Abstract
Alphaviruses productively infect a variety of vertebrate and insect cell lines. In vertebrate cells, Sindbis virus redirects cellular processes to meet the needs of virus propagation. At the same time, cells respond to virus replication by downregulating virus growth and preventing dissemination of the infection. The balance between these two mechanisms determines the outcome of infection at the cellular and organismal levels. In this report, we demonstrate that a viral nonstructural protein, nsP2, is a significant regulator of Sindbis virus-host cell interactions. This protein not only is a component of the replicative enzyme complex required for replication and transcription of viral RNAs but also plays a role in suppressing the antiviral response in Sindbis virus-infected cells. nsP2 most likely acts by decreasing interferon (IFN) production and minimizing virus visibility. Infection of murine cells with Sindbis virus expressing a mutant nsP2 leads to higher levels of IFN secretion and the activation of 170 cellular genes that are induced by IFN and/or virus replication. Secreted IFN protects naive cells against Sindbis virus infection and also stops viral replication in productively infected cells. Mutations in nsP2 can also attenuate Sindbis virus cytopathogenicity. Such mutants can persist in mammalian cells with defects in the alpha/beta IFN (IFN-α/β) system or when IFN activity is neutralized by anti-IFN-α/β antibodies. These findings provide new insight into the alphavirus-host cell interaction and have implications for the development of improved alphavirus expression systems with better antigen-presenting potential.
Alphaviruses are a group of significant human and animal pathogens. The nearly 30 members of this genus are transmitted by mosquitoes to higher vertebrates that serve as amplifying hosts (54). Alphaviruses cause different diseases but have similar replication strategies and life cycles. In insect vectors, they cause persistent lifelong infections and viral replication does not critically affect the viability of the hosts (51). These viruses also often establish persistent infection in cultured mosquito cells. In contrast, vertebrate hosts usually develop acute infection that often results in disease prior to virus clearance by the immune system (27, 31). The infection of susceptible vertebrate cells typically leads to rapid cytopathic effect (CPE) (54) and cell death.
Sindbis virus (SIN) is the prototype member of the Alphavirus genus. It can replicate productively in a wide variety of cell lines of insect and vertebrate origin and causes age-dependent encephalomyelitis in mice (28). As do all other alphaviruses, SIN enters the cells via receptor-mediated endocytosis. pH-dependent fusion of viral and endosomal membranes leads to release of nucleocapsids into the cytoplasm (12) followed by nucleocapsid disassembly and genome replication (64).
The SIN genome is a single RNA molecule of positive polarity, 11.7 kb in length. It contains a 5′ methylguanylate cap and a 3′ polyadenylate tract and is translated by host-cell machinery similar to cellular mRNAs (52). The 5′ two-thirds of the genome encodes the nonstructural proteins (nsPs), which comprise the viral components of the RNA replicase/transcriptase required for replication of the viral genome and transcription of subgenomic RNA. The 4.1-kb subgenomic RNA corresponds to the 3′ third of the genome and is translated into structural proteins that form virus particles. Replication of SIN is very rapid and leads to high-level accumulation of virus-specific RNAs and structural proteins. Finally, a large number of viral particles released by budding from the cell surface disseminate the infection. Viral replication profoundly affects cell metabolism, with downregulation of host cell protein synthesis playing the central role (31). Cells lose integrity and die within 24 to 48 h postinfection (p.i.). In many cell types, death is accompanied by apoptotic changes (37). Replication of viral RNA and/or accumulation of SIN nsPs is sufficient to cause translational shutoff and cell death. However, expression of viral structural proteins significantly accelerates the development of CPE (18).
In recent studies, we selected SIN self-replicating RNAs (replicons) that were capable of persisting in some vertebrate cell lines for an unlimited number of passages without causing CPE (1, 16). These noncytopathic replicons replicated less efficiently than the parent and contained point mutations in the gene encoding one of the nsPs, nsP2. One adaptive mutation, P726→L, was at the same position as that found in the SIN-1 variant that also exhibits reduced cytopathogenicity (11, 63). Subsequently another research group also detected adaptive mutations at nsP2-P726 (45). The nsP2-P726 is highly conserved among alphaviruses (16), and this remarkable convergence of mutations suggested that the noncytopathic phenotype of normally cytopathic, self-replicating SIN RNAs might be the result of a change in nsP2 function(s) rather than a simple downregulation of the RNA replication, which can be achieved by a wide variety of mutations in nsPs and cis-acting RNA elements (17, 43, 44).
nsP2 is one of four nsPs and is an essential component of the replicative complexes. It contains helicase (26) and RNA triphosphatase (60) activities, required for RNA synthesis, and is also a protease that orchestrates sequential cleavages of nonstructural polyprotein precursor P1234 (54). In the current model, the first cis-cleavage at the junction between nsP3 and nsP4 (referred to herein as the 3/4 site) creates a replicase efficient for minus-strand RNA synthesis. Further trans-cleavage at the 1/2 and 2/3 sites alters the activity of the replicase to create a complex efficient in plus-strand genomic and subgenomic RNA synthesis (34-36, 50). Because of the in-frame opal termination codon separating nsP3 and nsP4 (38) and the short half-life of nsP4 (7), nsP2 accumulates in infected cells in a 5- to 10-fold excess relative to nsP4, the catalytic subunit of the RNA-dependent RNA polymerase (RdRp) (53). Moreover, in Semliki Forest virus-infected cells, a significant fraction of the nsP2 is transported to the nucleus (47, 48). These observations suggest that nsP2 may possess an additional function(s) besides its roles in RNA replication.
We also noticed that noncytopathic replicons were able to persist in only a limited number of cell lines, such as BHK-21, CHO, and Vero cells (16). Other cell lines—including NIH 3T3, HeLa, L929, MDBK, MDCK, and others—could not support long-term replication and died during drug selection within a few days. In the present study, we have investigated the basis of cell-specific persistence of SIN. We show that nsP2 plays a key role in modulating virus-host interactions that determine the development of CPE and dissemination of the infection.
MATERIALS AND METHODS
Cell cultures and plaque assays.
BHK-21 cells (BHK-J) were kindly provided by Paul Olivo (Washington University, St. Louis, Mo.). NIH 3T3 cells were obtained from the American Type Tissue Culture Collection (Manassas, Va.). Both cell lines were maintained at 37°C in alpha minimum essential medium (αMEM) supplemented with 10% fetal bovine serum (FBS). L929 cells used for biological assay of alpha/beta interferon (IFN-α/β) were provided by Samuel Baron (University of Texas Medical Branch, Galveston). The IFN-α/βR−/− mouse embryonic fibroblasts (MEFs) were prepared from day-17 embryos of mice originally obtained from Michael Auget (59), as were wild-type (wt) MEFs from 129 Sv/Ev mice. Both MEFs were provided by Herbert W. Virgin (Washington University). The L929 cells and primary MEFs were propagated in Dulbecco's MEM supplemented with 10% FBS. Titers of viruses were determined by plaque assay on BHK-21 cells or NIH 3T3 monolayers as previously described (33).
Plasmid constructs.
Standard recombinant DNA techniques were used for all plasmid constructions. Maps and sequences are available from the authors upon request. A plasmid encoding the SIN replicon with a codon-optimized green fluorescent protein (GFP) gene under the control of the subgenomic promoter (SINrep/GFP) was kindly provided by Nicolas Ruggli (N. Ruggli and C. M. Rice, unpublished data). In a preliminary cloning step, the XbaI-NsiI fragment, derived from SINrep/GFP, with the NsiI site blunt-ended using T4 DNA polymerase (fragment A), was ligated with the BamHI-XhoI fragment (fragment B), which was derived from DH(26S)5′SIN (the BamHI site was filled in using T4 DNA polymerase), in pRS2 (a derivative of pUC19) digested with XbaI and XhoI. Fragment A contained the complete GFP gene, and fragment B contained the SIN subgenomic promoter and a complete cDNA of SIN TE12 subgenomic RNA. TE12, a SIN variant neurovirulent for mice, has been described elsewhere (40). This plasmid was named pGFP-SG. In order to replace the gene encoding puromycin acetyltransferase (PAC), the XbaI-XhoI fragment from pGFP-SG was cloned into pTSG/PAC, pTSG/nsP2-726G/PAC, and pTSG/44/PAC that had been digested with XbaI and XhoI. The pTSG/PAC and pTSG/nsP2-726G/PAC plasmids have been previously described elsewhere (16) and encode SIN replicons that differ in that nsP2-726 Pro (CCA) is replaced by Gly (GGA). The pTSG/44/PAC is identical to pTSG/PAC except for 44 silent mutations in the N-terminal part of the nsP1 coding region that destabilize the predicted secondary structure of the 51-nucleotide (51-nt) conserved sequence element (CSE) and downregulate SIN RNA replication (17). The final construct pwtSIN, derived from pTSG/PAC, contained the SP6 promoter followed by nt 1 to 7647 of the SIN genome; TCTAGAGCTTGCCGCCACC sequence and 720 nt representing the complete GFP gene (from ATG to TAA); AGCGG sequence and nt 11394 to 11448 of the SIN genome, followed by SIN nt 7335 to 11703 (containing the subgenomic promoter, subgenomic RNA with TE12 structural genes, and the 3′ untranslated region); and the poly(A) tail followed by EcoRI and XhoI restriction sites, useful for plasmid linearization prior to in vitro transcription. A schematic representation of these constructs is shown in Fig. 1. pSIN/G contained a single change, P726→G in nsP2, and pSIN44 had clustered silent mutations in nsP1, destabilizing the secondary structure of the 51-nt CSE (the replication enhancer).
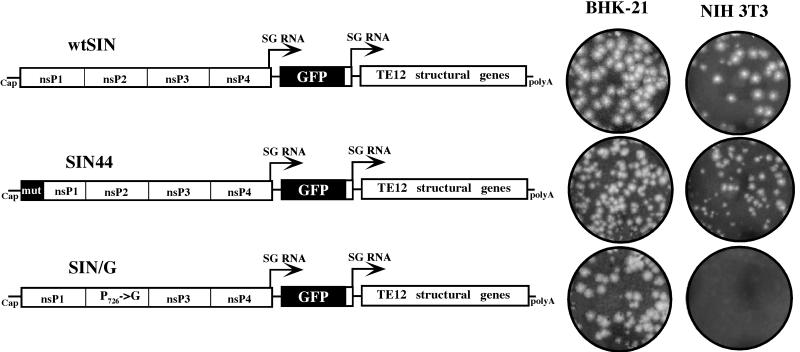
FIG. 1.
Schematic representation of the double subgenomic viral genomes and ability of viral mutants to form plaques on BHK-21 and NIH 3T3 cells. In all of the genomes, the first subgenomic promoter was driving the expression of GFP and the second one was driving the expression of SIN structural genes derived from the TE12 strain of SIN. The SIN/G variant was different from wtSIN by one amino acid in nsP2, P726→G. The SIN44 differed from wtSIN by clustered mutations in nsP1, which did not change the encoded protein sequence but destroyed the secondary structure of the replicational enhancer, the 51-nt CSE. Viral stocks generated by electroporation of in vitro-synthesized RNAs were titrated simultaneously on BHK-21 and NIH 3T3 cells. Plaques were allowed to develop for 48 h prior to fixation and staining. Stained, infected monolayers of both cell types correspond to the same dilutions of viruses.
RNA transcription.
Plasmids were purified by centrifugation in CsCl gradients. Prior to transcription, they were linearized with XhoI. RNAs were transcribed in the presence of cap analog using SP6 RNA polymerase. The yield and integrity of RNA transcripts were monitored by gel electrophoresis. Transcription reaction mixtures were aliquoted and stored at −80°C. For electroporation of BHK-21 cells, 4 μg of in vitro-synthesized RNA was used under previously described conditions (39). Viruses were harvested 24 h posttransfection.
Viruses recovered from pwtSIN, pSIN/G, and pSIN44 were named wtSIN, SIN/G, and SIN44, respectively.
RNA analysis.
The protocol used for labeling SIN-specific RNAs with [3H]uridine in the presence of dactinomycin is described in the legend to Fig. 2. RNAs were isolated from the cells by using TRIzol reagent as recommended by the manufacturer (Gibco-BRL), denatured with glyoxal in dimethyl sulfoxide, and analyzed by agarose gel electrophoresis using the conditions described elsewhere (16).
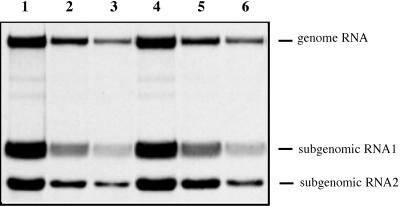
FIG. 2.
Analysis of the replication and transcription of SIN RNAs in infected cells. NIH 3T3 cells in six-well Costar plates were infected with different SIN viruses at an MOI of 20. At 1 h (lanes 1 to 3) and 5 h (lanes 4 to 6) p.i., medium in the wells was replaced by 1 ml of alpha MEM supplemented with 10% FBS, dactinomycin (2 μg/ml), and [3H]uridine (20 μCi/ml). After 3 h of incubation at 37°C, RNAs were isolated and analyzed by agarose gel electrophoresis as described in Materials and Methods. Lanes: 1 and 4, RNAs isolated from the cells infected with wtSIN; 2 and 5, RNAs from the cells infected with SIN44; 3 and 6, RNAs from the cells infected with SIN/G.
Analysis of protein synthesis.
NIH 3T3 cells were seeded into six-well Costar dishes at a concentration of 5 × 105 cells/well. Four hours later, the cells were infected with different viruses at a multiplicity of infection (MOI) of 20 PFU/cell in 200 μl of alpha MEM supplemented with 1% FBS at room temperature for 1 h. Medium was then replaced by alpha MEM with 10% FBS, and incubation continued at 37°C. At different times p.i., the cells were washed three times with phosphate-buffered saline (PBS) and then incubated for 30 min at 37°C in 1 ml of RPMI medium lacking methionine, supplemented with 1% FBS and 20 μCi of [35S]methionine. After the incubation, cells were scraped from the dish into PBS, pelleted by centrifugation, and dissolved in 200 μl of standard loading buffer. One-tenth of the samples was analyzed on sodium dodecyl sulfate-10% polyacrylamide gels. After electrophoresis, gels were dried, autoradiographed, and analyzed on a Storm 860 PhosphorImager (Molecular Dynamics).
IFN-α/β assay.
The concentrations of IFN-α/β in the media were measured by a previously described biological assay (56). Briefly, L929 cells were seeded in 100 μl of complete medium at a concentration of 5 × 104 cells/well in 96-well plates and incubated at 37°C for 4 to 6 h. Samples of media harvested from infected NIH 3T3 cells were treated with UV light for 1 h (no infectious virus could be detected after this treatment) and were serially diluted in twofold steps directly in the wells with L929 cells. After incubation for 24 h at 37°C, an additional 100 μl of medium with 2 × 105 PFU of vesicular stomatitis virus (VSV) was added to the wells, and incubation continued for 36 to 40 h. Then, cells were stained with crystal violet, and the end point was determined as the concentration of IFN-α/β required to protect 50% of the cells in monolayers from the VSV-specific CPE. The IFN-α/β standard for normalization of the results was received from ATCC.
Viral growth analysis.
Cells were seeded at a concentration of 5 × 105 cells/35-mm-diameter dish. After 4 h of incubation at 37°C, monolayers were infected with virus at the MOIs indicated (see figure legends) for 1 h at 37°C, washed three times with PBS, and overlaid with 1 ml of complete medium. At indicated times p.i., media were replaced by fresh media, and virus titers in the harvested samples were determined by plaque assay on BHK-21 (BHK-J) cells (33). The concentration of IFN in the samples was below a level that could possibly interfere with viral replication and plaque formation.
GeneChip expression analysis.
NIH 3T3 cells were seeded at a concentration of 5 × 106 cells per 150-mm-diameter dish. After 4 h of incubation at 37°C, cells were infected with wtSIN and SIN/G viruses at an MOI of 20 PFU/cell in 3 ml of MEM supplemented with 1% FBS. After 1 h of infection at room temperature with continuous shaking, media were replaced by 20 ml of alpha MEM containing 10% FBS, and cells were incubated at 37°C for 17 h. RNAs were isolated from 2 × 107 uninfected and infected cells by using TRIzol reagent as recommended by the manufacturer (Gibco-BRL). Samples contained very similar concentrations of total RNA. Poly(A)+ RNA was isolated from 100 μg of total RNA using Dynalbeads oligo(dT)25 by the procedure recommended by the manufacturer (Dynal). cDNA was synthesized using a T7(dT)24 primer and the SuperScript Choice system (Gibco-BRL). Biotin-labeled cRNA was synthesized on one-third of the cDNA using the RNA transcript labeling kit (ENZO). cRNA was purified and fragmented according to the Affymetrix protocol. Equal aliquots of cRNAs were hybridized to the Murine Genome U74A array (MG-U74A) in the GeneChip Core Facility (Alvin J. Siteman Cancer Center, Washington University). The data were analyzed using GeneChip Suite software (Affymetrix).
Virulence assay.
Two-day-old CD1 mice (Charles River) were inoculated intracranially with 103 PFU of wtSIN, SIN44, or SIN/G viruses. They were observed daily for mortality. At the indicated times p.i., mice were euthanized, brains were collected and homogenized in PBS, and homogenates were assayed on BHK-21 cells for plaque formation. These same brain homogenates were plated serially on L929 cells to assay IFN-α/β production as described above, except that virus was inactivated by overnight incubation at pH 2.0. Brain homogenates were neutralized before plating on L929 cells.
RESULTS
Recombinant SIN viruses.
In a previous study, we created a collection of SIN replicons and viruses with mutations in nsP2 that exhibited differences in cytopathogenicity and ability to persist in tissue culture (1, 16). Persistent replication was cell line dependent and correlated with lower levels of RNA replication (16). To further probe the mechanism of cell-specific persistence and to determine if the reduced cytopathogenicity of the replicons was a result of a lower level of virus-specific RNA replication or alterations in nsP2 functions, we created a set of recombinant SIN viruses (Fig. 1). All of the viral genomes encoded GFP, transcribed from an additional subgenomic promoter. GFP was used to monitor productive infection in tissue culture, particularly in the case of noncytopathic mutants with slow growth and low levels of CPE. A second subgenomic promoter drove the expression of viral structural genes. Since these experiments were performed in mouse cell lines and inbred mice, SIN envelope glycoprotein genes were derived from a mouse-adapted SIN variant, TE12 (40). The nsPs and cis-acting RNA elements were derived from Toto1101 (46).
Downregulation of viral RNA replication was achieved in two ways: (i) by mutation of nsP2 residue 726 (SIN/G) or (ii) by eliminating the 51-base CSE in the nsP1 coding region with a cluster of silent changes. wtSIN virus contained no mutations in the nonstructural genes or the 5′ and 3′ cis-acting elements required for RNA replication. The SIN/G variant contained a single nsP2 substitution (P726→G) that downregulated replication and cytopathogenicity. Compared to mutant viruses with other substitutions at this position (P726→V, S, and T) (16), SIN/G virus was the most stable, with minimal reversion or pseudoreversion to a cytopathic phenotype. This mutant caused partial CPE in BHK-21 cells, which was most likely dependent on the stage of cell cycle of each particular cell at the moment of infection, followed by persistent infection. The SIN44 variant expressed the wt nsPs but carried 44 silent mutations in the 51-base CSE near the 5′ end of the viral genome (17). These mutations significantly downregulated RNA replication.
All of the in vitro-synthesized RNAs were transfected into BHK-21 cells by electroporation, and viruses were harvested 24 h later. By that time, cells transfected by wtSIN and SIN44 RNAs had developed complete CPE; cells transfected with SIN/G had few morphological changes. Nevertheless, titers of all viral stocks were similar, approximately 109 PFU/ml.
Replication of SIN mutants in BHK-21 and NIH 3T3 cells.
First, we tested the ability of mutant viruses to form plaques on BHK-21 and NIH 3T3 cells. NIH 3T3 cells were previously shown not to support persistent replication of SIN replicons with mutated nsP2 (16). Viruses with the wt nsP2 (wtSIN and SIN44) were cytopathic and formed plaques with equal efficiency in both BHK-21 and NIH 3T3 cells (Fig. 1). The SIN/G virus was partially cytopathic for BHK-21 cells, as previously described for SIN with the same mutation (16), but was able to form plaques on this cell line. This can be explained by the reduced concentration of FBS in the agarose cover. In addition, the particular cell line that we used for the plaque assay, BHK-21 (BHK-J) cells, was previously found to develop better plaques in the case of not only SIN mutants but also other viral infections (I. Frolov, unpublished data). Plaque assays using NIH 3T3 cells led to formation of large foci of cells that transiently expressed GFP, but plaques did not develop (Fig. 1).
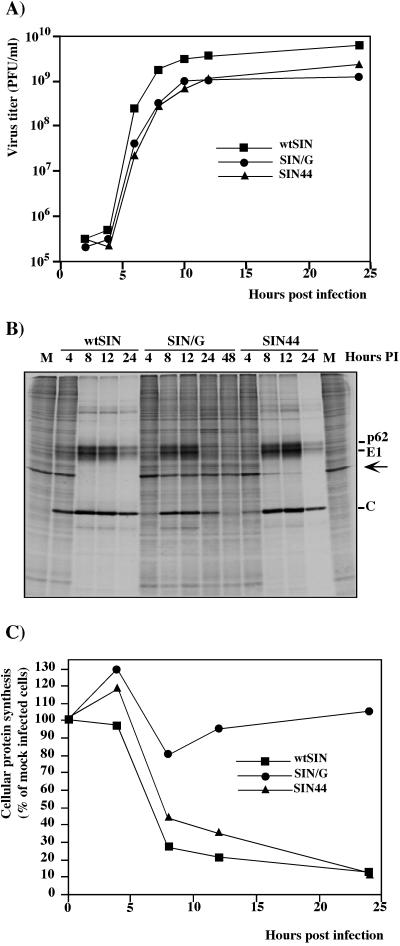
Next, monolayers of NIH 3T3 cells were infected with each virus at an MOI of 20 PFU/cell. wtSIN and SIN44 developed complete CPE within 24 to 36 h p.i.. SIN/G arrested cell growth for approximately 2 days (crisis period), after which cells recovered and resumed normal growth. To test the level of RNA replication, SIN RNAs were labeled with [3H]uridine in the presence of dactinomycin. Both in the early stage of infection (1 to 4 h p.i.) and later during the high virus release (5 to 8 h p.i.), SIN/G RNA replicated 7- to 10-fold less efficiently than wtSIN and 1.5-to 2-fold less efficiently than SIN44 (Fig. 2). SIN/G grew efficiently on NIH 3T3 cells, and virus was released with kinetics similar to those seen with SIN44 (Fig. 3A). Interestingly, structural proteins of SIN/G were produced almost as efficiently as the structural proteins of wtSIN (Fig. 3B), despite the lower levels of SIN-specific RNA synthesis (Fig. 2) and accumulation (data not shown). In contrast to wtSIN and SIN44, host cell protein synthesis was not shut off in cells infected with SIN/G (Fig. 3B and C). Within 24 h, NIH 3T3 cells began to suppress replication of the SIN/G variant, and by 48 h, the translation of SIN/G structural proteins was barely detectable (Fig. 3B). These differences in translational shutoff and CPE were not due to lower infectivity of SIN/G. Twelve hours p.i. with SIN/G, all cells were productively infected as monitored by GFP expression. For SIN variants with P726→S or P726→T mutations in nsP2, replication in NIH 3T3 cells also ceased 48 h p.i. (data not shown). These results show that NIH 3T3 cells infected with certain nsP2 mutants are able to develop an effective antiviral response that shuts down virus replication and precludes persistent infection.
FIG. 3.
Analysis of virus growth and protein synthesis in virus-infected cells. NIH 3T3 cells were infected with wtSIN, SIN/G, and SIN44 at an MOI of 20 PFU/cell. (A) At the indicated times, media were replaced and virus titers were determined as described in Materials and Methods. In other wells, proteins were pulse-labeled with [35S]methionine and analyzed on sodium dodecyl sulfate-10% polyacrylamide gel. Gels were dried and autoradiographed (B) or analyzed on a Storm 860 PhosphorImager (C). The levels of synthesis of cellular proteins were determined by measuring radioactivity in the protein band corresponding to actin (indicated by the arrow) and normalized to radioactivity in the actin band in uninfected cells.
Effects of SIN infection on gene expression in NIH 3T3 cells.
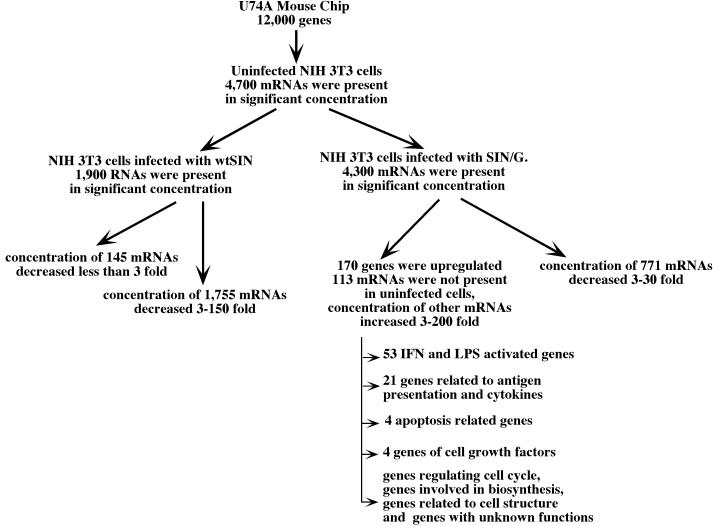
To examine possible differences in host cell response to virus infection, we compared host gene expression profiles in uninfected NIH 3T3 cells with cells infected by wtSIN or SIN/G. RNAs were isolated at 17 h p.i. and used for microarray analysis on Affymetrix U74A chips (Fig. 4). In uninfected NIH 3T3 cells, ∼4,700 mRNAs were detected. In cells infected with wtSIN, only 1,900 mRNAs were detected, and most of these RNAs were present at levels 3- to 150-fold lower than those found in uninfected cells. No cellular RNAs were upregulated in wt SIN-infected cells compared to uninfected cells. In contrast, 170 cellular mRNAs were upregulated after SIN/G virus infection, and 113 of these mRNAs were undetectable in uninfected cells. For the remainder, levels increased from 3- to 200-fold. Many upregulated genes were previously described as lipopolysaccharide or IFN-α/β activated, and 21 were cytokines or related to antigen presentation (9, 10). Eleven upregulated mRNAs were related to cell cycle, cell growth, or apoptosis. Others encode enzymes for cellular biosynthesis and cytoskeletal proteins. No function has been determined for 36 of the induced genes. These results show that wtSIN is capable of either suppressing or not inducing a cellular antiviral response in infected NIH 3T3 cells. The most-dramatic effect of wtSIN on host gene expression is the downregulation of many cellular mRNAs. SIN/G, on the other hand, induces and is incapable of suppressing transcription of IFN-α/β and IFN-inducible genes as well as other host genes. One or more of these host gene products presumably exerts an antiviral effect to downregulate SIN replication and allow cell survival.
FIG. 4.
The results of GeneChip analysis of mRNA expression in uninfected NIH 3T3 cells and the cells infected with wtSIN and SIN/G viruses at an MOI of 20 PFU/cell. The RNAs were isolated at 17 h p.i., and the analysis was performed as described in Materials and Methods. LPS, lipopolysaccharide.
Mutations in SIN nsP2 lead to an increase in IFN production.
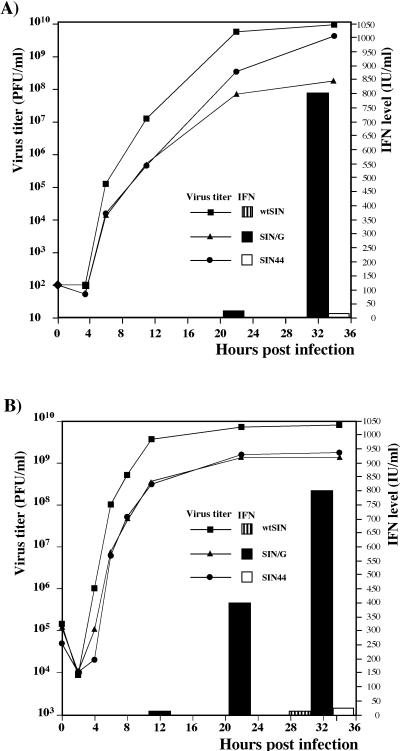
The results of the microarray analysis indicated that IFN-α/β could be an important factor in inhibiting SIN/G replication and promoting its clearance from infected NIH 3T3 cells. To examine IFN-α/β production directly, subconfluent NIH 3T3 cells were infected with wtSIN, SIN/G, or SIN44 at an MOI of 0.004 PFU/cell. We analyzed both virus growth and accumulation of IFN in the media (Fig. 5A). wtSIN virus caused complete CPE within 48 h p.i., and IFN-α/β was undetectable. SIN44 induced slower but complete CPE. A very low concentration of IFN was found in the media in the late stages of SIN44 infection. In contrast, SIN/G-infected cells released high levels of IFN, first detectable at 24 h; yielded almost 100-fold less virus than did wtSIN- or SIN44-infected cells; and did not induce significant CPE.
FIG. 5.
Virus growth and IFN-α/β production by the NIH 3T3 cells infected with wtSIN, SIN/G, and SIN44 variants at MOIs of 0.004 (A) and 20 (B) PFU/cell. Virus titers and concentrations of IFN-α/β were determined in the same samples harvested at the indicated time points. These data represent one of a series of repeated experiments which generated very reproducible data.
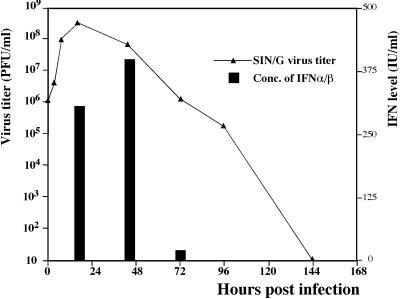
When NIH 3T3 cells were infected with virus at a high MOI (20 PFU/cell), the growth curves of SIN/G and SIN44 were indistinguishable (Fig. 5B), indicating that both viruses had similar efficiencies of intracellular replication. However, the profiles of IFN release were different. Only the SIN/G virus-infected cells released IFN, and it was detected in the medium significantly earlier than it was after infection with virus at a low MOI. The SIN/G mutant did not induce complete CPE, despite infecting all of the cells. The arrest of SIN/G replication and the decrease in virus release correlated with increased IFN-α/β in the medium (Fig. 6). SIN/G virus-infected cells secreted IFN until 72 h p.i., after which they resumed growth, but remained resistant to superinfection with wtSIN for at least 10 days (data not shown). These results strengthened the hypothesis that NIH 3T3 cells infected with SIN/G produce high levels of IFN-α/β that not only prevent the next round(s) of infection but also inhibit virus replication in previously infected cells.
FIG. 6.
Virus growth and IFN-α/β production by the NIH 3T3 cells infected with the SIN/G mutant at an MOI of 20 PFU/cell. Medium was replaced at the indicated time points; virus titers and concentrations of IFN-α/β were determined as described in Materials and Methods and demonstrate the accumulation of IFN and virus in the periods between the indicated time points. These data represent one of three repeated experiments which generated reproducible data.
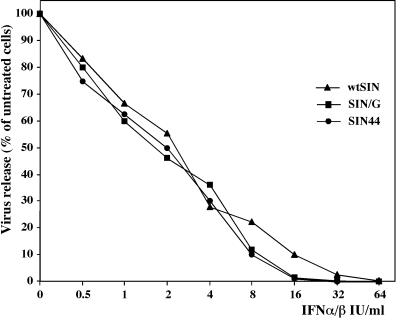
To test the sensitivities of viruses with wt or mutant nsP2 to IFN, we treated NIH 3T3 cells with serial dilutions of murine IFN-α/β for 24 h and then infected them with wtSIN, SIN/G, and SIN44 variants at an MOI of 20. Twenty-four hours later, replication of all three viruses (detected by GFP expression) was found in the cells treated with IFN at concentration of 16 IU/ml or less. Virus titers decreased with increases in IFN concentration at very similar rates (Fig. 7). These experiments indicate that wtSIN and the mutants, regardless of encoding wt or mutant nsP2, were equally sensitive to IFN-α/β.
FIG. 7.
IFN-α/β sensitivity assay. Monolayers of NIH 3T3 cells in 12-well plates were treated with the indicated concentrations of murine IFN-α/β for 24 h and then infected with wtSIN, SIN/G, and SIN44 variants at an MOI of 20. Titers of released viruses were measured 24 h p.i.. The results were normalized to titers of viruses released from the cells not treated with IFN-α/β.
Eliminating IFN-α/β action leads to persistent SIN/G replication.
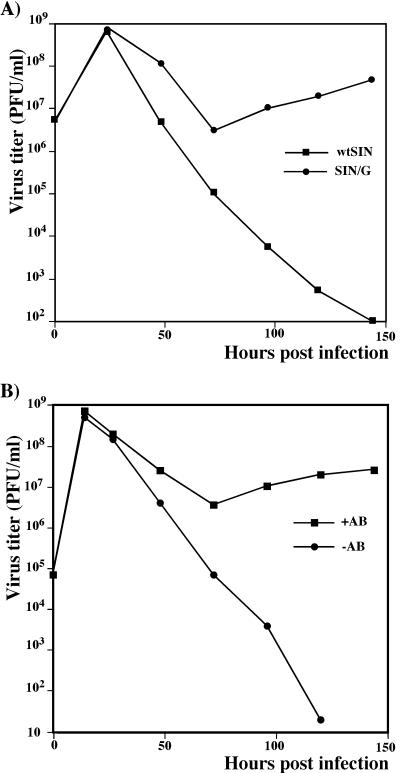
The significance of IFN release in the clearance of SIN/G was tested using MEFs from mice lacking the IFN-α/β receptor (IFN-α/βR). Cells were infected with the wtSIN and the SIN/G mutant viruses at an MOI of 20. Both viruses propagated in the IFN-α/βR−/− MEFs (Fig. 8A). The wtSIN-infected cells died within 24 h, but the SIN/G mutant caused only limited CPE. After 72 h of crisis period, all surviving cells remained infected, expressed GFP, released infectious virus, and grew without noticeable morphological changes. No persistent infection was detected upon SIN/G infection of MEFs derived from IFN-α/βR+/+ mice (data not shown).
FIG. 8.
(A) Growth of wtSIN and SIN/G viruses in MEFs derived from IFN-α/βR−/− mice; (B) growth of SIN/G virus in NIH 3T3 cells in the presence and absence of anti-IFN-α/β antibodies (+AB and −AB, respectively). Cells were infected at an MOI of 20 PFU/cell, and media were replaced at the indicated time points to determine viral titers. Sheep anti-mouse IFN-α/β AB was present in the medium at a concentration of 1,000 IFN neutralization units per ml. These data represent one of two repeated experiments.
This issue was also examined in NIH 3T3 cells using polyclonal antibodies that neutralize IFN-α/β. NIH 3T3 cells were infected with SIN/G in the absence or presence of anti-IFN-α/β antibodies. Neutralizing antibodies in the media prevented the clearance of SIN/G that usually occurred after 48 h p.i. (Fig. 8B). All of the cells remained infected and produced infectious virus, albeit less efficiently than during the first 24 h. These results show that IFN-α/β-induced gene expression is required for eliminating SIN/G replication but is not essential for cell survival or establishing persistent infection.
Induction of IFN-α/β in vivo.
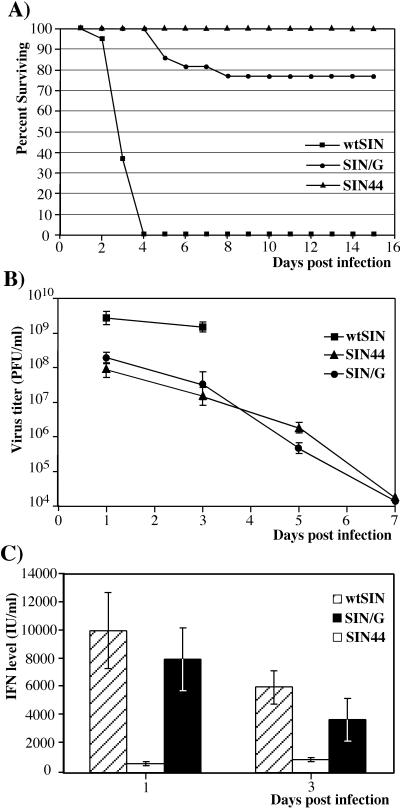
We also examined the effect of nsP2 and replication efficiency on pathogenicity and IFN production in vivo. Two-day-old CD1 mice were infected intracerebrally with 103 PFU of wtSIN, SIN44, or SIN/G. wtSIN caused 100% mortality within 4 days, but the virulence of SIN44 and SIN/G was significantly reduced (Fig. 9A). For the variants expressing wt nsP2 (wtSIN and SIN44), the level of secreted IFN correlated with virus propagation (Fig. 9B and C). SIN44 titers in mouse brains were 50-fold lower than those for the wtSIN, and SIN44 induced a proportionally lower level of IFN-α/β. SIN/G replication in mouse brains was very similar to SIN44 replication, but SIN/G induced IFN-α/β as efficiently as wtSIN. These data indicated that mutations in the nsP2 gene can lead to a higher level of IFN-α/β production by infected neuronal cells in vivo in spite of a lower level of virus propagation.
FIG. 9.
(A) Survival of mice infected with wtSIN, SIN/G, and SIN44; virus growth (B) and production of IFN-α/β (C) in mouse brains. Two-day-old CD1 mice were inoculated intracranially with 103 PFU of the indicated virus and observed for 15 days. Virus titers and concentrations of IFN-α/β were determined as described in Materials and Methods. Error bars, standard deviations.
DISCUSSION
Alphaviruses have simple genomes encoding four mature nsPs and three major structural proteins. These proteins are required for replication of the viral genetic material and its release from the cells in the form of infectious viral particles. Viral proteins may interact with host machinery to minimize activation of antiviral responses in infected cells. Hence, the outcome of SIN infection reflects the ability of the virus to redirect cellular processes for virus propagation while avoiding cellular antiviral responses. In the present study, we have initiated work aimed at understanding the mechanisms that SIN employs for suppressing cellular antiviral responses and the nature of cellular responses that can interfere with SIN replication.
Earlier work demonstrated that mutations in SIN nsP2 could be selected that allowed persistent noncytopathic replication in mammalian cells, but only for a limited spectrum of cell types (1, 16). Several possible explanations can be invoked for the mechanism of action of these mutations as well as their cell specificity. For example, CPE induction could be a quantitative effect (as it most certainly is), and the inability to cause CPE could result from lower levels of SIN nsPs or RNA production. Alternatively, mutations in nsP2 might block virus-host interactions critical for eliciting CPE. Restricted tropism of these adaptive changes might be due to cell-specific lethal defects in RNA replication or the activation cell defense mechanisms that arrest replication of the nsP2 mutants. To help distinguish between these possibilities, we created SIN variants that differed in the efficiency of genome RNA replication by incorporating mutations in nsP2 (SIN/G) or in the 51-nt CSE replication enhancer (SIN44).
The mutations in the nsP2, but not those in the 51-nt CSE, made SIN less cytopathic. The SIN/G mutant caused persistent infection with minor CPE in BHK-21 cells and noncytopathic infection in NIH 3T3 cells. These changes in cytopathogenicity appeared to result not from alterations in the level of nsP2, since mutant nsP2 accumulated to 80% of the wt level, and a significant fraction of this protein continued to be transported to the nucleus (data not shown). Furthermore, changes were not (or not only) associated with the lower levels of viral RNA replication, because SIN44 and SIN/G virus RNAs replicated with similar efficiency (Fig. 2). That concentration of nsP2 and other nsPs was sufficient to keep SIN/G virus growth at a level comparable to the growth of SIN44 and wtSIN (Fig. 3 and 5B). However, we cannot exclude the possibility that a lower level of replication could be a significant contributor in downregulation of the CPE. In any case, the point mutation in nsP2 affected not only functioning of RdRp, but also reduced the CPE of SIN replication in mammalian cells.
The most intriguing finding of this study was discovery that SIN/G replication in NIH 3T3 cells was not only noncytopathic but was suppressed beginning at 24 h p.i., becoming undetectable by 48 h. This suggested that cells infected with the mutant activated or allowed activation of defense mechanisms that suppressed viral replication. In contrast, wtSIN and SIN44 completely destroyed NIH 3T3 cells by 24 h p.i., indicating that these cells could not downregulate replication of viruses that expressed wt nsP2. Microarray analysis confirmed that biological changes differed greatly in cells infected with wtSIN and SIN/G. wtSIN infection caused a significant and global decrease in cellular mRNA levels, presumably due to inhibition of transcription. GeneChip expression analysis is based on isolation of total cellular RNA (both nuclear and cytoplasmic fraction), and this excludes an explanation of the differences in mRNA levels by alterations in their transport from the nucleus. Twelve hours p.i., transcription of rRNAs was strongly affected as well (data not shown). In contrast, SIN/G-infected cells activated synthesis of 170 mRNAs, including cytokines and IFN-induced genes and those involved in immunoproteosome information (10, 25, 30).
The microarray data were confirmed by direct measurements of IFN-α/β release from NIH 3T3 cells infected with the SIN variants. The nsP2 mutant, SIN/G, led to a strong induction of IFN-α/β secretion, and the kinetics of IFN release correlated with the shutoff of virus replication. Accumulation of IFN-α/β appeared to have an autocrine effect on virus replication and not only could protect uninfected cells (low-MOI experiments) but also could downregulate virus replication and clear virus from productively infected cells (high-MOI experiments). In contrast, infection of NIH 3T3 cells with wtSIN and SIN44, expressing wt nsP2, induced undetectable or barely detectable levels of IFN-α/β, respectively. However, in vivo experiments showed that wtSIN is clearly able to induce IFN production. Interestingly, all three viruses, wtSIN, SIN/G, and SIN44, were equally sensitive when cells were pretreated with IFN-α/β. This observation supports the hypothesis that shutoff of SIN/G replication was due in part to the inability of this mutant to downregulate IFN-α/β release. Therefore, lack of IFN induction by wtSIN in vitro is likely due to the rapid shutoff of host cell protein and RNA synthesis and the death of infected cells.
Additional experiments demonstrated that activation of IFN-α/β signaling pathways played a critical role in downregulation of SIN/G replication. MEFs derived from mice lacking IFN-α/β receptors could not eliminate the SIN/G mutant. SIN/G established persistent infection of IFN-α/βR−/− MEFs, with all the cells being productively infected. NIH 3T3 cells also could not stop replication of SIN/G in the presence of anti-IFN-α/β antibodies. Neutralization of IFN-α/β in the medium arrested virus clearance, and cells remained persistently infected for a number of days.
In both these experiments, blocking of IFN-α/β signaling made mouse fibroblasts unable to stop virus replication and caused persistent infection. This is a strong indication that the ability of SIN/G-infected cells to activate and respond to IFN-α/β signaling pathways played a key role in shutoff of SIN replication. It appears that SIN virus joins a growing number of infectious agents that have developed mechanisms to suppress IFN production. The best-known IFN-α/β antagonists of viral origin are NS1 protein of influenza A virus (20, 62), VP35 protein of Ebola virus (4), NSs protein of Rift Valley fever virus (5), hepatitis B virus open reading frame C product (65), and the C proteins of Sendai virus (21-23). It has also been shown that wt measles virus induces less IFN than an attenuated strain (42). In the case of SIN infection, rapid shutoff of cellular mRNA and rRNA synthesis could be a simple and efficient way to downregulate the production of IFN and other cytokines, thereby preventing activation of the antiviral state in both uninfected and already-infected cells. At first glance, the decrease in IFN production observed for wtSIN- and SIN44-infected cells could be the result of translational shutoff. However, the link may be not so simple. For instance, in VSV-infected cells, the accumulation of M protein, but not the inhibition of translation, was responsible for transcriptional shutoff in VSV-infected cells (2, 66).
There is no reason to expect that the expression of nsP2 during SIN infection in vivo leads to complete suppression of transcription in all infected cells and prevents IFN-α/β release. Upon SIN infection, animals respond with high levels of IFN-α/β (49, 55, 56), and this fact implies that SIN replicates differently in distinct tissues. In many cell types, SIN may not cause complete transcriptional shutoff like it does in NIH 3T3 cells. In our study, SIN variants expressing wt nsP2 (wt SIN and SIN44) induced IFN-α/β in proportion to the levels of virus replication in the brain and, probably, the number of infected cells. In contrast, the nsP2 mutant, SIN/G, induced IFN as efficiently as wtSIN, while replicating to a 50-fold lower titer, supporting the hypothesis that mutations in nsP2 affect the level of IFN-α/β release in vivo.
It was previously shown for different alphaviruses that the level of IFN induction in vivo depends on the virus strain and does not necessarily correlate with virus virulence (29). Previously studied natural isolates usually contained mutations in both nsPs and structural proteins, and the best-characterized mutants of Venezuelan equine encephalomyelitis virus strains TC-83 (32) and 230 (19) or SIN TRSBr114 (57) had mutations in structural proteins as well. Thus, it was difficult to distinguish if viral proteins could be directly involved in regulation of IFN production or if IFN level was determined by changes in virus tropism or in the rate of virus maturation and spread that led finally to different numbers of infected cells. Our experiments provided evidence that nsP2 is one of the proteins that determine the level of IFN-α/β release. These data correlate with recently published results on the replication of Semliki Forest virus mutants in mouse brains (14). These experiments indicated that mutations in a nuclear localization signal of nsP2 either make the virus more sensitive to interferon or lead to more-efficient induction of IFN-α/β.
In conclusion, we have shown that SIN nsP2 is a critical determinant of viral pathogenesis. It is not only an essential viral protease and an integral part of RdRp, but it also plays a significant role in the development of CPE and the suppression of the antiviral response in virus-infected cells. Mutations in nsP2 make SIN significantly less cytopathic and capable of persisting in cells with defects in IFN-α/β signaling. nsP2 mutations also affect the ability of SIN to downregulate IFN-α/β release and change the outcome of infection in mice. Not only does secreted IFN protect uninfected cells against SIN infection, but it can block replication of mutant virus in productively infected cells without leading to cell death. Vero and, possibly, BHK-21 cells have defects in IFN-α/β production (3, 6, 8, 13, 15, 58). This may explain why we were able to select adaptive SIN variants that could persist in these cells but failed in other cell types.
This information may be useful for improving alphavirus-based expression systems for vaccination. As professional antigen-presenting cells, dendritic cells are a primary target for immunization with SIN and Venezuelan equine encephalomyelitis virus vectors (24, 41, 49). Infection of dendritic cells with viruses expressing wt nsP2 or proteins with other functions devoted to preventing IFN-α/β induction and efficient antigen presentation may limit the effectiveness of alphavirus vectors. Indeed, a number of recent publications suggest that cytotoxic T lymphocyte responses to proteins expressed by alphavirus replicons were lower than might be expected based on the level of antigen expression (61). Inclusion of nsP2 mutations such as those described here may help to enhance antigen presentation, immunogenicity, and protection.
Acknowledgments
We thank Herbert W. Virgin for providing IFN-α/βR−/− and wt MEFs and Erik Barton for critical reading of the manuscript.
This work was supported by Public Health Service grants AI24134 (C.M.R), NS18596 (D.E.G.), and T32 AI07417 (S.H.C.) from the National Institutes of Health.
REFERENCES
- 1.Agapov, E. V., I. Frolov, B. D. Lindenbach, B. M. Prägai, S. Schlesinger, and C. M. Rice. 1998. Noncytopathic Sindbis virus RNA vectors for heterologous gene expression. Proc. Natl. Acad. Sci. USA 95:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed, M., and D. S. Lyles. 1998. Effect of vesicular stomatitis virus matrix protein on transcription directed by host RNA polymerases I, II, and III. J. Virol. 72:8413-8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andzhaparidze, O. G., N. N. Bogomolova, Y. S. Boriskin, M. S. Bektemirova, and I. D. Drynov. 1981. Comparative study of rabies virus persistence in human and hamster cell lines. J. Virol. 37:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Basler, C. F., X. Wang, E. Muhlberger, V. Volchkov, J. Paragas, H. D. Klenk, A. Garcia-Sastre, and P. Palese. 2000. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc. Natl. Acad. Sci. USA 97:12289-12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouloy, M., C. Janzen, P. Vialat, H. Khun, J. Pavlovic, M. Huerre, and O. Haller. 2001. Genetic evidence for an interferon-antagonistic function of Rift Valley fever virus nonstructural protein NSs. J. Virol. 75:1371-1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clarke, J. B., and R. E. Spier. 1983. An investigation into causes of resistance of a cloned line of BHK cells to a strain of foot-and-mouth disease virus. Vet. Microbiol. 8:259-270. [DOI] [PubMed] [Google Scholar]
- 7.de Groot, R. J., T. Rümenapf, R. J. Kuhn, E. G. Strauss, and J. H. Strauss. 1991. Sindbis virus RNA polymerase is degraded by the N-end rule pathway. Proc. Natl. Acad. Sci. USA 88:8967-8971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desmyter, J., J. L. Melnick, and W. E. Rawls. 1968. Defectiveness of interferon production and of rubella virus interference in a line of African green monkey kidney cells (Vero). J. Virol. 2:955-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Der, S. D., A. Zhou, B. R. Williams, and R. H. Silverman. 1998. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc. Natl. Acad. Sci. USA 95:15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Veer, M. J., M. Holko, M. Frevel, E. Walker, S. Der, J. M. Paranjape, R. H. Silverman, and B. R. Williams. 2001. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 69:912-920. [PubMed] [Google Scholar]
- 11.Dryga, S. A., O. A. Dryga, and S. Schlesinger. 1997. Identification of mutations in a Sindbis virus variant able to establish persistent infection in BHK cells: the importance of a mutation in the nsP2 gene. Virology 228:72-83. [DOI] [PubMed] [Google Scholar]
- 12.Edwards, J., E. Mann, and D. T. Brown. 1983. Conformational changes in Sindbis virus envelope proteins accompanying exposure to low pH. J. Virol. 45:1090-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emeny, J. M., and M. J. Morgan. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 43:247-252. [DOI] [PubMed] [Google Scholar]
- 14.Fazakerley, J. K., A. Boyd, M. L. Mikkola, and L. Kaariainen. 2002. A single amino acid change in the nuclear localization sequence of the nsP2 protein affects the neurovirulence of Semliki Forest virus. J. Virol. 76:392-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frey, T. K. 1994. Molecular biology of rubella virus. Adv. Virus Res. 44:69-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frolov, I., E. Agapov, T. A. Hoffman Jr., B. M. Prágai, M. Lippa, S. Schlesinger, and C. M. Rice. 1999. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J. Virol. 73:3854-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frolov, I., R. Hardy, and C. M. Rice. 2001. Cis-acting RNA elements at the 5′ end of Sindbis virus genome RNA regulate minus- and plus-strand RNA synthesis. RNA 7:1638-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frolov, I., and S. Schlesinger. 1994. Comparison of the effects of Sindbis virus and Sindbis virus replicons on host cell protein synthesis and cytopathogenicity in BHK cells. J. Virol. 68:1721-1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frolov, I. V., E. V. Agapov, A. A. Kolykhalov, S. V. Netesov, and L. S. Sandakhchiev. 1992. Effect of mutations in genes of Venezuelan equine encephalomyelitis virus structural proteins on its attenuation. Dokl. Akad. Nauk. 326:1078-1082. (In Russian.) [PubMed] [Google Scholar]
- 20.Garcia-Sastre, A., A. Egorov, D. Matassov, S. Brandt, D. E. Levy, J. E. Durbin, P. Palese, and T. Muster. 1998. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252:324-330. [DOI] [PubMed] [Google Scholar]
- 21.Garcin, D., J. Curran, M. Itoh, and D. Kolakofsky. 2001. Longer and shorter forms of Sendai virus C proteins play different roles in modulating the cellular antiviral response. J. Virol. 75:6800-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcin, D., J. Curran, and D. Kolakofsky. 2000. Sendai virus C proteins must interact directly with cellular components to interfere with interferon action. J. Virol. 74:8823-8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcin, D., P. Latorre, and D. Kolakofsky. 1999. Sendai virus C proteins counteract the interferon-mediated induction of an antiviral state. J. Virol. 73:6559-6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gardner, J. P., I. Frolov, S. Perri, Y. Ji, M. L. MacKichan, J. zur Megede, M. Chen, B. A. Belli, D. A. Driver, S. Sherrill, C. E. Greer, G. R. Otten, S. W. Barnett, M. A. Liu, T. W. Dubensky, and J. M. Polo. 2000. Infection of human dendritic cells by a Sindbis virus replicon vector is determined by a single amino acid substitution in the E2 glycoprotein. J. Virol. 74:11849-11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geiss, G., G. Jin, J. Guo, R. Bumgarner, M. G. Katze, and G. C. Sen. 2001. A comprehensive view of regulation of gene expression by double-stranded RNA-mediated cell signaling. J. Biol. Chem. 276:30178-30182. [DOI] [PubMed] [Google Scholar]
- 26.Gorbalenya, A. E., A. P. Donchenko, E. V. Koonin, and V. M. Blinov. 1989. N-terminal domains of putative helicases of flavi- and pestiviruses may be serine proteases. Nucleic Acids Res. 17:3889-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Griffin, D. E. 1989. Molecular pathogenesis of Sindbis virus encephalitis in experimental animals. Adv. Virus Res. 36:255-271. [DOI] [PubMed] [Google Scholar]
- 28.Griffin, D. E., B. Levine, W. R. Tyor, P. C. Tucker, and J. M. Hardwick. 1994. Age-dependent susceptibility to fatal encephalitis: alphavirus infection of neurons. Arch. Virol. Suppl. 9:31-39. [DOI] [PubMed] [Google Scholar]
- 29.Jahrling, P. B., E. Navarro, and W. F. Scherer. 1976. Interferon induction and sensitivity as correlates to virulence of Venezuelan encephalitis viruses for hamsters. Arch. Virol. 51:23-35. [DOI] [PubMed] [Google Scholar]
- 30.Johnston, C., W. Jiang, T. Chu, and B. Levine. 2001. Identification of genes involved in the host response to neurovirulent alphavirus infection. J. Virol. 75:10431-10445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kääriäinen, L., and M. Ranki. 1984. Inhibition of cell functions by RNA virus infections. Annu. Rev. Microbiol. 38:91-109. [DOI] [PubMed] [Google Scholar]
- 32.Kinney, R. M., G. J. Chang, K. R. Tsuchiya, J. M. Sneider, J. T. Roehrig, T. M. Woodward, and D. W. Trent. 1993. Attenuation of Venezuelan equine encephalitis virus strain TC-83 is encoded by the 5′-noncoding region and the E2 envelope glycoprotein. J. Virol. 67:1269-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lemm, J. A., R. K. Durbin, V. Stollar, and C. M. Rice. 1990. Mutations which alter the level or structure of nsP4 can affect the efficiency of Sindbis virus replication in a host-dependent manner. J. Virol. 64:3001-3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemm, J. A., and C. M. Rice. 1993. Assembly of functional Sindbis virus RNA replication complexes: requirement for coexpression of P123 and P34. J. Virol. 67:1905-1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lemm, J. A., and C. M. Rice. 1993. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J. Virol. 67:1916-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lemm, J. A., T. Rümenapf, E. G. Strauss, J. H. Strauss, and C. M. Rice. 1994. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus and plus-strand RNA synthesis. EMBO J. 13:2925-2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levine, B., Q. Huang, J. T. Isaacs, J. C. Reed, D. E. Griffin, and J. M. Hardwick. 1993. Conversion of lytic to persistent alphavirus infection by the bcl-2 cellular oncogene. Nature 361:739-742. [DOI] [PubMed] [Google Scholar]
- 38.Li, G., and C. M. Rice. 1989. Mutagenesis of the in-frame opal termination codon preceding nsP4 of Sindbis virus: studies of translational readthrough and its effect on virus replication. J. Virol. 63:1326-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liljeström, P., and H. Garoff. 1991. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology 9:1356-1361. [DOI] [PubMed] [Google Scholar]
- 40.Lustig, S., A. Jackson, C. S. Hahn, D. E. Griffin, E. G. Strauss, and J. H. Strauss. 1988. Molecular basis of Sindbis virus neurovirulence in mice. J. Virol. 62:2329-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacDonald, G. H., and R. E. Johnston. 2000. Role of dendritic cell targeting in Venezuelan equine encephalitis virus pathogenesis. J. Virol. 74:914-922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naniche, D., A. Yeh, D. Eto, M. Manchester, R. M. Friedman, and M. B. Oldstone. 2000. Evasion of host defenses by measles virus: wild-type measles virus infection interferes with induction of alpha/beta interferon production. J. Virol. 74:7478-7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Niesters, H. G. M., and J. H. Strauss. 1990. Defined mutations in the 5′ nontranslated sequence of Sindbis virus RNA. J. Virol. 64:4162-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niesters, H. G. M., and J. H. Strauss. 1990. Mutagenesis of the conserved 51 nucleotide region of Sindbis virus. J. Virol. 64:1639-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perri, S., D. A. Driver, J. P. Gardner, S. Sherrill, B. A. Belli, T. W. Dubensky, Jr., and J. M. Polo. 2000. Replicon vectors derived from Sindbis virus and Semliki forest virus that establish persistent replication in host cells. J. Virol. 74:9802-9807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rice, C. M., R. Levis, J. H. Strauss, and H. V. Huang. 1987. Production of infectious RNA transcripts from Sindbis virus cDNA clones: mapping of lethal mutations, rescue of a temperature-sensitive marker, and in vitro mutagenesis to generate defined mutants. J. Virol. 61:3809-3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rikkonen, M., J. Peranen, and L. Kaariainen. 1992. Nuclear and nucleolar targeting signals of Semliki Forest virus nonstructural protein nsP2. Virology 189:462-473. [DOI] [PubMed] [Google Scholar]
- 48.Rikkonen, M., J. Peranen, and L. Kaariainen. 1994. Nuclear targeting of Semliki Forest virus nsP2. Arch. Virol. Suppl. 9:369-377. [DOI] [PubMed] [Google Scholar]
- 49.Ryman, K. D., W. B. Klimstra, K. B. Nguyen, C. A. Biron, and R. E. Johnston. 2000. Alpha/beta interferon protects adult mice from fatal Sindbis virus infection and is an important determinant of cell and tissue tropism. J. Virol. 74:3366-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shirako, Y., and J. H. Strauss. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J. Virol. 68:1874-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stollar, V. 1980. Togaviruses in cultured arthropod cells, p. 584-621. In R. W. Schlesinger (ed.), The togaviruses—biology, structure, replication. Academic Press, Inc, New York, N.Y.
- 52.Strauss, E. G., C. M. Rice, and J. H. Strauss. 1984. Complete nucleotide sequence of the genomic RNA of Sindbis virus. Virology 133:92-110. [DOI] [PubMed] [Google Scholar]
- 53.Strauss, E. G., C. M. Rice, and J. H. Strauss. 1983. Sequence coding for the alphavirus nonstructural proteins is interrupted by an opal termination codon. Proc. Natl. Acad. Sci. USA 80:5271-5275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Strauss, J. H., and E. G. Strauss. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491-562. (Erratum, 58: 806.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trgovcich, J., J. F. Aronson, J. C. Eldridge, and R. E. Johnston. 1999. TNFα, interferon, and stress response induction as a function of age-related susceptibility to fatal Sindbis virus infection of mice. Virology 263:339-348. [DOI] [PubMed] [Google Scholar]
- 56.Trgovcich, J., J. F. Aronson, and R. E. Johnston. 1996. Fatal Sindbis virus infection of neonatal mice in the absence of encephalitis. Virology 224:73-83. [DOI] [PubMed] [Google Scholar]
- 57.Trgovcich, J., K. Ryman, P. Extrom, J. C. Eldridge, J. F. Aronson, and R. E. Johnston. 1997. Sindbis virus infection of neonatal mice results in a severe stress response. Virology 227:234-238. [DOI] [PubMed] [Google Scholar]
- 58.Truant, A. L., and J. V. Hallum. 1977. A persistent infection of baby hamster kidney-21 cells with mumps virus and the role of temperature-sensitive variants. J. Med. Virol. 1:49-67. [DOI] [PubMed] [Google Scholar]
- 59.van den Broek, M. F., U. Muller, S. Huang, R. M. Zinkernagel, and M. Aguet. 1995. Immune defence in mice lacking type I and/or type II interferon receptors. Immunol. Rev. 148:5-18. [DOI] [PubMed] [Google Scholar]
- 60.Vasiljeva, L., A. Merits, P. Auvinen, and L. Kaariainen. 2000. Identification of a novel function of the alphavirus capping apparatus. RNA 5′-triphosphatase activity of Nsp2. J. Biol. Chem. 275:17281-17287. [DOI] [PubMed] [Google Scholar]
- 61.Vidalin, O., A. Fournillier, N. Renard, M. Chen, E. Depla, D. Boucreux, C. Brinster, T. Baumert, I. Nakano, Y. Fukuda, P. Liljestrom, C. Trepo, and G. Inchauspe. 2000. Use of conventional or replicating nucleic acid-based vaccines and recombinant Semliki forest virus-derived particles for the induction of immune responses against hepatitis C virus core and E2 antigens. Virology 276:259-270. [DOI] [PubMed] [Google Scholar]
- 62.Wang, X., M. Li, H. Zheng, T. Muster, P. Palese, A. A. Beg, and A. Garcia-Sastre. 2000. Influenza A virus NS1 protein prevents activation of NF-κB and induction of alpha/beta interferon. J. Virol. 74:11566-11573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weiss, B., R. Rosenthal, and S. Schlesinger. 1980. Establishment and maintenance of persistent infection by Sindbis virus in BHK cells. J. Virol. 33:463-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wengler, G. 1984. Identification of a transfer of viral core protein to cellular ribosomes during the early stages of alphavirus infection. Virology 134:435-442. [DOI] [PubMed] [Google Scholar]
- 65.Whitten, T. M., A. T. Quets, and R. H. Schloemer. 1991. Identification of the hepatitis B virus factor that inhibits expression of the beta interferon gene. J. Virol. 65:4699-4704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yuan, H., S. Puckett, and D. S. Lyles. 2001. Inhibition of host transcription by vesicular stomatitis virus involves a novel mechanism that is independent of phosphorylation of TATA-binding protein (TBP) or association of TBP with TBP-associated factor subunits. J. Virol. 75:4453-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]