Abstract
ZEBRA protein converts Epstein-Barr virus (EBV) infection from the latent to the lytic state. The ability of ZEBRA to activate this switch is strictly dependent on the presence of serine or threonine at residue 186 of the protein (A. Francis, T. Ragoczy, L. Gradoville, A. El-Guindy, and G. Miller, J. Virol. 72:4543-4551, 1999). We investigated whether phosphorylation of ZEBRA protein at this site by a serine-threonine protein kinase was required for activation of an early lytic cycle viral gene, BMRF1, as a marker of disruption of latency. Previous studies suggested that phosphorylation of ZEBRA at S186 by protein kinase C (PKC) activated the protein (M. Baumann, H. Mischak, S. Dammeier, W. Kolch, O. Gires, D. Pich, R. Zeidler, H. J. Delecluse, and W. Hammerschmidt, J. Virol 72:8105-8114, 1998). Two residues of ZEBRA, T159 and S186, which fit the consensus for phosphorylation by PKC, were phosphorylated in vitro by this enzyme. Several isoforms of PKC (α, β1, β2, γ, δ, and ɛ) phosphorylated ZEBRA. All isoforms that phosphorylated ZEBRA in vitro were blocked by bisindolylmaleimide I, a specific inhibitor of PKC. Studies in cell culture showed that phosphorylation of T159 was not required for disruption of latency in vivo, since the T159A mutant was fully functional. Moreover, the PKC inhibitor did not block the ability of ZEBRA expressed from a transfected plasmid to activate the BMRF1 downstream gene. Of greatest importance, in vivo labeling with [32P]orthophosphate showed that the tryptic phosphopeptide maps of wild-type ZEBRA, Z(S186A), and the double mutant Z(T159A/S186A) were identical. Although ZEBRA is a potential target for PKC, in the absence of PKC agonists, ZEBRA is not constitutively phosphorylated in vivo by PKC at T159 or S186. Phosphorylation of ZEBRA by PKC is not essential for the protein to disrupt EBV latency.
ZEBRA, the protein product of the Epstein-Barr virus (EBV) BZLF1 gene, is a master switch between the latent and lytic life cycles of the virus (10, 11). ZEBRA is a member of the basic zipper (bZIP) group of transcriptional activators whose mammalian cellular members include c-Fos and c-Jun (14). ZEBRA and c-Fos/c-Jun both bind to an AP-1 site (TGAGTCA) (7). However, binding of this site is not sufficient for ZEBRA to disrupt latency. Chimeric proteins in which ZEBRA's basic DNA recognition region is replaced by that of c-Fos retain the ability to bind and activate transcription via AP-1 sites but are unable to disrupt EBV latency (24).
The crystal structure of c-Fos/c-Jun bound to an AP-1 site revealed that five amino acids within the basic domains of these proteins made direct contact with DNA (18). Four of these five residues are colinear in ZEBRA's basic domain. The fifth, a serine at position 186 in ZEBRA, is an alanine in c-Fos and c-Jun. These findings provoked the hypothesis that S186 of ZEBRA played a crucial role in the disruption of EBV latency.
The ZEBRA mutant Z(S186A) can bind to DNA and can activate transcription from viral early lytic cycle promoters present in reporter plasmid constructs but fails to disrupt latency from EBV itself (15, 16). The main defect in Z(S186A) is its inability to activate transcription of the viral gene BRLF1, which encodes a second transactivator, Rta (2, 16). ZEBRA and Rta collaborate to drive the expression of many downstream viral target genes (8, 17, 20, 22, 23, 31). The Z(S186A) mutant can be rescued by concomitant overexpression of Rta (2, 15). While the Z(S186A) mutant is deficient in its capacity to activate the promoter of BRLF1, it is fully competent to synergize with Rta on the promoter of an early gene, BMRF1, encoding the DNA polymerase processivity factor. BMRF1 is regulated by the combinatorial action of ZEBRA and Rta (15). Clues to the possible function of Z(S186) came from analysis of different amino acid substitutions at this position. Only a threonine substitution at S186 maintained the capacity of ZEBRA to disrupt latency. ZEBRA proteins with glycine, valine, or cysteine substitutions at S186 failed to activate transcription, although they were able to bind DNA (15). ZEBRA mutants with these other amino acid substitutions could not be rescued by Rta.
Since only serine or threonine at position 186 allowed the ZEBRA protein to activate early gene expression, a reasonable hypothesis was that S186 or S186T needed to be phosphorylated in order for ZEBRA to function. A corollary of this hypothesis was that phosphorylation of serine or threonine at amino acid 186 was not required for ZEBRA to synergize with Rta in the activation of downstream genes, since the Z(S186A) mutant, together with Rta supplied in trans, activated the early BMRF1 gene to wild-type levels (2, 15).
Considerable support existed for the idea that ZEBRA required phosphorylation at S186 in order to carry out its functions. ZEBRA is known to be a phosphoprotein in vivo (12). S186 was surrounded by basic residues which constitute a consensus sequence for phosphorylation by protein kinase C (PKC) (35). ZEBRA can be phosphorylated in vitro by PKC α, and mutation of residue S186 to alanine decreased in vitro phosphorylation by PKC (4). Baumann et al. reported that the phorbol ester tetradecanoyl phorbol acetate (TPA) markedly enhanced ZEBRA's ability to activate transcription (4). Since TPA is a potent PKC agonist, the implication of this result was that PKC-mediated phosphorylation of ZEBRA enhanced its behavior as a transcriptional activator. Support for the idea of PKC-mediated phosphorylation of ZEBRA came from the demonstration that TPA treatment of 293 cells in which ZEBRA was expressed induced the appearance of two additional ZEBRA phosphopeptides. These additional TPA-induced ZEBRA phosphopeptides were not present in cells transfected with the Z(S186A) mutant.
Our report addresses the question of whether phosphorylation of ZEBRA by PKC is obligatory for the protein to function in disruption of latency. More specifically, the experiments ask whether lack of phosphorylation of S186 by PKC accounts for the phenotype of the Z(S186A) mutant. We provide two lines of evidence indicating that phosphorylation of ZEBRA by PKC is not obligatory for its activity in the disruption of latency. A potent inhibitor of PKC that abolishes the ability of several isoforms of the enzyme to phosphorylate ZEBRA in vitro does not block the ability of ZEBRA to disrupt latency. The most compelling piece of evidence is the identical patterns of tryptic phosphopeptides of wild-type ZEBRA and ZEBRA mutated at its two PKC sites. Therefore, lack of phosphorylation by PKC does not appear to explain the phenotype of the ZEBRA S186A mutant.
MATERIALS AND METHODS
Cell lines and transient transfections.
Raji, an EBV-positive Burkitt's lymphoma cell line that is defective in lytic viral DNA replication (39), was maintained in RPMI 1640 medium containing 8% fetal bovine serum and antibiotics. For transient gene expression, 107 Raji cells were transfected with 10 μg of expression vector by electroporation (16).
The HKB5/Cl8 cell line (HKB for hybrid kidney B cell), a kind gift from Myung-Sam Cho, was generated by fusing the HH514-16 Burkitt lymphoma cell line with the 293 human embryonic kidney cell line. HKB5/Cl8 cells carry the EBV genome of HH514-16 cells. They were maintained in RPMI 1640 medium with 5% fetal bovine serum and antibiotics. The optimum transfection conditions of HKB5/Cl8 cells were achieved by using the DMRIE-C reagent (Invitrogen) and following the manufacturer's protocol. Briefly, 4 μg of plasmid DNA was mixed with 12 μl of DMRIE-C reagent and 1 ml of OPTI-MEM medium. The DNA and the DMRIE-C were left for 45 min at room temperature to form a complex; 2 × 106 HKB5/C18 cells in 0.2 ml of OPTI-MEM medium were added to the DNA-DMRIE-C complex and incubated for 5 h at 37°C in 5% CO2. After transfection, 2 ml of RPMI 1640 with 15% fetal calf serum was added to the cells.
ZEBRA and Rta expression systems.
EBV DNA from nucleotides 103181 to 102115, which encode genomic ZEBRA, was cloned into the pHD1013 plasmid under the control of the cytomegalovirus (CMV) immediate-early promoter (13). The Rta and Z(S186A) expression vectors were described previously (16, 31).
Additional point mutations in ZEBRA were generated by using the “Quick Change” site-directed mutagenesis system (Stratagene). pHD1013/genomic ZEBRA served as a template for eukaryotic expression vectors, and pET-22b/cDNA ZEBRA served as a template for bacterial expression constructs. The double mutant Z(T159A/S186A) was generated by using pHD1013/gZ(S186A) or pET-22b/cDNA Z(S186A) as a template. Mutations at position 186 were generated using the following mutagenic primer and its complementary strand: 5′-GAATCGGGTGGCTXXXAGAAAATGCCGG-3′, where XXX bases were changed to GCC to generate alanine, GAC to generate aspartate, or GAA to generate glutamate. To replace T159 with alanine, the mutagenic primer 5′-CCGGCACGACGCGCACGGAAACCACAACAGCC-3′ and its complementary strand were used. The alanine codon is underlined. All PCRs were carried out using Pfu turbo DNA polymerase; the parental DNA was digested with DpnI to select for the mutated DNA. The expression vectors encoding Z(T159A), Z(S186D), Z(S186E), and Z(T159A/S186A) were sequenced and were found to be identical to wild-type ZEBRA except where the point mutations were installed.
ZEBRA protein expression in E. coli and purification.
Escherichia coli (BL21) cells were transformed with pET22b expression vectors containing the cDNAs for wild-type ZEBRA, Z(T159A), Z(S186A), and Z(T159A/S186A). The bacteria were plated on Luria-Bertani (LB) agarose plates containing 100 μg of ampicillin/ml. A colony was used to inoculate 50 ml of LB medium plus ampicillin. After 3 h at 37°C with shaking, the culture was transferred to 450 ml of LB medium plus ampicillin and was grown to an optical density of 0.7 at 600 nm. Protein expression was induced by adding 5 ml of 100 mM IPTG (isopropyl-β-d-thiogalactopyranoside) to the culture and incubating it for 2 h at 37°C with shaking. The cells were collected by centrifugation at 5,000 rpm (Sorvall SLA 3000 rotor) for 10 min at 4°C and were suspended in 40 ml of nickel column binding buffer (5 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl, pH 7.9). The cells were sonicated, and the pellet was collected by centrifugation at 13,000 rpm (Sorvall SS34 rotor) for 15 min at 4°C. The pellet was washed twice with binding buffer and resuspended in binding buffer with 6 M urea. After sonication, the cell extract was centrifuged at 16,500 rpm (Sorvall SS34 rotor) for 30 min at 4°C, and the supernatant was loaded on a nickel column. The resin was washed with 15 ml of buffer (60 mM imidazole, 0.5 M NaCl, 20 mM Tris-HCl [pH 7.9], and 6 M urea). The retained proteins were eluted using a stepwise gradient of imidazole ranging from 0 to 1 M diluted in 0.25 M NaCl, 10 mM Tris-HCl (pH 7.9), and 6 M urea. The ZEBRA peak fractions, identified by Coomassie blue staining of a sodium dodecyl sulfate (SDS)-polyacrylamide gel, were pooled, and the protein was refolded by stepwise dialysis against buffer (15 mM HEPES [pH 7.5], 75 mM KCl, 0.2 mM EDTA, 1 mM dithiothreitol [DTT], and 15% glycerol) containing decreasing amounts of urea.
In vitro phosphorylation of ZEBRA.
The phosphorylation of ZEBRA by different PKC isoforms was analyzed by incubating 250 ng of recombinant ZEBRA expressed in and purified from E. coli with 25 ng of PKC and 1 μCi of [γ-32P]ATP in PKC phosphorylation buffer containing 20 mM HEPES (pH 7.4), 10 mM MgCl2, 0.1 mM CaCl2, 10 μg of phosphatidylserine, and 1 μg of diacylglycerol. The PKC isoforms were purchased from Upstate Biotechnology. The reaction mixtures (100 μl) were incubated at room temperature for 15 min. The reaction was stopped by adding trichloroacetic acid (TCA) to a final concentration of 10% and incubating the mixtures on ice for 30 min. The pellet formed after centrifugation at 14,000 rpm (Eppendorf model 5415C microcentrifuge) for 10 min at 4°C was washed with 70% ethanol to remove residual TCA. The phosphorylated proteins were separated by SDS-10% polyacrylamide gel electrophoresis (PAGE), dried, and autoradiographed.
EBV lytic cycle gene expression.
Twenty-four to 48 h after transfection, the cells were harvested, resuspended in SDS sample buffer, sonicated, boiled for 5 min, and spun for 1 min. Cell lysate corresponding to 3 × 106 cells was separated on an SDS-10% polyacrylamide gel by electrophoresis. The separated proteins were transferred to a nitrocellulose membrane, which was blocked with 5% nonfat milk. The filter was probed with rabbit polyclonal antiserum against ZEBRA (32) or Rta (31) at a dilution of 1:100 in 5% nonfat milk for 1 h at room temperature. Disruption of latency was detected by a monoclonal antibody directed against the BMRF1 product, early antigen-diffuse (EA-D) (1:1,000) (9). The EA-D antibody was first reacted with a rabbit anti-mouse antibody (1:400), which served as a bridge. The blots were washed with 10 mM Tris (pH 7.5), 200 mM NaCl, and 0.05% Tween 20, and the immunoreactive bands were detected with 1 μCi of 125I-protein A. All dilutions were carried out in 5% nonfat dry milk.
Metabolic labeling and immunoprecipitation.
HKB5/Cl8 cells or HH514-16 cells (1.6 × 107) were transfected with 32 μg of expression vectors for wild-type ZEBRA, Z(S186A), Z(T159A/S186A), or the empty vector (CMV). Twelve or 17 h after transfection, the cells were washed with either phosphate-free medium for 32P labeling or methionine- and cysteine-free medium for 35S labeling. Preliminary experiments showed that a minimal labeling period of 6 to 7 h was required to obtain 32P incorporation sufficient for phosphopeptide analysis. During the labeling period, the cells were incubated with 1.6 mCi of [γ-32P]orthophosphate or with 0.45 mCi of [35S]methionine and [35S]cysteine as the sole source of phosphate- or sulfur-containing amino acids. After being labeled, the cells were washed twice with Tris-buffered saline and resuspended in 200 μl of lysis buffer (50 mM Tris-HCl [pH 7.5], 0.25 M NaCl, 0.1% SDS, 0.1% Triton X-100, 5 mM EDTA, 50 mM NaF, 0.1 mM sodium vanadate, 1 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin per ml). The cell lysate was cleared by centrifugation at 14,000 rpm in an IEC microcentrifuge for 60 min at 4°C, and the supernatant was heated at 68°C for 5 min. Following heating, 800 μl of ice-cold buffer (50 mM Tris [pH 8], 1 M NaCl, and 1% NP-40) was added to the cell lysate, and ZEBRA was immunoprecipitated using a rabbit polyclonal antibody against exon 1 of ZEBRA (32) and protein A agarose beads (Invitrogen). The immunoprecipitated proteins were eluted by boiling the beads in 20 μl of SDS-PAGE sample buffer for 5 min, and the in vivo-labeled ZEBRA was detected by autoradiography following electrophoresis through SDS-8% PAGE.
Phosphopeptide mapping.
After in vitro or in vivo phosphorylation, the 32P-labeled ZEBRA band was excised from a dried gel. The gel slice was ground in 50 mM ammonium bicarbonate, 10% β-mercaptoethanol, and 0.2% SDS. The gel slurry was removed by centrifugation, and the eluted ZEBRA protein was precipitated from the supernatant with ice-cold 20% TCA plus 20 μg of bovine serum albumin as a carrier. The pellet was resuspended in 50 μl of 50 mM ammonium bicarbonate (pH 8.0 to 8.3) and digested with 10 μg of trypsin (Promega) for 24 h at 37°C. The peptides were washed twice with 400 μl of deionized water and dried by Speed-Vac. A final wash was carried out using 400 μl of the first-dimension pH 1.9 buffer (2.5% [vol/vol] formic acid and 7.8% [vol/vol] acetic acid). The dried peptides were resuspended in 5 μl of pH 1.9 buffer and separated on a thin-layer cellulose plate in two dimensions. The first dimension of separation was electrophoresis at 1,000 V for 30 min, and the second dimension was ascending chromatography for ∼10 h in the phosphochromatography buffer (32.5% [vol/vol] n-butanol, 25% [vol/vol] pyridine, and 7.5% [vol/vol] acetic acid). Following chromatography, the plates were dried and subjected to autoradiography.
Electrophoretic mobility shift assay.
DMRIE-C reagent was used to transfect 6 × 106 HKB5/Cl8 cells with 12 μg of empty vector (CMV) or expression vector for wild-type ZEBRA, Z(T159A), Z(S186A), Z(S186D), or Z(S186E). The cells were incubated for 48 h in 5% CO2 at 37°C. After incubation, the cells were harvested, washed with phosphate-buffered saline, and resuspended in 200 μl of lysis buffer (0.42 M NaCl, 20 mM HEPES [pH 7.5], 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, 25% glycerol, 1 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin/ml). The lysate was then centrifuged at 90,000 rpm for 15 min at 4°C using a TLA100 rotor (Beckman). The supernatant was collected and assayed for its protein content by the Bradford method and stored at −80°C (5). Annealed oligonucleotides containing a ZIIIB site (15) were labeled with 32P using polynucleotide kinase (Roche). Ten micrograms of protein from each cell extract was incubated with the 32P-labeled oligonucleotide for 10 min in the presence of a DNA binding buffer (10 mM HEPES [pH 7.5], 50 mM NaCl, 2 mM MgCl2, 2.5 μM ZnSO4, 0.5 mM EDTA, 1 mM DTT, and 15% glycerol) and 0.5 μg of poly[dI-dC]. Supershifts employed either a polyclonal antibody directed against the transactivation domain of ZEBRA (32) or a monoclonal antibody (BZ1) directed against the dimerization domain of ZEBRA (37); 2 μl of the antibody was added to the DNA binding reaction after the initial 10-min incubation of protein and DNA. The antibody was incubated with the protein-DNA complex for an additional 10 min at room temperature. The reaction mixtures were loaded on a 4% 0.5× Tris-borate-EDTA native polyacrylamide gel. The protein-DNA complexes were separated by electrophoresis at 200 V for 1.5 h, and the gel was dried and exposed to XAR films for autoradiography.
RESULTS
Two ZEBRA residues, T159 and S186, serve as PKC substrates in vitro.
Two amino acids of ZEBRA, threonine 159 and serine 186, fit the consensus site motif for phosphorylation by PKC (K/R X0-2 S/T X0-2 K/R, where X is any amino acid) (35). To determine whether ZEBRA was phosphorylated in vitro by PKC, His-tagged versions of wild-type ZEBRA and the mutants Z(T159A), Z(S186A), and Z(T159A/S186A) were expressed in E. coli and purified on nickel columns; 250 ng of each protein was phosphorylated in vitro by PKC α using [γ-32P]ATP as the phosphate source. The phosphorylated proteins were separated by SDS-10% PAGE, transferred to a nitrocellulose membrane, and autoradiographed (Fig. 1A). The single point mutation,T159A, reduced the level of phosphorylation slightly, the Z(S186A) mutation reduced phosphorylation markedly, and the T159A-S186A double mutation nearly abolished phosphorylation of ZEBRA by PKC α. Coomassie blue staining of the gel showed that equivalent amounts of ZEBRA protein had been loaded in all lanes (Fig. 1B).
FIG. 1.
Identification of two residues of ZEBRA phosphorylated by PKC α in vitro. (A) His-tagged wild-type ZEBRA and mutants Z(S186A), Z(T159A), and Z(T159A/S186A) were expressed in E. coli and purified on a nickel column; 250 ng of each protein was phosphorylated by PKC α using [γ-32P]ATP. The phosphorylated proteins were separated by SDS-10% PAGE, transferred to a nitrocellulose membrane, and autoradiographed. (B) Coomassie blue stain of the gel. (C) The 32P-labeled bands corresponding to ZEBRA were excised and digested with trypsin. Aliquots of each digest, containing 10,000 cpm, were applied to thin-layer cellulose plates and analyzed by two-dimensional thin-layer cellulose separation. a, wild type ZEBRA; b, Z(S186A); c, Z(T159A); d, mixture of the tryptic digests of Z(S186A) and Z(T159A).
The radioactive bands corresponding to ZEBRA were excised and digested with trypsin. The digests were subjected to two-dimensional separation on thin-layer cellulose plates (Fig. 1C). Two phosphopeptides were detected in wild-type ZEBRA. One of them, corresponding to S186, was more intensely labeled than the other, corresponding to T159 (Fig. 1C, image a). Mutating T159 or S186 to alanine resulted in only one major phosphopeptide. Pooling the peptides generated from each ZEBRA mutant protein resulted in two phosphopeptides with mobilities similar to those seen with wild-type ZEBRA (Fig. 1C, image d). However, the intensities of the two phosphopeptide spots generated from mixing the mutants (Fig. 1C, image d) were more nearly equal than those of the phosphopeptide spots from the wild-type protein (Fig. 1C, image a). These results indicated that T159 and S186 are both substrates for in vitro phosphorylation by PKC, but S186 is the preferred site.
One of the PKC substrate sites, T159A, is not required for disruption of latency.
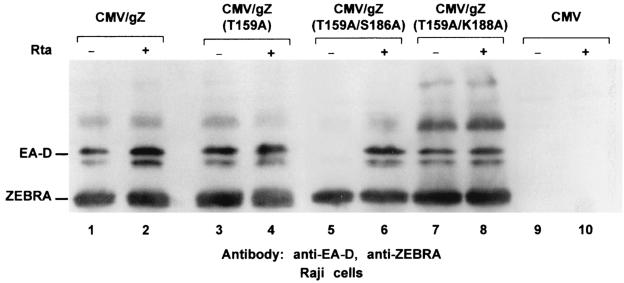
We previously reported that the ZEBRA point mutant Z(S186A) was unable to induce expression of lytic cycle mRNAs or proteins from the latent EBV genome but maintained its ability to synergize with Rta (15, 16). Therefore, we examined whether mutation of T159, the other PKC substrate site, to alanine also ablated the ability of ZEBRA, by itself or through synergy with Rta, to induce lytic cycle genes from the endogenous latent EBV genome. The EBV-positive Burkitt lymphoma cell line Raji was transfected with three constructs that expressed ZEBRA or ZEBRA mutants. Cells harvested 48 h after transfection were analyzed for the expression of the early lytic cycle gene BMRF1, an indicator of induction of the lytic cycle (Fig. 2). T159A was comparable to wild-type ZEBRA in its ability to activate BMRF1. When T159 and K188 were both mutated to alanine, the protein also behaved like the wild type. However, a ZEBRA protein that contained both the T159A and the S186A mutations was unable to disrupt latency but could synergize with Rta. These experiments showed that only one of the two major PKC sites within ZEBRA, S186, was required for the disruption of latency.
FIG. 2.
Z(T159A) maintains its ability to disrupt latency. Raji cells were transfected with 5 μg of expression vector of genomic ZEBRA (gZ), Z(T159A), Z(T159A/S186A), Z(T159A/K188A), or empty vector (CMV). Each transfection was carried out twice, once together with (+) 5 μg of the Rta expression vector and once with the empty vector (pRTS). Forty-eight hours after transfection, the cells were harvested and sonicated, and the cell extracts were analyzed by Western blotting. The membrane was probed with antibodies against ZEBRA and EA-D.
Bisindolylmaleimide I inhibits PKC isoforms that can phosphorylate ZEBRA in vitro.
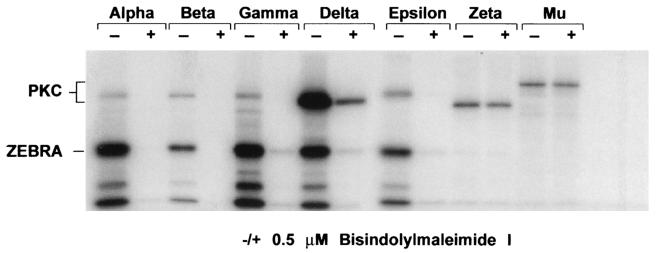
The PKC family comprises several groups of isoforms: conventional PKCs (α, β1, β2, and γ), novel (δ, ɛ, η, and θ), atypical (ι/λ and ζ), PKCμ/PKD, and the most recently identified isoform, PKC ν (21, 33). To study the differential activities of PKC isoenzymes on ZEBRA phosphorylation, purified ZEBRA was incubated at room temperature with eight different commercially available recombinant PKC isoforms, diacylglycerol, phosphatidyl serine, and [γ-32P]ATP for 10 min in the absence or presence of bisindolylmaleimide I, a specific inhibitor of PKC (34).
PKC α, β1, β2 (data not shown), γ, δ, and ɛ were competent to phosphorylate ZEBRA in vitro, while PKC ζ and μ were unable to do so (Fig. 3). All of the PKC isoforms that were found to phosphorylate ZEBRA in vitro were inhibited by bisindolylmaleimide I. The two PKC isoenzymes (ζ and μ) that could not phosphorylate ZEBRA were resistant to inhibition by bisindolylmaleimide I, since the levels of autophosphorylation of these two PKC isoforms were identical in the absence and the presence of the inhibitor (Fig. 3).
FIG. 3.
Phosphorylation of ZEBRA by different PKC isoforms in the absence (−) and presence (+) of bisindolylmaleimide I. In vitro phosphorylation reactions by different PKC isoforms (α, β1, β2, γ, ɛ, ζ, and μ) were performed using 250 ng of His-tagged ZEBRA and [γ-32P]ATP. The inhibitory effects of 0.5 μM bisindolylmaleimide I on different PKC isoforms were examined by the degree of autophosphorylation of each isoform in the absence or presence of the inhibitor. The reaction products were separated by SDS-10% PAGE, dried, and autoradiographed.
The ability of ZEBRA to induce the lytic cycle was not blocked by bisindolylmaleimide I.
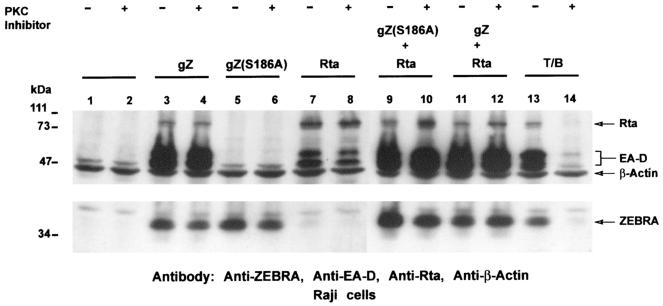
To explore whether those PKC isoforms that could phosphorylate ZEBRA in vitro were required for the ability of transiently expressed ZEBRA to induce the lytic cycle, the EBV-positive Burkitt lymphoma cell line Raji was transfected with ZEBRA or Rta expression vectors in the absence or presence of bisindolylmaleimide I.
As expected, wild-type ZEBRA was able to disrupt latency, as indicated by the expression of the BMRF1 product, EA-D (Fig. 4, lane 3), while the S186A mutant was deficient in activating the expression of EA-D from the dormant viral genome (Fig. 4, lane 5). Rta by itself caused a low level of BMRF1 activation (Fig. 4, lane 7). As previously reported, the S186A mutant was rescued to full wild-type level by coexpressing Rta in trans (Fig. 4, lane 9) (2, 15). Addition of bisindolylmaleimide I had no effect on the capacity of wild-type ZEBRA to activate the BMRF1 early lytic cycle gene (Fig. 4, lane 4), on the low level of activation of this gene by Rta (lane 8), or on the ability of Z(S186A) to synergize with Rta (Fig. 4, lane 10).
FIG. 4.
The ability of ZEBRA to disrupt latency is not blocked by bisindolylmaleimide I. Raji cells were transfected with the indicated expression vector(s) or treated with TPA-butyrate in the absence (−) or presence (+) of 10 μM bisindolylmaleimide I. Lanes 1 and 2, untreated cells; lanes 3 to 8, cells transiently transfected with 10 μg of plasmids expressing ZEBRA, Z(S186A), or Rta; lanes 9 to 12, cells cotransfected with 10 μg of vectors expressing ZEBRA or the mutant Z(S186A) together with a vector expressing Rta; lanes 13 and 14, cells treated with TPA and butyrate (T/B) to induce the lytic cycle. The cells were harvested 72 h after treatment, and cell extracts were separated by SDS-10% PAGE, transferred to a nitrocellulose membrane, probed with the indicated antibodies, and visualized by autoradiography. gZ, genomic ZEBRA.
To establish that the PKC inhibitor was active in Raji cells, we took advantage of the knowledge that TPA induces the lytic cycle in this cell background. Treating Raji cells with TPA activates PKC, which then leads to the expression of ZEBRA and consequently the expression of downstream targets such as EA-D (Fig. 4, lane 13). The addition of bisindolylmaleimide I inhibited PKC activation of ZEBRA and EA-D expression, as shown in Fig. 4, lane 14. Therefore, while bisindolylmaleimide I abrogated the induction of the lytic cycle by TPA and n-butyrate, the inhibitor had no effect on the disruption of latency by transiently transfected ZEBRA. This result showed that, while PKC isoforms inhibited by bisindolylmaleimide I play a significant role in activation of ZEBRA expression via TPA stimulation in Raji cells, these isoforms of PKC do not affect the activity of the ZEBRA protein in cells that have not been treated with TPA.
ZEBRA is not phosphorylated at the PKC recognition site motifs, T159 and S186, in vivo.
The experiments illustrated in Fig. 1 showed that ZEBRA is phosphorylated in vitro by PKC at two residues, T159 and S186. Moreover, Baumann et al. showed that ZEBRA was phosphorylated in vivo at S186 when 293 cells, transfected with a ZEBRA expression vector, were treated with a PKC agonist, TPA (4). Since transfected ZEBRA has repeatedly been shown to activate the lytic cycle in the absence of PKC agonists and since the presence of the EBV genome might have an effect on the phosphorylation state of ZEBRA, we examined the phosphorylation of ZEBRA in EBV-positive cell lines and compared the level of phosphorylation of wild-type ZEBRA with that of the mutant Z(T159A/S186A) in the absence of any PKC agonists.
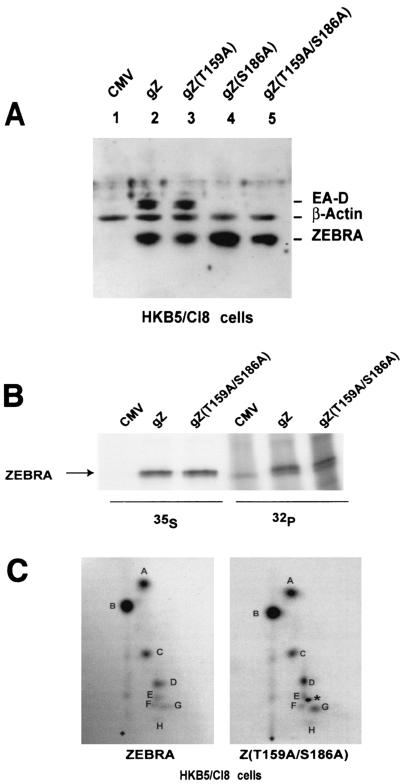
To study the phosphorylation of transfected ZEBRA, we initially used HKB5/Cl8 cells, which are hybrids between 293 cells and HH514-Cl16 cells. These cells are readily transfected, and they carry the EBV genome. To demonstrate that wild-type ZEBRA, Z(T159A), Z(S186A), and Z(T159A/S186A) maintained their phenotypes in this cell background, HKB5/Cl8 cells were transfected with the corresponding expression vectors and scored for induction of the EBV lytic cycle by immunoblotting for EA-D (Fig. 5A). Wild-type ZEBRA and Z(T159A) were competent to disrupt the latent state of EBV in HKB5/Cl8 cells, whereas Z(S186A) and Z(T159A/S186A) were crippled in activating the lytic cycle. Coexpression of Rta rescued the phenotype of the crippled Z(S186A) mutants in HKB5/Cl8 cells (data not shown). The mutant phenotypes reproduced those previously demonstrated in Raji cells (16).
FIG. 5.
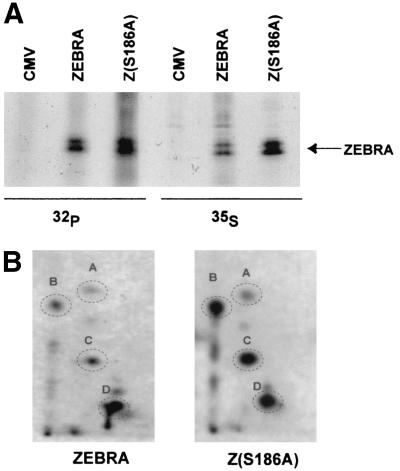
Alanine mutations at T159 and S186 do not affect the phosphorylation state of ZEBRA in vivo in HKB5/Cl8 cells. (A) Capacities of wild-type and mutant ZEBRA proteins to activate the EBV BMRF1 gene (EA-D) in HKB5/Cl8 cells. Cells (2 × 106) were transfected with 4 μg of plasmids expressing ZEBRA, Z(T159A), Z(S186A), or Z(T159A/S186A). The cells were harvested 24 h after transfection, and the cell lysates were separated by SDS-10% PAGE and immunoblotted using specific antibodies against the indicated proteins. gZ, genomic ZEBRA. (B) Immunoprecipitation of 35S- and 32P-labeled ZEBRA. HKB5/Cl-8 cells were transfected with empty vector (CMV) or vectors expressing wild-type ZEBRA (gZ) or Z(T159A/S186A). Twelve hours after transfection, the cells were incubated in medium containing both [35S]methionine and [35S]cysteine or [32P]orthophosphate for 7 h. The ZEBRA proteins were immunoprecipitated from the cell extracts and separated by SDS-8% PAGE. The gel was dried and autoradiographed. (C) Two-dimensional tryptic phosphopeptide maps of wild-type ZEBRA and Z(T159A/S186A). The 32P-labeled wild-type and mutant ZEBRA proteins were excised and extracted from the gel shown in panel B and digested with trypsin. The peptides generated by trypsin were separated in two dimensions on thin-layer cellulose. After autoradiography, eight phosphopeptides were detected, designated A to H. No difference was observed between the phosphopeptide map of wild-type ZEBRA and that of the double mutant. One extra dark spot (∗) among the Z(T159A/S186A) phosphopeptides between spots E and F is an autoradiography artifact.
To study phosphorylation of ZEBRA in vivo, HKB5/Cl8 cells were transfected with wild-type ZEBRA or with the Z(T159A/S186A) mutant and labeled with [35S]methionine and [35S]cysteine or with [32P]orthophosphate. The cells were labeled at two different intervals after transfection, 12 to 19 h (Fig. 5B and C) or 17 to 24 h (data not shown), with similar results. Immunoprecipitation of wild-type ZEBRA and the mutant indicated that the two proteins were expressed equally and were phosphorylated to the same level, as shown by the intensities of the 35S- and the 32P-labeled ZEBRA bands, respectively (Fig. 5B).
Since the overall level of phosphorylation of ZEBRA could not distinguish the presence or absence of one or two phosphorylation sites, we compared the phosphopeptide maps of wild-type ZEBRA with those of the T159A/S186A mutant (Fig. 5C). The 32P-labeled ZEBRA bands were excised, digested with trypsin, and loaded on a thin-layer cellulose plate. The phosphopeptide maps generated for wild-type ZEBRA and the mutant Z(T159A/S186A) were identical (Fig. 5C).
These experiments showed that ZEBRA was not phosphorylated at T159 or S186 in vivo. The experiment was repeated in a pure B-cell background, HH514-16 cells (Fig. 6). Again, wild-type ZEBRA and the mutant, Z(S186A), were both phosphorylated. 35S labeling indicated that the Z(S186A) mutant was expressed to a slightly greater level, and its phosphorylation state was correspondingly greater in this cell background (Fig. 6A). Nonetheless, the phosphopeptide maps of wild-type and mutant proteins were also identical (Fig. 6B). Although ZEBRA was phosphorylated in vitro by PKC at S186 as a major site and T159 as a minor site (Fig. 1), the in vivo experiments indicated that ZEBRA is not stably phosphorylated at either site at the times of maximal expression, 12 to 24 h after transfection (Fig. 5 and 6).
FIG. 6.
Phosphorylation states of ZEBRA and Z(S186A) in HH514-16 cells. (A) ZEBRA and the Z(S186A) mutant were immunoprecipitated from 35S- and 32P-labeled HH514-16 cells, resolved by SDS-8% PAGE, and visualized by autoradiography. The 32P-labeled ZEBRA was digested with trypsin, and the peptides were resolved on two-dimensional thin-layer cellulose plates by electrophoresis followed by ascending chromatography. (B) Autoradiography revealed identical phosphopeptide patterns for wild-type ZEBRA and the mutant.
A negative charge at position 186 abolishes the DNA binding activity of ZEBRA.
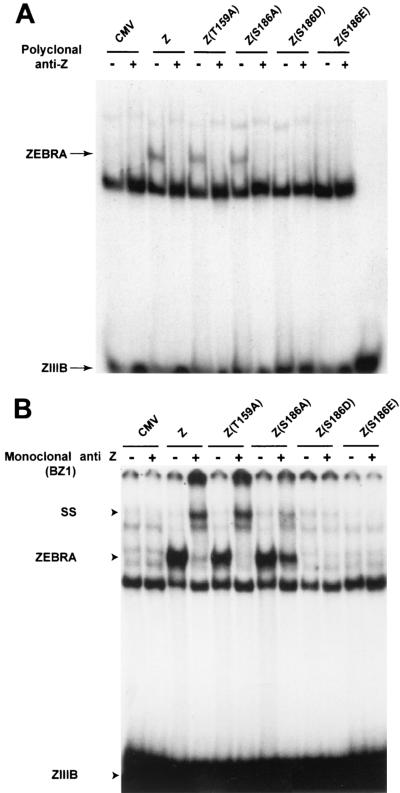
As a possible clue to how phosphorylation of ZEBRA at S186 might affect the function of ZEBRA, we mutated S186 into either aspartic acid or glutamic acid. Mutation of a specific serine or threonine residue into aspartate or glutamate is widely used to mimic the presence of a phosphate moiety. In previous experiments, we found that Z(S186D) and Z(S186E) were unable to activate Rta or EA-D expression, and this defect was not remedied by supplying Rta in trans (15). Total cell extracts prepared from HKB5/Cl8 cells expressing the ZEBRA mutants Z(S186D) and Z(S186E) failed to bind a ZIIIB site (Fig. 7). Z(T159A) and Z(S186A) bound to the same site with efficiencies equal or close to that of wild-type ZEBRA. These experiments showed that a negative charge at position 186 abrogated the disruption-of-latency function of ZEBRA, presumably by disrupting its DNA binding activity.
FIG. 7.
Effect of replacing S186 with a negatively charged amino acid on the ability of ZEBRA to bind DNA. Ten micrograms of total cell protein from HKB5/Cl8 cells transfected with the indicated plasmids were tested for their ability to bind a radiolabeled oligonucleotide containing a ZIIIB site by an electrophoretic mobility shift assay. The ZEBRA-DNA complex was identified by supershift (SS) analysis using a polyclonal antibody against ZEBRA (A) or the BZ1 monoclonal antibody (B). +, present; −, absent.
Alteration in the conformation of the Z(S186A) mutant.
In the course of carrying out these DNA binding experiments, we examined the abilities of two different antibodies to recognize ZEBRA and the Z(S186A) mutant when they were bound to a ZIIIB site. A polyclonal antibody to ZEBRA was competent to eliminate the ZEBRA-DNA complex, whether the source of the protein was wild-type ZEBRA or the T159A or S186A mutant (Fig. 7A). However, the BZ1 monoclonal antibody to ZEBRA was markedly deficient in its capacity to supershift the Z(S186A) mutant, even though the T159A and wild-type proteins were completely supershifted by the same antibody (Fig. 7B). The monoclonal antibody could recognize denatured Z(S186A) on an immunoblot and could immunoprecipitate the Z(S186A) mutant (unpublished data). This result indicated that the Z(S186A) mutant was altered in its conformation when bound to DNA. These results will be described in more detail in a subsequent manuscript (L. Heston, A. El-Guindy, Y. Endo, and G. Miller, unpublished data).
DISCUSSION
The experiments discussed in this report, addressing the question of whether PKC-mediated phosphorylation of ZEBRA is required for the protein to function in the disruption of EBV latency, can be divided into two groups: those which investigate PKC-mediated phosphorylation of ZEBRA in vitro and those which examine the potential role played by PKC in phosphorylation of ZEBRA in vivo.
By means of in vitro assays, we showed that ZEBRA can be phosphorylated by PKC α at two sites, T159 and S186. S186 appears to be the preferred site. The T159A mutant is phosphorylated at near-wild-type levels, whereas the S186A mutant is poorly phosphorylated (Fig. 1A). In the wild-type protein, the tryptic phosphopeptide containing S186 is more intense than the one containing T159 (Fig. 1C). ZEBRA can be phosphorylated in vitro by at least six isoforms of PKC (α, β1, β2, γ, δ, and ɛ), but it is not phosphorylated by PKC μ or ζ (Fig. 3). All isoforms of PKC that are competent to phosphorylate ZEBRA are inhibited by bisindolylmaleimide I; conversely, autophosphorylation of the two PKC isoforms that do not act on ZEBRA is not blocked by bisindolylmaleimide I.
In vivo studies demonstrated that mutation of only one of the two potential PKC substrate sites in ZEBRA exhibited a phenotype in disruption of latency. S186A was defective in disruption of latency, as previously described (15, 16). T159A behaved like wild-type ZEBRA (Fig. 2). The PKC inhibitor bisindolylmaleimide I had no effect on the capacity of transfected wild-type ZEBRA protein to disrupt latency or on the ability of the Z(S186A) mutant to synergize with Rta (Fig. 4). That the PKC inhibitor was active was shown by the failure of phorbol ester to induce ZEBRA and EA-D expression in the presence of the drug.
ZEBRA overexpressed from transfected plasmids was constitutively phosphorylated in human cells without a requirement for activation of PKC. Comparing wild-type ZEBRA with ZEBRA mutated at both potential PKC sites, we found that the overall levels of phosphorylation of the two proteins were identical (Fig. 5B). Of the greatest importance for the argument against constitutive phosphorylation of ZEBRA by PKC was the fact that the patterns of tryptic phosphopeptides of ZEBRA and the Z(S186A) and Z(T159A/S186A) mutants were also identical (Fig. 5C and 6B). Finally, two phosphomimetic mutants of ZEBRA, Z(S186D) and Z(S186E), which cannot disrupt latency, failed to bind DNA.
Taken together, the in vitro results show that ZEBRA T159 and S186 are potential substrates for many isoforms of PKC; however, the in vivo experiments fail to demonstrate phosphorylation of these residues in cells that have not been exposed to activators of PKC. The experiments exclude an obligatory role of PKC in the function of the ZEBRA protein.
Conflicting evidence concerning the functional importance of PKC-mediated phosphorylation of ZEBRA in vivo.
Only two previous studies have examined the phosphorylation state of ZEBRA in vivo. Daibata et al. were the first to show that ZEBRA was a phosphoprotein; they demonstrated that, following activation of the EBV lytic cycle by cross-linking of surface immunoglobulin in Akata cells, ZEBRA was phosphorylated at several serines (12). The addition of phorbol esters to activate PKC reduced the number of tryptic phosphopeptides. They proposed that, in a manner similar to its action on c-Jun, PKC activated a phosphatase which decreased the phosphorylation of ZEBRA and increased its capacity to bind DNA (12).
Baumann et al. showed that ZEBRA overexpressed in 293 cells was a phosphoprotein (4). Constitutive phosphorylation was evident from five tryptic phosphopeptides which were identical in wild-type ZEBRA and Z(S186A). Phorbol ester treatment of 293 cells that were transfected with BZLF1 induced two additional ZEBRA tryptic phosphopeptides. These additional phosphopeptides were not observed if the cells were transfected with the Z(S186A) mutant. Baumann et al. found that TPA treatment of BL41 and 293 cells markedly increased the capacity of transfected ZEBRA to activate a reporter consisting of the BHRF1 promoter linked to luciferase. TPA did not restore the capacity of the Z(S186A) mutant to activate this reporter. The importance of the PKC pathway in the EBV life cycle was inferred from the ability of the PKC inhibitor GF109203X (the same as bisindolylmaleimide I) to block the ability of transfected ZEBRA to induce replication of infectious green fluorescent protein-marked EBV from 293 cells. From these and other experiments, Baumann et al. argued that TPA-induced phosphorylation of ZEBRA (S186) augmented the DNA binding and transcriptional activities of the protein, presumably by mediating interactions with a cellular protein. This activation was required for ZEBRA to activate the latent EBV in 293 cells. In related studies, Baumann et al. found that the transcriptional activation domain of ZEBRA interacted with RACK1 (for receptor of activated C kinase) (3). However RACK1 did not activate ZEBRA or change its phosphorylation state (3).
Our studies argue against a requirement for PKC-mediated phosphorylation of Z(S186) in disruption of latency for the following reasons. First, ZEBRA is active at disruption of EBV latency in all cell backgrounds and does not require concomitant TPA treatment for its activity. Second, in some cell backgrounds in which ZEBRA is active, spontaneous PKC activity is not detected (19). Third, the PKC inhibitor bisindolylmaleimide I, which blocks the activity of PKC isoforms that can phosphorylate ZEBRA in vitro, does not block the activity of ZEBRA which has been introduced into cells by transfection (Fig. 4). Fourth, in cells that are not treated with activators of PKC but in which transfected ZEBRA is competent to disrupt latency, the levels of phosphorylation and the tryptic phosphopeptide maps of wild-type and Z(S186A) and Z(T159A/S186A) mutant proteins are the same (Fig. 5 and 6). With regard to the last two points, the experiments cannot rule out the possibility that the PKC inhibitor does not block the action of a PKC isoform which we were unable to test or that ZEBRA is only transiently phosphorylated at the PKC sites.
Lack of requirement for PKC activation in induction of the lytic cascade.
While these studies suggest that PKC-mediated phosphorylation of ZEBRA is not obligatory for its function, we have also considered whether PKC activation is obligatory for upstream events leading to the activation of ZEBRA expression. Again, our recent data argue that PKC activation is neither necessary nor sufficient for triggering the EBV lytic cascade (19). Although TPA, acting via PKC, is a potent inducer of the EBV lytic cycle in some cell backgrounds, in other cell backgrounds histone deacetylase (HDAC) inhibitors, such as sodium butyrate or trichostatin A, by themselves are sufficient to induce the EBV lytic cycle (6, 28, 36). In the cell backgrounds where the EBV lytic cycle is induced by HDAC inhibitors, lytic cycle gene expression is not blocked by the PKC inhibitor bisindolylmaleimide I (19). In the same cells, induction of the EBV lytic cycle by HDAC inhibitors is not accompanied by induction of PKC activity. That HDAC inhibitors can activate the entire lytic cascade, including ZEBRA expression, without activating PKC is a further argument against a requirement for PKC-mediated phosphorylation of ZEBRA in the disruption of latency. In yet other cell backgrounds, TPA treatment activates PKC, as expected, but TPA treatment is not associated with induction of the EBV lytic cascade.
What accounts for the phenotype of the Z(S186A) mutant?
The results presented indicate that ZEBRA is not constitutively phosphorylated in vivo at S186. Accordingly, the lack of phosphorylation of ZEBRA at this residue is not likely to account for the inability of the Z(S186A) mutant to disrupt latency. Since the Z(S186A) mutant can be rescued by overexpression of Rta, it is thought that the main deficit of the Z(S186A) mutant resides in its inability to activate expression of BRLF1, the gene for Rta. This deficiency may now be ascribed to one or a combination of the following deficits: (i) Z(S186A) has a relatively low affinity for the two ZEBRA binding sites in Rp, the promoter of BRLF1 (2, 15); (ii) the phenotype of Z(S186A) may reflect an inability to activate Rp in its chromatinized state, as the result of a loss of the capacity to reconfigure nucleosomes or to recruit a histone acetylase; (iii) a defect in specific protein-protein interactions; or (iv) an alteration in conformation of the mutant protein upon binding to DNA (Fig. 7B).
The affinity of wild-type ZEBRA for the two ZEBRA response elements ( ZREs) in Rp, ZIIIA and ZRE-R, is 2.4- and 8.5-fold higher, respectively, than the affinity of Z(S186A) (15). Although Z(S186A) is relatively impaired in its ability to bind to ZIIIA and ZRE-R, this defect is not reflected in an inability to activate reporter plasmids such as Rp (BRLF1p)-CAT (15). This result suggests that the mutant may be additionally deficient in some process required to activate the same promoter as it is configured on the latent endogenous virus.
One essential property of many transcriptional activators is the ability to rearrange nucleosomes (27). The bZIP regions of cFos/cJun have been shown in vitro to disrupt the structure of a nucleosome containing an AP-1 site (29). Nucleosome disruption was dependent on DNA binding activity and the concentrations of the AP-1 proteins and allowed other transcription factors to bind to sequences that were repressed by the nucleosome. In an analogous process, the involvement of the BRLF1 promoter in a nucleosome structure might be responsible for its inaccessibility to transcriptional activators (19). Wild-type ZEBRA may be able to disrupt this nucleosomal structure, whereas Z(S186A), with its impairment in DNA binding, may be hampered in its capacity to disrupt nucleosomes.
Alternatively, the Z(S186A) mutant may be unable to carry out additional protein-protein interactions that are essential for nucleosomal modification and thus disruption of latency. An example of such an interaction is the ability to recruit a histone acetylase (HAT) to the BRLF1 promoter. CBP has intrinsic HAT activity and functions as a coactivator for many transcription factors (26). CBP binds directly to ZEBRA and stimulates its transcriptional activity (1, 38); however, this interaction was maintained when S186 was mutated to alanine (1). New evidence suggests that specific HAT enzymes are required at some promoters. For example, promoters activated by the retinoic acid receptor and MyoD require the P/CAF and not the CBP acetyltransferase, even though both coactivators may be present on the promoter (26, 30). Different HAT enzymes may be acquired for different promoters due to nucleosome phasing or the presence of repressors. Therefore, the defect in the Z(S186A) mutant may be remedied by recruiting the appropriate HAT activity to the BRLF1 promoter.
How might such a defect in protein-protein interaction be conferred by the Z(S186A) mutant? A clue comes from comparing the interaction of a monoclonal antibody with the wild type and with Z(S186A) (Fig. 7B). This monoclonal antibody is directed against the dimerization domain of ZEBRA (37). ZEBRA present in cell extracts and bound to DNA is efficiently recognized by this monoclonal antibody, which induces a supershift of the ZEBRA-DNA complex. Z(S186A) is supershifted much less efficiently (Fig. 7B). In fact, the capacities of many mutants in ZEBRA's basic DNA recognition domain to be supershifted by this monoclonal antibody are markedly altered when they are bound to DNA (data not shown). This result suggests that basic domain mutations affect the conformation of the ZEBRA homodimerization domain. Crucial protein-protein interactions may be required for ZEBRA to carry out its many functions. These interactions, in turn, may depend on the correct conformation of ZEBRA when it is bound to DNA.
Role of phosphorylation in the function of ZEBRA.
Our data (Fig. 5 and 6) and those of others demonstrate that ZEBRA is a phosphoprotein (4, 12). Since there are four major ZEBRA tryptic phosphopeptides in both cell backgrounds, it is likely that ZEBRA is phosphorylated at multiple sites. One candidate site, S173, has previously been shown to be a substrate for casein kinase II phosphorylation in vitro (25). It has recently been found that this residue of ZEBRA is phosphorylated in vivo (A. S. El-Guindy and G. Miller, unpublished data). There are likely to be several different constitutively expressed kinases which phosphorylate ZEBRA. The identification of the sites on ZEBRA that are phosphorylated, the responsible kinases, and the functional significance of these phosphorylations is a fruitful area for future study.
Acknowledgments
This work was supported by grants CA12055 and CA16038 from the NIH.
We thank Jill Countryman, Pey-Jium Chang, Dan DiMaio, and Anthony Koleske for helpful discussions and critiques of the manuscript.
REFERENCES
- 1.Adamson, A. L., and S. Kenney. 1999. The Epstein-Barr virus BZLF1 protein interacts physically and functionally with the histone acetylase CREB-binding protein. J. Virol. 73:6551-6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamson, A. L., and S. C. Kenney. 1998. Rescue of the Epstein-Barr virus BZLF1 mutant, Z(S186A), early gene activation defect by the BRLF1 gene product. Virology 251:187-197. [DOI] [PubMed] [Google Scholar]
- 3.Baumann, M., O. Gires, W. Kolch, H. Mischak, R. Zeidler, D. Pich, and W. Hammerschmidt. 2000. The PKC targeting protein RACK1 interacts with the Epstein-Barr virus activator protein BZLF1. Eur. J. Biochem. 267:3891-3901. [DOI] [PubMed] [Google Scholar]
- 4.Baumann, M., H. Mischak, S. Dammeier, W. Kolch, O. Gires, D. Pich, R. Zeidler, H. J. Delecluse, and W. Hammerschmidt. 1998. Activation of the Epstein-Barr virus transcription factor BZLF1 by 12-O-tetradecanoylphorbol-13-acetate-induced phosphorylation. J. Virol. 72:8105-8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Chang, L. K., and S. T. Liu. 2000. Activation of the BRLF1 promoter and lytic cycle of Epstein-Barr virus by histone acetylation. Nucleic Acids Res. 28:3918-3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang, Y. N., D. L. Dong, G. S. Hayward, and S. D. Hayward. 1990. The Epstein-Barr virus Zta transactivator: a member of the bZIP family with unique DNA-binding specificity and a dimerization domain that lacks the characteristic heptad leucine zipper motif. J. Virol. 64:3358-3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chevallier-Greco, A., E. Manet, P. Chavrier, C. Mosnier, J. Daillie, and A. Sergeant. 1986. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 5:3243-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho, M. S., K. T. Jeang, and S. D. Hayward. 1985. Localization of the coding region for an Epstein-Barr virus early antigen and inducible expression of this 60-kilodalton nuclear protein in transfected fibroblast cell lines. J. Virol. 56:852-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Countryman, J., H. Jenson, R. Seibl, H. Wolf, and G. Miller. 1987. Polymorphic proteins encoded within BZLF1 of defective and standard Epstein-Barr viruses disrupt latency. J. Virol. 61:3672-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Countryman, J., and G. Miller. 1985. Activation of expression of latent Epstein-Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc. Natl. Acad. Sci. USA 82:4085-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daibata, M., R. E. Humphreys, and T. Sairenji. 1992. Phosphorylation of the Epstein-Barr virus BZLF1 immediate-early gene product ZEBRA. Virology 188:916-920. [DOI] [PubMed] [Google Scholar]
- 13.Davis, M. G., and E. S. Huang. 1988. Transfer and expression of plasmids containing human cytomegalovirus immediate-early gene 1 promoter-enhancer sequences in eukaryotic and prokaryotic cells. Biotechnol. Appl. Biochem. 10:6-12. [PubMed] [Google Scholar]
- 14.Farrell, P. J., D. T. Rowe, C. M. Rooney, and T. Kouzarides. 1989. Epstein-Barr virus BZLF1 trans-activator specifically binds to a consensus AP-1 site and is related to c-fos. EMBO J. 8:127-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francis, A., T. Ragoczy, L. Gradoville, A. El-Guindy, and G. Miller. 1999. Amino acid substitutions reveal distinct functions of serine 186 of the ZEBRA protein in activation of lytic cycle genes and synergy with the EBV Rta transactivator. J. Virol. 73:4543-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francis, A. L., L. Gradoville, and G. Miller. 1997. Alteration of a single serine in the basic domain of the Epstein-Barr virus ZEBRA protein separates its functions of transcriptional activation and disruption of latency. J. Virol. 71:3054-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giot, J. F., I. Mikaelian, M. Buisson, E. Manet, I. Joab, J. C. Nicolas, and A. Sergeant. 1991. Transcriptional interference between the EBV transcription factors EB1 and R: both DNA-binding and activation domains of EB1 are required. Nucleic Acids Res. 19:1251-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glover, J. N., and S. C. Harrison. 1995. Crystal structure of the heterodimeric bZIP transcription factor c-Fos-c-Jun bound to DNA. Nature 373:257-261. [DOI] [PubMed] [Google Scholar]
- 19.Gradoville, L., D. Kwa, A. El-Guindy, and G. Miller. 2002. Protein kinase C-independent activation of the Epstein-Barr virus lytic cycle. J. Virol. 76:5612-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruffat, H., N. Moreno, and A. Sergeant. 1990. The Epstein-Barr virus (EBV) ORI1yt enhancer is not B-cell specific and does not respond synergistically to the EBV transcription factors R and Z. J. Virol. 64:2810-2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayashi, A., N. Seki, A. Hattori, S. Kozuma, and T. Saito. 1999. PKCnu, a new member of the protein kinase C family, composes a fourth subfamily with PKCmu. Biochim. Biophys. Acta 1450:99-106. [DOI] [PubMed] [Google Scholar]
- 22.Holley-Guthrie, E. A., E. B. Quinlivan, E. C. Mar, and S. Kenney. 1990. The Epstein-Barr virus (EBV) BMRF1 promoter for early antigen (EA-D) is regulated by the EBV transactivators, BRLF1 and BZLF1, in a cell-specific manner. J. Virol. 64:3753-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kenney, S., E. Holley-Guthrie, E. C. Mar, and M. Smith. 1989. The Epstein-Barr virus BMLF1 promoter contains an enhancer element that is responsive to the BZLF1 and BRLF1 transactivators. J. Virol. 63:3878-3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolman, J. L., N. Taylor, L. Gradoville, J. Countryman, and G. Miller. 1996. Comparing transcriptional activation and autostimulation by ZEBRA and ZEBRA/c-Fos chimeras. J. Virol. 70:1493-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kolman, J. L., N. Taylor, D. R. Marshak, and G. Miller. 1993. Serine-173 of the Epstein-Barr virus ZEBRA protein is required for DNA binding and is a target for casein kinase II phosphorylation. Proc. Natl. Acad. Sci. USA 90:10115-10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Korzus, E., J. Torchia, D. W. Rose, L. Xu, R. Kurokawa, E. M. McInerney, T. M. Mullen, C. K. Glass, and M. G. Rosenfeld. 1998. Transcription factor-specific requirements for coactivators and their acetyltransferase functions. Science 279:703-707. [DOI] [PubMed] [Google Scholar]
- 27.Kuo, M. H., and C. D. Allis. 1998. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 20:615-626. [DOI] [PubMed] [Google Scholar]
- 28.Luka, J., B. Kallin, and G. Klein. 1979. Induction of the Epstein-Barr virus (EBV) cycle in latently infected cells by n-butyrate. Virology 94:228-231. [DOI] [PubMed] [Google Scholar]
- 29.Ng, K. W., P. Ridgway, D. R. Cohen, and D. J. Tremethick. 1997. The binding of a Fos/Jun heterodimer can completely disrupt the structure of a nucleosome. EMBO J. 16:2072-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Puri, P. L., V. Sartorelli, X. J. Yang, Y. Hamamori, V. V. Ogryzko, B. H. Howard, L. Kedes, J. Y. Wang, A. Graessmann, Y. Nakatani, and M. Levrero. 1997. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell 1:35-45. [DOI] [PubMed] [Google Scholar]
- 31.Ragoczy, T., L. Heston, and G. Miller. 1998. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol. 72:7978-7984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor, N., J. Countryman, C. Rooney, D. Katz, and G. Miller. 1989. Expression of the BZLF1 latency-disrupting gene differs in standard and defective Epstein-Barr viruses. J. Virol. 63:1721-1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Toker, A. 1998. Signaling through protein kinase C. Front. Biosci. 3:D1134-D1147. [DOI] [PubMed] [Google Scholar]
- 34.Toullec, D., P. Pianetti, H. Coste, P. Bellevergue, T. Grand-Perret, M. Ajakane, V. Baudet, P. Boissin, E. Boursier, F. Loriolle, et al. 1991. The bisindolylmaleimide GF 109203X is a potent and selective inhibitor of protein kinase C. J. Biol. Chem. 266:15771-15781. [PubMed] [Google Scholar]
- 35.Woodgett, J. R., K. L. Gould, and T. Hunter. 1986. Substrate specificity of protein kinase C. Use of synthetic peptides corresponding to physiological sites as probes for substrate recognition requirements. Eur. J. Biochem. 161:177-184. [DOI] [PubMed] [Google Scholar]
- 36.Yoshida, M., M. Kijima, M. Akita, and T. Beppu. 1990. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J. Biol. Chem. 265:17174-17179. [PubMed] [Google Scholar]
- 37.Young, L. S., R. Lau, M. Rowe, G. Niedobitek, G. Packham, F. Shanahan, D. T. Rowe, D. Greenspan, J. S. Greenspan, A. B. Rickinson, et al. 1991. Differentiation-associated expression of the Epstein-Barr virus BZLF1 transactivator protein in oral hairy leukoplakia. J. Virol. 65:2868-2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zerby, D., C. J. Chen, E. Poon, D. Lee, R. Shiekhattar, and P. M. Lieberman. 1999. The amino-terminal C/H1 domain of CREB binding protein mediates Zta transcriptional activation of latent Epstein-Barr virus. Mol. Cell. Biol. 19:1617-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang, C. X., G. Decaussin, J. Daillie, and T. Ooka. 1988. Altered expression of two Epstein-Barr virus early genes localized in BamHI-A in nonproducer Raji cells. J. Virol. 62:1862-1869. [DOI] [PMC free article] [PubMed] [Google Scholar]