Abstract
Porcine endogenous retroviruses (PERV) can infect human cell lines in vitro; hence, there is a presumed risk of viral exposure to a recipient when pig cells are transplanted into humans (xenotransplantation). Nonhuman primates (NHP) are considered a potential permissive animal model to study the risk of in vivo infection of PERV after xenotransplantation. We set out to determine whether PERV can infect and replicate in NHP primary cells or established cell lines from African green monkey, rhesus macaque, and baboon. We confirm that the NHP cell lines under investigation were infected with PERV as measured by detection of viral DNA and RNA by PCR and reverse transcription (RT)-PCR, respectively, indicating that a functional receptor must be present on the cell surface. However, the load of detectable viral DNA in infected NHP cells declined over time, and the cells never had detectable reverse transcriptase activity. Utilizing quantitative real-time TaqMan PCR we found detectable levels of unintegrated DNA intermediates, but the levels were approximately 100-fold lower compared to HEK 293 cells infected with PERV. Virions released from infected NHP cells could productively infect naïve human cell lines, HEK 293 and HeLa, as shown by RT-PCR and RT assay. However, naïve NHP cells remained negative in RT-PCR and RT assay after exposure to virions from infected NHP cells. Together our data demonstrate that NHP cells are not permissive to productive replication by PERV, presumably due to inefficient cell entry and replication. In light of these observations, the appropriateness of NHP as suitable animal models to study PERV infection in vivo needs to be reevaluated.
The increasing number of patients that would benefit from an organ transplant face a serious and worsening shortage of human donor organs. New technology and advances in the field of genetic modification of animals has opened the possibilities to use animals as an alternative source for cells, tissues, or organs, i.e., xenotransplantation. Xenotransplantation is defined by the Public Health Service as any procedure that involves transplantation, implantation, or infusion of live cells, tissues, or organs from a nonhuman animal source into a human recipient (PHS Guideline on Infectious Disease Issues in Xenotransplantation, 19 January 2001 [http://www.fda.gov/cber/xap/docs.htm]). A number of obstacles still remain, most notably immunological barriers. However, if successful, xenotransplantation could alleviate the shortage of human donor organs and tissues.
The pig has a number of advantages as a renewable source of donor tissue including a vast experience in its husbandry and health care, as well as the advancing technologies to engineer transgenic animals. A number of clinical trials are under way to test the feasibility of pig-derived tissues for xenotransplantation (7). Selective breeding and screening of the pig can reduce the risk of zoonoses, or animal-human infections, from porcine xenotransplantation (reviewed in reference 22). However, like many other mammalian species, pigs harbor endogenous retroviruses (porcine endogenous retrovirus [PERV]), which are encoded in their genomic DNA (33, 35) and assumed to be descendants of ancient viruses that became integrated into the host germ line.
Gammaretrovirus particles are released by pig cell lines (2, 3, 14), yet only recently have investigators looked into the potential risk of human infection by PERV. Two of the three identified receptor classes of PERV, distinguished by their envelope sequence and tropism, have been shown to be capable of replicating in human cells in vitro (18, 21, 25, 26, 27, 30, 38). Retrospective studies conducted to determine whether PERV was transmitted to recipients treated with pig cells, tissues, or organs have all shown negative results (9, 23, 24). One limitation of these studies is that these exposures of humans to pig tissues were generally very brief due to immunological rejection. In contrast, limited but not productive infection of mouse tissues with PERV has been observed after porcine islet xenotransplantation in NOD/SCID mice (6, 36). However, as advances in transplantation immunology, immunosuppression, source animal engineering, and even tolerance induction enhance success in xenotransplantation, subjects may be exposed to porcine cells in vivo for longer periods, increasing the risk of PERV transmission, and potentially leading to a public health risk. So far nothing is known about the potential pathogenicity of PERV in an in vivo setting. Therefore, the development of an animal model is critical to advancing our understanding of PERV infection and possible pathogenesis.
Nonhuman primates (NHP) have been used to study PERV in vivo. In studies using Old and New World monkeys exposed to porcine xenografts, tissue specimens from the tested animals tested negative for PERV infection (15, 19, 20, 28, 31). Some reports suggest that NHP cell lines are susceptible to PERV infection in vitro when analyzed by PCR or reverse transcriptase PCR (RT-PCR) for PERV sequences (4, 27, 31). In contrast, a study using PERV pseudotypes found that one NHP cell line examined was not susceptible to infection, based on lack of detection of a marker gene carried by the pseudotype (39). Since none of the studies have examined whether NHP cells are permissive to PERV replication or to productive infections, we set out to conduct a series of experiments using an isolate of PERV derived from NIH mini pigs, PERV-NIH (38), in both established NHP cell lines as well as several primary NHP cell cultures.
MATERIALS AND METHODS
Cell lines and virus.
The cell lines used in this study were obtained from the American Type Culture Collection unless otherwise indicated. Human embryonic kidney (HEK) 293 cells (ATCC CRL-1537), FRhK-4 rhesus monkey kidney (ATCC CRL-1688), and HeLa cervical carcinoma (ATCC CCL-2) were maintained in Dulbecco's modified Eagle's medium (Dulbecco's MEM) supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, 1 mM sodium pyruvate, penicillin (100 U/ml), and streptomycin (100 U/ml). FRhL-2 rhesus monkey lung (ATCC CL-160), BSC-1 rhesus macaque kidney (ATCC CCL-26), and RF/6A rhesus retinal endothelial cell line (ATCC CRL-1780) were maintained in essential MEM supplemented in the same manner as Dulbecco's MEM with the addition of 1% nonessential amino acids (Biofluids, Rockville, Md.). Primary cell cultures were developed from rhesus monkey lung, kidney, aorta, and umbilical veins. In brief, freshly harvested tissue was mechanically disrupted and digested with collagenase to yield single-cell suspensions after washing and filtration through a 60-gauge stainless steel mesh. The two primary epithelial cell lines were maintained in a supplemented commercial medium, REGM BulletKit (Cambrex, East Rutherford, N.J.), for no more than four passages before testing. The rhesus umbilical vein endothelial cells (UVEC) and rhesus aortic endothelial cells were maintained in EGM BulletKit medium (Cambrex) and used after at least four passages and no more than eight passages. 1 × 3731-6, a herpesvirus saimiri T-cell line, was maintained in RPMI-C (RPMI consisting of 2 mM glutamine, 1 mM sodium pyruvate, penicillin [100 U/ml], and streptomycin [100 U/ml], 1% MEM, 2.5% 1 M HEPES, 1% 1 N NaOH) supplemented with 15% interleukin-2 (Roche Molecular Biochemicals, Indianapolis, Ind.), 20% FBS, 2 mM glutamine, 1 mM sodium pyruvate, penicillin (100 U/ml), and streptomycin (100 U/ml). 8260, a herpesvirus papio B-cell line, was maintained in RPMI-C supplemented with 15% FBS, 2 mM glutamine, 1 mM sodium pyruvate, penicillin (100 U/ml), and streptomycin (100 U/ml). Both baboon cell lines were obtained from J. S. Allan (Southwest Foundation for Biomedical Research, San Antonio, Tex.). The 1 × 3731-6 and 8260 cell lines will be referred to as a baboon T-cell line or baboon B cell line, respectively.
HEK 293 cells productively infected with PERV-NIH were originally derived from activated PBMC of NIH minipigs (38). Virus produced by infected HEK 293 cells was named PERV-NIH-1°. Supernatant derived from PERV-NIH-1° was used to infect naïve HEK 293 cells to generate PERV-NIH-2° (39). PERV-NIH-2° was then used to infect naïve HEK 293 cells to generate PERV-NIH-3°, which has an approximately 10-fold higher infectious titer than PERV-NIH-2° as measured by 50% tissue culture infective dose assay (data not shown). The virus used for the experiments described in this paper is PERV-NIH-3° and will be referred to from here forward simply as PERV-NIH.
Derivation of viral pseudotypes.
293T cells were seeded at a density of 106 cells/10-cm2 dish 2 days before transfection. Expression vectors encoding a Moloney murine leukemia virus (Mo-MuLV)-based genome containing the packaging signal and the β-galactosidase gene coding sequence, Mo-MuLV gag-pol expression plasmid, and a plasmid encoding the vesicular stomatitis virus G protein (VSV-G) were transfected into 293T cells to generate virus pseudotypes as described previously (32). Culture supernatants were harvested 48 h posttransfection, filtered through a 0.45-μm-pore-size filter, adjusted to a 6-μg/ml final concentration of Polybrene, and used to superinfect HEK 293 cells or NHP cell lines infected with PERV-NIH. The resulting pseudovirions should carry PERV-NIH viral proteins surrounding the Mo-MuLV-based genome encoding β-galactosidase. Infectivity by these pseudovirions was monitored by immunohistochemical staining of target cells expressing β-galactosidase 48 to 72 h after infection as described previously (37).
Infectivity studies.
Supernatant infections were used to determine the infectivity of cell-free virus derived from different producer cell lines. Since freeze-thawing of viral supernatant resulted in >100-fold loss of infectious titer (data not shown), all experiments were done with freshly harvested supernatant from subconfluent cultures. One day prior to infection, target cells were seeded in 6- or 12-well dishes, varying from 3 × 104 to 105 cells per well depending on the cell type. On the day of the experiment, supernatant of subconfluent virus producer cells was harvested and filtered through a 0.45-μm-pore-size filter, adjusted to 6- to 8-μg/ml Polybrene, and used to replace culture medium of target cells. The day following infection, supernatant was replaced with fresh culture medium. A negative control that was not exposed to virus containing supernatant was maintained parallel to the infected cultures. When β-galactosidase expression was used to monitor infectivity, cells were fixed and histochemically stained 48 to 72 h after infection. Blue-staining focus-forming units (BFU) were counted under the microscope as previously reported (37). In other cases, cells were maintained by subpassage approximately twice per week and analyzed for evidence of infection as described below. In coculture assays, PERV-infected cell lines were plated into Transwell culture inserts (Costar, Cambridge, Mass.) at 2 × 104 to 5 × 104 cells per insert, and the “target” cells for infection were plated at 3 × 104 to 105 cells per well depending on the cell type. The inserts were removed after 1 week, and the exposed target cells were passed in culture approximately twice per week until testing.
PCR slot blot analysis for PERV DNA.
PCR was performed on DNA purified from cell preparations using a commercial genomic DNA isolation procedure (QIAamp blood kit; Qiagen, Valencia, Calif.) at a concentration of 1 μg per 50-μl reaction as previously described (23). PERV sequences were amplified with conserved pol sequence-derived primers: forward (5′ AGCTCCGGGAGGCCTACTC 3′) and reverse (5′ ACAGCCGTTGGTGTGGTCA 3′). Cycling parameters were 45 s at 64°C, 60 s at 72°C, and 30 s at 94°C for 35 cycles as previously described (36). Detection of PCR products was done by Southern slot blotting (Schleicher and Schuell, Keene, N.H.) of DNA using digoxigenin-labeled probes for full-length PERV pol and rabbit anti-digoxigenin conjugated to alkaline phosphatase (Boehringer Mannheim, Indianapolis, Ind.). The assay is sensitive enough to detect as few as 10 copies of PERV-specific DNA in a background of 105 cells.
VSV-G pseudotyping of RF/6A.
PERV-infected RF/6A or 293 cells in culture were transfected with a plasmid encoding the VSV-G to generate virus pseudotypes as described previously (32). Culture supernatants were collected 48 h later, filtered through a 0.45-μm-pore-size filter, adjusted to 8-μg/ml Polybrene, and used to replace culture medium of subconfluent monolayers of target cells.
Isolation and detection of unintegrated viral DNA.
HEK 293, FRhK-4, FRhL-2, and BSC-1 cells were seeded at a density of 5 × 106 cells in 150-cm2 flasks 1 day prior to infection. The following day, cells were exposed to supernatant containing PERV-NIH. Twenty-four hours later, cells were lysed and the low-molecular-weight DNA fraction was isolated (10). DNA concentration was determined by optical density at 260 nm, and 500-ng samples were used for PCR analysis. Circular DNA formation was detected by using long terminal repeat primers PERV-LTR1 (5′ CTGCGTGGTGTACGACTG 3′) and PERV-LTR2 (5′ GAGTCGGGACAGCTTTTATGG 3′) (94°C for 5 min, 35 cycles of 30 s at 94°C, 35 s at 57°C, and 30 s at 72°C, with a final extension step at 72°C for 7 min) specific to the long terminal repeat (LTR) sequence of PERV-C (GenBank accession no. AF038600). Quantitation of unintegrated PERV DNA was performed as described below.
Detection of viral RNA and RT activity.
Infection of target cells was measured with several methods. Samples were taken from supernatants of cultures at different time points after exposure to PERV to determine reverse transcriptase activity as previously described (38). Aliquots of samples from the same time points were also subject to RT-PCR analysis for detection of PERV RNA. RNA was extracted from cell culture supernatant using RNA-STAT 50-LS (Tel-Test “B,” Inc., Friendswood, Tex.) and coprecipitated with 5 μg of tRNA. RNA was reverse transcribed using Mo-MuLV reverse transcriptase RT (Life Technologies, Gaithersburg, Md.) using 50 ng of random primers (Life Technologies). The cDNA was amplified by PCR using primers specific to the pol coding sequence of PERV A (GenBank accession no. AF033259), the PCR products were fractionated on a 1% agarose gel, followed by Southern blotting and hybridization of the amplified products with an alkaline phosphatase-labeled oligonucleotide to PERV-pol as described previously (29).
Quantitative detection of PERV nucleic acid.
Total cellular RNA was extracted from infected cells using the Qiagen RNeasy kit according to the manufacturer's protocol. Viral RNA was isolated from cell supernatant as described above. Cellular DNA was prepared using the Qiagen DNeasy kit according to the manufacturer's protocol. The amplification primers used were Pol136FW (5′ TAACCCACTTCGTTTCTGGT 3′) and Pol293Rev (5′ TCATCTAATTGGAGGGTCAACA 3′), which correspond to a region of the PERV-pol gene (GenBank accession no. AF033259). The fluorogenic detection probe, Polprobe 162 (5′ TGAGTGCCCAGCACCCCTCTTAGGTA 3′) (GenBank accession no. AF033259) was labeled with the reporter dye FAM (6-carboxyfluorescein) at the 5′ end and a quencher dye TAMRA (6-carboxytetramethylrhodamine) at the 3′ end. The primers and probe were obtained from the core facility at the Center for Biologics Evaluation and Research (Bethesda, Md.). For DNA amplification the TaqMan Universal PCR master mix (PE Applied Biosystems, Foster City, Calif.) was used. A serial dilution of the PERV-pol plasmid was used as a standard for DNA PCR. In order to generate a standard for RT-PCR, the PERV-pol plasmid was linearized with KpnI and in vitro transcribed using the T7 RNA polymerase as described by the manufacturer (MAXIscript; Ambion, Austin, Tex.). The cRNA was phenol-chloroform extracted and ethanol precipitated. The integrity of the cRNA was verified by denaturing agarose gel-electrophoresis and visualization using ethidium bromide. The reaction conditions for a 25-μl PCR mix were as follows: 50°C for 2 min, 95°C for 10 min, and 50 cycles of 15 s at 95°C and 1 min at 60°C. RNA quantification was performed in a one-step RT-PCR. The reaction mixture for RT-PCR consisted of 1× TaqMan buffer; 0.15 mM (each) dATP, dCTP, dGTP, and dUTP; 0.3 μM of each Pol136FW and Pol293Rev; 2.5 mM manganese acetate; 0.1 μM TaqMan probe; and 1.0 μl of RTTH enzyme. The reaction was carried out in a 25-μl mix at 50°C for 2 min, 60°C for 30 min, 95°C for 2 min; then for 3 cycles at 95°C for 0.2 min, 60°C for 0.3 min; and then for 50 cycles at 95°C for 0.2 min, 60°C for 1 min. The PCR experiments were performed in the PRISM 7700 sequence detection system (PE Applied Biosystems) that gives a threshold cycle (CT) value for every sample. Quantification of DNA is achieved by comparing the CT value of the input DNA or RNA with the CT value of the standard template DNA or RNA, coamplified in the same run. All standard dilutions, controls and samples were run in duplicates and repeated at least two times. Standard curves for both DNA and RT-PCR were accepted when the coefficients of correlation (r2) were >0.9.
FACS detection of PERV infection by p10 Gag protein.
A rabbit polyclonal antisera was made with a peptide corresponding to the C-terminal 19 amino acids of the 10-kDa (p10) nucleocapsid portion of the PERV gag polyprotein (12) (kind gift from R. R. Toenjes). Cells were harvested, washed in phosphate-buffered saline, and then fixed in fresh 4% paraformaldehyde at room temperature for 20 min after which they were permeabilized with 0.05% saponin. Cells were blocked with human immunoglobulin G in 2% donkey serum and then incubated with either the rabbit polyclonal antisera or the preimmune sera as the negative control after which staining was detected with affinity-purified donkey anti-rabbit FITC secondary antibody at a 1:200 dilution (Jackson Labs, Bar Harbor, Maine). The staining detected with the preimmune negative control sera was used to set the fluorescent gain level after which specific staining was measured by the geometric median fluorescence intensity of the p10-stained cells as determined with a Becton Dickinson FACSCAN cytometer (Mansfield, Mass.).
RESULTS
Analysis of susceptibility of NHP primary cells and cell lines to PERV.
NHP cell lines representing two different species, rhesus macaque and African green monkey, were exposed to PERV-NIH pseudotype-containing supernatant, and infectious titers were determined after histochemical staining for β-galactosidase-expressing cells. As shown in Table 1, the relative susceptibility of FRhK-4, FRhL-2 and BSC-1 were approximately 1,000 to 10,000-fold lower when compared to HEK 293 cells.
TABLE 1.
Comparison of infectious titer of PERV-NIH virus pseudotypes on HEK 293 and NHP cells
| Cell line | Origin | Avg PERV-NIH/LacZa ±SEM (102 BFU/ml)b |
|---|---|---|
| HEK 293 | HuEK | 640 ± 31 |
| FrK-4 | Rhesus macaque kidney | 0.56 ± 0.02 |
| FrL-2 | Rhesus macaque lung | 0.142 ± 0.085 |
| BSC-1 | African green monkey kidney | 0.058 ± 0.006 |
Cells were exposed to supernatant containing viral pseudotypes encoding β-galactosidase as described in Materials and Methods.
BFU are as explained in Materials and Methods. Values shown are based on three independent experiments from triplicate wells.
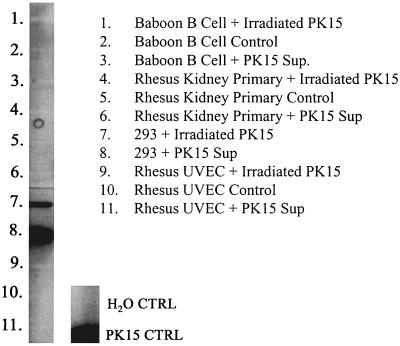
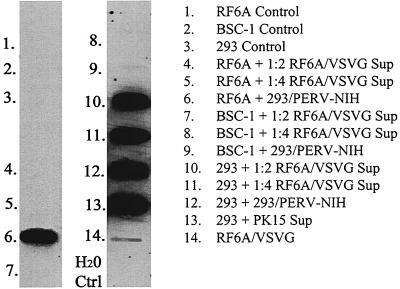
These results were confirmed and extended independently using the pig PK-15 PERV producer cell line (Fig. 1) and the same HEK 293/PERV-NIH producer cell line (data not shown) with a different but overlapping set of NHP cell lines, including primary rhesus derived from three tissues—B cell, kidney, and UVEC. Three to four passages after exposure to PERV, DNA was isolated from target cells and subject to DNA PCR slot blots. The DNA PCR slot blot assay system has a sensitivity of detecting 10 copies of PERV in a background of 105 HEK 293 or human T cells (data not shown). Figure 1 shows the representative results from one of three experiments. There is no evidence of PERV DNA detected in an EBV-transformed baboon B-cell line, a primary rhesus kidney and a primary rhesus UVEC line exposed to PK-15 supernatants. Identical studies with primary rhesus lung and aortic endothelial cell cultures were also negative (data not shown). As shown in Fig. 5, after exposure to PERV-NIH, viral DNA was detected after six passages in RF/6A (lane 6), a rhesus retinal endothelial cell line, and was barely detectable in the African green monkey kidney cell line, BSC-1 (lane 9). In contrast, supernatants from PERV DNA-positive RF/6A were repeatedly unable to transfer infection to RF/6A or BSC-1 cells, though they did consistently transfer infection to HEK 293 targets (data not shown).
FIG. 1.
Detection of PERV DNA in primary and established NHP cells and HEK 293 cells. Cells were exposed to supernatant from pig PK-15 PERV producer cells (PK15 sup) or cocultured with these cells (Irradiated PK15). Three to four passages after exposure, DNA was isolated, and PERV-specific sequences were amplified by PCR and hybridized by slot blotting.
FIG. 5.
Detection of PERV DNA in primary and established NHP cells and HEK 293 cells. Cells were exposed to supernatant from PERV-NIH (labeled 293/PERV-NIH) or to supernatant from RF6A cells exposed to PERV-NIH and then pseudotyped with VSVG (labeled PERV-RF6A/VSVG) two times and cultured for six passages before DNA was isolated, amplified by PCR, and subjected to DNA slot blotting (as described in Materials and Methods). Lanes 1 to 3 are results from negative control cells not exposed to any virus-containing supernatant. Lane 14 is from RF6A cells exposed to VSVG pseudotyped PERV-NIH.
Viral DNA is detected in extrachromosomal DNA from NHP cells exposed to PERV.
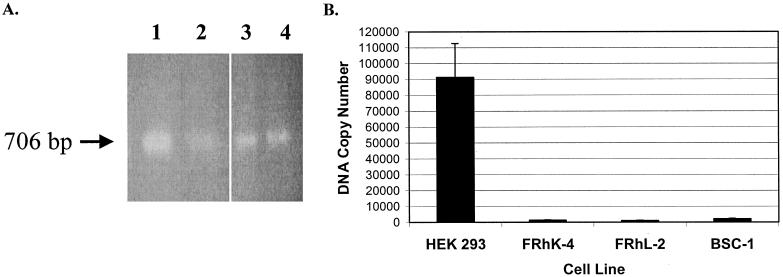
In order to determine if the inefficient infection of NHP cells by PERV is due to an entry block or a block in reverse transcription, we analyzed HEK 293 and NHP cell lines exposed to PERV-NIH for the presence of unintegrated viral DNA as described in Materials and Methods. Primers were designed to the PERV LTR that would amplify across LTR-LTR junctions of circular DNA intermediates, using an approach described by Towers and coworkers (34). A band corresponding to the expected size of 706 bp was obtained for all cell lines under study (Fig. 2A). To confirm the identity of this band as a PERV LTR, the amplified products were cloned into a TA cloning vector (Invitrogen, Carlsbad, Calif.) and sequenced. Sequence analysis showed 100% nucleotide identity to the PERV-C-LTR (GenBank accession no. AF038600) (data not shown). In order to quantitate the total amount of unintegrated, reverse-transcribed viral DNA, we performed PCR using TaqMan technology with primers directed to PERV-pol, as described in Materials and Methods. Copy numbers of unintegrated viral DNA detected were approximately 100-fold lower for FRhK-4 (1311.67 copies of viral DNA) and FRhL-2 (999.63 copies of viral DNA), and about 50-fold lower for BSC-1 (2,129 copies of viral DNA) compared to HEK 293 cells (91,268 copies of viral DNA) (Fig. 2B).
FIG. 2.
Detection of PERV-NIH DNA in extrachromosomal fraction isolated from NHP or HEK 293 cells exposed to PERV. (A) Extrachromosomal DNA was isolated (10) and subjected to PCR analysis that would amplify across LTR-LTR junctions of circular DNA intermediates (see Materials and Methods). An expected 706-bp fragment was observed for HEK 293 (lane 1), FRhK-4 (lane 2), FRhL-2 (lane 3), and BSC-1 (lane 4). (B) Unintegrated viral DNA was quantitated using TaqMan PCR with primers directed toward PERV-pol. A plasmid encoding PERV-pol was used as a standard. All samples were run in duplicate and repeated at least three times. Error bars represent the standard error of the mean.
Exposure of NHP cell lines to PERV-NIH repeatedly leads to PERV infection.
To determine whether NHP cells are permissive to PERV replication, we passaged the cells after initial exposure to PERV-NIH containing supernatant for more than 8 weeks and periodically collected supernatant for RT and RT-PCR analysis. At 2 weeks postexposure, BSC-1 and FRhL-2, but not FRhK-4 or baboon T and B cells, showed a positive signal for PERV-pol in RT-PCR, but all remained negative by RT assay (data not shown). After additional passaging (>4 weeks) of the exposed cells, all cells were negative for PERV-pol by RT-PCR. In an attempt to increase the number of virions entering the cells, the NHP cells were exposed to supernatant collected from PERV-NIH cells two to four times over a period of 2 weeks. Exposed NHP cells were continuously passaged for a period of 12 to 16 weeks and supernatant and cell pellets were collected periodically for analysis. Table 2 shows the results from the analysis of one representative experiment for NHP cells exposed to PERV-NIH. Analyses included quantitation of DNA copy number by real-time PCR as described in Materials and Methods, detection of viral RNA by RT-PCR, and detection of viral RT activity. HEK 293 and HeLa cells served as positive controls. While all NHP cells tested positive after exposure to PERV for PERV expression by RT-PCR, none had detectable levels of RT activity over the course of the experiment. We repeated these experiments three times and could not detect measurable RT-activity during culture periods up to 16 weeks postexposure to PERV. Quantitative analysis of DNA copy number revealed that the PERV DNA copy number was comparable in PERV-exposed NHP cell lines compared to HEK 293 or HeLa cells at the 2-week time point after exposure, but that the copy numbers detected in the NHP cultures quickly declined at subsequent time points, while the DNA copy number increased over time in the cultures of HEK 293 and HeLa cells.
TABLE 2.
Analysis of NHP, HEK 293, and HeLa cells after exposure to PERV-NIH
| Cell line | Time postexposure (wk)a | DNA copy no/ngb | RT-PCR resultc | [3H]TTPd (cpm) |
|---|---|---|---|---|
| RF6A | 2 | NDe | ND | Bkgd |
| 6 | ND | ND | Bkgd | |
| FrK-4 | 2 | 15.6 | ND | Bkgd |
| 4 | 1.3 | + | Bkgd | |
| 12 | 0.3 | + | Bkgd | |
| >16 | 0.06 | ND | ND | |
| FrL-2 | 2 | 3.91 | ND | Bkgd |
| 4 | 1.96 | + | Bkgd | |
| 12 | 1.0 | + | Bkgd | |
| >16 | 0.9 | ND | ND | |
| BSC-1 | 2 | 2.97 | ND | Bkgd |
| 4 | 3.78 | + | Bkgd | |
| 12 | 3.14 | + | Bkgd | |
| >16 | 1.65 | ND | ND | |
| Baboon B | 2 | 0.6 | ND | Bkgd |
| 4 | 0.2 | + | Bkgd | |
| 12 | 0.1 | + | Bkgd | |
| Baboon T | 2 | 0.32 | ND | Bkgd |
| 4 | 0.01 | + | Bkgd | |
| 12 | ND | + | Bkgd | |
| HeLa | 2 | 386 | + | 5,227 ± 920 |
| 4 | 705 | + | 26,268 ± 2,360 | |
| 12 | ND | + | 21,404 ± 140 | |
| HEK 293 | 2 | 858 | + | 15,052 ± 330 |
| 4 | 1,152 | + | 51,129 ± 5,283 | |
| 12 | ND | + | 77,062 ± 301 |
Cells were exposed to supernatant from PERV-NIH.
DNA copy numbers were determined using real-time quantitative PCR for detection of PERV pol sequences from genomic DNA, as described in Materials and Methods. Numbers represent the values obtained per nanogram of genomic DNA analyzed.
RT-PCR analysis for PERV pol sequences was performed as described in Materials and Methods. +, signal was observed either on the gel after agarose gel electrophoresis or after hybridization with a PERV pol-specific probe as described.
Values shown reflect the [3H]TTP incorporated in an RT assay as described previously (40) with background values subtracted. The background value is obtained from a matched mock infected control assayed at the same time point as the culture exposed to the virus. If the value observed for the virus-exposed culture is <2-fold greater than the background value, it is reported as “Bkgd.”
ND, not done.
Analysis of susceptibility of HEK 293 and HeLa cells to supernatant derived from NHP/PERV-NIH.
In order to determine whether NHP cells previously exposed to PERV-NIH (referred to here as NHP/PERV-NIH) produce infectious virions, we exposed HEK 293 and HeLa cells to supernatant derived from PERV-NHP that was RT-negative but RT-PCR positive for PERV. Cultures were passaged for 10 weeks, and supernatant and cell pellets were collected periodically for analysis. HEK 293 became RT positive by 4 weeks postexposure to supernatant derived from BSC-1/PERV-NIH and FRhL-2/PERV-NIH and stayed positive for the remainder of the experiment (Fig. 3A). In contrast, RT activity was not detected in HEK 293 cells exposed to NHP/PERV-NIH obtained from FRhK-4, baboon T, or baboon B cells. HeLa cells only became RT positive after exposure to supernatant derived from BSC-1 cells (Fig. 3B) but not any of the other NHP/PERV-NIH-derived supernatants. The experiment was repeated three times, yielding the same results.
FIG. 3.
Analysis of RT activity in HEK 293 (A) and HeLa cells (B) exposed to FRhK-4/PERV-NIH-containing supernatant (squares), FRhL-2/PERV-NIH-containing supernatant (triangle), BSC-1/PERV-NIH-containing supernatant (upside down triangles), baboon B/PERV-NIH-containing supernatant (diamonds), or baboon T/PERV-NIH-containing supernatant. Data points represent [3H]TTP incorporated (y axis) in an RT assay measured in cell supernatants sampled at time points after NHP/PERV-NIH exposure, as indicated on the x axis.
To determine whether the different results could be due to differences in viral load in the initial supernatants, we measured RNA copy number of the supernatants used for infecting the HEK 293 and HeLa cell cultures. In all cases, the NHP/PERV-NIH supernatants carried >100-fold-lower levels of PERV RNA compared to HEK 293/PERV-NIH supernatants (Table 3). Interestingly, of the samples analyzed, BSC-1 and FrL-2 consistently had higher RNA copies of PERV present than the other cell lines correlating with the resulting RT activity detected in HEK 293 cells exposed to those cultures.
TABLE 3.
Quantitative assessment of NHP/PERV-NIH viral RNA copy number
| NHP/PERV-NIHa | PERV RNA copy no.b (mean ± SEM) |
|---|---|
| FRhK-4 | 90 ± 15 |
| FRhL-2 | 427 ± 114 |
| BSC-1 | 582 ± 106 |
| Baboon B-cell line | 72 ± 27 |
| Baboon T-cell line | 224 ± 7 |
| HEK 293/PERV-NIH | 70,428 ± 2,723 |
Supernatants were harvested from each of the indicated NHP cell lines. These cell lines were previously shown to be positive for PERV RNA and DNA sequences (Table 2) but negative for RT activity.
RNA copy numbers were measured using the TaqMan-based RT-PCR assay for detection and quantitation of PERV pol RNA, as described in Materials and Methods. Values are based on results obtained for duplicate samples run in two assays.
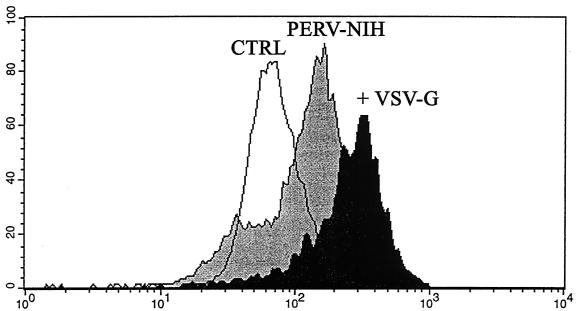
To determine if we could increase the productivity of infection with RF/6A-produced PERV virions by VSV-G pseudotyping we did a transient VSV-G transfection of the PERV DNA-positive RF/6A cell line. Our hypothesis was that increasing the initial viral load over some threshold might allow a productive infection to be developed subsequently in NHP cells. To validate the strategy, we used flow cytometry and a polyclonal anti-p10 PERV Gag antisera. Figure 4 depicts representative staining of RF6A, 3 days after exposure to a 1:2 dilution of RF6A-derived PERV-NIH after VSV-G pseudotyping. The results compared to the same preparation of RF6A-derived PERV-NIH without pseudotyping demonstrate the higher levels of p10 Gag expression on the PERV VSV-G infected cells (geometric mean fluorescent intensities [MFIs] = 29 for Control/RF6A, 39 for PERV-NIH, and 161 for PERV-NIH+VSV-G). Moreover, by 2 weeks of culture (three passages), p10 expression could only be detected on the PERV VSV-G infected RF6A (geometric MFIs = 25 for Control/RF6A, 26 for PERV-NIH and 112 for PERV-NIH+VSV-G). However, as shown in Fig. 5, minimal dilutions of culture supernatants from the VSV-G pseudotyped, PERV-NIH DNA-positive RF/6A cell line readily infected HEK 293 controls (lanes 10 and 11) but not fresh RF/6A (lanes 4 and 5) or BSC-1 (lanes 7 and 8) when cells were studied after six passages (4 weeks). In sum, these results are consistent with early infection of NHP cells with PERV but no evidence of productive infection. Increasing the initial viral exposure using VSV-G pseudotyping does appear to increase short-term infection levels but this also fails to lead to a productive infection when retested after 4 weeks of culture.
FIG. 4.
Analysis of PERV infection by expression of p10 Gag. We developed a flow cytometry assay (see Materials and Methods) to quantify the expression of the p10 nucleocapsid Gag protein recognized by a polyclonal antisera. The control RF6A (CTRL) were naïve cells never exposed to PERV-NIH, and gates were set for analysis using a preimmune serum. This histogram compares the p10 staining of control RF6A to RF6A 3 days after exposure to PERV-NIH or PERV-NIH pseudotyped with VSV-G. The relative fluorescent intensity is shown on the x axis, and the cell number is on the y axis. The geometric MFI for each curve is given in the text.
Analysis of susceptibility of NHP and HEK 293 cell lines to NHP/PERV-NIH pseudotypes.
We generated viral pseudotypes expressing β-galactosidase by superinfecting NHP/PERV-NIH cells with retrovirus vector-containing supernatant as described in Materials and Methods to assess whether infectious particles could be rescued. HEK 293, FRhK-4, FRhL-2, and BSC-1 cells were exposed to supernatant of FRhK-4/PERV-NIH and BSC-1/PERV-NIH pseudotypes and infectious titers were determined after histochemical staining 72 h after infection. Very low levels of β-galactosidase positive cells were observed on HEK 293 cells whereas no β-galactosidase positive cells were observed on NHP cells tested (Table 4).
TABLE 4.
Infectious titers on HEK 293 and NHP cells of NHP/PERV-NIH-LacZ virus pseudotypes
| Cell line | Avg BFU/ml ± SEMa for pseudotype
|
|
|---|---|---|
| FrK-4/PERV-NIH-LacZ (16.2 ± 6.4)b | BSC-1/PERV-NIH-LacZ (230 ± 31) | |
| HEK 293 | 3.4 ± 0.8 | 16.5 ± 5.4 |
| FrK-4 | NDc | ND |
| FrL-2 | ND | ND |
| BSC-1 | ND | ND |
BFU are as explained in Materials and Methods. Cells were exposed to supernatant containing viral pseudotypes encoding β-galactosidase as described in Materials and Methods. Values shown are averages of three independent experiments from triplicate wells.
Data in parentheses are input RNA copy numbers as measured by TaqMan RT-PCR and described in Materials and Methods. Values are reported as average of three independent experiment ± standard error of the mean.
ND, none detected.
Infection of naïve NHP cells by supernatant derived from NHP/PERV-NIH.
Since NHP/PERV-NIH supernatant contained infectious particles, as measured by productive infection of HeLa and HEK 293 cells and by ability to rescue the MuLV vector genome, we wanted to determine if the same supernatant derived from NHP/PERV-NIH could productively infect naive NHP cells. Target cells were exposed singly or multiple times to supernatant collected from NHP/PERV-NIH and then passaged over a period of 12 weeks. Supernatant and cell pellets were collected periodically for RT and RT-PCR analysis. In a total of 4 experiments, all target cells remained negative for RT activity (data not shown). RT-PCR on RNA isolated from the culture supernatant and TaqMan PCR on isolated cellular DNA did not show a positive result for any of the cell lines tested.
DISCUSSION
Previous reports about the susceptibility of NHP cell lines to PERV have produced contradictory results. The results presented in this report clearly demonstrate that while NHP cells allow for PERV infection, they do not support PERV replication in NHP cell cultures well enough to create productive infections. The difference in susceptibility to infection vs. productive infection may explain the apparent inconsistency of previous studies. Most authors measured infection by directly analyzing target cells with PCR or RT-PCR for viral nucleic acid sequences (4, 27, 31). The enhanced sensitivity of PCR-based assays may yield a positive result indicating viral infection of the cells despite insufficient levels of viral replication to produce infectious supernatants of high enough titer to create productive infections.
Using PERV-NIH-LacZ pseudotypes, we found that the NHP cell lines FRhK-4, FRhL-2, and BSC-1 are susceptible to PERV infection, but the apparent titer on these cell lines is 1,000- to 10,000-fold lower when compared to the apparent titer on HEK 293 cells. Previous results using LacZ pseudotypes (39) were negative for PERV infection of FRhK-4 cells. The different results likely reflect the use of the LacZ pseudotypes made with PERV-NIH-3° in this study that had an approximately 10-fold higher infectious titer on HEK 293 cells than the pseudotypes used in the previous report. The increased titer of infectious pseudotypes, therefore, increases the sensitivity of detecting virus infection in NHP cell lines.
Since receptor-mediated blocks to infection usually result in a >6-log decrease in infectivity, we suspected a postentry-mediated inhibition of infection (1). In particular, the 1,000- to 10,000-fold reduction in titer is similar to that observed in murine cells expressing the Fv-1 restriction to certain strains of MuLV (8). Since MuLV infection in Fv-1 restricted cells is blocked at the formation of circular DNA intermediates, but not in the formation of linear viral DNA (5, 11, 40), we examined whether reverse-transcribed circular DNA intermediates were present in the NHP cell lines by analyzing for the presence of LTR-LTR junctions. Detection of these intermediates in NHP cells after exposure to PERV, suggests that an Fv-1-like restriction does not account for the reduction in infection of these cells. However, the quantitative analysis of total unintegrated viral DNA revealed that there is a greater than 100-fold lower DNA copy number in NHP cells, suggesting that entry, reverse transcription, or both are occurring less efficiently than in human HEK 293 cells. This observation is similar to the one made by Towers and colleagues that certain NHP and human cell lines mediated an Fv-1-like restriction that manifested as a block to formation of any linear DNA (34).
The observed decrease in unintegrated DNA copy number (100-fold) is not as great as the observed decrease in the infectivity titer of LacZ pseudotypes (1,000- to 10,000-fold). The observed differences may reflect the fact that our assay measured circular DNA intermediates that are not integration-competent, and thus may not accurately reflect the number of DNA intermediates available for integration. Alternatively, we suspected that additional steps in viral replication might be compromised. A simple reduction in viral entry efficiency into NHP cells, for example by relatively low viral receptor expression levels, was excluded by our experiments with pseudotyping RF/6A PERV DNA-positive cells. The resulting supernatants were still not capable of carrying infection over to fresh RF/6A or BSC-1, though their capability to infect HEK 293 remained evident (Fig. 5). Moreover, analysis of viral-containing supernatant of NHP cells after exposure to PERV-NIH failed to detect RT activity, in spite of positive results from DNA PCR and RT-PCR for viral sequences (Table 2). A similar result has been observed for other cell lines (feline cell line, AK-D, FC3TG; human colorectal cancer cell line, Caco-2, human Jurkat and K56 cells) (39). Quantitation of viral DNA copy number with TaqMan PCR at different time points after exposure to PERV-NIH strongly suggested that the NHP cell lines were not permissive to PERV replication. Whereas the copy number stayed constant or increased for HEK 293 and HeLa cells, it decreased in NHP cells during the 16 weeks of culture after exposure to PERV, suggesting that the virus was not spreading in the cultured NHP cells.
In spite of an apparent block to PERV replication in NHP cells, these cells do release infectious particles. Exposure of naïve HEK 293 cells to supernatant collected from BSC-1/PERV-NIH, FRhL-2/PERV-NIH, and RF6A/PERV-NIH resulted in a productive infection. RT activity was detectable at 4 weeks postexposure and remained positive for the remainder of the experiment. HeLa cells were not quite as sensitive to infection by NHP-derived PERV, as only supernatant derived from BSC-1/PERV-NIH resulted in a productive infection of HeLa cells. None of the other NHP cell lines, FRhK-4, baboon T, or baboon B, that were positive for PERV by RT-PCR were able to productively infect either HEK 293 or HeLa cells. Nor were any of the NHP/PERV-NIH supernatants able to transmit virus to naïve NHP cells. The absence of transmission of PERV by some NHP/PERV-NIH but not all is correlated to the viral RNA copy number measured by TaqMan RT-PCR of the input supernatant. All of the NHP/PERV-NIH RNA copy numbers measured were significantly lower compared to PERV-NIH from HEK 293 cells. PERV RNA copy numbers were consistently low (<250 [Table 3]) for the supernatants from NHP/PERV-NIH, FRhK-4, or baboon T or baboon B cells that did not result in infection of HEK 293 or HeLa cells. In contrast, the RNA levels were always greater than 350 copies in the three NHP/PERV-NIH supernatants that did transmit PERV to HEK 293 cells. These reproducible results suggest that there may be a threshold number of particles required to productively infect HEK 293 or HeLa cells with HeLa cells requiring a slightly higher number of particles (FRhL-2 supernatants with slightly lower values than BSC-1 did not transmit virus to HeLa). Since susceptibility of NHP to PERV is at least 100-fold lower compared to HEK 293 cells and the number of virions released by NHP/PERV-NIH is 2 to 3 logs lower than to HEK 293/PERV-NIH, the correlation between RNA copy number and ability to initiate a productive infection can be further extrapolated to explain the inability of NHP/PERV-NIH to transmit virus to other NHP cells.
Our data, together with previous studies (4, 27, 31), provide evidence that a number of different NHP cell lines examined express a functional cell surface receptor for PERV. However, the early steps of infection prior to the formation of double-stranded DNA intermediates in the cytoplasm are compromised—occurring 100-fold less efficiently than in HEK 293 control cells. In addition, the decrease in DNA copy number in genomic DNA of NHP cells during culture suggests that there may be multiple blocks to PERV infection in NHP cells. For example, a number of cellular factors in addition to the receptor and coreceptor have been demonstrated to play a role in the inability of HIV to replicate in murine cells. Inefficient tat-mediated transactivation of the HIV LTR can be rescued by expression of human cyclin T1, due to incompatibility of murine cyclin T1 (for example, see reference 13). However, additional cellular factors appear to be necessary to fully reconstitute efficient HIV replication in murine cells. Murine cells transfected with the receptor, coreceptor and human cyclin T1, can support HIV reverse transcription, integration, and expression at levels comparable to those in human permissive cells, but assembly is defective resulting in formation of virions with insufficient p24 gag (17). In fact, this observation results in a phenomenon similar to the one reported here, where a low level of infectious virions are released when assayed upon a permissive human cell line, but that the levels are too low to generate replication and spread throughout the murine cell culture (17). Subsequent analysis of human-mouse heterokaryons have demonstrated that the block to accurate HIV assembly in murine cells is due to absence of a cellular factor in murine cells (16). It remains to be seen whether the block to PERV replication in NHP cells is due to the lack of cellular factors present in HEK 293 but absent in NHP cells.
In summary, we have examined five different cell lines representing three different primates (rhesus macaques, African green monkey, and baboon) and four different tissue types (kidney, lung, B, and T cells) and found that all restrict PERV replication. In addition, we found that primary cells derived from rhesus kidney or umbilical vein and aortic endothelial cells were also resistant to PERV infection. While this is not a comprehensive analysis of all different types of tissues that may be found in vivo, it suggests that there may be a common restriction to PERV replication in NHP cells. In addition, our study demonstrates the need to use multiple methods to assess cell-specific susceptibility to retroviral infection vs. permissiveness to replication. Finally, these results indicate that NHP may not provide a good animal model for examining the potential transmission and pathogenicity of PERV in vivo, and that the significance of previous studies where negative results have been reported may need to be reevaluated in light of these data.
Acknowledgments
We thank Jonathan Allan for the gift of the baboon B- and T-cell lines, Ralf Toenjes for the gift of the anti-PERV antibody, and Takele Argaw for designing and testing the TaqMan assay used in this study. We also thank Susan Wong and Winston Colon-Moran for providing expert technical assistance. Nancy Markovitz and Andrew Byrnes provided critical review of the manuscript.
This work was supported by NIH grant R01 AI 45494 to D. R. Salomon and a JDRF fellowship award to L. J. W. van der Laan.
REFERENCES
- 1.Albritton, L. M., L. Tseng, D. Scadden, and J. M. Cunningham. 1989. A putative murine ecotropic receptor gene encodes a multiple membrane-spanning protein and confers susceptibility to virus infection. Cell 57:659-666. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, J. A., J. S. Porterfield, and A. T. de Madrid. 1971. C-type virus particles in pig kidney cell lines. J. Gen. Virol. 10:195-198. [DOI] [PubMed] [Google Scholar]
- 3.Benveniste, R. E., and G. J. Todaro. 1975. Evolution of type C viral genes: preservation of ancestral murine type C viral sequences in pig cellular DNA. Proc. Natl. Acad. Sci. USA 72:4090-4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blusch, J. H., C. Patience, Y. Takeuchi, C. Templin, C. Roos, K. van der Helm, G. Steinhoff, and U. Martin. 2000. Infection of nonhuman primate cells by pig endogenous retrovirus. J. Virol. 74:7687-7690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chinsky, J., and R. Seiro. 1981. Fv-1 host restriction of Friend leukemia virus: analysis of unintegrated proviral DNA. J. Virol. 40:45-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng, Y.-M., B. E. Tuch, and W. D. Rawlinson. 2000. Transmission of porcine endogenous retroviruses in severe combined immunodeficient mice xenotransplanted with fetal porcine pancreatic cells. Transplantation 70:1010-1016. [DOI] [PubMed] [Google Scholar]
- 7.Fink, J. S., J. M. Schumacher, S. L. Ellias, E. P. Palmer, M. Saint-Hilaire, K. Shannon, R. Penn, P. Starr, C. VanHorne, H. S. Kott, P. K. Dempsey, A. J. Fischman, R. Raineri, C. Manhart, J. Dinsmore, and O. Isacson. 2000. Porcine xenografts in Parkinson's disease and Huntington's disease patients: preliminary results. Cell Transplant 9:273-278. [DOI] [PubMed] [Google Scholar]
- 8.Hartley, J. W., W. P. Rowe, and R. J. Huebner. 1970. Host-range restrictions of murine leukemia viruses in mouse embryo cell cultures. J. Virol. 5:221-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heneine, W., A. Tibell, W. M. Switzer, P. Sandstrom, G. V. Rosales, A. Mathews, O. Korsgren, L. E. Chapman, T. M. Folks, and C. G. Groth. 1998. No evidence of infection with porcine endogenous retrovirus in recipients of porcine islet-cell xenografts. Lancet 352:695-699. [DOI] [PubMed] [Google Scholar]
- 10.Hirt, B. 1967. Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26:365-369. [DOI] [PubMed] [Google Scholar]
- 11.Jolicoer, P., and E. Rassart. 1980. Effect of Fv-1 gene product on synthesis of linear and supercoiled viral DNA in cells infected with murine leukemia virus. J. Virol. 33:183-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krach, U., N. Fischer, F. Czauderna, R. Kurth, and R. Tonjes. 2000. Generation and testing of a highly specific anti-serum directed against porcine endogenous retrovirus nucleocapsid. Xenotransplantation 7:221-229. [DOI] [PubMed] [Google Scholar]
- 13.Kwak, Y. T., D. Ivanov, J. Guo, E. Nee, and R. B. Gaynor. 1999. Role of the human and murine cyclin T proteins in regulating HIV-1 tat-activation. J. Mol. Biol. 288:57-69. [DOI] [PubMed] [Google Scholar]
- 14.Lieber, M. M., C. J. Sherr, R. E. Benveniste, and G. J. Todaro. 1975. Biologic and immunologic properties of porcine type C viruses. Virology 66:616-619. [DOI] [PubMed] [Google Scholar]
- 15.Loss, M., H. Arends, M. Winkler, M. Przemeck, G. Steinhoff, S. Rensing, F.-J. Kaup, H. J. Hedrich, M. E. Winkler, and U. Martin. 2001. Analysis of potential porcine endogenous retrovirus (PERV) transmission in a whole-organ xenotransplantation model without interfering microchimerism. Transplant. Int. 14:31-37. [DOI] [PubMed] [Google Scholar]
- 16.Mariani, R., B. A. Rasala, G. Rutter, K. Wiegers, S. M. Brandt, H. G. Krausslich, and N. R. Landau. 2001. Mouse-human heterokaryons support efficient human immunodeficiency virus type 1 assembly. J. Virol. 75:3141-3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mariani, R., G. Rutter, M. E. Harris, T. J. Hope, H. G. Krausslich, and N. R. Landau. 2000. A block to human immunodeficiency virus type 1 assembly in murine cells. J. Virol. 74:3859-3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martin, U., V. Kiessig, J. Blusch, A. Haverich, K. van der Helm, T. Herden, and G. Steinhoff. 1998. Expression of pig endogenous retrovirus by primary porcine endothelial cells and infection of human cells. Lancet 352:692-694. [DOI] [PubMed] [Google Scholar]
- 19.Martin, U., G. Steinhoff, V. Kiessig, M. Chikobava, M. Anssar, T. Morschheuser, B. Lapin, and A. Haverich. 1999. Porcine endogenous retrovirus is transmitted neither in vivo nor in vitro from porcine endothelial cells to baboons. Transplant. Proc. 31:913-914. [DOI] [PubMed] [Google Scholar]
- 20.Martin, U., G. Steinhoff, V. Kiessig, M. Chikobava, M. Anssar, T. Morschheuser, B. Lapin, and A. Haverich. 1998. Porcine endogenous retrovirus (PERV) was not transmitted from transplanted porcine endothelial cells to baboons in vivo. Transplant. Int. 11:247-251. [DOI] [PubMed] [Google Scholar]
- 21.Martin, U., M. E. Winkler, M. Id, H. Radeke, L. Arseniev, Y. Takeuchi, A. R. Simon, C. Patience, A. Haverich, and G. Steinhoff. 2000. Productive infection of primary human endothelial cells by pig endogenous retrovirus (PERV). Xenotransplantation 7:138-142. [DOI] [PubMed] [Google Scholar]
- 22.Onions, D. E., and C. J. Witt. 2000. Xenotransplantation: an overview of microbiological risks and potentials for risk management. Rev. Sci. Technol. 19:289-301. [DOI] [PubMed] [Google Scholar]
- 23.Paradis, K., G. Langford, Z. Long, W. Heneine, P. Sandstrom, W. Switzer, L. Chapman, C. Lockey, D. Onions, T. X. S. Group, and E. Otto. 1999. Search for cross-species transmission of porcine endogenous retrovirus in patients treated with living pig tissue. Science 285:1236-1241. [DOI] [PubMed] [Google Scholar]
- 24.Patience, C., G. S. Patton, Y. Takeuchi, R. A. Weiss, M. O. McClure, L. Rydberg, and M. E. Breimer. 1998. No evidence of pig DNA or retroviral infection in patients with short-term extracorporeal connection to pig kidneys. Lancet 352:699-701. [DOI] [PubMed] [Google Scholar]
- 25.Patience, C., Y. Takeuchi, and R. A. Weiss. 1997. Infection of human cells by an endogenous retrovirus of pigs. Nat. Med. 3:282-286. [DOI] [PubMed] [Google Scholar]
- 26.Specke, V., S. Rubant, and J. Denner. 2001. Productive infection of human primary cells and cell lines with porcine endogenous retroviruses. Virology 285:177-180. [DOI] [PubMed] [Google Scholar]
- 27.Specke, V., S. J. Tacke, K. Boller, J. Schwendemann, and J. Denner. 2001. Porcine endogenous retroviruses: in vitro host range and attempts to establish small animal models. J. Gen. Virol. 82:837-844. [DOI] [PubMed] [Google Scholar]
- 28.Switzer, W. M., R. E. Michler, V. Shanmugam, A. Matthews, A. I. Hussain, A. Wright, P. Sandstrom, L. E. Chapman, C. Weber, S. Safley, R. R. Denny, A. Navarro, V. Evans, A. J. Norin, P. Kwiatkowski, and W. Heneine. 2001. Lack of cross-species transmission of porcine endogenous retrovirus infection to nonhuman primate recipients of porcine cells, tissues, or organs. Transplantation 71:959-969. [DOI] [PubMed] [Google Scholar]
- 29.Takefman, D. M., S. Wong, T. Maudru, K. Peden, and C. A. Wilson. 2001. Detection and characterization of porcine endogenous retrovirus in porcine plasma and porcine factor VIII. J. Virol. 75:4551-4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeuchi, Y., C. Patience, S. Magre, R. A. Weiss, P. T. Banerjee, P. L. Tissier, and J. P. Stoye. 1998. Host range and interference studies of three classes of pig endogenous retrovirus. J. Virol. 72:9986-9991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Templin, C., C. Schroder, A. R. Simon, G. Laaff, J. Kohl, M. Chikobava, B. Lapin, G. Steinhoff, and U. Martin. 2000. Analysis of potential porcine endogenous retrovirus transmission to baboon in vitro and in vivo. Transplant. Proc. 32:1163-1164. [DOI] [PubMed] [Google Scholar]
- 32.Ting, Y.-T., C. A. Wilson, K. B. Farrell, G. J. Chaudry, and M. V. Eiden. 1998. Simian sarcoma-associated virus fails to infect Chinese hamster cells despite the presence of functional gibbon ape leukemia virus receptors. J. Virol. 72:9453-9458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Todaro, G. J., R. E. Benveniste, M. M. Lieber, and C. J. Sherr. 1974. Characterization of a type C virus released from the porcine cell line PK(15). Virology 58:65-74. [DOI] [PubMed] [Google Scholar]
- 34.Towers, G., M. Bock, S. Martin, Y. Takeuchi, J. P. Stoye, and O. Danos. 2000. A conserved mechanism of retrovirus restriction in mammals. Proc. Natl. Acad. Sci. USA 97:12295-12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tristem, M., P. Kabat, L. Lieberman, S. Linde, A. Karpas, and F. Hill. 1996. Characterization of a novel murine leukemia virus-related subgroup within mammals. J. Virol. 70:8241-8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van der Laan, L. J. W., C. Lockey, B. C. Griffeth, F. S. Frasier, C. A. Wilson, D. E. Onions, B. J. Hering, Z. Long, E. Otto, B. E. Torbett, and D. R. Salomon. 2000. Infection by porcine endogenous retrovirus after islet xenotransplantation in SCID mice. Nature 407:90-94. [DOI] [PubMed] [Google Scholar]
- 37.Wilson, C., and M. Eiden. 1991. Viral and cellular factors governing hamster cell infection by murine and gibbon ape leukemia viruses. J. Virol. 65:5975-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilson, C., S. Wong, J. Muller, C. Davidson, T. Rose, and P. Burd. 1998. Type C retrovirus released from porcine primary peripheral blood mononuclear cells infects human cells. J. Virol. 72:3082-3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilson, C. A., S. Wong, M. V. Brocklin, and M. J. Federspiel. 2000. Extended analysis of the in vitro tropism of porcine endogenous retrovirus. J. Virol. 74:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang, W. K., J. O. Kiggans, D.-M. Yang, C.-Y. Ou, R. W. Tennant, A. Brown, and R. H. Bassin. 1980. Synthesis and circularization of N- and B-tropic retroviral DNA in Fv-1 permissive and restrictive mouse cells. Proc. Natl. Acad. Sci. USA 77:2994-2998. [DOI] [PMC free article] [PubMed] [Google Scholar]