Abstract
The teschoviruses constitute a recently defined picornavirus genus. Most of the genome sequence of the porcine teschovirus-1 (PTV) Talfan and several other strains is known. We now demonstrate that initiation of protein synthesis occurs at nucleotide (nt) 412 on the PTV Talfan RNA and that nt 1 to 405 contains an internal ribosome entry site (IRES) that functions efficiently in vitro and within mammalian cells. In comparison with other picornavirus IRES elements, the PTV IRES is relatively short and lacks a significant polypyrimidine tract near the 3′ end. Expression of an enterovirus 2A protease, which induces cleavage of eIF4G within the translation initiation complex eIF4F, has little effect on the PTV IRES activity within BHK cells. The PTV IRES has a unique set of properties and represents a new class of picornavirus IRES element.
The Teschovirus genus within the family Picornaviridae was recently established (19) to include a collection of porcine enteroviruses (PEVs) that differ significantly in their genome organization from the other picornaviruses (e.g., poliovirus) that constitute the Enterovirus genus. Some of these PEVs cause significant disease; for example, the Teschen and Talfan strains of PEV-1 were isolated from outbreaks of polioencephalomyelitis in swine (14, 25). The Talfan strain is now the reference strain for the Teschovirus genus and has been renamed porcine teschovirus (PTV-1) Talfan. The sequences of the major portion of the genome of the PTV-1 F65 strain (9) and the PTV-1 Talfan virus (15) have been determined. However, the 5′ terminal sequences of these genomes, upstream of a possible poly(C) tract (9), have not yet been cloned or sequenced.
All picornaviruses have a positive-sense RNA genome of about 8 kb. The RNA has a poly(A) tail at the 3′ terminus and includes a single large open reading frame. In contrast to eukaryotic cellular mRNAs, the viral RNA does not have a cap structure (m7GpppN…) at its 5′ terminus but the genomic RNA is linked to a small virus-encoded peptide termed VPg (or 3B). The 5′ untranslated regions (5′UTRs) vary from about 600 nucleotides (nt) to 1,300 nt among different picornaviruses and include multiple AUG codons that are not used for the initiation of protein synthesis. It is now well established that picornavirus RNAs contain an element in their 5′UTRs, termed an internal ribosome entry site (IRES), which directs the cap-independent initiation of protein synthesis on the viral RNA (2, 3). To date, three distinct types of picornavirus IRES elements have been recognized. The rhino- and enterovirus RNAs contain one type of element, which functions poorly in the rabbit reticulocyte lysate (RRL) in vitro translation system unless it is supplemented with extracts from HeLa cells (7, 10). The cardio- and aphthovirus IRES elements (e.g., from encephalomyocarditis virus [EMCV] and foot-and-mouth disease virus [FMDV]) constitute a second type of element with a predicted secondary structure that is completely different from that derived for the rhino- and enterovirus IRES elements. The cardio- and aphthovirus IRES elements function very efficiently in the RRL translation system. The third type of picornavirus IRES element that has been identified is that found within the hepatitis A virus (HAV) genome (8). This has another different predicted secondary structure, and its activity within the RRL system is not enhanced by HeLa cell factors, but it has been reported that liver cell extracts can stimulate activity (13). The HAV IRES has low activity compared to the EMCV IRES in vitro and within cells (26). A unique feature of the HAV IRES is its requirement for an intact cap-binding complex, eIF4F (1, 4, 26). The translation initiation factor eIF4F complex (12) comprises eIF4E (which binds to the cap structure on mRNAs), eIF4A (an RNA helicase), and eIF4G. The latter component is believed to have a scaffold function that enables the eIF4F complex to bridge between the mRNA and the ribosome. The eIF4G interacts with several other proteins, e.g., eIF3 (on the small ribosomal subunit), the poly(A) binding protein, and Mnk-1 (an eIF4E kinase). When eIF4G is cleaved, a process induced by the 2A protease of the entero- and rhinoviruses, the cap-binding complex is inactivated for the initiation of translation on capped mRNAs. However, the modified complex is still competent to initiate translation on the entero- and rhinovirus and cardio- and aphthovirus IRES elements (for a review, see reference 2). Indeed, in certain cell types the activity of the entero- and rhinovirus IRES elements is stimulated by the expression of the 2A protease (5, 22). In contrast, the HAV IRES is inhibited under these conditions (1, 6, 26).
Following sequence analysis of the PTV-1 strain F65, it was suggested that initiation of protein synthesis on the PTV RNA could occur at one of two different AUG codons (9). However, comparison with other PTV sequences indicated that the first of these AUGs (nt 336 in the F65 sequence) was not conserved (despite an extremely high level of sequence identity between different strains of PTV within the 5′UTR [28]). In the PTV Talfan sequence in place of the AUG codon at position 316 (equivalent to position 336 in the F65 sequence), there is a GUG codon (15). It has, therefore, been proposed (but not shown) that the next, in-frame AUG (at nt 432 in the F65 sequence), which is adjacent to a region of complementarity to 18S rRNA, is the authentic initiation codon (28).
Identification of the initiation codon on PTV Talfan RNA.
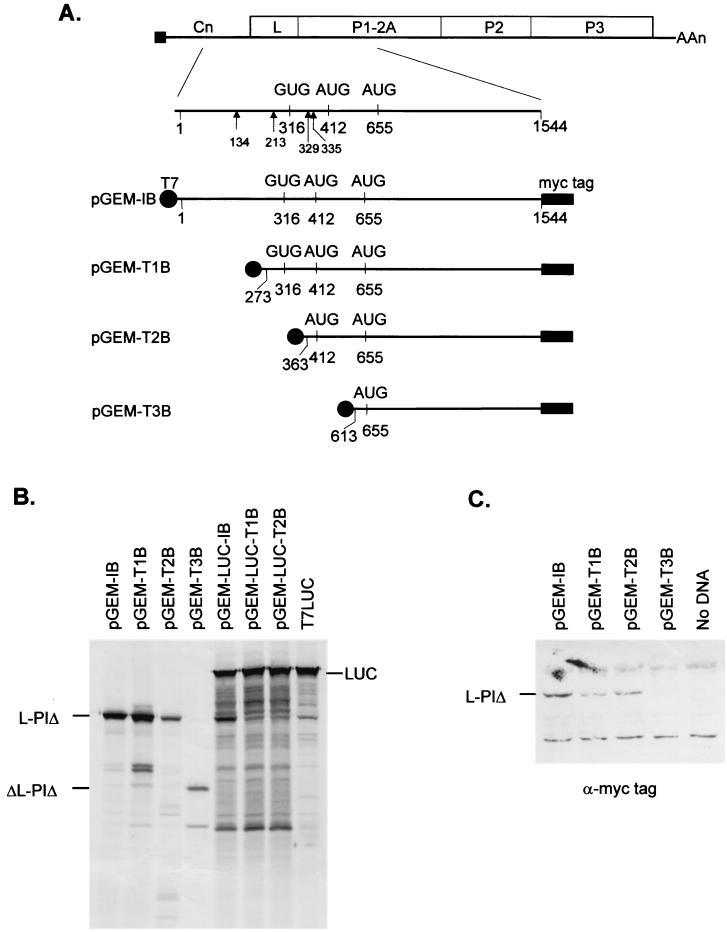
In order to identify the location of the initiation codon within the PTV Talfan RNA sequence, a series of reporter plasmids were constructed. By using PTV Talfan cDNA as a template, the known 5′UTR plus a portion of the viral coding region (encoding the L protein and part of the capsid precursor P1, termed P1Δ) was amplified by PCR with the primers XhoI+Talfan 1F and HindIII+Talfan1544R (Table 1) to generate a fragment of about 1.6 kbp. This was digested with XhoI and HindIII, and the purified fragment was ligated into similarly digested pBAD/myc-HisB (Invitrogen) by using standard methods (24). The vector adds the coding sequence for a C-terminal myc-epitope tag (to facilitate detection of protein expression) together with a termination codon. Using this plasmid (pBAD-I-mycB) as a template, another PCR was performed with primers pBADF and pBADR (Table 1). The product was digested with BamHI and SphI, and the fragment, including the PTV 5′UTR and myc-tagged coding sequence, was isolated and ligated into similarly digested pGEM3Z (Promega) to create pGEM-IB (Fig. 1). This plasmid directs the expression of the PTV RNA (nt 1 to 1544) under control of the T7 promoter. Derivatives of this plasmid which expressed shorter regions of the viral RNA that were truncated from the 5′ terminus were prepared. The cDNAs were generated by PCR using the plasmid pBAD-I-mycB as a template, with the forward primers TST1, TST2, and TST3 (Table 1) in conjunction with the pBADR primer. The fragments, of about 1.4, 1.3, and 1.05 kbp, respectively, were purified, digested with BamHI and SphI, and then ligated into similarly digested pGEM3Z to generate pGEM-T1B, pGEM-T2B, and pGEM-T3B, respectively (Fig. 1). Each of these plasmids expresses, from a T7 promoter, a monocistronic mRNA encoding a myc-tagged portion of the PTV coding region. The RNAs differ in their 5′ terminal start sites (Fig. 1A), which were selected to test the possible use of different AUG initiation codons. The plasmids were used to program in vitro TNT reactions (Promega) containing RRL with [35S]-methionine, and the products were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (17) and autoradiography (Fig. 1B). Plasmids pGEM-IB, pGEM-T1B, and pGEM-T2B, which contain Talfan cDNA sequences starting at nt 1, 273, and 363, respectively, predominantly expressed a product of about 48 kDa. This corresponds to the myc-tagged L-P1Δ product encoded by the open reading frame that commences at nt 412 and was terminated at nt 1544 in the PTV Talfan sequence by the addition of the myc-tag sequence and termination codon. Plasmid pGEM-T3B, which expresses an RNA that starts at nt 613, produced a truncated product (ΔL-P1Δ, about 33 kDa) presumably resulting from initiation at the first AUG codon within this transcript at nt 655 (this is in the correct reading frame).
TABLE 1.
PCR primers
| Primer | Sequence (5′-3′)a | Location (nt) on PTV RNAb |
|---|---|---|
| XhoI+Talfan 1F | ATACTGCAGCCC TCTGGACTTGTAACTGGTAA | 1-20 |
| HindIII+Talfan1544R | TGTAAGCTTGACAGGGTTGCTGAAGAATTTGT | 1544c-1522 |
| pBADF | ATCGGATCCGGAATTAACCTTGGATCCG | |
| pBADR | TGTGCATGCAAAACAGCCAAGCTGGAG | |
| TST1 | AGCGGATCCATATCCCTAGGCACCTATTG | 273-292 |
| TST2 | AGCGGATCCAGGGTCGCGGCTGGCCGTCT | 363-382 |
| TST3 | AGCGGATCCCCTTTAGATCCAACTTCTTT | 613-632 |
| TAL2R | CCCGGATCCTTTCAACTGACTATACAAAGT | 405-385 |
| TAL3R | CCCGGATCCGCTCAAAATCACAAACCAAAGA | 648-628 |
Sequences within the oligonucleotides that introduce restriction enzyme sites are indicated in italics.
The numbering system is that used for the PTV Talfan sequence (15) (Genbank accession number AB038528). The sequences of the reverse primers (suffixed R) are complementary to the locations indicated.
nt 1544 has been changed from a G to a C to prevent the formation of a stop codon with the HindIII site sequence.
FIG. 1.
Identification of the initiation codon on PTV Talfan RNA. (A) Genome organization of the PTV Talfan RNA. The presence of the virus-encoded peptide VPg (also called 3B), attached to the 5′ end of the genome, is depicted by a black square. The locations of potential initiation codons are indicated, and the sites of additional AUG codons that are out of frame with the polyprotein-coding sequence, are shown by arrowheads. The structures of plasmids which express, from the T7 promoter (black circles), portions of the PTV genome (within nt 1 to 1544) followed by sequences encoding a myc-epitope tag plus a termination codon (indicated by solid rectangles) are illustrated. (B) The indicated plasmids were used to program in vitro TNT reactions containing RRL and [35S]-methionine. The products were analyzed by SDS-PAGE and autoradiography. (C) Plasmids expressing monocistronic transcripts were transfected into vTF7-3-infected BHK cells. After 20 h, cell lysates were prepared and analyzed by SDS-PAGE and immunoblotting using antibodies specific for the c-myc epitope tag (9E10). Products were detected with peroxidase-labeled antimouse antibodies and chemiluminescence reagents on X-ray film.
The same monocistronic plasmids were also assayed by transfection using Lipofectin (Life Technologies) into BHK cells infected with the recombinant vaccinia virus vTF7-3 (11), which expresses the T7 RNA polymerase, as described previously (22). The products were analyzed by SDS-PAGE and detected by immunoblotting with an anti-myc tag antibody (MAb 9E10; Santa Cruz Biotechnology Inc.) and peroxidase-labeled antimouse antibodies (Amersham) with chemiluminescence reagents (Amersham). Consistent with the TNT results, a single myc-tagged-L-P1Δ species was observed within extracts of cells transfected with plasmids pGEM-IB, pGEM-T1B, and pGEM-T2B (Fig. 1C). However, no product was detected from pGEM-T3B; perhaps the recognition of the AUG codon at nt 655 was poor in this system or the product was unstable within the cells.
Identification of an IRES within the PTV Talfan RNA.
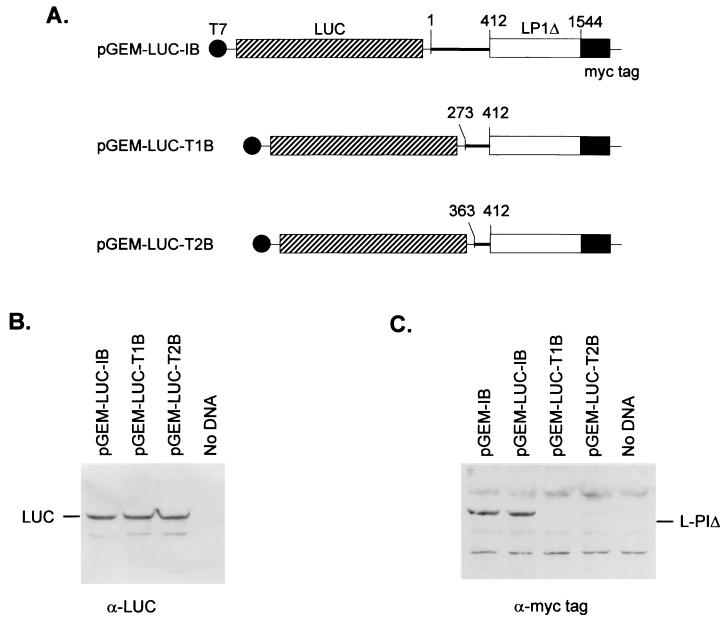
The standard method for identifying IRES elements is the construction and assay of plasmids that express dicistronic mRNAs. In order to preserve the normal linkage between the 5′UTR of the PTV Talfan RNA and its coding sequence, the dicistronic constructs were produced by introducing the coding sequence for firefly luciferase (LUC) sequence (BamHI-XhoI fragment from pGEM-luc [Promega]) between the T7 promoter and the PTV 5′UTR sequences. Thus, the myc-tagged L-P1Δ Talfan coding sequences are the second cistrons in these constructs (Fig. 2A). Derivatives of the monocistronic plasmids pGEM-IB, pGEM-T1B, and pGEM-T2B, which each expressed the correct L-P1Δ product (Fig. 1) were prepared and termed pGEM-LUC-IB, pGEM-LUC-T1B, and pGEM-LUC-T2B, respectively (Fig. 2).
FIG. 2.
Identification of an IRES element within the PTV Talfan RNA. (A) Plasmids that express, from a T7 promoter, dicistronic mRNAs were constructed. They contain the LUC coding sequence between the T7 promoter and the indicated PTV Talfan sequences. (B and C) Plasmids were used to transfect vTF7-3-infected BHK cells, and cell extracts were prepared and analyzed as described for Fig. 1C by using antibodies directed against LUC (B) and the c-myc tag (C). Detection was achieved by using appropriate peroxidase-labeled antispecies antibodies and chemiluminescence reagents. Plasmid pGEM-IB (Fig. 1) served as a positive control for the expression of the myc-tagged L-P1Δ.
These dicistronic plasmids were assayed by using TNT assays (Fig. 1B) and within the transient expression assay system within vTF7-3-infected BHK cells (Fig. 2B and C). In the TNT assays, each of the plasmids produced the LUC product (Fig. 1B) but only the pGEM-LUC-IB construct produced any detectable L-P1Δ product (note that the generation of truncated forms of LUC within the TNT system may have masked low-level synthesis of this product). Within cells, each of the dicistronic plasmids expressed the upstream cistron, LUC, with a similar efficiency (Fig. 2B), as expected, but only the plasmid pGEM-LUC-IB (with PTV RNA sequence starting at nt 1) expressed the myc-tagged L-P1Δ product from the downstream cistron (Fig. 2C), consistent with the TNT assay results. The expression of the L-P1Δ product in cells from the dicistronic plasmid pGEM-LUC-IB and from the parental monocistronic plasmid (pGEM-IB) was similar. These results indicated that a functional IRES element is contained within nt 1 to 1544 of the PTV Talfan sequence which comprises nt 1 to 412 from the 5′UTR followed by part of the polyprotein coding region. Deletion from the 5′ terminus (nt 1) to nt 273 within the PTV RNA 5′UTR (as in pGEM-LUC-T1B) or beyond (to nt 363 in pGEM-LUC-T2B) abolished IRES function (Fig. 2C).
Characterization of the PTV Talfan IRES element.
The next step was to determine whether the PTV Talfan IRES required any viral RNA coding sequences for activity. It has been shown that the hepatitis C virus (HCV) IRES can require sequences from within the coding sequence for activity (20). Plasmids were constructed in which cDNA corresponding to the PTV Talfan 5′UTR alone or with the first part (ca. 240 nt) of the coding sequence was inserted into the dicistronic reporter plasmid pGEM-CAT/LUC (Fig. 3A), which has been described previously (22). This plasmid expresses a dicistronic mRNA encoding chloramphenicol acetyltransferase (CAT) and LUC (Fig. 3); each cistron has its own initiation and termination codons. Regions of the PTV-1 Talfan cDNA corresponding to the 5′UTR were generated by PCR with primers pBADF and either TAL2R or TAL3R (Table 1) with the pBAD-I-mycB plasmid as a template. The fragments obtained were purified and digested with BamHI, and the products of 445 and 688 nt were ligated into BamHI-linearized and phosphatased pGEM-CAT/LUC. Plasmids containing the inserts in both possible orientations were isolated and named pGEM-CAT/TAL2F/LUC (sense orientation), pGEM-CAT/TAL2R/LUC (antisense orientation), pGEM-CAT/TAL3F/LUC, and pGEM-CAT/TAL3R/LUC, respectively (Fig. 3). Derivatives containing cDNA corresponding to the IRES elements from FMDV and the coxsackievirus B4 (CB4) have been described previously (22). The reading frame of the PTV sequence initiated at nt 412 was maintained into the LUC coding sequence in the pGEM-CAT/TAL3F/LUC plasmid so that a fusion protein, ΔL-LUC, should be expressed from a functional IRES. These reporter plasmids were used to program TNT assays (Fig. 3B). Efficient expression of CAT from each plasmid was observed. LUC was also efficiently expressed from pGEM-CAT/TAL2F/LUC and from pGEM-CAT/FMD/LUC (a positive control) but not from pGEM-CAT/TAL2R/LUC or pGEM-CAT/LUC (negative control). These results indicated that the PTV sequence from nt 1 to 405 functions as an IRES element only when expressed in the sense orientation. The PTV IRES clearly functioned well within the RRL translation system, as did the FMDV IRES. Furthermore, efficient expression of the ΔL-LUC fusion protein was detected from pGEM-CAT/TAL3F/LUC but not from pGEM-CAT/TAL3R/LUC (Fig. 3B) with the IRES in the inverse orientation. No evidence for a requirement of the PTV IRES for any coding sequence was apparent from these assays, since the expressions of LUC from pGEM-CAT/TAL2F/LUC and of ΔL-LUC from pGEM-CAT/TAL3F/LUC were similar.
FIG. 3.
Characterization of the PTV Talfan IRES element. (A) Derivatives of the reporter plasmid pGEM-CAT/LUC (22) were prepared by the insertion of PTV Talfan cDNA (flanked by BamHI sites) into the unique BamHI site (indicated by a B) of the parental vector. The orientations of the PTV sequences are shown by arrowheads, and the presence of PTV coding sequences is depicted by the open rectangle. The limits of the PTV sequences used are indicated. (B) Reporter plasmids were used to program in vitro TNT reactions as for Fig. 1B. (C) The plasmids shown were transfected into vTF7-3-infected BHK cells as for Fig. 1C, and the products were analyzed by immunoblotting with antibodies specific for CAT and LUC as described in the legend for Fig. 2.
When these reporter plasmids were assayed within the transient expression assay system within cells, the upstream cistron, CAT, was again efficiently expressed from each plasmid as expected (Fig. 3C), which indicated that the plasmids were transfected and transcribed with similar efficiencies. The efficient expression of LUC was observed from plasmid pGEM-CAT/TAL2F/LUC and from the positive control pGEM-CAT/FMD/LUC, while the ΔL-LUC fusion protein was expressed from pGEM-CAT/TAL3F/LUC (Fig. 3C). The levels of expression of LUC and ΔL-LUC, as judged by immunoblotting, were very similar, indicating that the presence of coding sequence had little effect on the efficiency of the IRES within cells also. No expression of LUC-related proteins was detected when the PTV 5′UTR sequences were present in the reverse orientation (Fig. 3C). The production of the ΔL-LUC fusion protein from pGEM-CAT/TAL3F/LUC was fully consistent with the use of the AUG codon at nt 412 as the initiation codon. The expression of LUC was also detected by LUC activity assays (Promega) using a luminometer, and the relative levels of LUC expression detected were fully consistent with the Western blot analyses (data not shown). The plasmid pGEM-CAT/TAL2F/LUC (containing the PTV IRES in the sense orientation) directed the synthesis of a >200-fold-higher level of LUC activity than the parental pGEM-CAT/LUC vector alone. In four parallel transfections, the mean activity of the PTV IRES was 96% of the FMDV IRES activity in BHK cells. Much lower levels of LUC activity were detected in extracts that contained the ΔL-LUC product than in those containing the native LUC protein. Thus, it seemed that the fusion protein was much less active than the native protein since the immunoblot detection indicated that the levels of protein expression were similar.
Influence of enterovirus 2A protease expression on PTV IRES activity in BHK cells.
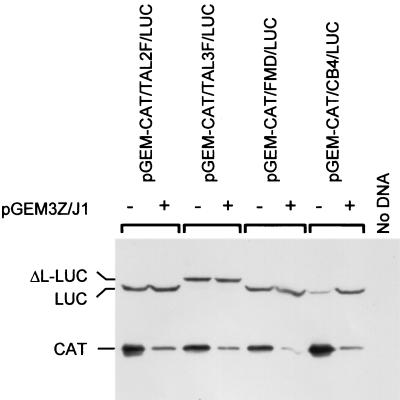
Expression of an enterovirus 2A protease strongly inhibits cap-dependent protein synthesis but has different effects on the activities of distinct picornavirus IRES elements (5, 22). The cardio- and aphthovirus IRES elements are little affected by this protease, but within BHK cells, the expression of an enterovirus 2A strongly stimulates the activity of entero- and rhinovirus IRES elements. In contrast, the HAV IRES is inhibited under similar conditions. To analyze the effect of the enterovirus 2A expression on the PTV IRES activity, the dicistronic reporter plasmids pGEM-CAT/TAL2F/LUC and pGEM-CAT/TAL3F/LUC were transfected into BHK cells alone or with plasmid pGEM3Z/J1, which expresses the swine vesicular disease virus 1D-2A coding region (23). For comparison, the reporter plasmids pGEM-CAT/FMD/LUC and pGEM-CAT/CB4/LUC, which express dicistronic mRNAs containing the FMDV and CB4 IRES elements, respectively, were assayed in parallel. As expected, each of the plasmids efficiently expressed CAT when assayed alone but this was severely inhibited in the presence of the 2A protease, since cap-dependent initiation of protein synthesis was blocked (Fig. 4). Expression of LUC directed by the PTV and FMDV IRES elements was high in both the presence and absence of the 2A protease. This demonstrates that the PTV sequence directs cap-independent internal initiation of protein synthesis. In contrast, the CB4 IRES-directed expression of LUC was low in the absence of 2A but was strongly stimulated by the 2A protease as shown previously (22, 23). The results from the Western blot analysis were entirely consistent with LUC assays performed in parallel (data not shown).
FIG. 4.
PTV Talfan IRES activity is unaffected by the coexpression of an enterovirus 2A protease. vTF7-3-infected BHK cells were transfected with the indicated reporter plasmids alone or with pGEM3Z/J1 (this plasmid encodes the swine vesicular disease virus 2A protease). Cell extracts were prepared after 20 h and analyzed by SDS-PAGE and immunoblotting with antibodies specific for CAT and LUC as indicated.
The studies presented here demonstrate that the 5′UTR of the PTV Talfan RNA contains an IRES element that directs the initiation of protein synthesis from nt 412. This codon corresponds to the AUG at position nt 432 in the PEV-1 F65 sequence (9) and is consistent with the initiation site proposed previously (28). The identification of this start site was made by the functional analysis of mRNAs with different lengths of PTV RNA sequence and was also confirmed by the production of a fusion protein (ΔL-LUC) between the PTV sequences and the LUC-coding sequence (Fig. 3). The AUG codon at nt 412 is not preceded by a significant polypyrimidine tract; this contrasts with the conserved presence of such a tract (typically 8 to 9 nt long with a core motif of UUUC) about 20 nt upstream of an AUG codon, at the 3′ end of the IRES elements of all other picornaviruses (see reference 18 for a review). It should be noted that the entero- and rhinovirus RNAs have a polypyrimidine tract upstream of an AUG codon that is not the initiation codon but is believed to be the point of ribosome entry from which ribosome scanning occurs. The spacer region on the entero- and rhinovirus RNAs between the ribosome entry point and the initiation site of protein synthesis is always free of AUG codons (for a review, see reference 2). It should be mentioned that a short, conserved CUUU motif is present just over 20 nt upstream of the nt 412 start site in the PTV sequence; but whether this can be considered a vestigial polypyrimidine tract is open to debate. A more significant pyrimidine tract (UUUCUCU) is located further upstream in the PTV RNA, some 13 nt upstream of the GUG codon at nt 316, and it should be noted that in some strains of PTV this codon is an AUG codon (28). In the light of the very high sequence conservation between the 5′UTRs of different PTV strains (28), this particular difference in sequence at nt 316 is noteworthy, but its significance is not known. However, it does not seem likely that ribosome entry takes place near nt 316 on the PTV Talfan RNA since there are two additional AUG codons (not in frame with the polyprotein) between this site and the authentic initiation site at nt 412. These would be expected to decrease the efficiency of any scanning ribosomes correctly initiating protein synthesis. Furthermore, ribosome entry near nt 316 would indicate that the PTV IRES is extremely short compared to other picornavirus IRES elements. It should be noted that the importance of the polypyrimidine tract for the picornavirus IRES elements is not entirely clear (2). For example, although this feature is conserved among all other picornaviruses, when the polypyrimidine tract of EMCV was converted to a tract of A residues it was found that this IRES element still functioned at about 70% of wild-type efficiency (16).
In general, the picornavirus IRES elements have been shown to be about 450 nt in length (2) and do not require any significant part of the coding sequence for activity. The PTV Talfan IRES is also contained entirely within the 5′UTR but is significantly shorter, at most 405 nt. We have shown that when the 5′UTR is truncated from the 5′ terminus (nt 1) to nt 273 the residual sequence lacks IRES activity, but the boundaries of this element (both 5′ and 3′) have yet to be established. The PTV IRES is clearly shorter than other picornavirus elements, but it may still be significantly longer than the IRES element from HCV (a Hepacivirus within the family Flaviviridae), which is about 300 to 340 nt in length. The HCV IRES can be optimally active when linked to a portion (at least 40 nt) of the viral coding sequence (20); however, this may reflect a requirement for an unstructured region of RNA following the IRES to prevent perturbation of the IRES structure (21).
The 2A protease from enteroviruses induces the cleavage of the translation initiation factor eIF4G (thereby inhibiting cap-dependent protein synthesis) and stimulates the activity of the entero- and rhinovirus IRES elements within cell types (e.g., BHK cells) in which these IRES elements display submaximal levels of activity (5, 22). In BHK cells, the activity of the cardio- and aphthovirus IRES elements is little affected by the 2A protease. The results presented here indicate that the activity of the PTV Talfan IRES is also maintained, but not stimulated, in the presence of the 2A protease (cf. the CB4 IRES). The PTV IRES also functions efficiently within the RRL in vitro translation system. Thus, the biological properties of the PTV IRES are most similar to the FMDV and EMCV IRES elements. However, computer prediction of the secondary structure of the PTV IRES sequence shows no apparent resemblance to the cardio- and aphthovirus IRES elements (27).
In summary, the PTV IRES element has a unique set of properties. These are as follows: (i) the IRES functions efficiently in the RRL system and within cells using the vaccinia virus/T7 expression system; (ii) it is, at most, 405 nt in length; (iii) IRES activity is unaffected by the coexpression of an enterovirus 2A protease and the cleavage of eIF4G; (iv) it lacks a significant polypyrimidine tract associated with an AUG codon; and (v) the predicted RNA secondary structure is distinct from other picornavirus IRES elements. We conclude that the PTV RNA contains a fourth class of picornavirus IRES element.
Acknowledgments
Financial support from the Ministry of Education, Culture, Sports, Science and Technology of Japan and the BBSRC (United Kingdom) is gratefully acknowledged.
REFERENCES
- 1.Ali, I. K., L. McKendrick, S. J. Morley, and R. J. Jackson. 2001. Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J. Virol. 75:7854-7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belsham, G. J., and R. J. Jackson. 2000. Translation initiation on picornavirus RNA, p. 869-900. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Monograph 39. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 3.Belsham, G. J., and N. Sonenberg. 1996. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol Rev. 60:499-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borman, A. M., and K. M. Kean. 1997. Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology 237:129-136. [DOI] [PubMed] [Google Scholar]
- 5.Borman, A. M., P. le Mercier, M. Girard, and K. M. Kean. 1997. Comparison of picornaviral IRES-driven internal initiation in cultured cells of different origins. Nucleic Acids Res. 25:925-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borman, A. M., Y. M. Michel, and K. M. Kean. 2001. Detailed analysis of the requirements of hepatitis A virus internal ribosome entry segment for the eukaryotic initiation factor complex eIF4F. J. Virol. 75:7864-7871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown, B., and E. Ehrenfeld. 1979. Translation of poliovirus RNA in vitro: changes in cleavage pattern and initiation sites by ribosomal salt wash. Virology 97:396-405. [DOI] [PubMed] [Google Scholar]
- 8.Brown, E. A., A. J. Zajac, and S. M. Lemon. 1994. In vitro characterization of an internal ribosome entry site (IRES) present within the 5′ nontranslated region of hepatitis A virus RNA: comparison with the IRES of encephalomyocarditis virus. J. Virol. 68:1066-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doherty, M., D. Todd, N. McFerran, and E. M. Hoey. 1999. Sequence analysis of a porcine enterovirus serotype 1 isolate: relationships with other picornaviruses. J. Gen. Virol. 80:1929-1941. [DOI] [PubMed] [Google Scholar]
- 10.Dorner, A. J., B. L. Semler, R. J. Jackson, R. Hanecak, E. Duprey, and E. Wimmer. 1984. In vitro translation of poliovirus RNA: utilization of internal initiation sites in reticulocyte lysate. J. Virol. 50:507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gingras, A.-C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 13.Glass, M. J., and D. F. Summers. 1993. Identification of a trans-acting activity from liver that stimulates hepatitis A virus translation in vitro. Virology 193:1047-1050. [DOI] [PubMed] [Google Scholar]
- 14.Harding, J. D. J., J. T. Done, and G. F. Kershaw. 1957. A transmissible polio-encephalomyelitis of pigs (Talfan disease). Vet. Rec. 69:824-832. [Google Scholar]
- 15.Kaku, Y., A. Sarai, and Y. Murakami. 2001. Genetic reclassification of porcine enteroviruses. J. Gen. Virol. 82:417-424. [DOI] [PubMed] [Google Scholar]
- 16.Kaminski, A., G. J. Belsham, and R. J. Jackson. 1994. Translation of encephalomyocarditis virus RNA: parameters influencing the selection of the internal initiation site. EMBO J. 13:1673-1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 18.Meerovitch, K., and N. Sonenberg. 1993. Internal initiation of picornavirus RNA translation. Semin. Virol. 4:217-227. [Google Scholar]
- 19.Pringle, C. R. 1999. Virus taxonomy at the XIth International Congress of Virology, Sydney, Australia, 1999. Arch. Virol. 144:2065-2070. [DOI] [PubMed] [Google Scholar]
- 20.Reynolds, J. E., A. Kaminski, H. J. Kettinen, K. Grace, B. E. Clarke, A. R. Carroll, D. J. Rowlands, and R. J. Jackson. 1995. Unique features of internal initiation of hepatitis-C virus-RNA translation. EMBO J. 14:6010-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rijnbrand, R., P. J. Bredenbeek, P. C. Haasnoot, J. S. Keift, W. J. M. Spaan, and S. M. Lemon. 2001. The influence of downstream protein-coding sequence on internal ribosome entry on hepatitis C virus and other flavivirus RNAs. RNA 7:585-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roberts, L. O., R. A. Seamons, and G. J. Belsham. 1998. Recognition of picornavirus internal ribosome entry sites within cells; influence of cellular and viral proteins. RNA 4:520-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakoda, Y., N. Ross-Smith, T. Inoue, and G. J. Belsham. 2001. An attenuating mutation in the 2A protease of swine vesicular disease virus, a picornavirus, regulates cap- and internal ribosome entry site-dependent protein synthesis. J. Virol. 75:10643-10650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Trefny, L. 1930. Massive illness of swine in Teschen area. Zveroleki Obzori 23:235-236. [Google Scholar]
- 26.Whetter, L. E., S. P. Day, O. Elroy-Stein, E. A. Brown, and S. M. Lemon. 1994. Low efficiency of the 5′ nontranslated region of hepatitis A virus RNA in directing cap-independent translation in permissive monkey kidney cells. J. Virol. 68:5253-5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Witwer, C., S. Rauscher, I. L. Hofacker, and P. F. Stadler. 2001. Conserved RNA secondary structures in Picornaviridae genomes. Nucleic Acids Res. 29:5079-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zell, R., M. Dauber, A. Krumbholz, A. Henke, E. Birch-Hirschfeld, A. Stelzner, D. Prager, and R. Wurm. 2001. Porcine teschoviruses comprise at least eleven distinct serotypes: molecular and evolutionary aspects. J. Virol. 75:1620-1631. [DOI] [PMC free article] [PubMed] [Google Scholar]