that HeV and NiV may use the same receptor on the surfaces of the target cells. Earlier data had already suggested that HeV may be using a cell surface protein as its receptor for fusion and viral entry (4). The broad species tropism demonstrated by HeV and NiV in the cell fusion assay is a unique biological property that is not common to other paramyxoviruses. Moreover, the large number of species that contain receptor-positive cells may play an important role in the cross-species transmission of these viruses from animals to humans. As in previous studies with HeV F and G, NiV F and G were unable to mediate fusion with the pig kidney cell line used in this study. As previously discussed, more pig cell lines need to be tested to further support the notion that our in vitro host cell tropism results correlate with natural infections.
FIG. 2.
Quantitation of NiV envelope glycoprotein-mediated cell fusion. HeLa cells were infected with recombinant vaccinia viruses encoding either NiV F, NiV G, both NiV F and G, neither (none), or both HeV F and G, along with a recombinant vaccinia virus encoding T7 RNA polymerase (effector cells). Each designated target cell type was infected with the reporter vaccinia virus vCB21R, encoding E. coli lacZ. NiV or HeV glycoprotein-expressing cells (105) were mixed with each target cell type (105) in duplicate wells of a 96-well plate. After 3 h at 37°C, Nonidet P-40 was added and β-Gal activity was quantitated. Key: target cells only, level of background β-Gal activity in target cell populations alone; none, β-Gal activity from target cells mixed with HeLa partner cells infected with T7 RNA polymerase-encoding vaccinia virus only and no recombinant vaccinia viruses encoding NiV or HeV glycoproteins. The level of background β-Gal activity in effector cell populations alone is labeled “effector cells” on the x axis. (A) Species tropism of NiV-mediated cell fusion. (B) NiV-mediated cell fusion, compared to HeV-mediated fusion, with human T-cell and neuroblastoma cell lines.
Specificity of HeV- and NiV-mediated fusion activity.
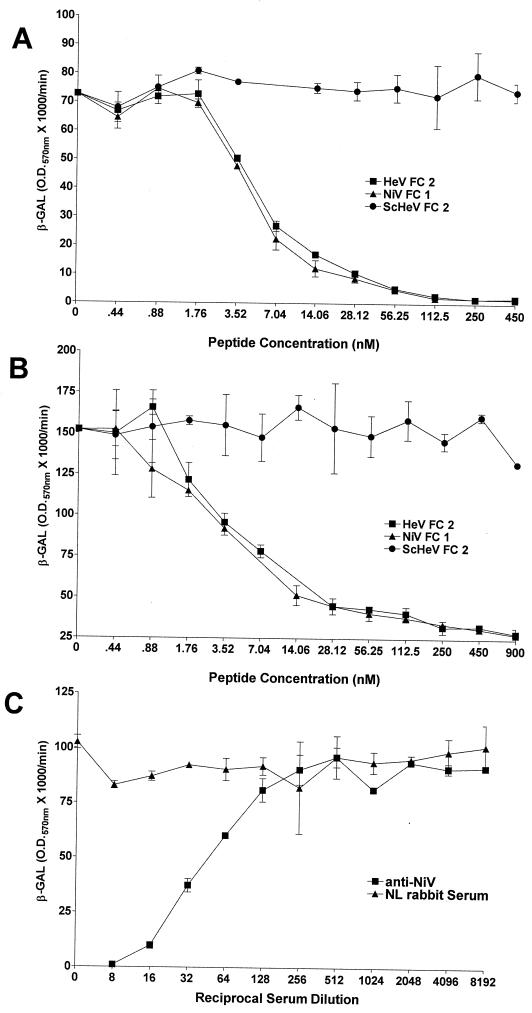
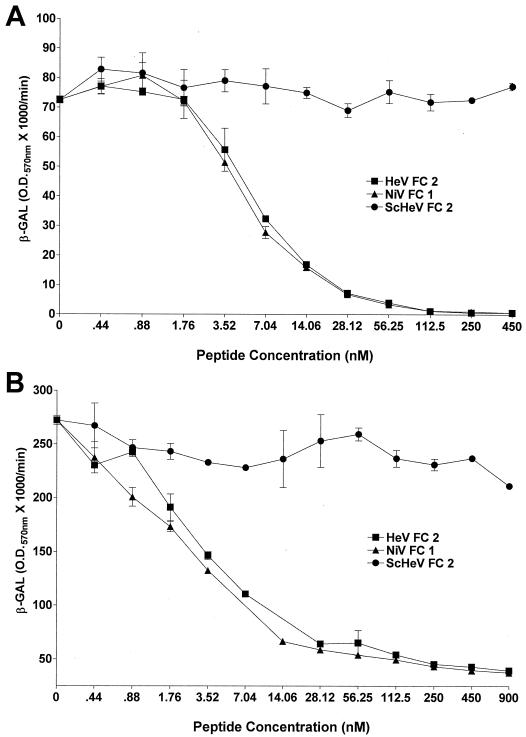
Major advances have been made recently in the understanding of the structural requirements and potential mechanisms involved in the fusion of the membranes of enveloped viruses with their host cell membranes (reviewed in references 14, 16, 46, and 54). Current evidence from a number of groups supports a model indicating that the formation of a trimer-of-hairpins structure whose oligomeric coiled-coil formation is mediated by the two α-helical heptad repeat domains of the fusion protein is coupled to membrane fusion. Peptides derived from either of the α-helical heptad repeat regions of enveloped viral fusion proteins have previously been shown to be potent inhibitors of the fusion process for a number of viruses, including several paramyxoviruses, when present during the fusion process (24, 28, 30, 45, 55, 58, 60). Both HeV and NiV have two putative heptad repeat domains in F, one proximal to the F1 fusion peptide (N terminus) and the other very close to the predicted transmembrane domain (C terminus). Helical wheel analysis of HeV F revealed a high degree of sequence homology of important functional residues of the heptad repeats with those of SV5 F, and synthetic C-terminal HeV peptides inhibited HeV-mediated fusion (4). To determine if these domains played an important role in NiV-mediated fusion, a 42-amino-acid peptide analogous to the NiV F C-terminal heptad repeat was synthesized (NiV FC1) and tested for its ability to interfere with NiV-mediated fusion. Since there were three amino acid differences within the C-terminal heptad repeat of HeV and NiV, a second peptide, corresponding to the HeV F C-terminal heptad repeat, was also synthesized (HeV FC2). A scrambled version of HeV FC2 (ScHeV) was synthesized and used as a negative control. Shown in Fig. 3A and B are the results obtained in the presence of these peptides for both HeV- and NiV-mediated fusion. HeV FC2 and NiV FC1 could inhibit both HeV- and NiV-mediated fusion in a dose-dependent manner and were completely inhibitory in the nanomolar range, with 50% inhibitory concentrations (IC50) between 5.2 and 5.8 nM, respectively. ScHeV FC2 had no inhibitory effect on HeV- or NiV-mediated fusion. These data suggest that HeV and NiV have similar mechanisms of virion-cell membrane fusion and that these mechanisms are likely comparable to those proposed for other viral fusion systems, where a trimer of hairpins has been hypothesized to form. There were no significant differences between the abilities of HeV FC2 and NiV FC1 to neutralize either HeV- or NiV-mediated fusion. The conservative Y450F and K479R amino acid substitutions in NiV F did not affect the ability of NiV FC1 to inhibit HeV-mediated fusion or the ability of HeV FC2 to inhibit NiV-mediated fusion. These results are further supported by helical wheel analysis, which revealed that none of these amino acids fall in the proposed functional points of the putative C-terminal α-helix of HeV and NiV F thought to be involved in protein-protein interactions leading to the formation of the trimer-of-hairpins fusogenic conformation. Neither NiV FC1, HeV FC2, nor ScHeV FC2 had any effect on cell fusion mediated by the envelope glycoproteins of MeV or CDV (data not shown), further demonstrating the specificity of this recombinant HeV- and NiV-mediated membrane fusion system.
FIG. 3.
Specificity of NiV- and HeV-mediated fusion. Effector cells were prepared as described in the legend to Fig. 2. Human U373 cells were infected with the reporter vaccinia virus vCB21R encoding E. coli lacZ (target cells). Peptides or a rabbit polyclonal anti-NiV serum were diluted and added to the glycoprotein-expressing cells (105) in a 96-well plate, and U373 cells were then added (105). Each peptide and serum concentration was tested in duplicate in 96-well-plate format. After 3 h at 37°C, Nonidet P-40 was added and β-Gal activity was quantitated. (A) Inhibition of HeV-mediated fusion by synthetic C-terminal F peptides. (B) Inhibition of NiV-mediated fusion by synthetic C-terminal F peptides. (C) Inhibition of NiV-mediated fusion by an anti-NiV antiserum.
To further evaluate the specificity of NiV-mediated fusion, a polyclonal rabbit anti-NiV antiserum and a normal rabbit serum were compared for their abilities to inhibit NiV-mediated cell fusion. Both sera were serially diluted and added to envelope glycoprotein-expressing effector cell populations just prior to the addition of target cells. The normal rabbit serum lowered NiV-mediated fusion slightly, but by no more than ∼15% at the highest serum concentration; conversely, the NiV-specific antiserum could block cell fusion by >90% at a 1:8 dilution, and there was approximately 50% inhibition at a 1:50 dilution (Fig. 3C). The NiV-specific antiserum was also able to block HeV-mediated cell fusion, but to a lesser extent (data not shown). This is probably due to the polyclonal nature of the anti-NiV antiserum and the high level of antigenic relatedness between HeV and NiV.
Heterologous fusion activity of the HeV and NiV F and G glycoproteins.
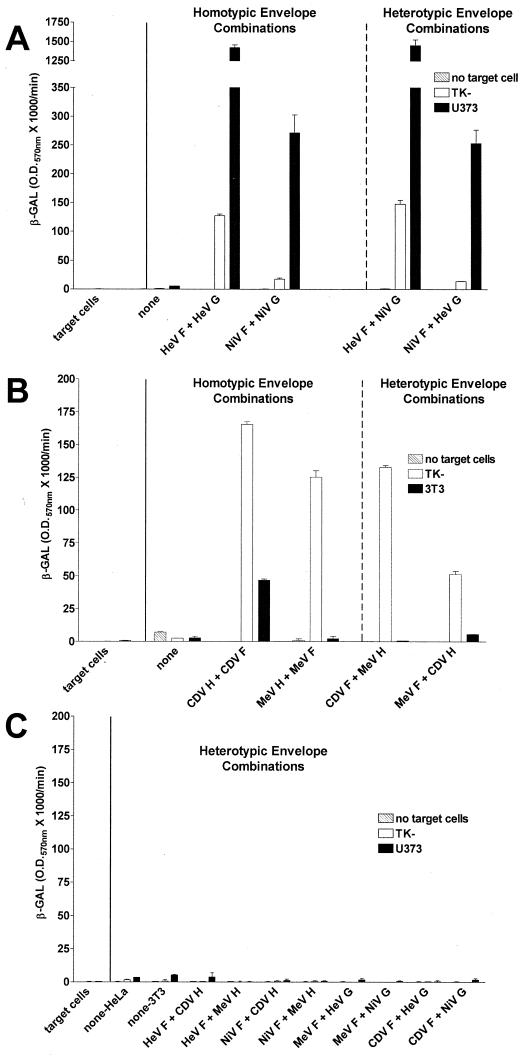
Since the cellular tropism and fusion requirements of HeV and NiV appeared to be very similar to one another yet distinct from those of other paramyxoviruses, we explored whether the envelope glycoproteins from these henipaviruses could function in the context of heterologous combinations of the fusion and attachment envelope glycoproteins. For most paramyxoviruses, including HeV and NiV, efficient membrane fusion requires the presence of both the fusion and attachment envelope glycoproteins, although there is considerable evidence that fusion mediated by F alone can be readily measured for SV5 (reviewed in reference 46). It is also clear that methods to facilitate close membrane-to-membrane contact with an F-alone fusion system can also enhance membrane fusion in the absence of the homotypic attachment protein, which in the case of SV5 is the HN protein (43). Among the paramyxoviruses, members of the genus Morbillivirus are most closely related to HeV and NiV (52), and previously, two morbilliviruses, MeV and CDV, were examined and heterologous function with different combinations of the MeV and CDV envelope glycoproteins was demonstrated (40). Here, in a similar fashion, we examined the abilities of the HeV and NiV envelope glycoproteins to function in heterologous combinations in a syncytium formation assay. Shown in Fig. 4 are the syncytium formation results for U373 target cells mixed with HeLa effector cells expressing several HeV and NiV heterologous envelope glycoprotein combinations. The human U373 cells were chosen because of the high level of cell fusion observed with both HeV and NiV, presumably due to expression of high levels of the virus receptor. Homotypic glycoprotein combinations are shown in Fig. 4C and D for HeV and NiV, respectively, and efficient cell fusion with the U373 target cell was evident for both. No syncytia were observed with HeLa effector cells expressing only the F glycoprotein of either HeV or NiV (Fig. 4A and B, respectively). Effector cells expressing heterotypic mixes of the F and G glycoproteins of HeV and NiV were clearly capable of mediating cell fusion with U373 target cells (Fig. 4E and F). It was also evident that HeV-mediated cell fusion resulted in somewhat larger and more numerous syncytia than NiV-mediated cell fusion.
FIG. 4.
Syncytium formation mediated by homotypic and heterotypic NiV and HeV envelope glycoprotein combinations. HeLa cells were infected with recombinant vaccinia viruses encoding either HeV F (A), NiV F (B), HeV F and HeV G (C), NiV F and NiV G (D), HeV F and NiV G (E), or NiV F and HeV G (F) (effector cells). Partner U373 cells were detached by using EDTA and washed three times with PBS. The effector cells (105) were mixed with the U373 partner cells (105) in duplicate wells of a 96-well plate and incubated at 37°C. After 18 h, photographs were taken at ×400 magnification.
In light of these results indicating functional compatibility of the HeV and NiV glycoproteins as well as the enhanced cell fusion mediated by HeV F compared to NiV F, we sought to examine these cell fusion processes in a quantitative manner in order to define any subtle differences between homologous and heterologous envelope combinations. Shown in Fig. 5 are quantitative cell fusion results mediated by effector cell populations expressing various combinations of the HeV, NiV, MeV, and CDV envelope glycoproteins. The NiV and HeV envelope glycoproteins could efficiently mediate fusion in heterologous envelope combinations with each other (Fig. 5A). Further, the fusion signal observed with either heterologous combination correlated quite well to the fusion level seen with the homologous combination which included the same F glycoprotein. These fusion results were also in agreement with the syncytium formation results shown in Fig. 4. Because HeV and NiV were so efficient in supporting heterotypic envelope glycoprotein-mediated fusion, we wanted to examine whether they could also support a heterotypic fusion reaction with glycoproteins derived from other related viruses that were available and suitable for our cell fusion system. In parallel, we reassessed our heterotypic fusion results using the fusion and attachment glycoproteins from the morbilliviruses MeV and CDV (Fig. 5B). As expected, heterotypic combinations of MeV and CDV envelope glycoproteins were capable of mediating fusion, although less efficiently than the homologous envelope combinations. Here, human TK− cells are permissive for fusion with MeV, while murine 3T3 cells are not, owing to the absence of a functional MeV receptor, whereas CDV can mediate fusion with both target cell types. We then tested whether coexpression of NiV and HeV F or G with morbillivirus F or H from MeV or CDV could result in any heterotypic fusion activity; however, no fusion was detectable with any of these glycoprotein combinations (Fig. 5C). Here, the human TK− and U373 target cells were chosen because they were the most proficient cells for fusion observed for all four viruses. The fact that fusion with heterotypic glycoprotein combinations of these related morbilliviruses is much less efficient than that observed for HeV and NiV is consistent with the degree of close genetic relatedness between the latter virus pair and the notion that HeV and NiV likely use the same cell surface receptor while MeV and CDV, although closely related, do not. The fusion specificity observed with the HeV and NiV heterotypic functional activity was verified by using HeV FC2 and NiV FC1, as was done for homologous envelope combinations, and results are shown in Fig. 6. HeV F- and NiV G-mediated fusion was completely inhibited by HeV FC2 or NiV FC1, and the dose-dependent curve closely resembled that seen with HeV F and HeV G. NiV F- and HeV G-mediated fusion was also completely inhibited by HeV FC2 or NiV FC1 and resembled that seen with NiV F and NiV G. The IC50 of HeV FC2 and NiV FC1 for fusion mediated by all combinations of HeV and NiV envelope glycoproteins are summarized in Table 1.
FIG. 5.
Quantitation of cell fusion mediated by homotypic and heterotypic NiV and HeV envelope glycoprotein combinations. HeLa or 3T3 cells were infected with recombinant vaccinia viruses encoding various combinations of the envelope glycoproteins HeV F, HeV G, NiV F, NiV G, MeV F, MeV H, CDV H, and CDV F, along with a recombinant vaccinia virus encoding T7 RNA polymerase (effector cells). HeLa cells were used as effector cells for expression of HeV and NiV G envelope combinations, and 3T3 cells were used as effector cells for expression of MeV and CDV HA envelope combinations. TK−, U373, and 3T3 target cells were infected with the reporter vaccinia virus vCB21R encoding E. coli lacZ (target cells). Glycoprotein-expressing cells (105) were mixed with each target cell type (105) in duplicate wells of a 96-well plate. After 3 h at 37°C, Nonidet P-40 was added and β-Gal activity was quantitated. Key: no target cells, level of background β-Gal activity in effector cell populations alone. The level of background β-Gal activity in target cell populations alone is labeled “target cells” on the x axis, and that from target cells mixed with effector cells infected only with a vaccinia virus encoding T7 RNA polymerase is labeled “none.” (A) HeV and NiV envelope combinations; (B) MeV and CDV envelope combinations; (C) henipavirus and morbillivirus envelope combinations.
FIG. 6.
Specificity of heterotypic envelope function. Exactly the same procedures as those described in the legend to Fig. 3 were followed. (A) Inhibition of HeV F- and NiV G-mediated fusion by synthetic C-terminal F peptides. (B) Inhibition of NiV F- and HeV G-mediated fusion by synthetic C-terminal F peptides.
TABLE 1.
IC50 of peptides derived from the fusion glycoproteins of HeV and NiV
| Envelope glycoprotein combination | IC50 (nM) of:
|
|
|---|---|---|
| HeV FC2a | NiV FC1b | |
| HeV F + HeV G | 5.8 | 5.2 |
| HeV F + NiV G | 6.5 | 5.9 |
| NiV F + NiV G | 5.3 | 5.8 |
| NiV F + HeV G | 2.5 | 2.9 |
HeV FC2 is specific to the C-terminal heptad repeat of HeV F.
NiV FC1 is specific to the C-terminal heptad repeat of NiV F.
DISCUSSION
The results presented here have established the requirements for NiV-mediated fusion and have defined some functional similarities and differences between the NiV and HeV envelope glycoproteins. These two viruses emerged in two geographically isolated countries 5 years apart. However, they appear to have the same reservoir in nature, namely, certain fruit bat species found in the Australasian region, commonly known as flying foxes (10, 19, 20, 32, 57, 59). As a group, these animals have a large range which encompasses much of Southeast Asia, spreading as far west as the eastern coast of Africa. It is also of interest that these two viruses independently spread, almost certainly from flying foxes, to different animal species and from there to cause fatal disease episodes in humans. In light of the evidence for the existence of additional and distinct Hendra-like viruses in various pteropid bat species, these observations suggest that additional emerging viruses may yet appear as other animal species serve as amplifying hosts. It is important to understand the mechanisms which underlie the transmission of such new infectious agents in nature to animals and ultimately to humans. In the initial outbreak HeV caused a fatal respiratory disease in 14 horses, and although only 2 human cases were diagnosed at that time, an unknown disease in such a large number of horses led to an investigation and the eventual discovery of the virus. In contrast, it appeared at first that NiV did not cause fatal disease in pigs, but the large outbreak of human cases of encephalitis necessitated an investigation into the causative agent, which was then traced to infected pigs in close contact with humans. However, before the recognition of the disease outbreak occurring in both humans and pigs, there were animal deaths from viral pneumonia and encephalitis which were attributed to swine fever but are now recognized to have been caused by NiV (35). The diseases of animals and humans caused by these two viruses are different, but their genetic makeup and some related biological properties show their relatedness. Here we report studies detailing several functional differences and commonalities between NiV and HeV envelope glycoproteins, the viral proteins which influence tropism and facilitate virus entry.
The NiV envelope glycoproteins were cloned into vaccinia virus shuttle vectors, and recombinant vaccinia viruses were made. Protein expression was verified through metabolic labeling and immunoprecipitation using a polyclonal anti-NiV antiserum. The molecular size of the recombinant-virus-expressed NiV G was comparable to that of HeV G, migrating at a slightly smaller apparent size of ∼72 to 75 kDa, which, however, was larger than its predicted molecular size of ∼67 kDa. The molecular size of the recombinant-virus-expressed NiV F0 precursor was ∼61 kDa, that of the processed F1 subunit was ∼49 kDa, and that of the processed F2 subunit was ∼19 kDa. The recombinant-virus-expressed NiV F glycoprotein appeared to be processed to a greater extent than the HeV F glycoprotein, in agreement with observations made with infectious viruses (52). Based on the similarity in molecular weights in comparison to those of other members of the Paramyxoviridae, the F and G glycoproteins of NiV are undoubtedly N glycosylated at one or more sites.
The functional activity and cell fusion species tropism for NiV were examined and compared to those for HeV. Both viruses were found to use the same receptor recognition pattern among the cell lines we examined, and those cell lines that supported the highest level of HeV-mediated fusion also supported the highest level of NiV-mediated fusion. These data suggest that HeV and NiV are likely using the same receptor for entry into receptive host cells. Previous work had also suggested that HeV may be using a cell surface protein as its receptor (4). As with HeV, NiV also requires both F and G to mediate membrane fusion, which has been observed for all paramyxoviruses with the exception of SV5, where F-mediated fusion in the absence of HN is detectable (42). The human cell line U373 supported the highest level of membrane fusion for both NiV and HeV. This finding is of interest because U373 is a human cell line of neural origin, and it suggests that related cell types may be important targets in the central nervous system in NiV or HeV infections of humans.
For most paramyxoviruses, membrane fusion mediated by the fusion and attachment glycoproteins is a type-specific event, and functional activity derived from mixing the fusion and attachment glycoproteins of different viruses (heterotypic) has only rarely been observed. The heterotypic fusion activity that has been measured is considerably lower than a virus's homotypic fusion activity (2, 5, 40, 51). In prior work, Nussbaum et al. were the first to demonstrate heterotypic fusion with the mixing of the F and H glycoproteins of the morbilliviruses MeV and CDV and to show that CDV-mediated fusion on CD46-expressing cells could be rescued upon coexpression of MeV H with CDV F (40). The heterotypic fusion activity in that system was not as potent or efficient as the fusion obtained with homotypic MeV or CDV F and H. However, this heterotypic activity was bidirectional, i.e., fusion could occur with either heterotypic combination, which was unlike the efficient heterotypic fusion results observed with Sendai virus F combined with hPIV-1 HN, in contrast to the inability of Sendai virus HN to functionally complement hPIV-1 F (5). Given the percent similarities of the F and HN glycoproteins of these viruses compared to those of MeV and CDV, this would not have been expected. We examined the heterotypic functional activities of the HeV and NiV F and G glycoproteins and found that they are highly functionally compatible in a bidirectional manner. Indeed, the level of fusion measured with either heterotypic mix was the same as that measured for the homotypic combination containing the same F glycoprotein; that is, the fusion potency correlated to the species of F glycoprotein, a finding that has not been observed with any other paramyxovirus system. Evidence of a physical association between the paramyxovirus fusion and attachment glycoproteins has been observed with only limited success and only with NDV (13, 49), hPIV (56), and most recently with MeV (44). Whether the observed highly efficient heterotypic interactions between HeV and NiV reported here translate into efficient measurable physical interactions remains to be determined.
Another observation of interest from our studies was that HeV-mediated fusion was consistently more potent than NiV-mediated fusion. The basis of the observed fusogenic differences between HeV and NiV is not clear and does not appear to be related to the levels of envelope glycoproteins expressed. Since we believe that HeV and NiV may share the same receptor, the difference in the potency of fusion may be attributable to structural and/or functional differences in the F and G envelope glycoproteins or to differences in the ways the two proteins engage one another. The apparent difference in processing efficiency observed here between recombinant vaccinia virus-expressed NiV F and HeV F is in agreement with observations on infectious NiV (52), but whether this distinction directly affects NiV-mediated fusion activity is unknown at present. Even though NiV and HeV are quite closely related on a genetic basis, the cleavage recognition site of the HeV F precursor polypeptide contains a lysine (K) residue in the P1 position, whereas the NiV F precursor contains an arginine (R) in that position, which is similar to the residue in that position for all other fusion glycoproteins among Paramyxoviridae members and across several virus families including the Orthomyxoviridae, Flaviviridae, Togaviridae, and Retroviridae (25). Although this is a conservative amino acid substitution, it may be important for proteolytic cleavage and activation of F. Mutagenesis studies are under way to determine what role this cleavage site distinction plays in F0 processing and the subsequent fusion rates seen for HeV and NiV F. Indeed, the fusion rates demonstrated here in the heterologous mixing experiments support the notion that the F envelope glycoprotein is the more important component in the mechanism affecting the rate of fusion. This notion is supported by the observation that either G glycoprotein is utilized equally well by either F, and differences in receptor recognition are a less likely explanation. Indeed, we examined the fusion specificities for homologous and heterologous envelope combinations by using peptides derived from the C-terminal α-heptad repeat from either HeV or NiV F. However, we observed no significant differences in the IC50 (Table 1) for either peptide in either the homologous or heterologous envelope combinations. These data, demonstrating inhibition of the fusion processes of HeV and NiV, offer an attractive avenue for the development of therapeutics, which has met with promising success with human immunodeficiency virus type 1, and these peptides do inhibit infectious HeV and NiV entry (B. T. Eaton, unpublished data).
Although processing of F0 is necessary for its activation and there are apparent functional differences between HeV and NiV F, it is possible that the observed functional differences between the efficiencies of the two F glycoproteins in mediating fusion could be related to the ways in which the HeV and NiV F glycoproteins interact with or engage the G glycoprotein in the fusion process. Delineation of the regions in both F and G that are involved in their interaction in mediating membrane fusion should aid in our understanding of the mechanism of paramyxovirus fusion in general. Preliminary studies in our laboratory have revealed an N-linked glycosylation site deletion mutant of HeV G that is no longer capable of supporting efficient HeV-mediated fusion (C. C. Broder and K. N. Bossart, unpublished results). This kind of posttranslational modification may be critical in determining the native structure of G or may play an important role in the interaction between HeV F and G and/or act to stabilize the proposed fusogenic conformation of HeV F. The HeV and NiV G glycoproteins share only 83% amino acid identity, yet they are identical in the location and number of seven extracellular potential N-linked glycosylation sites (22, 52), suggesting that certain sites may be critical for proper folding or function of the glycoprotein.
In summary, we have established a recombinant system to express and characterize the F and G membrane glycoproteins of NiV and HeV. This system has afforded the opportunity to examine these glycoproteins on a functional level in a quantitative manner and will also serve as a useful tool in future experiments aimed at exploring the interactions between the F and G glycoproteins. We have also demonstrated that efficient NiV-mediated membrane fusion requires both the F and G glycoproteins, as was observed for HeV. NiV-mediated fusion has demonstrated a broad species tropism, similar to results obtained with HeV. In addition, the results presented here have indicated that HeV-mediated fusion is more potent or efficient than that of NiV. The membrane fusion mechanism shown here by NiV as well as HeV can be specifically inhibited with either an antiserum or targeted peptides, and this system may prove useful as a surrogate assay for measuring immune-system-based inhibition of virus infection outside of BSL-4 containment. Finally, the NiV and HeV glycoproteins are capable of highly efficient heterotypic functional activity among themselves, but no heterotypic activity was observed with two related morbilliviruses. Taken together, these functional studies have laid the foundation for a variety of approaches which may be taken for reagent development and for exploring the fusion and attachment glycoprotein functions of these interesting and unique emerging paramyxoviruses.
Acknowledgments
We thank Yan-Ru Feng and Joseph Isaac for viruses and cells, and Paul Selleck and Chris Morrissy for Hendra and Nipah virus-specific rabbit antisera.
This study was supported by USUHS grant R073IL to C.C.B.
REFERENCES
- 1.Alkhatib, G., C. C. Broder, and E. A. Berger. 1996. Cell type-specific fusion cofactors determine human immunodeficiency virus type 1 tropism for T-cell lines versus primary macrophages. J. Virol. 70:5487-5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bagai, S., and R. A. Lamb. 1995. Quantitative measurement of paramyxovirus fusion: differences in requirements of glycoproteins between simian virus 5 and human parainfluenza virus 3 or Newcastle disease virus. J. Virol. 69:6712-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berger, E. A., O. Nussbaum, and C. C. Broder. 1995. HIV envelope glycoprotein/CD4 interactions: studies using recombinant vaccinia virus vectors, p. 123-145. In J. Karn (ed.), HIV: a practical approach, vol. 2. Oxford University Press, Cambridge, United Kingdom.
- 4.Bossart, K. N., L. F. Wang, B. T. Eaton, and C. C. Broder. 2001. Functional expression and membrane fusion tropism of the envelope glycoproteins of Hendra virus. Virology 290:121-135. [DOI] [PubMed] [Google Scholar]
- 5.Bousse, T., T. Takimoto, W. L. Gorman, T. Takahashi, and A. Portner. 1994. Regions on the hemagglutinin-neuraminidase proteins of human parainfluenza virus type-1 and Sendai virus important for membrane fusion. Virology 204:506-514. [DOI] [PubMed] [Google Scholar]
- 6.Broder, C. C., and E. A. Berger. 1995. Fusogenic selectivity of the envelope glycoprotein is a major determinant of human immunodeficiency virus type 1 tropism for CD4+ T-cell lines vs. primary macrophages. Proc. Natl. Acad. Sci. USA 92:9004-9008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Broder, C. C., and P. L. Earl. 1999. Recombinant vaccinia viruses. Design, generation, and isolation. Mol. Biotechnol. 13:223-245. [DOI] [PubMed] [Google Scholar]
- 8.Carroll, M. W., and B. Moss. 1995. E. coli beta-glucuronidase (GUS) as a marker for recombinant vaccinia viruses. BioTechniques 19:352-354, 356. [PubMed] [Google Scholar]
- 9.Chong, H. T., S. R. Kunjapan, T. Thayaparan, J. Tong, V. Petharunam, M. R. Jusoh, and C. T. Tan. 2002. Nipah encephalitis outbreak in Malaysia, clinical features in patients from Seremban. Can. J. Neurol. Sci. 29:83-87. [DOI] [PubMed] [Google Scholar]
- 10.Chua, K. B., C. Lek Koh, P. S. Hooi, K. F. Wee, J. H. Khong, B. H. Chua, Y. P. Chan, M. E. Lim, and S. K. Lam. 2002. Isolation of Nipah virus from Malaysian Island flying-foxes. Microbes Infect. 4:145-151. [DOI] [PubMed] [Google Scholar]
- 11.Chung, M., K. Kizhatil, L. M. Albritton, and G. N. Gaulton. 1999. Induction of syncytia by neuropathogenic murine leukemia viruses depends on receptor density, host cell determinants, and the intrinsic fusion potential of envelope protein. J. Virol. 73:9377-9385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crennell, S., T. Takimoto, A. Portner, and G. Taylor. 2000. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Biol. 7:1068-1074. [DOI] [PubMed] [Google Scholar]
- 13.Deng, R., Z. Wang, P. J. Mahon, M. Marinello, A. Mirza, and R. M. Iorio. 1999. Mutations in the Newcastle disease virus hemagglutinin-neuraminidase protein that interfere with its ability to interact with the homologous F protein in the promotion of fusion. Virology 253:43-54. [DOI] [PubMed] [Google Scholar]
- 14.Dimitrov, D. S. 2000. Cell biology of virus entry. Cell 101:697-702. [DOI] [PubMed] [Google Scholar]
- 15.Eaton, B. T. 2001. Introduction to current focus on Hendra and Nipah viruses. Microbes Infect. 3:277-278. [DOI] [PubMed] [Google Scholar]
- 16.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 17.Feng, Y., C. C. Broder, P. E. Kennedy, and E. A. Berger. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872-877. [DOI] [PubMed] [Google Scholar]
- 18.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halpin, K., P. L. Young, H. Field, and J. S. Mackenzie. 1999. Newly discovered viruses of flying foxes. Vet. Microbiol. 68:83-87. [DOI] [PubMed] [Google Scholar]
- 20.Halpin, K., P. L. Young, H. E. Field, and J. S. Mackenzie. 2000. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J. Gen. Virol. 81:1927-1932. [DOI] [PubMed] [Google Scholar]
- 21.Harcourt, B. H., A. Tamin, K. Halpin, T. G. Ksiazek, P. E. Rollin, W. J. Bellini, and P. A. Rota. 2001. Molecular characterization of the polymerase gene and genomic termini of Nipah virus. Virology 287:192-201. [DOI] [PubMed] [Google Scholar]
- 22.Harcourt, B. H., A. Tamin, T. G. Ksiazek, P. E. Rollin, L. J. Anderson, W. J. Bellini, and P. A. Rota. 2000. Molecular characterization of Nipah virus, a newly emergent paramyxovirus. Virology 271:334-349. [DOI] [PubMed] [Google Scholar]
- 23.Harrington, R. D., and A. P. Geballe. 1993. Cofactor requirement for human immunodeficiency virus type 1 entry into a CD4-expressing human cell line. J. Virol. 67:5939-5947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Joshi, S. B., R. E. Dutch, and R. A. Lamb. 1998. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology 248:20-34. [DOI] [PubMed] [Google Scholar]
- 25.Klenk, H. D., and W. Garten. 1994. Host cell proteases controlling virus pathogenicity. Trends Microbiol. 2:39-43. [DOI] [PubMed] [Google Scholar]
- 26.Krueger, D. K., S. M. Kelly, D. N. Lewicki, R. Ruffolo, and T. M. Gallagher. 2001. Variations in disparate regions of the murine coronavirus spike protein impact the initiation of membrane fusion. J. Virol. 75:2792-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamb, R. A. 1993. Paramyxovirus fusion: a hypothesis for changes. Virology 197:1-11. [DOI] [PubMed] [Google Scholar]
- 28.Lamb, R. A., S. B. Joshi, and R. E. Dutch. 1999. The paramyxovirus fusion protein forms an extremely stable core trimer: structural parallels to influenza virus haemagglutinin and HIV-1 gp41. Mol. Membr. Biol. 16:11-19. [DOI] [PubMed] [Google Scholar]
- 29.Lamb, R. A., and D. Kolakofsky. 2001. Paramyxoviridae: the viruses and their replication, p. 1305-1340. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 30.Lambert, D. M., S. Barney, A. L. Lambert, K. Guthrie, R. Medinas, D. E. Davis, T. Bucy, J. Erickson, G. Merutka, and S. R. Petteway, Jr. 1996. Peptides from conserved regions of paramyxovirus fusion (F) proteins are potent inhibitors of viral fusion. Proc. Natl. Acad. Sci. USA 93:2186-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim, C. C., K. E. Lee, W. L. Lee, P. A. Tambyah, C. C. Lee, Y. Y. Sitoh, A. P. Auchus, B. K. Lin, and F. Hui. 2002. Nipah virus encephalitis: serial MR study of an emerging disease. Radiology 222:219-226. [DOI] [PubMed] [Google Scholar]
- 32.Mackenzie, J. S., K. B. Chua, P. W. Daniels, B. T. Eaton, H. E. Field, R. A. Hall, K. Halpin, C. A. Johansen, P. D. Kirkland, S. K. Lam, P. McMinn, D. J. Nisbet, R. Paru, A. T. Pyke, S. A. Ritchie, P. Siba, D. W. Smith, G. A. Smith, A. F. van Den Hurk, L. F. Wang, and D. T. Williams. 2001. Emerging viral diseases of Southeast Asia and the Western Pacific. Emerg. Infect. Dis. 7:497-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michalski, W. P., G. Crameri, L. Wang, B. J. Shiell, and B. Eaton. 2000. The cleavage activation and sites of glycosylation in the fusion protein of Hendra virus. Virus Res. 69:83-93. [DOI] [PubMed] [Google Scholar]
- 34.Middleton, D. J., H. A. Westbury, C. J. Morrissy, B. M. van der Heide, G. M. Russell, M. A. Braun, and A. D. Hyatt. 2002. Experimental nipah virus infection in pigs and cats. J. Comp. Pathol. 126:124-136. [DOI] [PubMed] [Google Scholar]
- 35.Mohd Nor, M. N., C. H. Gan, and B. L. Ong. 2000. Nipah virus infection of pigs in peninsular Malaysia. Rev. Sci. Tech. 19:160-165. [DOI] [PubMed] [Google Scholar]
- 36.Morrison, T., and A. Portner. 1991. Structure, function, and intracellular processing of the glycoproteins of Paramyxoviridae, p. 347-382. In D. W. Kingsbury (ed.), The Paramyxoviruses. Plenum, New York, N.Y.
- 37.Morrison, T. G. 1988. Structure, function, and intracellular processing of paramyxovirus membrane proteins. Virus Res. 10:113-135. [DOI] [PubMed] [Google Scholar]
- 38.Murray, K., B. Eaton, P. Hooper, L. Wang, M. Williamson, and P. Young. 1998. Flying foxes, horses, and humans: a zoonosis caused by a new member of the Paramyxoviridae, p. 43-58. In W. M. Scheld, D. Armstrong, and J. M. Hughes (ed.), Emerging infections. ASM Press, Washington, D.C.
- 39.Nussbaum, O., C. C. Broder, and E. A. Berger. 1994. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J. Virol. 68:5411-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nussbaum, O., C. C. Broder, B. Moss, L. B. Stern, S. Rozenblatt, and E. A. Berger. 1995. Functional and structural interactions between measles virus hemagglutinin and CD46. J. Virol. 69:3341-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pastey, M. K., and S. K. Samal. 1997. Analysis of bovine respiratory syncytial virus envelope glycoproteins in cell fusion. J. Gen. Virol. 78:1885-1889. [DOI] [PubMed] [Google Scholar]
- 42.Paterson, R. G., M. L. Johnson, and R. A. Lamb. 1997. Paramyxovirus fusion (F) protein and hemagglutinin-neuraminidase (HN) protein interactions: intracellular retention of F and HN does not affect transport of the homotypic HN or F protein. Virology 237:1-9. [DOI] [PubMed] [Google Scholar]
- 43.Paterson, R. G., C. J. Russell, and R. A. Lamb. 2000. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology 270:17-30. [DOI] [PubMed] [Google Scholar]
- 44.Plemper, R. K., A. L. Hammond, and R. Cattaneo. 2001. Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J. Biol. Chem. 276:44239-44246. [DOI] [PubMed] [Google Scholar]
- 45.Rapaport, D., M. Ovadia, and Y. Shai. 1995. A synthetic peptide corresponding to a conserved heptad repeat domain is a potent inhibitor of Sendai virus-cell fusion: an emerging similarity with functional domains of other viruses. EMBO J. 14:5524-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Russell, C. J., T. S. Jardetzky, and R. A. Lamb. 2001. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 20:4024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russell, R., R. G. Paterson, and R. A. Lamb. 1994. Studies with cross-linking reagents on the oligomeric form of the paramyxovirus fusion protein. Virology 199:160-168. [DOI] [PubMed] [Google Scholar]
- 48.Santoro, F., P. E. Kennedy, G. Locatelli, M. S. Malnati, E. A. Berger, and P. Lusso. 1999. CD46 is a cellular receptor for human herpesvirus 6. Cell 99:817-827. [DOI] [PubMed] [Google Scholar]
- 49.Stone-Hulslander, J., and T. G. Morrison. 1997. Detection of an interaction between the HN and F proteins in Newcastle disease virus-infected cells. J. Virol. 71:6287-6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takikawa, S., K. Ishii, H. Aizaki, T. Suzuki, H. Asakura, Y. Matsuura, and T. Miyamura. 2000. Cell fusion activity of hepatitis C virus envelope proteins. J. Virol. 74:5066-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tong, S., and R. W. Compans. 1999. Alternative mechanisms of interaction between homotypic and heterotypic parainfluenza virus HN and F proteins. J. Gen. Virol. 80:107-115. [DOI] [PubMed] [Google Scholar]
- 52.Wang, L., B. H. Harcourt, M. Yu, A. Tamin, P. A. Rota, W. J. Bellini, and B. T. Eaton. 2001. Molecular biology of Hendra and Nipah viruses. Microbes Infect. 3:279-287. [DOI] [PubMed] [Google Scholar]
- 53.Wang, L. F., W. P. Michalski, M. Yu, L. I. Pritchard, G. Crameri, B. Shiell, and B. T. Eaton. 1998. A novel P/V/C gene in a new member of the Paramyxoviridae family, which causes lethal infection in humans, horses, and other animals. J. Virol. 72:1482-1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weissenhorn, W., A. Dessen, L. J. Calder, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1999. Structural basis for membrane fusion by enveloped viruses. Mol. Membr. Biol. 16:3-9. [DOI] [PubMed] [Google Scholar]
- 55.Wild, T. F., and R. Buckland. 1997. Inhibition of measles virus infection and fusion with peptides corresponding to the leucine zipper region of the fusion protein. J. Gen. Virol. 78:107-111. [DOI] [PubMed] [Google Scholar]
- 56.Yao, Q., X. Hu, and R. W. Compans. 1997. Association of the parainfluenza virus fusion and hemagglutinin-neuraminidase glycoproteins on cell surfaces. J. Virol. 71:650-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yob, J. M., H. Field, A. M. Rashdi, C. Morrissy, B. van der Heide, P. Rota, A. bin Adzhar, J. White, P. Daniels, A. Jamaluddin, and T. Ksiazek. 2001. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg. Infect. Dis. 7:439-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Young, J. K., D. Li, M. C. Abramowitz, and T. G. Morrison. 1999. Interaction of peptides with sequences from the Newcastle disease virus fusion protein heptad repeat regions. J. Virol. 73:5945-5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Young, P. L., K. Halpin, P. W. Selleck, H. Field, J. L. Gravel, M. A. Kelly, and J. S. Mackenzie. 1996. Serologic evidence for the presence in Pteropus bats of a paramyxovirus related to equine morbillivirus. Emerg. Infect. Dis. 2:239-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao, X., M. Singh, V. N. Malashkevich, and P. S. Kim. 2000. Structural characterization of the human respiratory syncytial virus fusion protein core. Proc. Natl. Acad. Sci. USA 97:14172-14177. [DOI] [PMC free article] [PubMed] [Google Scholar]