Abstract
The role of the gamma-2 herpesvirus open reading frame (ORF) 73 gene product has become the focus of considerable interest. It has recently been shown that the Kaposi's sarcoma-associated herpesvirus (KSHV) latency-associated nuclear antigen (LANA) is expressed during a latent infection and can modulate both viral and cellular gene expression. The herpesvirus saimiri (HVS) ORF 73 gene product has some sequence homology to LANA; however, the role of HVS ORF 73 is unknown. We have previously demonstrated that HVS ORF73 is expressed in a stably transduced human carcinoma cell line, where HVS genomes persist as nonintegrated circular episomes. This implies that there may be some functional homology between these proteins. To further investigate the role of the HVS ORF 73 protein, the yeast two-hybrid system was employed to identify interacting cellular proteins. We demonstrate that ORF 73 interacts with the cellular protein p32 and triggers the accumulation of p32 in the nucleus. Using reporter gene-based transient-transfection assays, we demonstrate that ORF 73 can transactivate a number of heterologous promoter constructs and also upregulate its own promoter. Moreover, ORF 73 and p32 act synergistically to transactivate these promoters. The binding of ORF 73 to p32 is mediated by an amino-terminal arginine-rich domain, which contains two functionally distinct nuclear localization signals. The p32 binding domains are required for ORF 73 transactivating abilities and for ORF 73 to induce nuclear accumulation of p32. These results suggest that ORF 73 can function as a regulator of gene expression and that p32 is involved in ORF 73-dependent transcriptional activation.
Herpesvirus saimiri (HVS) is the prototype gamma-2 herpesvirus, originally isolated from its natural host the squirrel monkey (Saimiri sciureus), in which it causes a persistent asymptomatic infection. However, on experimental transfer to other New World primates, HVS infection results in a number of lymphoproliferative diseases within a few weeks (19). HVS subgroups A and C possess the ability to immortalize common marmoset T lymphocytes to interleukin-2-independent proliferation (17, 49). Moreover, subgroup C viruses are capable of transforming human, rabbit, and rhesus monkey lymphocytes in vitro (5-8). HVS can also stably transduce a variety of human cell lines, where the viral genome persists as multiple nonintegrating circular episomes (22, 23, 46). HVS has significant homology to a number of other herpesviruses with oncogenic potential, including the gamma-2 herpesviruses Kaposi's sarcoma-associated herpesvirus (KSHV) and murine gammaherpesvirus 68 and the gamma-1 herpesvirus Epstein-Barr virus (EBV). Their genomes are predominantly colinear, with homologous genes in approximately the same locations and orientation. However, conserved gene blocks are separated by genes unique to each virus (1, 37, 45, 53).
The role of the open reading frame (ORF) 73 gene products encoded by gamma-2 herpesviruses has become the focus of considerable interest. Analysis of latent KSHV gene transcription in the PEL cell line showed that a region spanning ORFs 71 to 73 is expressed and that separate mRNAs encoding ORF 73 and ORF 72 are generated from a common latency-specific promoter (18). This observation is further supported by the work of Sarid et al. (45a) and Talbot et al. (49a), who detected two transcripts of approximately 6.0 and 2.0 kb in the BC-1 and PEL cell lines, respectively. The larger transcript encodes the ORF 73, ORF 72, and ORF K13 products, while the smaller transcript encodes only the ORF 72 and ORF K13 products. We have recently analyzed gene expression in an HVS persistently infected human lung carcinoma cell line, A549, in which the HVS DNA is stably maintained as multiple nonintegrated circular episomes (23). Virus production can be reactivated using chemical inducing agents, including tetradecanoyl phorbol acetate (TPA) and n-butyrate, suggesting that the infection in A549 cells may be latent. Analysis of viral gene expression using Northern blotting demonstrated that similar adjacent sets of genes encoding the ORF 71 to 73 products are expressed in this stably transduced cell line. Moreover, these genes are transcribed as a polycistronic mRNA species produced from a common promoter upstream of ORF 73.
In both HVS and KSHV, ORF 71 and ORF 72 encode an anti-apoptosis protein, v-FLIP, and v-cyclin D homologue, respectively (12, 29, 51). However, there is limited homology between the HVS and KSHV ORF 73 proteins. KSHV ORF 73 encodes the latency-associated nuclear antigen (LANA). LANA is a large (222- to 234-kDa) protein exhibiting a distinctive nuclear speckling pattern (30, 31, 42). It interacts with a number of cellular proteins, including RING3, a homologue of the Drosophila female sterile homeotic gene product (39), p53 (20), retinoblastoma protein (41), and proteins of the mSin3 corepressor complex (33). Moreover, in cells harboring KSHV episomes, LANA and KSHV DNA colocalize at discrete points in interphase nuclei and along mitotic chromosomes. This suggests that LANA is involved in episomal maintenance by tethering viral genomes to host cell chromosomes (2, 3, 15). In support of this hypothesis is the observation that LANA can bind the histone H1 protein in immunoprecipitation assays (15). These results have led to the suggestion that although unrelated by sequence homology, KSHV LANA is a functional homologue of the EBV EBNA-1 protein, which is essential for episomal maintenance and also functions as a transcriptional regulator (26, 40, 48). To date, no similar function of HVS ORF 73 has been reported.
In this study we have further investigated the role of the HVS ORF 73 protein. The yeast two-hybrid system was employed to identify interacting cellular proteins, using HVS ORF 73 as bait. We demonstrate that ORF 73 interacts with the cellular protein, p32. First isolated associated with the splicing factor SF2/ASF (32), p32 interacts with a number of additional cellular and viral proteins, including human immunodeficiency virus type 1 (HIV-1) Rev (34, 50) and Tat (56, 57), herpes simplex virus type 1 (HSV-1) ORF P (10) and ICP27 (11), adenovirus core protein V (35), rubella virus capsid protein (4), and EBV EBNA-1 protein (13, 52, 54). Although the function of p32 is as yet undetermined, it has been suggested to play roles in splicing (11, 32, 38, 50), nucleocytoplasmic transport (9, 11, 35), maintenance of oxidative phosphorylation (36), transcriptional activation (52, 54, 56, 57), and possibly latent-cycle DNA replication (13, 52). To address the implications of this interaction, we demonstrate that ORF 73 and p32 have a synergistic effect on the transactivation of a number of heterologous promoters and its own promoter. In addition, ORF 73 mutants lacking the p32 binding domain were defective for reporter gene upregulation. These results suggest that ORF 73 can function as a regulator of gene expression and that p32 is involved in ORF 73-dependent transcriptional activation.
MATERIALS AND METHODS
Yeast two-hybrid screen for ORF 73-interacting proteins.
The Gal4-based yeast two-hybrid system screening technique (14) was used to identify proteins that interacted with the ORF 73 protein. The “bait” plasmid was constructed by cloning the ORF 73 coding region, previously excised from p73GFP (23) as a BamHI-XhoI fragment, into pGBT9 (GAL41-147DNA-BD, TRP1) (Clontech) to derive the GAL4 DNA-binding domain (DBD) fusion, pDBD73. A human kidney cDNA-GAL4 activation domain fusion library in the vector pACT2 (GAL4768-881AD, LEU2) (Clontech) was used to identify ORF 73-interacting proteins. The bait plasmid was transformed into Saccharomyces cerevisiae strain HF7c (MATα ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3,112 gal4-542 gal80-538 LYS2::GAL1UAS-GAL1TATA-HIS3 URA3::GAL417-MERS(x3)-CYC1TATA-lacZ) (Clontech). Clones were selected on minimal synthetic dropout medium in the absence of tryptophan. Yeast clones harboring the bait plasmids were then sequentially transformed with the “prey” library. Positive clones potentially harboring ORF 73-interacting species were identified both by their ability to grow on medium without tryptophan, leucine, and histidine and by the detection of β-galactosidase activity, as specified by the manufacturer (Clontech). Plasmids were isolated from positive yeast clones, selecting for pACT2 cDNA library plasmids by transformation of the leucine auxotroph into Escherichia coli strain HB101. Clones were then grouped by size and restriction analysis.
To confirm the specificity of the interactions, pACT-2 library plasmids were transformed in yeast strains harboring no plasmid, yeast containing pGBT9 vector only, yeast containing pLAM5 (a GAL4 human lamin C fusion), or pDBD73. Only library plasmids demonstrating a requirement for the pDBD73 plasmid for induced expression of HIS3 or lacZ reporter genes were considered further and selected for DNA sequencing.
Viruses, cell culture, and transfections.
HVS-GFP (23), based on strain A11, was propagated in owl monkey kidney (OMK) cells which were maintained in Dulbecco modified Eagle medium (Life Technologies) supplemented with 10% fetal calf serum. Cos-7 cells were also maintained in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. Plasmids used in the transfections were prepared using Qiagen Plasmid kits as specified by the manufacturer. Cells were seeded at 5 × 105 cells per 35-mm-diameter petri dish 24 h prior to transfection. Transfections were performed using Lipofectamine (Life Technologies) as described by the manufacturer, with 2 μg of the appropriate DNAs.
Plasmid constructs.
The yeast two-hybrid ORF 73 deletion series was produced by excising the ORF 73 amino and carboxy termini from p73Δ1-6GFP (24) as BamHI-XhoI fragments and ligated into pGBT9 (Clontech), to obtain the GAL4 DBD fusion, pDBD73Δ1-3. The ORF 73 bacterial expression constructs were constructed by a similar procedure, cloning into pGEX4T-3, to yield the expression constructs pGST73, pGST73N, pGST73ΔAcidic, and pGST73C, respectively.
The amino-terminal ORF 73 deletion bacterial expression constructs were produced by a PCR-based method using full-length mutants of pORF73 p73ΔNLS1, p73ΔNLS2, and p73ΔNLS1+2 (24) as templates. PCR amplication was performed using the forward primer 5′-CGC GGA TCC ATG GCG CCC AGA AGA AGA AAA GCG and the reverse primer 5′-CCG GAA TTC GTC ATC GTC GCC TTG AGG CTT CAG TTT TCG TCT TCT CTT GCG. These oligonucleotides incorporated BamHI and EcoRI restriction sites, respectively, which facilitated the cloning of the PCR product into pGEX-2T, to yield pGST73NLSΔ1, pGST73NLSΔ2, and pGST73NLSΔ1+2.
pSV40-CAT and pRSV-CAT, containing the simian virus 40 and Rous sarcoma virus minimal promoters upstream of the chloramphericol acetyltransferase (CAT) reporter genes, were kindly provided by G. E. Blair, University of Leeds. The ORF 73 promoter-CAT reporter construct has been previously described (23). The p32 eukaryotic expression vector was produced by amplification of the complete p32 coding region using the forward and reverse primers 5′-GCA GAT CTA TGC TGC CTC TGC TGC GCT GCG TG and 5′-GTG AAT TCT ACT GGC TCT TGA CAA AAC TCT, respectively. These oligonucleotides incorporated BglII and EcoRI restriction sites, respectively, to facilitate cloning into pIRESGFP in order to derive p32IRESGFP.
GST pull-down assays.
The ORF 73 deletion series were expressed as glutathione S-transferase (GST) fusion proteins in E. coli DH5α. A fresh overnight culture of transformed E. coli was diluted 1:20 with Luria-Bertari medium containing ampicillin (100 μg/ml). After growth at 37°C for 2 h, the culture was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and grown at 37°C for a further 4 h. The cells were harvested by centrifugation and resuspended in 0.1 volume of lysis buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Triton X-100). They were then sonicated and stored on ice for 30 min, and cellular debris was pelleted. The recombinant protein was purified from crude lysates by incubation with glutathione-Sepharose 4B affinity beads as specified by the manufacturer (Pharmacia Biotech). The protein-bound beads were then incubated for 16 h at 4°C with Cos-7 cell extracts previously lysed with lysis buffer (0.3 M NaCl, 50 mM Tris-HCl [pH 8.0], 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS]) containing protease inhibitors (AEBSF; Roche). The beads were then pelleted, washed four times in lysis buffer, and resuspended in Laemmli buffer, and the precipitated polypeptides were resolved on an SDS-12% polyacrylamide gel. The proteins were then transferred to nitrocellulose membranes by electroblotting and probed by immunoblot analysis.
Coimmunoprecipitation assays.
OMK cells were left uninfected, transfected using 2 μg of the appropriate DNAs, or infected with HVS at multiplicity of infection of 1. After 48 h, the cells were harvested and lysed with lysis buffer (0.3 M NaCl, 1% Triton X-100, 50 mM and HEPES buffer [pH 8.0] containing the protease inhibitors leupeptin and phenylmethylsulfonyl fluoride). For each immunoprecipitation 50 μl of the ORF 73 polyclonal antiserum (24) was incubated with protein A-Sepharose beads (Pharmacia Biotech) for 16 h at 4°C. The beads were then pelleted, washed, and incubated with each respective cell lysate for 16 h at 4°C. The beads were then pelleted and washed, and precipitated polypeptides were resolved on an SDS-12% polyacrylamide gel and analysed by immunoblot analysis.
p32 and glucose oxidase column pull-down assays.
Bacterially expressed p32 was purified by charge affinity chromatography and coupled to activated Sepharose to generate p32 columns as previously described (35). A control matrix was prepared by the same method using 5 mg of purified glucose oxidase (GO) (Sigma). Pull-down assays were performed with 50 μl of p32 or GO column material and 200 μl of infected or transfected cell extract as previously described (35). After being washed, bound proteins were resuspended in Laemmli buffer and precipitated polypeptides were resolved on an SDS-12% polyacrylamide gel. The proteins were then transferred to nitrocellulose membranes by electroblotting and probed by immunoblot analysis.
Immunoblot analysis.
Immunoblot analysis was performed with the pull-down samples. Precipitated polypeptides were resolved on an SDS-12% polyacrylamide gel and then soaked for 10 min in transfer buffer (25 mM Tris, 192 mM glycine, 20% [vol/vol] methanol, 0.1% SDS). The proteins were transferred to nitrocellulose membranes by electroblotting for 3 h at 250 mA. After the transfer, the membranes were soaked in phosphate-buffered saline (PBS) and blocked by preincubation with 2% (wt/vol) nonfat milk powder for 2 h at 37°C. The membranes were then incubated with either a 1:400 dilution of affinity-purified p32 polyclonal antiserum (35), a 1:500 dilution of monoclonal p32 antiserum (11), a 1:1,000 dilution of green fluorescent protein monoclonal antiserum (Clontech), or a 1:2,000 dilution of GST monoclonal antiserum (Sigma), washed with PBS, and incubated for 1 h at 37°C with a 1:1,000 dilution of anti-rabbit or anti-mouse immunoglobulin conjugated with horseradish peroxidase (Dako) in blocking buffer. After five washes with PBS, the nitrocellulose membranes were developed using enhanced chemiluminescence (Pierce) as specified by the manufacturer.
Immunofluorescence analysis.
Cells were fixed with 4% formaldehyde in PBS, washed in PBS, and permeabilized in 0.5% Triton X-100 for 5 min. The cells were rinsed in PBS and blocked by preincubation with 1% (wt/vol) nonfat milk powder for 1 h at 37°C. A 1:100 dilution of goat p32 polyclonal antiserum (Santa Cruz), was layered over the cells, which were then incubated for 1 h at 37°C. Texas red-conjugated anti-goat immunoglobulin (Dako) (1:200 dilution) was then added for 1 h at 37°C. After each incubation step, the cells were washed extensively with PBS. The p73GFP deletion series contained a carboxy-terminal green fluorescent protein fusion tag, which allowed direct visualization of these proteins, as previously described (24). Samples were visualized using a Leica TCS SP confocal microscope with a PlanApo 100× UV oil immersion lens, collecting data from the three channels sequentially.
CAT assay.
Cell extracts were prepared 48 h after transfection and incubated with [14C]chloramphenicol in the presence of acetyl coenzyme A as described previously (21). The percent acetylation of chloramphenicol was quantified using a Bio-Rad FX molecular phosphorimager.
RESULTS
p32 interacts with the HVS ORF 73 gene product.
To identify ORF 73-interacting cellular proteins, 1.5 × 106 independent cDNA clones of a human kidney cDNA library fused to the GAL4 activation domain were screened. A total of 48 clones were identified which activated histidine and β-galactosidase reporter gene expression in the presence of the ORF 73-DBD fusion protein. The specificity of the interaction was confirmed by transforming the putative ORF 73-interacting cellular clones into yeast strains harboring no plasmid, yeast containing pGBT9 vector only, yeast containing pLAM5 (a GAL4 human lamin C fusion), or pDBD73. Only library plasmids demonstrating a requirement of pDBD73 for induced expression of histidine and β-galactosidase reporter genes were considered further. The sequences of clones that fulfilled all these criteria were determined and searched against the EMBL/GenBank database using the Blast tool. Analysis revealed that four ORF 73-interacting clones corresponded to the cellular protein p32.
p32 and HVS ORF73 interact in vitro.
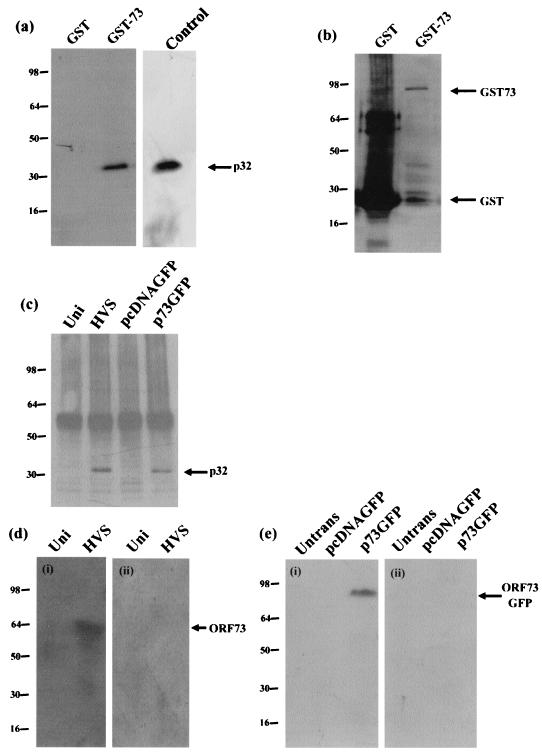
To confirm whether this interaction could also be observed in vitro, GST pull-down experiments were performed. The full-length ORF 73 coding region was cloned into the expression vector pGEX4T-3, allowing expression of the ORF 73 product as a GST fusion protein. Protein extracts prepared from untransfected Cos-7 cells were incubated with GST alone or ORF73GST protein bound to glutathione-Sepharose beads. Bound proteins were then separated by SDS-polyacrylamide gel electrophoresis (PAGE), and the presence of p32 was detected by Western blotting using a p32 polyclonal antibody (35) (Fig. 1a). The expression of the GST-ORF 73 fusion protein was confirmed by Western blotting with a mouse monoclonal anti-GST antibody (Fig. 1b). The results demonstrated that p32 bound to the ORF 73-GST fusion protein but not GST alone. In addition, a control lane of whole-cell lysate was used to estimate that approximately 50% of cellular p32 interacts with GST-ORF 73.
FIG. 1.
The ORF 73 protein interacts with the cellular protein p32. (a) Control GST alone and GST-ORF 73 were expressed in E. coli and purified from crude lysate by incubation with glutathione-Sepharose 4B affinity beads. Protein extracts of untransfected cells were incubated with GST or GST-ORF 73 proteins. Bound proteins were resolved by SDS-PAGE (12% polyacrylamide), and p32 was detected by Western blotting using p32 antiserum. A control lane of whole-cell lysate is shown to estimate the level of interaction between p32 and GST-73. (b) To confirm the expression of GST and GST-ORF 73 proteins, Western blot analysis was performed using a mouse monoclonal GST antibody. (c) Mock-infected (Uni), HVS-infected, pcDNAGFP-transfected, or p73GFP-transfected cell extracts were incubated with ORF 73 polyclonal antiserum bound to protein A-Sepharose beads. The beads were then pelleted and washed, and precipitated polypeptides were resolved on an SDS-12% polyacrylamide gel and analyzed by immunoblot analysis with p32 antiserum. (d) Mock-infected OMK (Uni) and HVS-infected cell extracts were incubated with rp32-Sepharose (i) or rGO-Sepharose (ii). After being washed and eluted, the cells were separated by SDS-PAGE and analyzed by Western blotting with ORF 73 polyclonal antisera. (e) Untransfected Cos-7 and pcDNAGFP-transfected or p73GFP-transfected cell extracts were incubated with rp32-Sepharose (i) or rGO-Sepharose (ii). After being washed and eluted, the cells were separated by SDS-PAGE and analyzed by Western blotting with GFP monoclonal antisera.
In addition, to confirm the GST pull-down analysis, coimmunoprecipitation studies were performed. Control untransfected OMK cells were compared with cells transfected with p73GFP or HVS-infected cells (multiplicity of infection, 1). After 24 h, the cells were harvested and cell lysates were utilized in coimmunoprecipitation analysis with an ORF 73 polyclonal antiserum (24). Polypeptides precipitated from untransfected, transfected, and infected cellular extracts were then resolved by SDS-PAGE and transferred to a nitrocellulose membrane. Immunoblot detection was performed using antiserum specific for p32. The results demonstrate that in both ORF 73-transfected and HVS-infected cells, ORF 73 specifically interacts with p32 (Fig. 1c).
HVS ORF 73 interacts with rMp32-Sepharose.
Potentially the interaction between cellular p32 and bacterially expressed GST-ORF 73 fusion proteins could be mediated by minor bacterial contaminants from GST-purified proteins or could be a result of abnormal folding of the ORF 73 fusion protein. Therefore we used bacterially expressed p32 (which is purified by a different protocol from that of GST-ORF 73) to affinity purify ORF 73 expressed in ORF 73-transfected cells or from HVS-infected cells. Recombinant p32 (35) coupled to Sepharose beads was incubated with extracts from mock-infected or HVS-infected OMK cells, untransfected Cos-7 cells, or cells transfected with pcDNAGFP or p73GFP. After a washing step, bound proteins were eluted from the Sepharose beads and separated by SDS-PAGE. Western blotting revealed that ORF 73 was bound to recombinant p32 in both infected (Fig. 1d, i) and p73GFP-transfected (Fig. 1e, i) cells. The p32 protein is highly acidic with a strong net negative charge (pI 4.2). Potentially the interaction between p32 and ORF 73 could be the result of an indiscriminate charge-based interaction. To determine whether the interaction was specific, a protein with a similar pI to p32, GO, was used in similar experiments. Recombinant GO coupled to Sepharose beads was incubated with extracts from mock-infected, HVS-infected, or p73GFP-transfected cells. Western blotting revealed no interaction between ORF 73 and recombinant GO in HVS-infected (Fig. 1d, ii) or ORF 73-transfected (Fig. 1e, ii) cells. This suggests that the interaction between HVS ORF 73 and p32 is not indiscriminate and that the proteins can also bind in the absence of any other viral proteins.
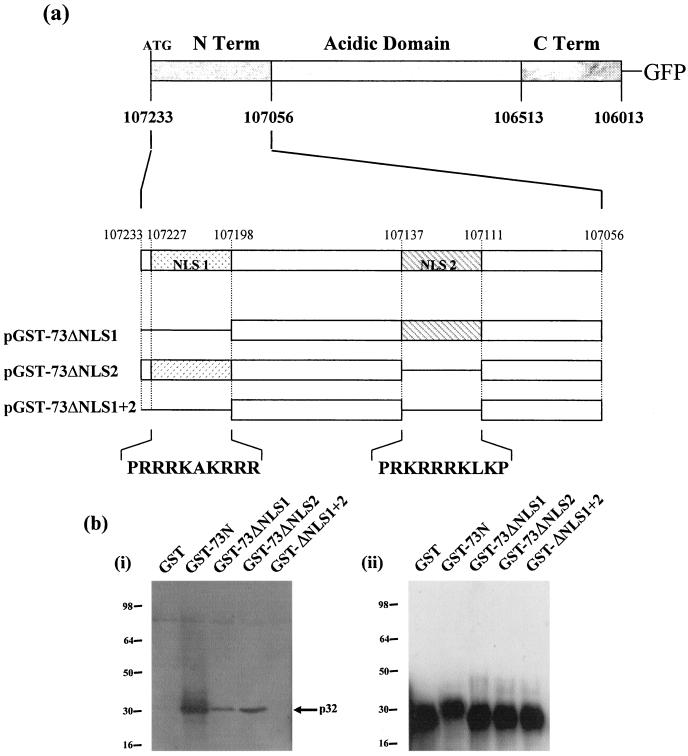
The ORF 73 amino-terminal arginine-rich domain is required for p32 binding.
To map the domain within ORF 73 required for its specific interaction with p32, a series of ORF 73 deletions were expressed in yeast as fusions with GAL4-DBD (Fig. 2a). Competent yeast strain HF7c was cotransformed with p32AD, pDBD73Δ1, pDBD73Δ2, and pDBD73Δ3 and assessed for the ability to grow on selective medium and activate β-galactosidase. The results indicated that the amino terminus of ORF 73 was sufficient for its interaction with p32 (Fig. 2a).
FIG. 2.
The ORF 73 amino terminus is sufficient for p32 binding. (a) A series of ORF 73 deletions were inserted into pGBT9. Each deletion plasmid was cotransformed into HF7c with p32AD and assessed for the ability to grow on selective medium and activiate β-galactosidase expression. +, results of a positive interaction; −, no interaction. (b) Control GST alone, GST-73N, GST-ΔAcidic, and, GST-73C were expressed in E. coli and purified from crude lysate by incubation with glutathione-Sepharose 4B affinity beads. Protein extracts of untransfected cells were incubated with GST or GST-ORF 73 deletion proteins. (i) Bound proteins were resolved by SDS-PAGE (12% polyacrylamide), and p32 was detected by Western blotting using p32 antiserum. (ii) To confirm the expression of GST and GST-ORF 73 deletion fusion proteins, Western blot analysis was performed using a mouse monoclonal GST antibody.
To determine whether the ORF 73 amino terminus was required for the interaction with p32, a GST pull-down analysis was performed. The amino and carboxy termini of ORF 73 were expressed as GST fusion proteins. Protein extracts prepared from untransfected Cos-7 cells were incubated with GST alone or GST-ORF73 deletion proteins bound to glutathione-Sepharose beads. Bound proteins were then separated by SDS-PAGE, and the presence of p32 was detected by Western blotting using a p32 polyclonal antibody (Fig. 2b, i). As a control, the expression of the GST-ORF 73 deletion fusion proteins was confirmed by Western blotting with a mouse monoclonal anti-GST antibody (Fig. 2b, ii). Results demonstrated that p32 specifically bound to the ORF 73 amino terminus, confirming the analysis in the yeast two-hybrid assay.
To delineate the sequences within the ORF 73 amino terminus required for p32 binding, further amino-terminal deletions were expressed as GST fusion proteins (Fig. 3a). Since it is thought that arginine- and lysine-rich regions of the adenovirus minor core protein V are responsible for mediating an interaction with p32 (D. A. Matthews, unpublished observations), we speculated that the interaction between HVS ORF 73 and p32 may be mediated by one or both of the arginine-rich nuclear localization signal (NLS) regions present in the HVS ORF 73 amino terminus (24). Therefore, amino-terminal mutant proteins lacking either one or both of the NLS regions were expressed as GST fusion proteins and pull-down analysis was performed using untransfected Cos-7 cellular lysates. Western blotting with a p32 polyclonal antibody indicated that the complete amino-terminal region showed a strong interaction with p32, as described above. Moreover, results demonstrate that deletion of either amino-terminal arginine-rich region reduced p32 binding to approximately 30% of the levels observed using the complete ORF 73 amino terminus. Furthermore, deletion of both arginine-rich domains completely abolished p32 binding (Fig. 3b, i). As a control, expression of the GST-ORF 73 deletion fusion proteins was confirmed by Western blotting with a mouse monoclonal anti-GST antibody (Fig. 3b, ii). This suggests that both amino-terminal arginine-rich domains are required for the specific interaction with p32.
FIG. 3.
The ORF 73 amino-terminal arginine-rich domains are required for p32 binding. (a) A further series of amino-terminal ORF 73 deletions were expressed as GST fusion proteins. (b) Control GST alone, GST-73N, GST-73ΔNLS1, GST-73ΔNLS2, and GST-73ΔNLS1+2 were expressed in E. coli and purified from crude lysate by incubation with glutathione-Sepharose 4B affinity beads. Protein extracts of untransfected cells were incubated with GST or GST-ORF 73 amino deletion proteins. (i) Bound proteins were resolved by SDS-PAGE (12% polyacrylamide), and p32 was detected by Western blotting using p32 antiserum. (ii) To confirm the expression of GST and GST-ORF 73 amino deletion fusion proteins, Western blot analysis was performed using a mouse monoclonal GST antibody.
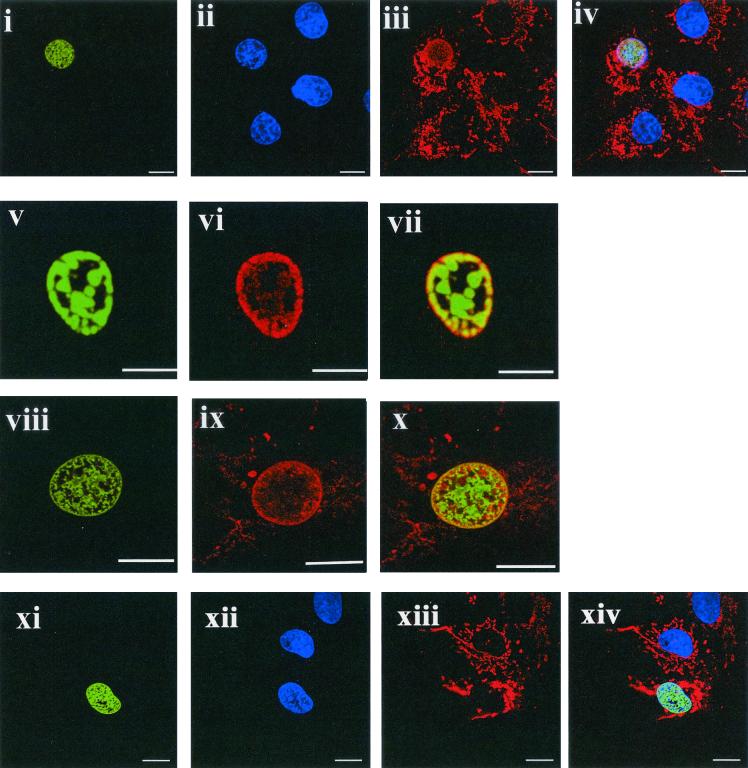
HVS ORF 73 triggers an accumulation of p32 in the nucleus.
p32 is localized predominantly to the mitochondria of many different cell types but has also recently been observed in the nucleus at low levels (9, 44). To ascertain the effect of ORF 73 expression on the subcellular localization of p32, indirect immunofluorescence was performed. p32 subcellular localization was compared in mock- or HVS-infected cells and Cos-7 cells transfected with a range of ORF 73-GFP deletion expression constructs (24). In both untransfected and pcDNAGFP-transfected cells, p32 was predominantly cytoplasmic in a mitochondrial pattern as previously described (9, 16, 35, 36, 47). In contrast, p73GFP-transfected cells (Fig. 4i to vii) and HVS-infected cells (Fig. 4viii to x) demonstrated an accumulation of p32 in the nucleus. Results suggested that the accumulation of p32 in the nucleus might be dose dependent, whereby more p32 colocalized with ORF 73 in cells that were expressing ORF 73 to higher levels (data not shown). We noted that there is some degree of similarity in the localization of ORF 73 and p32, particularly in ORF 73-GFP-transfected cells at the nuclear periphery (Fig. 4v and vii). However, in the infected cells this is less obvious (Fig. 4viii and x); potentially this slight difference may be a result of the presence of other virus proteins in the infected cells. Similar p32 nuclear accumulation results have also been observed using an ORF 73-myc tagged protein in both Cos-7 and OMK cells (data not shown).
FIG. 4.
The ORF 73 protein accumulates p32 in the nucleus. In all color images, ORF73-GFP or ORF 73 in infected cells stains green, 4; 6-diamidino-2-phenylindole (DAPI) stains blue, and p32 stains red. Images represent a single 0.36-μm focal plane approximately halfway through the cell. Bars, 10 μm. Cells were transfected with p73GFP (i and v), infected with HVS (viii), or transfected with p73ΔNLS1GFP (xi). Each experiment resulted in nuclear localization of ORF73 (or the GFP fusion protein), as shown by DAPI costaining (ii and xii). For infected cells, ORF73 was detected by ORF73 polyclonal antiserum and an anti-rabbit fluorescein thiocyanate conjugate (viii). In all cases, p32 was detected using an p32 antiserum (Santa Cruz) and an anti-goat Texas red conjugate (iii, vi, ix, and xiii). Images iv, vii, x, and xiv are an overlay of the three channels (i to iii), (v to vi), (viii to ix), and (xi to xiii), respectively.
In addition, to determine whether the amino-terminal arginine-rich domains were required for p32 nuclear accumulation, p32 redistribution was assessed in p73NLSΔ1-transfected cells (Fig. 4xi to xiv) and p73NLSΔ2-transfected cells (data not shown). Results demonstrate that when either one of the arginine-rich domains was removed, p32 did not accumulate in the nucleus. This indicates that both ORF 73 amino-terminal arginine-rich domains, which are required for p32 binding, are necessary for the accumulation of p32 in the nucleus.
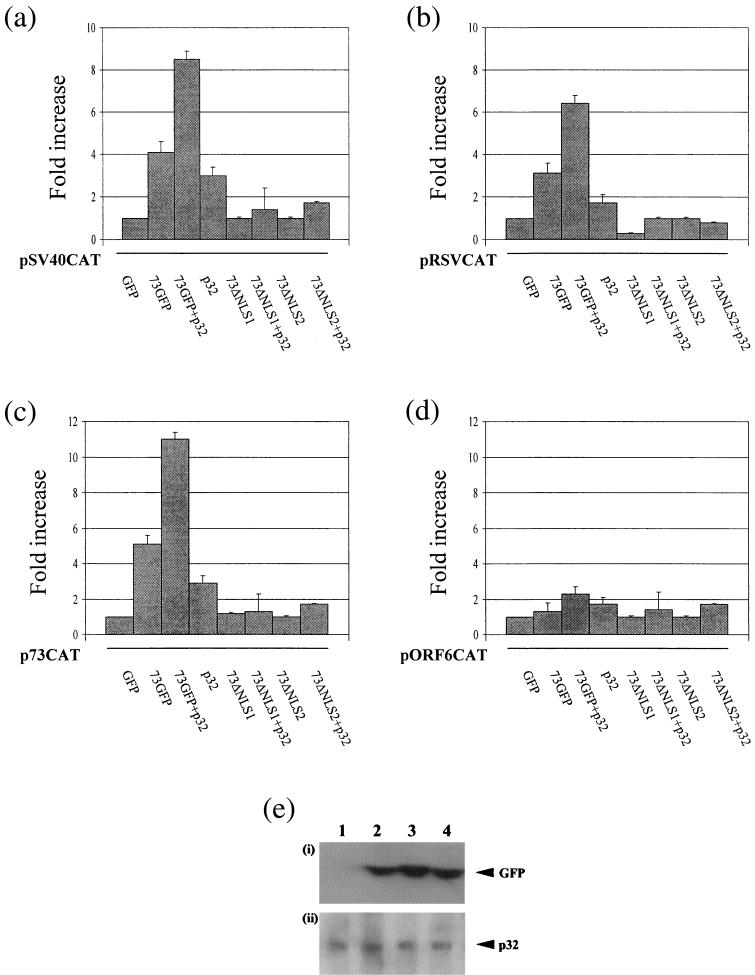
The p32-interacting domains of ORF 73 are required for ORF 73-dependent transactivation.
It has previously been shown that p32 interacts with the EBV EBNA-1 protein (13, 52, 54). p32 has been implicated in the activation of transcription by EBNA-1 or in EBV genome replication and/or maintenance. To determine whether p32 plays a role in ORF 73-mediated transactivation, we first confirmed whether ORF 73 augments transcription from a set of reporter plasmids. A series of transient-transfection assays were performed to assess the effect of the ORF 73 protein on the heterologous simian virus 40 and Rous sarcoma virus promoters, driving expression of the CAT reporter gene. Cos-7 cells were transiently transfected with either pSV40CAT or pRSVCAT in the absence or presence of pcDNAGFP or p73GFP (24). They were harvested at 48 h posttransfection, and the cell extracts were assayed for CAT activity. The results shown in Fig. 5 indicate that ORF 73 has the ability to transactivate both heterologous promoter constructs approximately three- to fourfold (Fig. 5), suggesting that ORF 73 can function as a transcriptional activator. Furthermore, the coexpression of p32 and ORF 73 resulted in a six- to eightfold increase in CAT activity, suggesting that p32 can cooperate with ORF 73 to stimulate transcription synergistically, since limited activation was observed with p32 alone. To confirm that p32 is implicated in ORF 73-dependent transactivation, the transient-transfection experiments were repeated with ORF 73 mutants lacking the amino-terminal arginine-rich p32-binding site. Transient transfections were repeated in the absence or presence of p73NLSΔ1GFP or p73NLSΔ2GFP. The results demonstrated that deletion of the amino-terminal arginine-rich p32-interacting domains abrogated the ability of ORF 73 to activate the heterologous promoters (Fig. 5). As a control, the transfection efficiency was monitored by calculating the number of GFP-expressing cells. In addition, equivalent expression of p73GFP, ORF 73 deletions, and p32 was confirmed by Western blot analysis (Fig. 5e). The results demonstrate that the wild-type ORF 73 construct was expressed to levels similar to those of the p32-binding domain deletions. Therefore, these results suggest that p32 binding to ORF 73 is required for ORF 73-dependent transactivation.
FIG. 5.
The p32-binding domain is required for ORF73-dependent transactivation. Cos-7 cells were transfected with pSV40CAT (a), pRSVCAT (b), p73Δ4 (c), or pAWCAT1 (d) in the presence of pGFP, p73GFP, p73GFP+p32, p32, p73ΔNLS1, p73ΔNLS1+p32, p73ΔNLS2, or p73ΔNLS2+p32. The cells were harvested at 48 h posttransfection, and cell extracts were assayed for CAT activity. Results are shown in graphical form, where CAT activity is presented as fold induction over background levels. The variations between two replicated assays are indicated. (e) (i) Cos-7 cell extracts, untransfected (lane 1) or transfected with p73GFP (lane 2), p73ΔNLS1 (lane 3), or p73ΔNLS2 (lane 4), were resolved by SDS-PAGE, and GFP was detected by Western blotting using GFP antiserum. (ii) Cos-7 cell extracts transfected with p73GFP (lane 1), p73GFP+p32 (lane 2), p73ΔNLS1+p32 (lane 3), or p73ΔNLS2+p32 (lane 4) were resolved by SDS-PAGE, and p32 was detected by Western blotting using p32 antiserum.
To elucidate whether p32 binding is required for ORF 73-mediated transactivation of any HVS promoters, we determined whether ORF 73 regulated its own viral promoter or an ORF 50-responsive DE ORF 6 promoter. Recent analysis has demonstrated that LANA positively autoregulates its own promoter (27, 43). A series of transient-transfection assays were performed to assess the effect of the ORF 73 protein on p73Δ4, a plasmid consisting of an ORF 73 639-bp promoter fragment upstream of the CAT reporter gene (23), and pAWCAT1, a plasmid containing the DE ORF 6 promoter upstream of CAT (55). Cos-7 cells were transiently transfected with either p73Δ4CAT or pAWCAT1 in the absence or presence of pcDNAGFP, p73GFP, and p32IRESGFP. Cells were harvested at 48 h posttransfection, and the cell extracts were assayed for CAT activity. The results demonstrate that ORF 73 has the ability to positively autoregulate its promoter approximately fivefold. Moreover, the coexpression of ORF 73 and p32 has a synergistic effect on the transactivation of the ORF 73 promoter, as observed with the heterologous promoters (Fig. 5c). Also, deletion of the amino-terminal arginine-rich p32-interacting domains abrogated the ability of ORF 73 to activate its own promoter. In contrast, neither ORF 73 nor p32 has any effect on the transcriptional upregulation of the DE ORF 6-ORF 50-responsive promoter. This suggests that the ORF 73-p32 interaction does not function as a general transcriptional transactivator. Thus, ORF73 affects a specific subset of promoters and its interaction with p32 augments these effects.
DISCUSSION
In this study we have utilized the yeast two-hybrid system and in vitro pull-down analysis to demonstrate that HVS ORF 73 interacts with the host cellular protein p32. Our data also indicate that the association between p32 and ORF 73 is charge based since p32 is highly acidic and the binding requires basic regions on ORF 73. This charge-based interaction is not indiscriminate, however, since ORF 73 does not interact with a similarly charged acidic protein, GO.
Also known as SF2-associated protein, TAP, or gC1qR, p32 was originally isolated from HeLa cells by copurification with the nuclear pre-mRNA splicing factor ASF/SF2 (32). The protein is synthesized as a pro-protein of 282 amino acids that resides in the cytoplasm before undergoing cleavage of amino acids 1 to 73 to generate the mature form of the protein, which is located in the mitochondria (25). However, recent analysis has shown that p32 accumulates in the nuclei of transfected cells after treatment with actinomycin D, leptomycin B, or TPA, suggesting that p32 can shuttle between the mitochondria and the nucleus (9, 44).
Here we show that expression of ORF 73 is responsible for the accumulation of p32 in the nucleus. Similar observations have been demonstrated for other p32-interacting viral proteins, including EBV EBNA-1, HSV-1 ICP27, and adenovirus protein V (11, 35, 52, 54). At present, the mechanism of p32 nuclear redistribution is unknown. Potentially, viral proteins bind to nascent p32 in the cytoplasm before p32 interacts with the mitochondrial protein import machinery. This might augment the levels of extra mitochondrial p32 available for nuclear-cytoplasmic shuttling. Indeed, ORF 73 may also increase the rate of nuclear import of p32. For example, p32 may bind ORF 73 protein at one of the NLS regions and the complex may be imported via the second NLS on ORF 73. On the other hand, the accumulation of p32 in the nucleus might result from a failure of nuclear transport pathways required for p32 export. However, mutants of ORF 73 lacking only one NLS are still transported to the nucleus but are unable to trigger nuclear accumulation of p32. Thus, it is difficult to envisage how reinstatement of the missing second NLS leads to deficient export of p32 from the nucleus. The fluorescent-image data do not demonstrate unequivocally that p32 and ORF 73 exclusively colocalize in either infected or transfected cells, although they appear to be in similar locations. This may indicate that the interaction between p32 and ORF 73 in the nucleus is dynamic or that the p32-ORF 73 complex (perhaps containing additional factors) is not readily detectable in situ by the anti-p32 serum used here.
Although the exact physiological function of p32 remains unknown, it has been shown to be important in maintaining oxidative phosphorylation in yeast mitochondria (36). The highly asymmetric distribution of aspartic and glutamic acids on the surface of the p32 trimer is a feature reminiscent of the Ca2+ storage protein calsequestrin (28). Through its association with the splicing factors ASF/SF2 and SR30pc, p32 has also been implicated in control of gene expression at a posttranscriptional level (38). This specific function of p32 has been implicated in the mechanism by which both HIV-1 Rev and HSV-1 ICP27 inhibit cellular splicing (11, 34, 50).
In addition to its role in splicing, p32 functions as a transcriptional activator and is implicated in the regulation of both HIV-1 and EBV gene expression. HIV-1 Tat associates with p32, and in this context p32 is thought to function as a coactivator of viral gene expression (56, 57). Deletion analysis of p32 identified a highly acidic carboxy terminus that acts as a strong transactivating domain, which is required for the binding of p32 to the general transcription factor, TFIIB (56). Therefore, these results suggest that p32 bridges Tat to the cellular transcription machinery via TFIIB (56). However, deletion mutations of p32 may also have affected the more recently reported ability of p32 to multimerize, which may complicate the interpretation of the data.
EBV EBNA-1 can function as a transactivator of viral latency promoters but lacks a detectable activation domain (26, 40, 48). EBNA-1 interacts with p32 through two basic arginine-glycine-rich domains and is a possible coactivator of EBNA-1-dependent transactivation (52, 54). Here we show that the HVS ORF 73 protein interacts with p32 via basic regions, ORF 73 can function as a transcriptional activator, and p32 acts synergistically with ORF 73 to upregulate transcription. Moreover, we have demonstrated that deletion of the amino-terminal arginine-rich p32 interacting domain abrogated the ability of ORF 73 to activate the heterologous promoters. This suggests that p32 binding to HVS ORF 73 plays a key role in ORF 73-dependent transactivation. To the best of our knowledge, this is the first observation of HVS ORF 73 protein functioning as a transcriptional activator and suggests some functional similarities to EBNA-1 and KSHV LANA, both of which have transactivating properties. Additionally, our data suggest that ORF 73 activates promoters from other virus families and its own promoter, indicating that there could be a broad range of responsive promoters. However, it does not activate the expression of HVS ORF 6, implying that it is, at least, restricted in what type of HVS promoter it modulates. At present we have only preliminary analysis demonstrating the transactivating capability of HVS ORF 73 on its own regulatory region. The mechanism by which HVS ORF 73 transactivates gene expression remains to be elucidated, and it will be of interest to determine whether p32 bridges ORF 73 to the cellular transcriptional machinery in a similar manner to HIV-1 Tat (56). Experiments are under way to determine if ORF 73 (or ORF 73-p32 complex) binds promoters directly to regulate transcription or if transcription regulation occurs via an indirect method. It is also interesting that reporter gene assays have demonstrated that KSHV LANA can exert a positive or negative effect on gene expression (27, 33, 43). It also remains to be elucidated whether HVS modulates similar viral and cellular gene expression and whether LANA-dependent transactivation is also dependent on an interaction with p32.
In summary, we have demonstrated that the HVS ORF 73 protein functions as a transcriptional activator and interacts with the cellular protein p32, leading to its accumulation in the nucleus. An amino-terminal arginine-rich domain, which contains two distinct NLSs, is required for this interaction. Further analysis suggests a role for p32 in ORF 73-dependent transcriptional activation.
Acknowledgments
We thank the MMU Confocal User Group for assistance with confocal microscopy and G. E. Blair (University of Leeds) for providing the heterologous promoter constructs.
This work was supported in parts by grants to A.W. from Yorkshire Cancer Research, Medical Research Council, and the Wellcome Trust. D.A.M. and M.S.G. are recipients of MRC Non-Clinical and Wellcome Trust Clinical Research Training Fellowships, respectively.
REFERENCES
- 1.Albrecht, J. C., and B. Fleckenstein. 1992. New member of the multigene family of complement control proteins in herpesvirus saimiri. J. Virol. 66:3937-3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballestas, M. E., P. A. Chatis, and K. M. Kaye. 1999. Efficient persistence of extrachromosomal KSHV DNA mediated by latency-associated nuclear antigen. Science 284:641-644. [DOI] [PubMed] [Google Scholar]
- 3.Ballestas, M. E., and K. M. Kaye. 2001. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 75:3250-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beatch, M. D., and T. C. Hobman. 2000. Rubella virus capsid associates with host cell protein p32 and localizes to mitochondria. J. Virol. 74:5569-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berend, K. R., J. U. Jung, T. J. Boyle, J. M. DiMaio, S. A. Mungal, R. C. Desrosiers, and H. K. Lyerly. 1993. Phenotypic and functional consequences of herpesvirus saimiri infection of human CD8+ cytotoxic T lymphocytes. J. Virol. 67:6317-6321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biesinger, B., I. Muller-Fleckenstein, B. Simmer, G. Lang, S. Wittmann, E. Platzer, R. C. Desrosiers, and B. Fleckenstein. 1992. Stable growth transformation of human T lymphocytes by herpesvirus saimiri. Proc. Natl. Acad. Sci. USA 89:3116-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biesinger, B., J. J. Trimble, R. C. Desrosiers, and B. Fleckenstein. 1990. The divergence between two oncogenic Herpesvirus saimiri strains in a genomic region related to the transforming phenotype. Virology 176:505-514. [DOI] [PubMed] [Google Scholar]
- 8.Broker, B. M., A. Y. Tsygankov, I. Muller-Fleckenstein, A. H. Guse, N. A. Chitaev, B. Biesinger, B. Fleckenstein, and F. Emmrich. 1993. Immortalization of human T cell clones by Herpesvirus saimiri. Signal transduction analysis reveals functional CD3, CD4, and IL-2 receptors. J. Immunol. 151:1184-1192. [PubMed] [Google Scholar]
- 9.Brokstad, K. A., K. H. Kalland, W. C. Russell, and D. A. Matthews. 2001. Mitochondrial protein p32 can accumulate in the nucleus. Biochem. Biophys. Res. Commun. 281:1161-1169. [DOI] [PubMed] [Google Scholar]
- 10.Bruni, R., and B. Roizman. 1996. Open reading frame P—a herpes simplex virus gene repressed during productive infection encodes a protein that binds a splicing factor and reduces synthesis of viral proteins made from spliced mRNA. Proc. Natl. Acad. Sci. USA 93:10423-10427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bryant, H. E., D. A. Matthews, S. Wadd, J. E. Scott, J. Kean, S. Graham, W. C. Russell, and J. B. Clements. 2000. Interaction between herpes simplex virus type 1 IE63 protein and cellular protein p32. J. Virol. 74:11322-11328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, Y., P. S. Moore, S. J. Talbot, C. H. Boshoff, T. Zarkowska, K. Godden, H. Paterson, R. A. Weiss, and S. Mittnacht. 1996. Cyclin encoded by KS herpesvirus. Nature 382:410. [DOI] [PubMed] [Google Scholar]
- 13.Chen, M. R., J. F. Yang, C. W. Wu, J. M. Middeldorp, and J. Y. Chen. 1998. Physical association between the EBV protein EBNA-1 and P32/TAP/hyaluronectin. J. Biomed. Sci. 5:173-179. [DOI] [PubMed] [Google Scholar]
- 14.Chien, C. T., P. L. Bartel, R. Sternglanz, and S. Fields. 1991. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc. Natl. Acad. Sci. USA 88:9578-9582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cotter, M. A., Jr., and E. S. Robertson. 1999. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology 264:254-264. [DOI] [PubMed] [Google Scholar]
- 16.Dedio, J., W. Jahnen-Dechent, M. Bachmann, and W. Muller-Esterl. 1998. The multiligand-binding protein gC1qR, putative C1q receptor, is a mitochondrial protein. J. Immunol. 160:3534-3542. [PubMed] [Google Scholar]
- 17.Desrosiers, R. C., D. P. Silva, L. M. Waldron, and N. L. Letvin. 1986. Nononcogenic deletion mutants of herpesvirus saimiri are defective for in vitro immortalization. J. Virol. 57:701-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dittmer, D., M. Lagunoff, R. Renne, K. Staskus, A. Haase, and D. Ganem. 1998. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 72:8309-8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fleckenstein, B., G. W. Bornkamm, C. Mulder, F. J. Werner, M. D. Daniel, L. A. Falk, and H. Delius. 1978. Herpesvirus ateles DNA and its homology with herpesvirus saimiri nucleic acid. J. Virol. 25:361-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Friborg, J., Jr., W. Kong, M. O. Hottiger, and G. J. Nabel. 1999. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature 402:889-894. [DOI] [PubMed] [Google Scholar]
- 21.Gorman, C. M., L. F. Moffat, and B. H. Howard. 1982. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol. Cell. Biol. 2:1044-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grassmann, R., and B. Fleckenstein. 1989. Selectable recombinant herpesvirus saimiri is capable of persisting in a human T-cell line. J. Virol. 63:1818-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hall, K. T., M. S. Giles, D. J. Goodwin, M. A. Calderwood, I. M. Carr, A. J. Stevenson, A. F. Markham, and A. Whitehouse. 2000. Analysis of gene expression in a human cell line stably transduced with herpesvirus saimiri. J. Virol. 74:7331-7337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall, K. T., M. S. Giles, D. J. Goodwin, M. A. Calderwood, A. F. Markham, and A. Whitehouse. 2000. Characterization of the herpesvirus saimiri ORF73 gene product. J. Gen. Virol. 81:2653-2658. [DOI] [PubMed] [Google Scholar]
- 25.Honore, B., P. Madsen, H. H. Rasmussen, J. Vandekerckhove, J. E. Celis, and H. Leffers. 1993. Cloning and expression of a cDNA covering the complete coding region of the P32 subunit of human pre-mRNA splicing factor SF2. Gene 134:283-287. [DOI] [PubMed] [Google Scholar]
- 26.Horner, D., M. Lewis, and P. J. Farrell. 1995. Novel hypotheses for the roles of EBNA-1 and BHRF1 in EBV-related cancers. Intervirology 38:195-205. [DOI] [PubMed] [Google Scholar]
- 27.Jeong, J., J. Papin, and D. Dittmer. 2001. Differential regulation of the overlapping Kaposi's sarcoma-associated herpesvirus vGCR (orf74) and LANA (orf73) promoters. J. Virol. 75:1798-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang, J., Y. Zhang, A. R. Krainer, and R. M. Xu. 1999. Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein. Proc. Natl. Acad. Sci. USA 96:3572-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jung, J. U., M. Stager, and R. C. Desrosiers. 1994. Virus-encoded cyclin. Mol. Cell. Biol. 14:7235-7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kedes, D. H., M. Lagunoff, R. Renne, and D. Ganem. 1997. Identification of the gene encoding the major latency-associated nuclear antigen of the Kaposi's sarcoma-associated herpesvirus. J. Clin. Investig. 100:2606-2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kellam, P., C. Boshoff, D. Whitby, S. Matthews, R. A. Weiss, and S. J. Talbot. 1997. Identification of a major latent nuclear antigen, LNA-1, in the human herpesvirus 8 genome. J. Hum. Virol. 1:19-29. [PubMed] [Google Scholar]
- 32.Krainer, A. R., A. Mayeda, D. Kozak, and G. Binns. 1991. Functional expression of cloned human splicing factor SF2: homology to RNA-binding proteins, U1 70K, and Drosophila splicing regulators. Cell 66:383-394. [DOI] [PubMed] [Google Scholar]
- 33.Krithivas, A., D. B. Young, G. Liao, D. Greene, and S. D. Hayward. 2000. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J. Virol. 74:9637-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luo, Y., H. Yu, and B. M. Peterlin. 1994. Cellular protein modulates effects of human immunodeficiency virus type 1 Rev. J. Virol. 68:3850-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matthews, D. A., and W. C. Russell. 1998. Adenovirus core protein V interacts with p32—a protein which is associated with both the mitochondria and the nucleus. J. Gen. Virol. 79:1677-1685. [DOI] [PubMed] [Google Scholar]
- 36.Muta, T., D. Kang, S. Kitajima, T. Fujiwara, and N. Hamasaki. 1997. p32 protein, a splicing factor 2-associated protein, is localized in mitochondrial matrix and is functionally important in maintaining oxidative phosphorylation. J. Biol. Chem. 272:24363-24370. [DOI] [PubMed] [Google Scholar]
- 37.Neipel, F., J. C. Albrecht, and B. Fleckenstein. 1997. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J. Virol. 71:4187-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petersen-Mahrt, S. K., C. Estmer, C. Ohrmalm, D. A. Matthews, W. C. Russell, and G. Akusjarvi. 1999. The splicing factor-associated protein, p32, regulates RNA splicing by inhibiting ASF/SF2 RNA binding and phosphorylation. EMBO J. 18:1014-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Platt, G. M., G. R. Simpson, S. Mittnacht, and T. F. Schulz. 1999. Latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus interacts with RING3, a homolog of the Drosophila female sterile homeotic (fsh) gene. J. Virol. 73:9789-9795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polvino-Bodnar, M., and P. A. Schaffer. 1992. DNA binding activity is required for EBNA 1-dependent transcriptional activation and DNA replication. Virology 187:591-603. [DOI] [PubMed] [Google Scholar]
- 41.Radkov, S. A., P. Kellam, and C. Boshoff. 2000. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 6:1121-1127. [DOI] [PubMed] [Google Scholar]
- 42.Rainbow, L., G. M. Platt, G. R. Simpson, R. Sarid, S. J. Gao, H. Stoiber, C. S. Herrington, P. S. Moore, and T. F. Schulz. 1997. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 71:5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Renne, R., C. Barry, D. Dittmer, N. Compitello, P. O. Brown, and D. Ganem. 2001. Modulation of cellular and viral gene expression by the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 75:458-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Robles-Flores, M., E. Rendon-Huerta, H. Gonzalez-Aguilar, G. Mendoza-Hernandez, S. Islas, V. Mendoza, M. V. Ponce-Castaneda, L. Gonzalez-Mariscal, and F. Lopez-Casillas. 2002. p32 (gC1qBP) is a general protein kinase C (PKC)-binding protein; interaction and cellular localization of P32-PKC complexes in ray hepatocytes. J. Biol. Chem. 277:5247-5255. [DOI] [PubMed] [Google Scholar]
- 45.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45a.Sarid, R., J. S. Wiezorek, P. S. Moore, and Y. Chang.1999. Characterization and cell cycle regulation of the major Kaposi’s sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J. Virol. 73:1438-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simmer, B., M. Alt, I. Buckreus, S. Berthold, B. Fleckenstein, E. Platzer, and R. Grassmann. 1991. Persistence of selectable herpesvirus saimiri in various human haematopoietic and epithelial cell lines. J. Gen. Virol. 72:1953-1958. [DOI] [PubMed] [Google Scholar]
- 47.Soltys, B. J., D. Kang, and R. S. Gupta. 2000. Localization of P32 protein (gC1q-R) in mitochondria and at specific extramitochondrial locations in normal tissues. Histochem. Cell Biol. 114:245-255. [DOI] [PubMed] [Google Scholar]
- 48.Sugden, B., and N. Warren. 1989. A promoter of Epstein-Barr virus that can function during latent infection can be transactivated by EBNA-1, a viral protein required for viral DNA replication during latent infection. J. Virol. 63:2644-2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Szomolanyi, E., P. Medveczky, and C. Mulder. 1987. In vitro immortalization of marmoset cells with three subgroups of herpesvirus saimiri. J. Virol. 61:3485-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49a.Talbot, S. J., R. A. Weiss, P. Kellam, and C. Boshoff. 1999. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology 257:84-94. [DOI] [PubMed] [Google Scholar]
- 50.Tange, T. O., T. H. Jensen, and J. Kjems. 1996. In vitro interaction between human immunodeficiency virus type 1 Rev protein and splicing factor ASF/SF2-associated protein, p32. J. Biol. Chem. 271:10066-10072. [DOI] [PubMed] [Google Scholar]
- 51.Thome, M., P. Schneider, K. Hofmann, H. Fickenscher, E. Meinl, F. Neipel, C. Mattmann, K. Burns, J. L. Bodmer, M. Schroter, C. Scaffidi, P. H. Krammer, M. E. Peter, and J. Tschopp. 1997. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature 386:517-521. [DOI] [PubMed] [Google Scholar]
- 52.Van Scoy, S., I. Watakabe, A. R. Krainer, and J. Hearing. 2000. Human p32: a coactivator for Epstein-Barr virus nuclear antigen-1-mediated transcriptional activation and possible role in viral latent cycle DNA replication. Virology 275:145-157. [DOI] [PubMed] [Google Scholar]
- 53.Virgin, H. W., IV, P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, Y., J. E. Finan, J. M. Middeldorp, and S. D. Hayward. 1997. P32/TAP, a cellular protein that interacts with EBNA-1 of Epstein-Barr virus. Virology 236:18-29. [DOI] [PubMed] [Google Scholar]
- 55.Whitehouse, A., I. M. Carr, J. C. Griffiths, and D. M. Meredith. 1997. The herpesvirus saimiri ORF50 gene, encoding a transcriptional activator homologous to the Epstein-Barr virus R protein, is transcribed from two distinct promoters of different temporal phases. J. Virol 71:2550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu, L., P. M. Loewenstein, Z. Zhang, and M. Green. 1995. In vitro interaction of the human immunodeficiency virus type 1 Tat transactivator and the general transcription factor TFIIB with the cellular protein TAP. J. Virol. 69:3017-3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu, L., Z. Zhang, P. M. Loewenstein, K. Desai, Q. Tang, D. Mao, J. S. Symington, and M. Green. 1995. Molecular cloning and characterization of a cellular protein that interacts with the human immunodeficiency virus type 1 Tat transactivator and encodes a strong transcriptional activation domain. J. Virol. 69:3007-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]