Abstract
A 14-amino-acid spacer peptide termed SP1 that separates the capsid (CA) and nucleocapsid (NC) sequences plays an active role in the assembly of human immunodeficiency virus type 1. This activity of SP1 involves its amino-terminal residues that, together with adjacent CA residues, constitute a putative α-helical structure spanning Gag residues from positions 359 to 371. In this study, we have determined that the virus assembly determinants within this putative α-helix were residues H359, K360, A361, L364, A367, and M368, of which K360 and A367 contribute to virus production to lesser extents. Notably, changes of the two basic amino acids H359 and K360 to arginine (R) impaired virus production, whereas mutations L364I and M368I, in contrast to L364A and M368A, generated near-wild-type levels of virus particles. This suggests that within Gag complexes, amino acids H359 and K360 are involved in stricter steric interactions than L364 and M368. Since L364 and M368 are separated by four residues and thus presumably located on the same side of the helical surface, they may initiate synergistic hydrophobic interactions to stabilize Gag association. Further analysis in the context of the protease-negative mutation D185H confirmed the key roles of amino acids H359, A361, L364, and M368 in virus assembly. Importantly, when transfected cells were subjected to Dounce homogenization and the cell lysates were treated by ultracentrifugation at 100,000 × g, Gag molecules containing each of the H359A, A361V, L364A, and M368A mutations were found mainly in the supernatant fraction (S100), whereas approximately 80% of wild-type Gag proteins were found in the pellet. Therefore, these four mutations must have prevented Gag from generating large complexes.
Particle formation of human immunodeficiency virus type 1 (HIV-1) is driven by Gag proteins (20). Gag molecules multimerize within host cells and form immature particles along the plasma membrane. In the meantime, other components essential for the construction of infectious particles, such as viral genomic RNA, Gag-Pol polyproteins, envelope proteins, Vpr, and a number of cellular proteins, are incorporated mainly via interactions with distinct regions of Gag. The immature particles are further converted to mature forms by virtue of proteolytic processing of Gag mediated by the viral protease during or shortly after virus budding from the host cell. This maturation process involves conformational transitions of the capsid shell from a spherical form to a conical form (for reviews, see references 12, 17, and 18). Cleavage of Gag generates matrix (MA), capsid (CA), SP1 (previously termed p2), nucleocapsid (NC), SP2 (previously termed p1), and p6 proteins (25). Of these proteins, MA, CA, and NC are the major structural components of mature virus particles.
HIV-1 Gag multimerization occurs at a number of levels, and accordingly, involves distinct sequences. The MA sequences can direct the formation of trimeric Gag complexes (26, 36). Two dimeric interfaces have been located within CA (3, 15, 31, 34, 52). SP1 is needed for a higher-order multimerization of Gag (35). The NC sequences promote Gag-Gag interactions largely through high affinity for RNA (5-7, 9, 10, 23, 45). Of the protein sequences involved in Gag assembly, SP1 is the least-characterized sequence, mainly because of a lack of structural information regarding this region (52). Targeting of Gag to the plasma membrane is mediated by a fatty acid, myristate, that is attached to the glycine residue at the amino terminus of MA and by the basic residues that are present within the first 31 amino acids of MA (10, 20, 22, 46, 56). Release of HIV-1 particles from the host cells is facilitated by cellular factors, including TSG101, that bind to a conserved four-residue stretch, P(T/S)AP, at the amino terminus of p6 (13, 19, 21, 33, 49).
The SP1 region regulates HIV-1 assembly in two distinct fashions. First, the SP1 sequence is essential for the generation of virus particles. Deletion of the SP1 sequence, especially of the residues at the amino terminus, virtually abolishes virus production (1, 27). Computer modeling predicts an α-helical structure across the CA-SP1 boundary (1). Although this putative α-helix awaits verification by structural studies, insertion of either glycine or proline, two α-helix breakers, into the SP1 sequences that constitute the carboxy-terminal portion of this helix lead to decreased levels of virus particle production (1). On the other hand, SP1 must be removed from the carboxy terminus of CA during virus maturation for the spherical capsid shell to be converted to a mature conical core (24, 51). Conceivably, SP1 removal disrupts the relevant α-helix and thus leads to rearrangement of CA-CA interactions. These conformational transitions switch the state of virus particle from an “assembly mode” to a “disassembly mode” (24). On the basis of these findings, it is hypothesized that an assembly domain exists at the carboxy terminus of CA and further extends into the SP1 region (1, 24, 27, 51). The predicted α-helix at the CA-SP1 boundary may represent the key structural element of this assembly domain.
Largely because of the lack of structural information regarding the position of this putative helix in the context of CA sequences and its interactions with other domains of Gag, the mechanisms underlying its role in virus assembly are unclear. In an attempt to shed light on this subject, we have performed a genetic analysis to identify the key amino acids within this helical structure that are essential for virus assembly.
Gag residues H359, A361, L364, and M368 are essential for HIV-1 production.
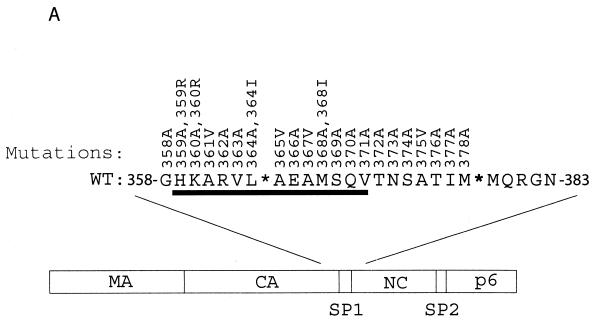
Mutagenesis studies were performed to change each of the 21 residues from positions 358 to 378, which cover the carboxy terminus of CA and the SP1 region, in the context of wild-type HIV-1 cDNA BH10 (Fig. 1A and Table 1). Except for amino acids A361, A365, A367, and A375, which were changed to valine (V), the rest were substituted by alanine (A) (Fig. 1A and Table 1). None of the mutations thus constructed disrupts the α-helix at the CA-SP1 boundary on the basis of folding patterns predicted by the PHD program (41-44) (Table 2).
FIG. 1.
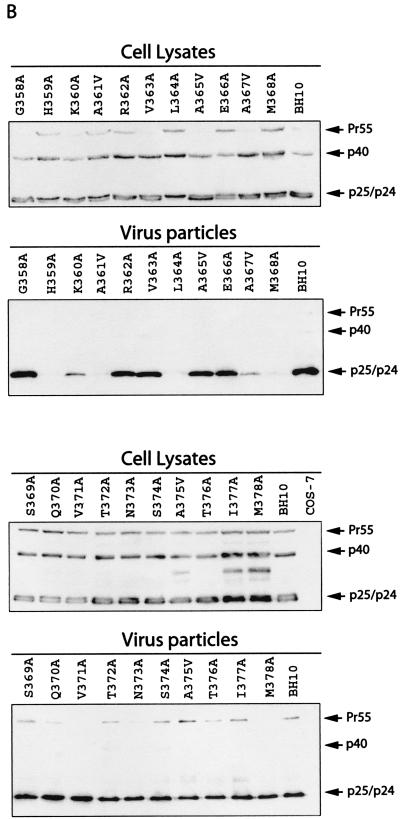
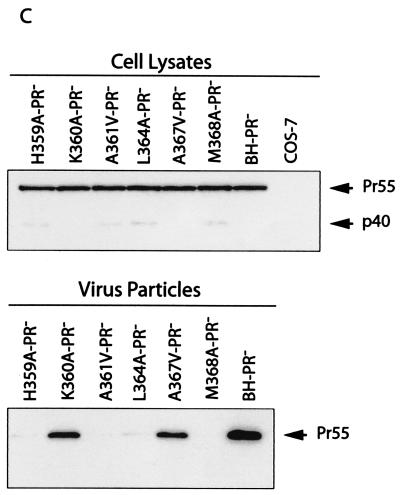
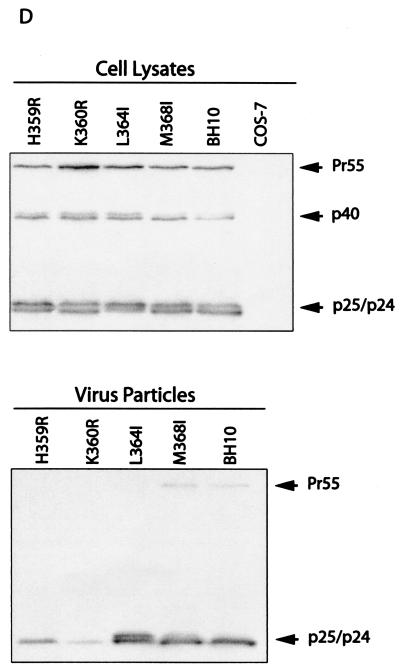
Effects of the mutations in SP1 and the carboxy terminus of CA on virus production. (A) Amino acids from positions 358 to 378 were substituted with alanine except that A361, A365, A367, and A375 were changed to valine. Amino acids H359 and K360 were further changed to arginine, and L364 and M368 were changed to isoleucine. These mutations were generated through use of a PCR strategy as described previously (40). The primers employed are listed in Table 1. The underlined residues from positions 359 to 371 may form an α-helix as predicted by the PHD program (1, 41-44). The asterisks denote viral protease cleavage sites. Numbering of amino acids starts from the first residue of Gag. WT, wild type. (B) Analysis of viral proteins expressed in cells or associated with virus particles. Either COS-7 or HeLa cells were transfected with mutated or wild-type DNA constructs. Forty-eight hours after transfection, cells were lysed in NP-40 lysis buffer containing 50 mM Tris-Cl (pH 7.4), 150 mM NaCl, 1% NP-40, 0.02% sodium azide, and protease inhibitor cocktails (Roche, Laval, Quebec, Canada). The lysates were clarified in a Beckman GR-6S centrifuge at 3,000 rpm for 30 min at 4°C and then assessed by Western blotting using monoclonal antibodies (MAbs) against CA (p24) (ID Lab, Inc., London, Ontario, Canada). Mock-transfected cells were also used in Western blots as negative controls. Virus particles in culture fluids were concentrated by ultracentrifugation at 210,000 × g for 1 h and then analyzed by Western blotting using the same antibodies. The positions of Gag precursor Pr55 and its derivatives are shown to the right of the gels. Similar results from Western blots were obtained from transfection of COS-7 and HeLa cells, and results from COS-7 are shown. The quantities of Gag proteins that were associated with either the cell lysates or virus particles were further determined by quantitative ELISA (Vironostika HIV-1 Antigen Microelisa System; Organon Teknika Corporation, Durham, N.C.). The data were used to calculate the efficiency of virus production for each of the mutated and wild-type DNA constructs. The results are summarized in Table 2. (C) Effects of mutations H359A, K360A, A361V, L364A, A367V, and M368A on virus production in the context of a protease-negative virus. These mutated DNA constructs were tested in both COS-7 and HeLa cells. Similar results from Western blots were obtained in both cases, and results from COS-7 cells are shown. The Pr55 precursor is the major form of Gag protein that was detected by Western blotting using MAbs against CA (p24). The quantities of Pr55 associated with cell lysates and virus particles were measured by quantitative ELISA, and the data were used to calculate the efficiency of virus production. The results are summarized in Table 3. (D) Effects of mutations H359R, K360R, L364I, and M368I on virus production. Either COS-7 or HeLa cells were transfected with the mutated or wild-type constructs. Gag proteins associated with the cell lysates or virus particles were assessed by Western blotting using MAbs against CA (p24). Similar results were obtained for both cell lines, and results from COS-7 cells are shown. The efficiency of virus production by the mutated or wild-type constructs was calculated on the basis of quantitative ELISA. The results are summarized in Table 2 FIG. 1—Continued..
TABLE 1.
Sequences of primers that were used for mutagenesis
| Mutation | Sequence of primera | Positionb |
|---|---|---|
| G358A | 5′-CAGGGAGTAGGAGGACCCGCCCATAAGGCAAGAG-3′ | 1840-1873 |
| H359A | 5′-GGAGTAGGAGGACCCGGCgcTAAGGCAAGAG-3′ | 1842-1873 |
| H359R | 5′-GGACCCGGCCgTAAGGCAAGAG-3′ | 1852-1873 |
| K360A | 5′-GTAGGAGGACCCGGCCATgcGGCAAGAGTTTTGG-3′ | 1846-1879 |
| K360R | 5′-GGACCCGGCCATagGGCAAGAGTTTTGG-3′ | 1852-1878 |
| A361V | 5′-GACCCGGCCATAAGGtAAGAGTTTTGG-3′ | 1853-1879 |
| R362A | 5′-GACCCGGCCATAAGGCAgcAGTTTTGGCTCAAG-3′ | 1853-1885 |
| V363A | 5′-GCCATAAGGCAAGAGcTTTGGCTGAAGC-3′ | 1859-1886 |
| L364A | 5′-GCCATAAGGCAAGAGTTgcGGCTGAAGCAATGAGC-3′ | 1859-1893 |
| L364I | 5′-CCCATAAGGCAAGAGTTaLcGCTGAAGCAATGAGCC-3′ | 1859-1894 |
| A365V | 5′-GGCAAGAGTTTTGGtTGAAGCAATGAG-3′ | 1866-1892 |
| E366A | 5′-GAGTTTTGGCTGcAGCAATGAGCC-3′ | 1871-1894 |
| A367V | 5′-GCAAGAGTTTTGGCTGAAGtAATGAGCCAAG-3′ | 1867-1897 |
| M368A | 5′-GAGTTTTGGCTGAAGCAgcGAGCCAAGTAAC-3′ | 1871-1901 |
| M368I | 5′-GGCTGAAGCAATcAGCCAAGTAAC-3′ | 1878-1901 |
| S369A | 5′-GCTGAAGCAATGgcCCAAGTAACAAATTC-3′ | 1879-1907 |
| Q370A | 5′-GAAGCAATGAGCgcAGTAACAAATTC-3′ | 1882-1907 |
| V371A | 5′-GCAATGAGCCAAGcAACAAATTCAGC-3′ | 1885-1910 |
| T372A | 5′-GAGCCAAGTAgCAAATTCAGC-3′ | 1880-1910 |
| N373A | 5′-GACCCAAGTAACAgcTTCAGCTACCATAATG-3′ | 1880-1920 |
| S374A | 5′-CAAGTAACAAATgCAGCTACCATAATG-3′ | 1884-1920 |
| A375V | 5′-GTAACAAATTCAGtTACCATAATGATGC-3′ | 1897-1924 |
| I377A | 5′-CAAATTCAGCTACCgcAATGATGCAGAG-3′ | 1901-1928 |
| M378A | 5′-CAAATTCAGCTACCATAgcGATGCAGAGAGGC-3′ | 1901-1932 |
Mutated nucleotides are denoted in lowercase type.
Nucleotide positions refer to the HIV-1 cDNA sequence.
TABLE 2.
Effects of various mutations on virus production and virus infectiousnessa
| Mutation | Virus productionb
|
Infectivityc (Jurkat) | |||
|---|---|---|---|---|---|
| COS-7
|
HeLa
|
||||
| p24 | RT | p24 | RT | ||
| G358A | 105 | 102 | 168 | 118 | ++++ |
| H359A | 0.86 | 0.88 | 3.4 | 2.3 | − |
| H359R | 5.4 | 5.6 | 13 | 18 | − |
| K360A | 7.7 | 7.3 | 36 | 34 | − |
| K360R | 2.0 | 2.5 | 14 | 18 | − |
| A361V | 0.75 | 0.65 | 4.3 | 2.8 | − |
| R362A | 35 | 38 | 62 | 42 | + |
| V363A | 56 | 48 | 118 | 89 | ++ |
| L364A | 1.3 | 1.0 | 2.4 | 2.9 | − |
| L364I | 39 | 44 | 44 | 51 | − |
| A365V | 49 | 50 | 52 | 49 | ++++ |
| E366A | 55 | 51 | 78 | 112 | ++ |
| A367V | 3.4 | 2.2 | 9.3 | 8.3 | − |
| M368A | 0.69 | 0.56 | 2.1 | 1.6 | − |
| M368I | 137 | 126 | 96 | 124 | +++ |
| S369A | 132 | 86 | 102 | 109 | ++++ |
| Q370A | 136 | 117 | 121 | 95 | ++++ |
| V371A | 35 | 34 | 52 | 56 | ++ |
| T372A | 145 | 174 | 110 | 114 | ++++ |
| N373A | 147 | 146 | 143 | 137 | ++++ |
| S374A | 383 | 292 | 146 | 122 | ++++ |
| A375V | 238 | 183 | 91 | 93 | +++ |
| T376A | 128 | 157 | 116 | 176 | ++++ |
| I377A | 119 | 102 | 104 | 134 | ++++ |
| M378A | 39 | 35 | 41 | 42 | ++ |
| BH10 | 100 | 100 | 100 | 100 | ++++ |
All viruses tested had the CA-SP1 α-helix.
The yield of virus particles was measured either by p24 determination or by RT assays. The efficiency of virus production for each construct was calculated on the basis of p24 levels associated with the cell lysates and the virus particles. The levels of virus production by BH10 are set as 100. The data are the average of two to three independent transfection experiments.
The degree of virus infectivity on Jurkat cells is indicated as follows: −, not infective; +, 5% infective; ++, 10 to 25% infective; +++, 50% infective; ++++, 100% infective.
When the mutated DNA constructs were transfected into COS-7 cells, the mutated Gag proteins were expressed at levels similar to those seen with wild-type BH10 Gag (Fig. 1B). Of the mutations analyzed, L364A led to an accumulation of the p25 intermediate and A375V, T377A, and M378A delayed the processing of the CA-SP1-NC intermediate (Fig. 1B). These defects in Gag processing are presumably due to amino acid changes within the relevant protease cleavage sites (1, 25, 51). In contrast to the wild-type levels of Gag expression within the cytoplasm, the mutated constructs H359A, K360A, A361V, L364A, A367V, and M368A yielded dramatically lower quantities of extracellular virus particles than did wild-type BH10 (Fig. 1B).
We next measured p24 quantities associated with the pelleted virus particles and in cell lysates by quantitative enzyme-linked immunosorbent assay (ELISA). These data were then used to calculate the efficiency of virus production by the mutated HIV-1 cDNA (Table 2). In comparison to wild-type BH10, mutations H359A, A361V, L364A, and M368A decreased levels of virus production by more than 100-fold, and mutations K360A and A367V led to 20- to 30-fold reductions (Table 2). A moderate decrease in this regard was observed with mutations R362A, V363A, A365V, E366A, V371A, and M378A (Table 2). Similar observations were made on the basis of results obtained from reverse transcriptase (RT) assays (Table 2).
To determine whether the assembly defects seen with the aforementioned mutations were cell type specific, we also transfected the mutated DNA constructs into HeLa cells. Virus production was assessed by either Western blotting, quantitative ELISA, or RT assays, and the data were summarized in Table 2. The results revealed 20- to 30-fold reductions in virus production for mutations H359A, A361V, L364A, and M368A, an ≈10-fold decrease for A367V, and two- to threefold reductions for mutations K360A, R362A, A365V, V371A, and M378A. These results are, in general, consistent with those obtained by transfection of COS-7 cells, although the levels of reduction observed in HeLa cells were not the same as those seen in COS-7 cells. This may be attributable to different levels of Gag expression in COS-7 and HeLa cells (data not shown).
The mutations H359A, A361V, L364A, and M368A delay Gag processing at certain stages (Fig. 1B). To address whether the assembly defects associated with these mutations were caused by aberrant Gag processing, we combined a D185H substitution, which mutates the protease active-site D185 and thus inactivates protease activity (37), with each of these four mutations. The constructs thus generated were termed H359A-PR−, A361V-PR−, L364A-PR−, and M368A-PR−. Each of these four Gag mutations led to severely diminished virus production in both COS-7 and HeLa cells (Fig. 1C and Table 3). Therefore, the H359A, A361V, L364A, and M368A mutations must have directly suppressed the assembly of Pr55Gag precursors. We also assessed the effects of the K360A and A367V mutations on virus production in the presence of the protease-negative mutation D185H. The results showed that these two mutations only modestly affected virus production (Fig. 1C and Table 3).
TABLE 3.
Efficiency of virus productiona
| Virus | Virus production in cells
|
|
|---|---|---|
| COS-7 | HeLa | |
| H359A-PR− | 3.4 | 7.0 |
| K360A-PR− | 32 | 77 |
| A361V-PR− | 4.9 | 5.0 |
| L364A-PR− | 5.9 | 12 |
| A367V-PR− | 25 | 56 |
| M368A-PR− | 5.2 | 12 |
| BH-PR− | 100 | 100 |
The amounts of virus particles were measured by p24 determination. The level of virus production by BH-PR− is set at 100. The data are the average of two to three independent transfection experiments.
Taken together, mutations H359A, A361V, L364I, and M368A nearly abolish virus production from both COS-7 and HeLa cells, and mutations K360A and A367V affect virus production to lesser extents. Notably, these six key residues are all located within the putative CA-SP1 α-helix (Fig. 1A).
Of the six mutations that display severe deficits in virus production, H359A and K360A alter two basic amino acids, and L364A and M368A change two hydrophobic residues. We speculate that the basic feature of H359 and K360 and the hydrophobic nature of L364 and M368 may have accounted for the importance of these residues in virus production. To test this possibility, we further changed the former two amino acids to arginine (R) and the latter two to isoleucine (I). The mutations thus generated were termed H359R, K360R, L364I, and M368I, respectively. The last two mutations have been tested by other groups (1, 51), and the results obtained in the present study were consistent with their results. Regardless of the wild-type levels of Gag expression within the cytoplasm of COS-7 cells, mutations H359R and K360R but not L364I and M368I decreased virus production by 20- to 40-fold (Fig. 1D and Table 2). Similarly, a five- to sevenfold virus reduction was observed for mutations H359R and K360R in HeLa cells, whereas mutations L364I and M368I did not show adverse effects in this regard (Table 2). These results suggest that their basic feature alone cannot explain the critical roles of H359 and K360 in virus assembly; instead, strict steric interactions must have also been involved. As for residues L364 and M368, their key contribution to virus assembly is the development of hydrophobic interactions with other residues of Gag.
The H359A, A361V, L364A, and M368A mutations attenuate the formation of large Gag complexes.
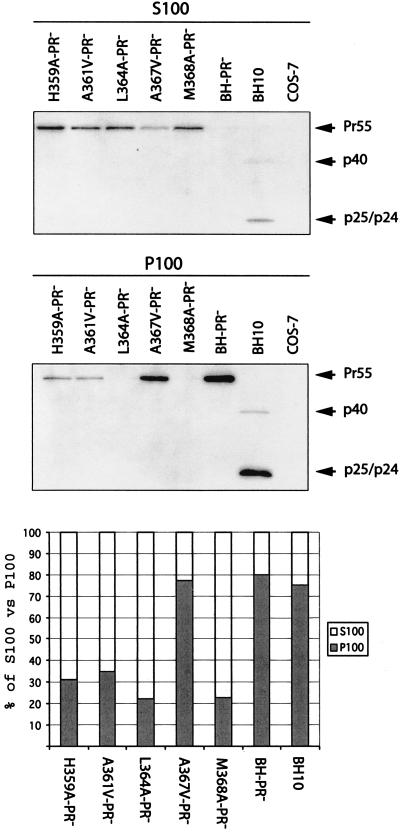
Gag assembly leads to the formation of a series of complexes of variable size that can be isolated by velocity sedimentation. These complexes serve as sequential intermediates in the process of virus construction (29, 30, 32, 35, 48). We speculate that the mutations in SP1 and in the carboxy terminus of CA may have blocked Gag assembly at a certain stage and, consequently, suppressed virus production. To test this possibility, transfected COS-7 cells were subjected to Dounce homogenization in the absence of detergents and then centrifugation at 100,000 × g. The majority of wild-type Gag molecules generated by either BH-PR− or BH10 were recovered in the P100 fraction (Fig. 2). In contrast, the mutated Gag molecules with H359A, A361V, L364A, and M368A mutations were mainly found in the S100 fraction (Fig. 2). This suggests that these four mutations must have restricted Gag multimerization to early stages when only low-molecular-weight complexes have been formed. The A367V-PR− Gag was mostly pelleted at 100,000 × g (Fig. 2), which explains the observation that this mutated Gag can efficiently produce virus particles (Fig. 1C).
FIG. 2.
Analysis of Gag complexes within the cytoplasm by velocity sedimentation. COS-7 cells were transfected with mutated or wild-type DNA constructs containing the protease-negative mutation D185H. Cells were then treated by Dounce homogenization in ice-cold TNE buffer containing protease inhibitor cocktails (Roche). After the supernatants were clarified in a Beckman GR-6S centrifuge at 3,000 rpm for 30 min at 4°C, they were subjected to ultracentrifugation at 100,000 × g for 1 h at 4°C through a 20% sucrose cushion. The supernatants (termed S100) and pelleted materials (termed P100) were analyzed for Gag proteins either by anti-p24 Western blotting or by quantitative anti-p24 ELISA. The relative quantities of Gag proteins in the S100 and P100 fractions for each construct are shown in the bar graph at the bottom of the figure.
Effects of the various mutations on production of infectious virus particles in Jurkat cells.
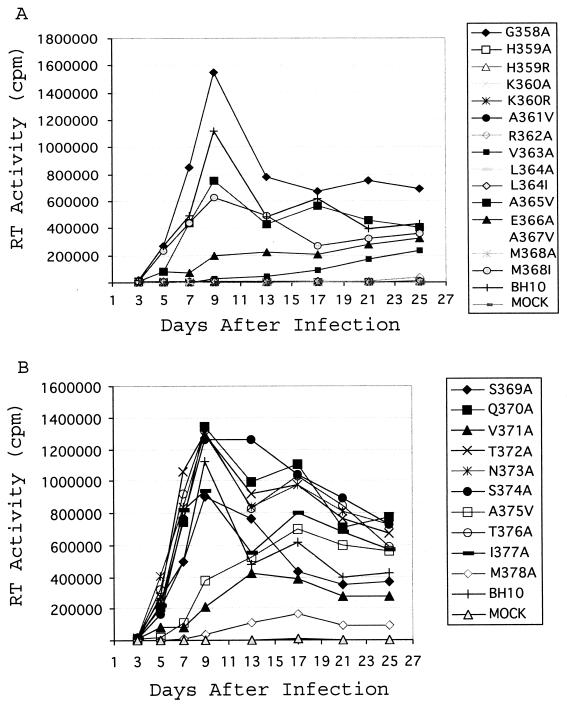
Next, we assessed the infectiousness of the viruses containing each of the mutations illustrated in Fig. 1A. Jurkat cells (106) were transfected by 1 μg of the mutated or wild-type HIV-1 cDNA. Viral growth was then monitored by measuring RT activity in the culture fluids. Consistent with their severely diminished ability to generate virus particles, mutations H359A, H359R, K360A, K360R, A361V, L364A, A367V, and M368A failed to produce any infectious virus (Fig. 3 and Table 2). The L364I mutation impaired processing of CA-SP1 (p25) (Fig. 1D) (1, 51), an event that has been shown to be essential for morphogenesis of a mature conical core (51). Therefore, even though the L364I mutated construct was able to yield wild-type levels of virus particles (Fig. 1D and Table 2), the viruses thus generated were noninfectious (Fig. 3). Of the other mutations, R362A, V363A, E366A, V371A, A375V, and M378A delayed viral replication kinetics to various extents (Fig. 3). These replication defects may have also been the results of diminished virus production (Table 2). Moreover, mutations of amino acids from positions 369 to 377, which cover 70% of the SP1 sequences, gave minor effects on viral replication. This observation supports the results of a previous study showing that deletion of amino acids 370 to 374 was relatively well tolerated with respect to viral replication (1).
FIG. 3.
Effects of the various mutations on viral replication. Jurkat cells were transfected with mutated or wild-type viral cDNA. Viral growth was monitored by measuring levels of RT activity in culture fluids at various times. For clarity, the growth curves are presented in two separate graphs (A and B).
Conclusions.
Our data, together with those of other groups (1, 24, 27, 51), support the hypothesis that a continuous assembly domain exists at the CA-SP1 boundary. Mutations within the SP1 sequence can suppress virus production, result in aberrant particle morphology, and alter the morphogenesis of Gag assembly (1, 22, 24, 27, 38, 50, 51). Interestingly, a continuous α-helical structure has been predicted at the CA-SP1 boundary (1). In support of the importance of this putative α-helix in virus assembly, insertions of glycine or proline, two α-helix breakers, disrupt this α-helical structure and virtually eliminate virus production (1). We have now localized the key residues for virus assembly within this predicted helix to H359, K360, A361, L364, A367, and M368 (Fig. 1 and Table 2).
Understanding of the mechanisms underlying the involvement of this putative α-helix in virus assembly is hampered by limited structural information regarding the location of this α-helix in the context of CA sequences. This is mainly because the last 11 residues at the carboxy terminus of CA and the SP1 sequence are disordered in the crystal form of the CA146-231-SP1 molecule (52). To counter this disadvantage, we have performed genetic analysis and identified six key residues within this predicted α-helix that are essential for virus assembly (Fig. 1 and Table 2). On this basis, it is possible to gain insights into the types of potential molecular interactions by which this putative α-helix binds to other portions of Gag or to unknown cellular factors. Substitutions L364A and M368A nearly eliminate virus production; however, mutations L364I and M368I, which retain the hydrophobic nature of both L364 and M368, display marginal effects in this regard (Table 2). This strongly suggests that residues L364 and M368 promote Gag assembly by establishing hydrophobic interactions. Since each turn of an α-helix consists of 3.5 amino acids, it follows that residues L364 and M368, which are four residues apart, must be closely located on the same side of the helical surface. This spatial proximity may allow them to form a hydrophobic center and to develop synergistic hydrophobic interactions. On the other hand, the basic feature of residues H359 and K360 is not sufficient to explain their critical roles in virus assembly, since mutations H359R and K360R also significantly inhibit virus production (Fig. 1D). Conceivably, strict steric interactions of H359 and K360 with other residues are needed for virus assembly.
We have tried to determine the stages at which our mutations affect virus assembly. Virus production involves Gag multimerization, targeting of Gag to the plasma membrane, formation of the spherical bud, and release of virus particles from host cells. The results of our sedimentation studies showed that the mutant Gag H359A, A361V, L364A and M368A were mainly found within the S100 fraction (Fig. 2). This suggests that these four mutations impede Gag multimerization and thus prevent formation of large Gag complexes at wild-type levels. Our observations are consistent with a previous report showing that deletion of the SP1 sequence blocks Gag oligomerization at the trimer stage (35). Electron microscopy (EM) studies revealed large electron-dense patches underneath the plasma membrane for assembly-defective HIV-1 mutants lacking SP1 sequences (1, 27). These EM data indicate that the mutant Gag can aggregate but fail to generate the spherical capsids seen with wild-type Gag. At this stage, it is difficult to compare these EM data obtained with HIV-1 mutants in which SP1 sequences had been deleted with the results of sedimentation experiments performed with our substitution mutations. We speculate that although our mutant Gag proteins were detected in the P100 fraction at levels lower than the level for the wild-type Gag, this minor portion of mutant Gag may have bound to the plasma membrane and can be visualized by EM as electron-dense plaques underneath the plasma membrane. Additional EM studies of the mutants H359A, A361V, L364A, and M368A will help to address this issue.
The roles for the CA-SP1 boundary region in virus assembly need further evaluation in the context of two other important assembly domains: the carboxy-terminal domain of CA and the amino-terminal domain of NC. CA consists of two domains: an amino-terminal domain (residues 1 to 146) and a carboxy-terminal domain (residues 148 to 231) (14 15 16). In general, mutations within the carboxy-terminal third of CA inhibit virus assembly, whereas mutations of the amino-terminal sequences of CA lead to the production of noninfectious virus particles with aberrant morphology (8, 11, 39, 46, 47). More strikingly, removal of CA sequences amino terminal to the major homology region is still permissive for virus production (2, 4). Structural determination of CA146-231 reveals four helices that adopt an ovoid fold (15). Of these helices, helix 2 comprises an interface to mediate CA dimerization. Notably, a short stretch of GP-rich amino acids 353-GVGGPG-358 is located close to the carboxy terminus of CA. This short sequence is fairly conserved in the Gag proteins of primate lentiviruses (28). Its GP-rich feature implicates structural flexibility and may result in the separation of the putative α-helix at the CA-SP1 boundary from the upstream CA structures. It remains to be elucidated whether this CA-SP1 boundary region acts independently in virus assembly or functions together with the carboxy-terminal sequences of CA.
NC plays a crucial role in mediating Gag-Gag interactions and has thus been termed the interaction (I) domain. This NC function is mainly determined by its amino-terminal residues (45, 53). NC acts in Gag association largely by virtue of its RNA binding activity (5-7, 9, 10, 23, 45). Accordingly, its roles in Gag assembly can be performed by foreign protein sequences, such as the Bacillus subtilis MtrB protein domain and the leucine zipper domain of the Saccharomyces cerevisiae transcription factor GCN4 (2, 54, 55). These findings suggest that NC does not initiate direct interactions with other regions of Gag during virus assembly. Thus, it is unlikely that the putative α-helix at the CA-SP1 boundary represents an element of the I domain within NC, although the former structure is located proximal to the amino terminus of NC.
Acknowledgments
We thank Shan Cen and Rabih Halwani for valuable suggestions and Mervi Detorio and Maureen Oliveira for technical assistance.
This research was supported in part by grants from the Canadian Institutes of Health Research (CIHR) and by the Fonds de la Recherche en Sante du Quebec (FRSQ). Rodney S. Russell is a recipient of a CIHR fellowship. Chen Liang is a Chercheur-Boursier of the FRSQ.
REFERENCES
- 1.Accola, M. A., S. Hoglund, and H. G. Gottlinger. 1998. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J. Virol. 72:2072-2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Accola, M. A., B. Strack, and H. G. Gottlinger. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395-5402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berthet-Colominas, C., S. Monaco, A. Novelli, G. Sibai, F. Mallet, and S. Cusack. 1999. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab. EMBO J. 18:1124-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borsetti, A., A. Ohagen, and H. G. Gottlinger. 1998. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J. Virol. 72:9313-9317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burniston, M. T., A. Cimarelli, J. Colgan, S. P. Curtis, and J. Luban. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 73:8527-8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell, S., and A. Rein. 1999. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 73:2270-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell, S., and V. M. Vogt. 1995. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 69:6487-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chazal, N., C. Carriere, B. Gay, and P. Boulanger. 1994. Phenotypic characterization of insertion mutants of the human immunodeficiency virus type 1 Gag precursor expressed in recombinant baculovirus-infected cells. J. Virol. 68:111-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cimarelli, A., S. Sandin, S. Hoglund, and J. Luban. 2000. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 74:3046-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craven, R. C., and L. J. Parent. 1996. Dynamic interactions of the Gag polyprotein. Curr. Top. Microbiol. Immunol. 214:65-94. [DOI] [PubMed] [Google Scholar]
- 11.Dorfman, T., A. Bukovsky, A. Ohagen, S. Hoglund, and H. G. Gottlinger. 1994. Functional domains of the capsid protein of human immunodeficiency virus type 1. J. Virol. 68:8180-8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed, E. O. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 13.Freed, E. O. 2002. Viral late domains. J. Virol. 76:4679-4687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gamble, T. R., F. F. Vajdos, S. Yoo, D. K. Worthylake, M. Houseweart, W. I. Sundquist, and C. P. Hill. 1996. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell 87:1285-1294. [DOI] [PubMed] [Google Scholar]
- 15.Gamble, T. R., S. Yoo, F. F. Vajdos, U. K. von Schwedler, D. K. Worthylake, H. Wang, J. P. McCutcheon, W. I. Sundquist, and C. P. Hill. 1997. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science 278:849-853. [DOI] [PubMed] [Google Scholar]
- 16.Ganser, B. K., S. Li, V. Y. Klishko, J. T. Finch, and W. I. Sundquist. 1999. Assembly and analysis of conical models for the HIV-1 core. Science 283:80-83. [DOI] [PubMed] [Google Scholar]
- 17.Garnier, L., J. B. Bowzard, and J. W. Wills. 1998. Recent advances and remaining problems in HIV assembly. AIDS 12(Suppl. A):S5-S16. [PubMed] [Google Scholar]
- 18.Garoff, H., R. Hewson, and D. J. Opstelten. 1998. Virus maturation by budding. Microbiol. Mol. Biol. Rev. 62:1171-1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garrus, J. E., U. K. von Schwedler, O. W. Pornillos, S. G. Morham, K. H. Zavitz, H. E. Wang, D. A. Wettstein, K. M. Stray, M. Cote, R. L. Rich, D. G. Myszka, and W. I. Sundquist. 2001. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107:55-65. [DOI] [PubMed] [Google Scholar]
- 20.Gheysen, D., E. Jacobs, F. de Foresta, C. Thiriart, M. Francotte, D. Thines, and M. De Wilde. 1989. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 59:103-112. [DOI] [PubMed] [Google Scholar]
- 21.Göttlinger, H. G., T. Dorfman, J. G. Sodroski, and W. A. Haseltine. 1991. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 88:3195-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Göttlinger, H. G., J. G. Sodroski, and W. A. Haseltine. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc. Natl. Acad. Sci. USA 86:5781-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gross, I., H. Hohenberg, and H.-G. Kräusslich. 1997. In vitro assembly properties of purified bacterially expressed capsid proteins of human immunodeficiency virus. Eur. J. Biochem. 249:592-600. [DOI] [PubMed] [Google Scholar]
- 24.Gross, I., H. Hohenberg, T. Wilk, K. Wiegers, M. Grattinger, B. Muller, S. Fuller, and H.-G. Kräusslich. 2000. A conformational switch controlling HIV-1 morphogenesis. EMBO J. 19:103-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henderson, L. E., R. C. Sowder, T. D. Copeland, S. Oroszlan, and R. E. Benveniste. 1990. Gag precursors of HIV and SIV are cleaved into six proteins found in the mature virions. J. Med. Primatol. 19:411-419. [PubMed] [Google Scholar]
- 26.Hill, C. P., D. Worthylake, D. P. Bancroft, A. M. Christensen, and W. I. Sundquist. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc. Natl. Acad. Sci. USA 93:3099-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kräusslich, H.-G., M. Facke, A. M. Heuser, J. Konvalinka, and H. Zentgraf. 1995. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J. Virol. 69:3407-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuiken, C., B. Foley, et al. 1999. Human retroviruses and AIDS: a compilation and analysis of nucleic acid and amino acid sequences. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, Los Alamos, N.Mex.
- 29.Lee, Y. M., B. Liu, and X. F. Yu. 1999. Formation of virus assembly intermediate complexes in the cytoplasm by wild-type and assembly-defective mutant human immunodeficiency virus type 1 and their association with membranes. J. Virol. 73:5654-5662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, Y. M., and X. F. Yu. 1998. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T cells. Virology 243:78-93. [DOI] [PubMed] [Google Scholar]
- 31.Li, S., C. P. Hill, W. I. Sundquist, and J. T. Finch. 2000. Image reconstructions of helical assemblies of the HIV-1 CA protein. Nature 407:409-413. [DOI] [PubMed] [Google Scholar]
- 32.Lingappa, J. R., R. L. Hill, M. L. Wong, and R. S. Hegde. 1997. A multistep, ATP-dependent pathway for assembly of human immunodeficiency virus capsids in a cell-free system. J. Cell Biol. 136:567-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin-Serrano, J., T. Zang, and P. D. Bieniasz. 2001. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat. Med. 7:1313-1319. [DOI] [PubMed] [Google Scholar]
- 34.Momany, C., L. C. Kovari, A. J. Prongay, W. Keller, R. K. Gitti, B. M. Lee, A. E. Gorbalenya, L. Tong, J. McClure, L. S. Ehrlich, M. F. Summers, C. Carter, and M. G. Rossmann. 1996. Crystal structure of dimeric HIV-1 capsid protein. Nat. Struct. Biol. 3:763-770. [DOI] [PubMed] [Google Scholar]
- 35.Morikawa, Y., D. J. Hockley, M. V. Nermut, and I. M. Jones. 2000. Roles of matrix, p2, and N-terminal myristoylation in human immunodeficiency virus type 1 Gag assembly. J. Virol. 74:16-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morikawa, Y., W. H Zhang, D. J. Hockley, M. V. Nermut, and I. M. Jones. 1998. Detection of a trimeric human immunodeficiency virus type 1 Gag intermediate is dependent on sequences in the matrix protein, p17. J. Virol. 72:7659-7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morin, N., E. Cherry, X. Li, and M. A. Wainberg. 1998. Cotransfection of mutated forms of human immunodeficiency virus type 1 Gag-Pol with wild-type constructs can interfere with processing and viral replication. J. Hum. Virol. 1:240-247. [PubMed] [Google Scholar]
- 38.Pettit, S. C., M. D. Moody, R. S. Wehbie, A. H. Kaplan, P. V. Nantermet, C. A. Klein, and R. Swanstrom. 1994. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J. Virol. 68:8017-8027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reicin, A. S., S. Paik, R. D. Berkowitz, J. Luban, I. Lowy, and S. P. Goff. 1995. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J. Virol. 69:642-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rong, L., R. S. Russell, J. Hu, Y. Guan, L. Kleiman, C. Liang, and M. A. Wainberg. 2001. Hydrophobic amino acids in the human immunodeficiency virus type 1 p2 and nucleocapsid proteins can contribute to the rescue of deleted viral RNA packaging signals. J. Virol. 75:7230-7243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rost, B. 1996. PHD: predicting one-dimensional protein structure by profile based neural networks. Methods Enzymol. 266:525-539. [DOI] [PubMed] [Google Scholar]
- 42.Rost, B., and C. Sander. 1993. Improved prediction of protein secondary structure by use of sequence profiles and neural networks. Proc. Natl. Acad. Sci. USA 90:7558-7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rost, B., and C. Sander. 1993. Prediction of protein secondary structure at better than 70% accuracy. J. Mol. Biol. 232:584-599. [DOI] [PubMed] [Google Scholar]
- 44.Rost, B., and C. Sander. 1994. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins 19:55-72. [DOI] [PubMed] [Google Scholar]
- 45.Sandefur, S., R. M. Smith, V. Varthakavi, and P. Spearman. 2000. Mapping and characterization of the N-terminal I domain of human immunodeficiency virus type 1 Pr55Gag. J. Virol. 74:7238-7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spearman, P., J. J. Wang, N. Vander Heyden, and L. Ratner. 1994. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J. Virol. 68:3232-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang, S., T. Murakami, B. E. Agresta, S. Campbell, E. O. Freed, and J. G. Levin. 2001. Human immunodeficiency virus type 1 N-terminal capsid mutants that exhibit aberrant core morphology and are blocked in initiation of reverse transcription in infected cells. J. Virol. 75:9357-9366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tritel, M., and M. D. Resh. 2000. Kinetic analysis of human immunodeficiency virus type 1 assembly reveals the presence of sequential intermediates. J. Virol. 74:5845-5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.VerPlank, L., F. Bouamr, T. J. LaGrassa, B. Agresta, A. Kikonyogo, J. Leis, and C. A. Carter. 2001. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55Gag. Proc. Natl. Acad. Sci. USA 98:7724-7729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Von Poblotzki, A., R. Wagner, M. Niedrig, G. Wanner, H. Wolf, and S. Modrow. 1993. Identification of a region in the Pr55gag-polyprotein essential for HIV-1 particle formation. Virology 193:981-985. [DOI] [PubMed] [Google Scholar]
- 51.Wiegers, K., G. Rutter, H. Kottler, U. Tessmer, H. Hohenberg, and H.-G. Kräusslich. 1998. Sequential steps in human immunodeficiency virus particle maturation revealed by alterations of individual Gag polyprotein cleavage sites. J. Virol. 72:2846-2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Worthylake, D. K., H. Wang, S. Yoo, W. I. Sundquist, and C. P. Hill. 1999. Structures of the HIV-1 capsid protein dimerization domain at 2.6 A resolution. Acta Crystallogr. D Biol. Crystallogr. 55:85-92. [DOI] [PubMed] [Google Scholar]
- 53.Zabransky, A., E. Hunter, and M. Sakalian. 2002. Identification of a minimal HIV-1 gag domain sufficient for self-association. Virology 294:141-150. [DOI] [PubMed] [Google Scholar]
- 54.Zhang, Y., and E. Barklis. 1997. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J. Virol. 71:6765-6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang, Y., H. Qian, Z. Love, and E. Barklis. 1998. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 72:1782-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou, W., L. J. Parent, J. W. Wills, and M. D. Resh. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J. Virol. 68:2556-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]