Abstract
Transcription of the six Epstein-Barr virus (EBV) EBNA genes is coordinately regulated, being driven by either the Cp promoter, which is encoded within the unique region just upstream of the EBV major internal repeat (IR-1), or by the Wp promoter, which is encoded within the IR-1 repeat and thus present in multiple copies. Previous analyses of Cp- and Wp-initiated transcription have identified a shared cis-regulatory element mapping to the region extending from −169 to −369 bp upstream of the Wp transcription initiation site (M. T. Puglielli, N. Desai, and S. H. Speck, J. Virol. 71:120-128, 1997). To assess the impact of this regulatory region on Cp and Wp activity in the context of the viral genome, we attempted to delete this regulatory region upstream of the first copy of Wp (Wp1). While 10 recombinant viruses were obtained in which this deletion was incorporated in the interior of the IR-1 repeat, only a single lymphoblastoid cell line (LCL) immortalized by a recombinant EBV harboring the deletion upstream of Wp1 was recovered. In contrast, using a control targeting vector in which the Wp regulatory sequences were intact but which contained a sequence tag within the W0 exon, we demonstrated that of the five recombinant viruses analyzed in which the crossover event had occurred upstream of the Wp sequence tag, four had incorporated the tagged sequences into Wp1 of the virus. Taken together, these results indicate that deletion of the regulatory sequences from −369 to −169 bp upstream of Wp1 is unfavorable for EBV-driven B-cell immortalization but is tolerated within the interior of the IR-1 repeat. Analysis of promoter usage in the clone 9-60 LCL, in which the W enhancer sequences were deleted upstream of Wp1, revealed the following: (i) the level of Cp-initiated transcription was significantly diminished compared to that of wild-type LCLs; (ii) the decreased Cp-initiated transcription was not efficiently compensated by transcription initiation from Wp1; and (iii) transcription initiation from downstream Wp promoters was detectable. This is the first report of an LCL in which transcription initiation from a Wp downstream of Wp1 has been documented.
Epstein-Barr virus (EBV) immortalization of primary B cells requires the expression of five nuclear antigens (EBNA1, EBNA2, EBNA3a, EBNA3c, and EBNA4 [also referred to as EBNA-LP]) and latency-associated membrane protein 1 (LMP1) (reviewed in reference 11). Transcription of the genes encoding the EBNAs is driven from one of two promoters, Cp and Wp, located near the left end of the viral genome (reviewed in reference 21). Wp is encoded within the major internal repeat (IR-1) of EBV and as such is present in multiple copies. During the initial stages of infection of B cells, Wp is exclusively used to drive EBNA gene expression, followed by upregulation of transcription from Cp and a concomitant downregulation of transcription initiation from Wp (15, 17).
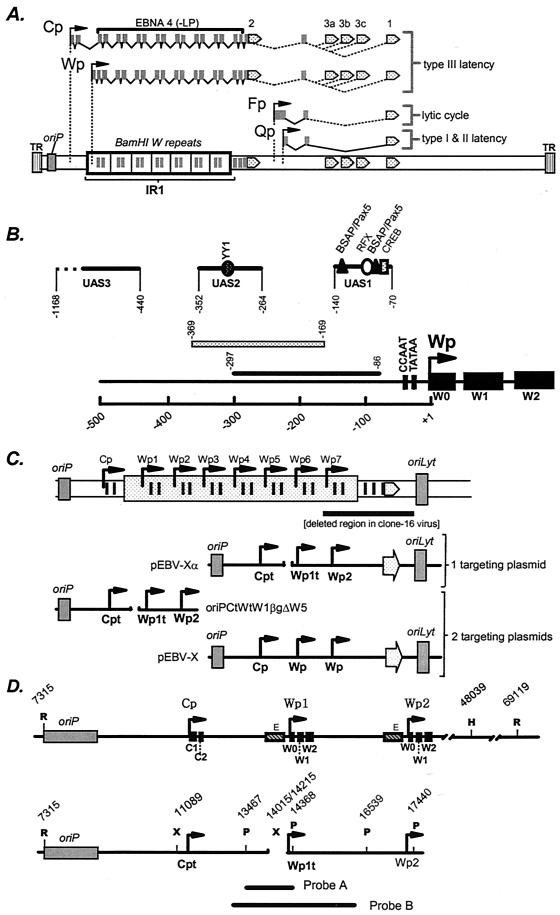
Sequences upstream of Wp have been shown to be important for Wp-initiated transcription (3, 7, 8, 10, 12, 16) and, in some cases, Cp-initiated transcription (8). Ricksten et al. (10) mapped an enhancer in the region from −297 to −86 bp upstream of Wp that exhibits B-cell-specific activity (Fig. 1). The analysis of cis elements in this region has been further characterized by Bell et al. (3), who have identified the presence of three regulatory regions: UAS1 exhibits B-cell-specific activity, while UAS2 and UAS3 exhibit cell lineage-independent activity (Fig. 1). Tierney et al. (12) have subsequently shown that UAS1 binds the cellular transcription factors CREB/ATF, BSAP, and RFX (Fig. 1). Whether UAS1 is the critical regulatory element that leads to Wp-initiated transcription during the initial stages of B-cell infection remains to be determined.
FIG. 1.
(A) Schematic illustration of EBNA gene transcription arising from Cp and Wp during immortalizing latency (type III latency) or restricted latency where EBNA1 transcription is driven from the Qp promoter (types I and II latency). During virus replication, EBNA1 is transcribed at low levels from the Fp promoter. (B) Schematic illustration of the regulatory sequences that have been identified upstream of Wp. Puglielli et al. (8) have identified a regulatory region mapping from −169 to −369 bp upstream of Wp that influences both Wp-initiated and Cp-initiated transcription. Ricksten et al. (10) identified an enhancer in the region from −86 to −297 bp upstream of Wp that regulates Wp-initiated transcription, in the absence of extensive upstream regulatory sequences, in EBV-negative B-cell lines. Further analyses of cis elements that regulate Wp activity in the absence of any EBV latency-associated antigens have identified three distinct regulatory regions (UAS1, UAS2, and UAS3) (3). Subsequent analyses have shown that UAS1 exhibits B-cell-specific activity, while UAS2 and UAS3 exhibit cell lineage-independent activity (3, 7, 12). Binding sites for the cellular transcription factors YY1, BSAP/Pax5, RFX, and CREB have been identified, and their locations are indicated (7, 12). (C) Schematic illustrations of the single and W plasmid-targeting strategies used to generate recombinant EBV harboring a deletion in regulatory sequences upstream of Wp. pEBV-X is modified from vector p135 (5) and contains EBV genomic sequences spanning the region from the EcoRI site in BamHI C (position 7315) to the SalI site in BamHI F (position 56081) cloned into pBR322 (note that only two copies of the BamHI W repeat are present in this plasmid). pEBV-XαWT is a modified form of pEBV-X in which the C1 exon and the first W0 exon are tagged (15). OriPCtWtW1BgΔW5 contains the region from oriP to the second copy of Wp, except with a deletion of the sequences from −169 to −369 bp upstream of the first copy of Wp (positions 14015 to 14215). The C1 exon and the first W0 exon are tagged as previously described (8, 15). pEBV-Xα and oriPΔW5orilyt are similar to pEBV-XαWT and oriPWTorilyt, respectively, except that they harbor a deletion of the sequences from −169 to −369 bp upstream of the first copy of Wp. (D) Locations of restriction endonuclease cleavage sites used to determine the presence and location of the deletion in the EBV genome. The probes used for the Southern blots are also shown (Probe A was used to hybridize to the PstI-digested DNA as shown in Fig. 2A, while Probe B was used to hybridize to the Southern blot shown in Fig. 2B). Note that the genomic coordinates indicated for the HindIII and EcoRI sites downstream of the IR-1 repeat are based on the presence of 10 copies of the BamHI W repeat. The XbaI site indicated upstream of Cp was engineered into the targeting construct for cloning purposes (replaces a SacII site) and is present in all of the recombinant viruses analyzed (Fig. 4). Cpt, Cp in which a sequence tag has been introduced into the C1 exon; Wpt, Wp with a sequence tag in the W0 exon, allowing transcription from Wpt to be distinguished from that from downstream copies of Wp; R, EcoRI; H, HindIII; X, XbaI; P, PstI.
While the above-mentioned studies have attempted to define cis elements involved in regulating Wp activity in the absence of EBV gene products, and thus presumably important during the initial stages of virus infection, parallel analyses have been carried out to identify cis elements involved in regulating Cp and Wp in established EBV-immortalized lymphoblastoid cell lines (LCLs) (8, 9, 14-17). It has previously been shown that the region from −169 to −369 bp upstream of Wp is important for both Cp- and Wp-initiated transcription in established LCLs (8). Here, we examine the impact of deleting this regulatory region on LCL formation and on the steady-state levels of Cp- and Wp-initiated transcripts in an established LCL.
Employing two targeting constructs to generate EBV recombinants lacking the sequences from −169 to −369 bp upstream of Wp.
To generate a recombinant EBV lacking the regulatory sequences from −169 to −369 bp upstream of Wp1, we utilized the previously described strategy of employing two targeting constructs (13, 20) (Fig. 1C). One targeting construct (EBV-X) contained the EBNA2-coding exon, as well as the exons encoding the carboxy terminus of EBNA4, along with flanking sequences, allowing the rescue of immortalizing virus from the P3HR-1 clone 16 Burkitt's LCL. The second targeting construct (oriPCtWtW1BgΔW5) (8) was designed to introduce the deletion upstream of Wp1. This strategy has previously been used to delete the EBNA2 enhancer upstream of Cp, and it was observed that ca. 3 to 5% of the immortalizing virus recovered had also incorporated the EBNA2 enhancer deletion (20).
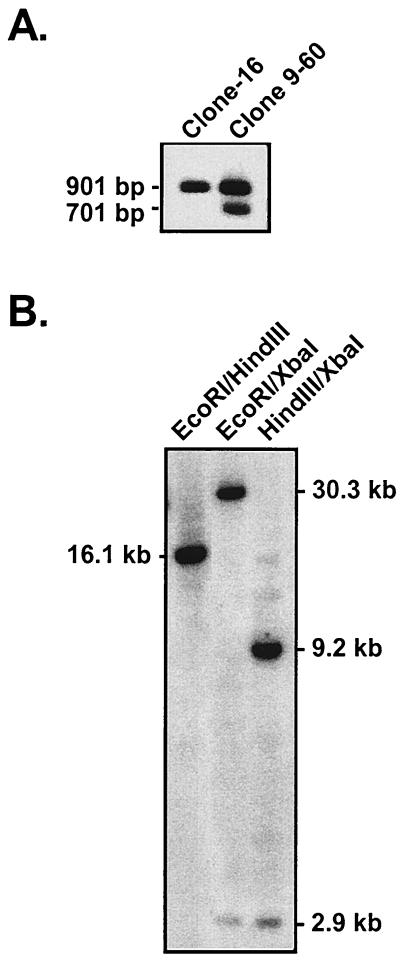
The P3HR-1 clone 16 cell line was transfected with the two targeting plasmids, along with a BZLF1 gene expression vector (pSV40-BZLF1) to induce virus replication (20), and the resulting heterogeneous virus stock was used to infect primary B cells. All LCLs which arose from this infection were screened by PCR for the presence of the enhancer deletion. We screened 253 clonal LCLs generated by this approach and recovered a single LCL, clone 9-60, which had incorporated the Wp enhancer deletion. Thus, the efficiency of generating a recombinant virus containing the enhancer deletion upstream of Wp1 was quite low (<0.5%), raising the possibility that this mutation is unfavorable for B-cell immortalization (see below). A Southern blot of PstI-digested genomic DNA revealed the presence of both the enhancer deletion (701 bp) and wild-type (901 bp) fragments in DNA isolated from clone 9-60, while only the 901-bp wild-type PstI fragment was present in DNA isolated from the parental P3HR-1 clone 16 cell line (Fig. 1D and 2A). We anticipated observing both wild-type and mutant-length PstI fragments in the clone 9-60 cell line, since there should be multiple copies of the BamHI W repeat present in this mutant viral genome.
FIG. 2.
(A) Southern blot of DNA isolated from the P3HR-1 clone 16 cell line and the clone 9-60 LCL, demonstrating the presence of the deletion of Wp regulatory sequences in the clone 9-60 LCL. Total cellular DNA was digested with PstI and probed with the PstI fragment of BamHI W (see probe A shown in Fig. 1D). (B) Southern blot of DNA isolated from the 9-60 LCL to determine the size of the IR-1 repeat and the location of the deletion within IR-1. Clone 9-60 LCL DNA was digested with EcoRI and HindIII, EcoRI and XbaI, or XbaI and HindIII and probed with the entire BamHI W fragment (see probe B shown in Fig. 1D). For Southern analyses, 5 μg from each sample was digested with the specified restriction enzyme(s) and the digested fragments were separated on an agarose gel. Subsequently, the gel was denatured and the DNA was transferred to a nylon membrane (Nytran membrane; Schleicher & Schuell), using the Turboblot transfer system (Schleicher & Schuell). Membranes were prehybridized in 6× SSPE (1× SSPE is 0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]), 5× Denhardt's solution, 0.5% sodium dodecyl sulfate, 150 μg of salmon sperm/ml, and 50% formamide solution for 2.5 h at 42°C, followed by the addition of 106 cpm of 32P-labeled probe/μl.
To determine the size of the IR-1 repeat, as well as the location of the enhancer deletion within the IR-1 repeat, further Southern blotting was carried out. Digestion of clone 9-60 DNA with EcoRI and HindIII, which cut in the unique regions upstream and downstream of IR-1, respectively, revealed a 16.1-kb band indicating the presence of three copies of the BamHI W repeat and, by implication, three copies of Wp (Fig. 1D and 2B). The incorporation of XbaI sites at the point of the enhancer deletion and upstream of Cp at bp 11089 in the viral genome (in the generation of the targeting construct, the SacII site at bp 11089 was replaced with an XbaI site [8]) allowed, in conjunction with either EcoRI or HindIII digestion, the determination of the location of the enhancer deletion (Fig. 1D). The detection of a 2.9-kb XbaI fragment indicates that the enhancer deletion is located upstream of the first copy of Wp (Wp1). Consistent with this interpretation, digestion with HindIII (which cuts in the unique region just downstream of the IR-1 repeat) in conjunction with XbaI revealed the presence of a 9.2-kb fragment which, based on the size of the IR-1 repeat determined by EcoRI and HindIII digestion, confirmed that the enhancer deletion is located upstream of Wp1 (Fig. 2B). As discussed below, it should be noted that the introduction of sequence tags within the C1 and W0 exons and the unique XbaI restriction endonuclease site upstream of Cp had no discernible impact on the utilization of Cp or Wp1 as assessed with “wild-type” recombinant viruses harboring these sequence tags.
Characterization of EBNA gene promoter usage in the clone 9-60 LCL.
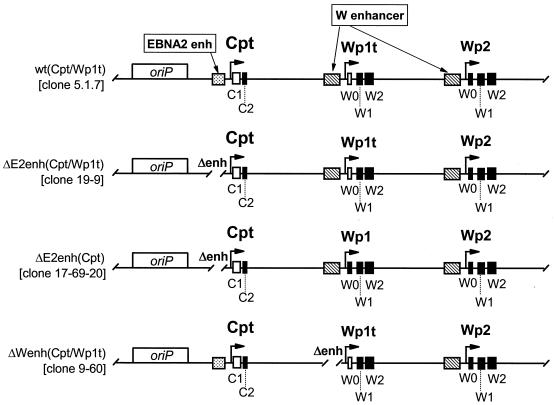
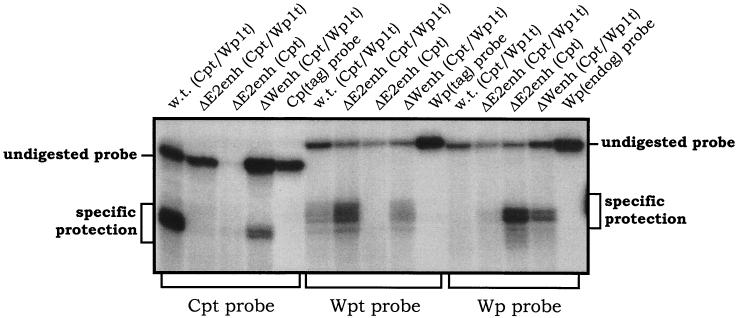
S1 nuclease protection analyses were carried out to quantitatively determine the steady-state levels of Cp- and Wp-initiated transcripts in the 9-60 LCL. As controls, RNA was prepared from several LCLs harboring recombinant EBV genomes with specific mutations and/or sequence tags (Fig. 3 shows schematic illustrations of the Cp and Wp regions in these recombinant viruses). As previously shown with LCLs immortalized with wild-type recombinant viruses (20), the LCL harboring a wild-type EBV recombinant containing sequence-tagged C1 and W0 exons [w.t. (Cpt/Wp1t)] exhibited high levels of Cp-initiated transcripts, a very low level of Wp1-initiated transcripts, and no detectable transcripts arising from the copies of Wp downstream of Wp1 (Fig. 4). Thus, in wild-type EBV-infected LCLs, EBNA gene transcription is heavily biased toward utilization of Cp (15-17, 20). In contrast, as previously shown (20), LCLs harboring virus with the Cp EBNA2 enhancer deleted [ΔE2enh (Cpt/Wp1t) and ΔE2enh (Cpt)] did not contain detectable levels of Cp-initiated transcripts. The LCL immortalized with ΔE2enh (Cpt/Wp1t) exhibited high levels of Wp1-initiated transcripts and only very low levels of transcripts initiating from downstream copies of Wp (Fig. 4). The LCL immortalized with ΔE2enh (Cpt) did not have a tagged W0 exon, so no transcript was detected with the probe specific for the Wp with a sequence tag in the W0 exon (Wpt), but high levels of transcripts were detected with the probe specific for untagged Wp-initiated transcripts [equivalent to the levels of Wp1-initiated transcripts detected in the LCL immortalized with ΔE2enh (Cpt/Wp1t) virus]. Presumably, these Wp-initiated transcripts arose from Wp1. Thus, as has been shown previously (20), absence of the Cp EBNA2 enhancer leads to LCLs that are heavily biased toward utilization of Wp1 for driving EBNA gene transcription. Notably, analysis of EBNA transcripts in the 9-60 LCL [ΔWpenh (Cpt/Wp1t)] revealed low levels of Cp- and Wp1-initiated transcripts compared to those in the appropriate control cell lines. However, in contrast to the other recombinant LCLs, there were significant steady-state levels of EBNA gene transcripts arising from downstream copies of Wp (Fig. 4). Thus, deletion of the regulatory sequences from −169 to −369 bp upstream of Wp1 appears to lead to impaired Cp and Wp1 activity, which is compensated by upregulation of transcription from downstream copies of Wp. Whether the latter activity exclusively arises from Wp2 or represents the sum of activity from Wp2 and Wp3 (there are only three copies of Wp in the 9-60 viral genome) is unclear. This is the first instance in which transcription from downstream copies of Wp has been documented. Overall, these data are consistent with the results from previous analyses carried out using transient transfection of reporter constructs into established LCLs. However, these results must be interpreted with caution since they are based on the behavior of a single mutant LCL.
FIG. 3.
Schematic illustrations of the structures of the Cp and Wp regions of the recombinant viral genomes present in the LCLs analyzed (Fig. 4). The wild-type recombinant (clone 5.1.7) and Cp EBNA2 enhancer deletion mutants (clone 19-9 and clone 17-69-20) were generated as previously described (20). Cpt, Cp in which a sequence tag has been introduced into the C1 exon; Wpt, Wp with a sequence tag in the W0 exon, allowing transcription from Wpt to be distinguished from that from downstream copies of Wp; W enhancer, region from −169 to −369 bp upstream of Wp (Fig. 1B shows a more detailed summary of identified regulatory elements upstream of Wp). Exons containing introduced sequence tags are depicted as open rectangles, while exons lacking sequence tags are depicted as filled rectangles.
FIG. 4.
S1 nuclease protection analyses of RNA prepared from the indicated LCLs. The structural features of the recombinant EBV harbored by each LCL are summarized in Fig. 3. S1 nuclease protection assays were carried out as previously described (8, 9, 15, 20), using 32P-labeled single-stranded oligonucleotide probes which specifically detect transcription arising from Cpt, Wpt, or Wp. The migration of undigested and specifically protected fragments is indicated. RNA from the indicated cell lines was isolated by the single-step guanidinium thiocyanate-phenol-chloroform method as described previously (4). The S1 nuclease protection assay was performed as previously described (9), using 40 μg of total cellular RNA for the analyses of Cp- and Wp-initiated transcripts. Gels were exposed to PhosphorImager screens (Molecular Dynamics) for ca. 20 h, and signals were quantitated. The sequences of the S1 nuclease probes were as previously described (20). w.t. (Cpt/Wp1t), a wild-type recombinant EBV in which distinct sequence tags have been introduced into the C1 and first W0 exons; ΔE2enh (Cpt/Wp1t), a recombinant EBV in which the Cp EBNA2 enhancer has been deleted and sequence tags have been introduced into the C1 and first W0 exons; ΔE2enh (Cpt), a recombinant EBV in which the Cp EBNA2 enhancer has been deleted and a sequence tag has been introduced into the C1 exon; ΔWenh (Cpt/Wp1t), the 9-60 LCL which lacks the regulatory sequences from −169 to −369 bp upstream of Wp1 and also contains sequence tags that have been introduced into the C1 and first W0 exons.
Utilization of a single targeting vector to generate LCLs harboring recombinant EBV containing the deletion of the region from −169 to −369 bp upstream of Wp. We were unsatisfied with the mutant generation strategy used to generate the 9-60 LCL because of its poor efficiency. In an attempt to generate additional LCLs harboring virus with the enhancer deletion upstream of Wp1, we altered the mutagenesis strategy. We reasoned that at least part of the inefficiency of our initial approach was due to the use of two separate targeting plasmids, one harboring the EBNA2 gene and the other harboring the enhancer deletion (Fig. 1C). Thus, we engineered a single plasmid-targeting construct (EBV-Xα) containing both the EBNA2 gene and the enhancer deletion (Fig. 1C). As a control, we used a related plasmid (EBV-XαWT) in which the Wp regulatory sequences were intact. Both EBV-Xα and EBV-XαWT contained two complete copies of the BamHI W repeat, along with a sequence tag in the W0 exon of the first (Cp-proximal) Wp (Wp1). The latter allowed determination of (i) the site of recombination within the IR-1 repeat and (ii) transcription initiation from the tagged copy of Wp (see the discussion of S1 nuclease analyses above).
Using the single targeting vector, we were able to isolate 10 LCLs immortalized with recombinant viruses containing the enhancer deletion. However, analyses of these recombinant viruses demonstrated that the mutation was never located upstream of the first copy of Wp (Table 1). The recovery of mutants in which recombination had occurred within the interior of the IR-1 repeat could reflect either (i) that recombination within IR-1 occurs at a higher frequency than recombination within the unique sequences upstream of IR-1 or (ii) that recombinants that introduce the mutation upstream of Wp1 are impaired in their ability to form LCLs. Analysis of five LCLs generated with the wild-type targeting vector (EBV-XαWT), which contained the tagged W0 exon but lacked the enhancer deletion, demonstrated that in four of these the tagged W0 exon was present in the first copy of Wp (Table 1). The latter result indicates that recombination upstream of the IR-1 repeat sequences is favored over recombination within the interior of IR-1 and thus that the failure to recover mutant LCLs harboring the enhancer deletion upstream of Wp1 likely reflects an impairment of this mutant virus in LCL generation.
TABLE 1.
Location of Wp tags and mutations
| Clone | Location of Wp tag and/or mutation | IR-1 repeat size (no. of copies) |
|---|---|---|
| Wild-type recombinants | ||
| 5.1.7 | First repeat | 2 |
| 5.1.8 | First repeat | 2 |
| 10-26W | First repeat | 2 |
| 11-1 | First repeat | 3 |
| 5.1.2 | Seventh repeat | 9 |
| W enhancer mutants | ||
| 2.1.8 | Second repeat | 3 |
| 11-17a | Second repeat | 3 |
| 2.3.12 | Fourth repeat | 4 |
| 2.4.10 | Fourth repeat | 5 |
| 2.1.3 | Fourth repeat | 5 |
| 11-19a | Fourth repeat | 5 |
| 2.1.18 | Fourth repeat | 6 |
| 2.1.4 | Fifth repeat | 6 |
| 2.2.10 | Fifth repeat | 7 |
| 2.4.1 | Sixth repeat | 7 |
Discussion.
In established LCLs, the most powerful regulator of Wp activity appears to be Cp itself (8, 9, 15, 17, 20). Analysis of a panel of 15 long-established LCLs for endogenous Cp and Wp activity found that 11 lines exclusively used Cp while only 4 cell lines used Wp exclusively (2, 16). For two of the Wp-using cell lines, it has been shown that Cp is deleted from the viral genome (17, 18), while the basis for exclusive Wp utilization in the other two LCLs remains unknown. More recent analyses of low-passage LCLs have demonstrated the presence of both Cp- and Wp-initiated EBNA gene transcripts, with activity being strongly biased toward Cp-initiated transcripts in the majority of these cell lines (20). Consistent with the observed dominance of Cp in established LCLs, we have shown by transient transfection employing reporter plasmids containing Cp and Wp, in the context of extensive flanking sequences, that Wp-initiated transcripts are only detected when the upstream Cp was deleted or inverted (8). This suggests that transcription from the distal promoter, Cp, interferes with transcription initiation from the proximal promoter, Wp. Transcriptional interference has been proposed to explain the expression of a number of cellular genes whose transcription is driven by multiple distinct promoters (1). Further evidence supporting the proposed interference by Cp-initiated transcription of Wp activity has been provided using the conditional EBNA2 LCL, er/eb 2-5, in which EBNA2 function is dependent on the presence of β-estradiol (6). It has previously been shown that upon withdrawal of β-estradiol the steady-state level of Cp-initiated transcripts decreased more than threefold over 48 h and that there was a concomitant increase of approximately fivefold in the levels of Wp-initiated transcripts (19). The latter experiment appears to conclusively demonstrate that diminished Cp activity leads directly to increased levels of Wp-initiated transcripts, as predicted by the transcription interference model.
In this paper, we have extended these analyses to demonstrate that the regulatory region from −169 to −369 bp upstream of Wp1 is important for both Cp- and Wp1-initiated transcription. Analysis of the single LCL recovered that harbors virus in which this regulatory region has been deleted upstream of Wp1 demonstrated decreased steady-state levels of Cp- and Wp1-initiated transcripts and greatly enhanced steady-state levels of transcripts arising from downstream copies of Wp. The detection of EBNA transcripts arising from downstream copies of Wp is unique and is consistent with the hypothesis that transcription initiation at the upstream promoters Cp and Wp1 normally suppresses transcription initiation from downstream copies of Wp.
That we were unable to identify any recombinants by using the single targeting vector, in which the deletion of Wp regulatory sequences was inserted upstream of Wp1, even though a control targeting vector preferentially recombined upstream of Wp1, suggests that insertion of this mutation upstream of Wp1 is unfavorable for LCL generation. Notably, we did identify recombinants in which this mutation was inserted in the interior of the IR-1 repeat, indicating that the mutation is tolerated at other positions within the repeat. It is possible that since the targeting vector contains only two copies of the BamHI W repeat, and thus only two copies of Wp, insertion of the deletion upstream of Wp1 would force the generation of a recombinant containing only two copies of the BamHI W repeat. Previous analyses of LCLs harboring wild-type recombinants, or Cp EBNA2 enhancer mutant recombinants, generated from the rescue of the P3HR-1 clone 16 virus, has shown that EBV recombinants containing only two copies of the BamHI W repeat are quite common and do not appear to be impaired in B-cell immortalization (19, 20). However, it is possible that deletion of the regulatory sequences upstream of Wp1 are not well tolerated in the context of only two copies of the BamHI W repeat but would be tolerated if more copies of the BamHI W repeat (and thus more copies of Wp) were present. Notably, however, when we employed two targeting plasmids, which should not restrict the number of copies of BamHI W repeats present in the recombinants, we were able to recover only a single mutant (clone 9-60) out of 253 LCLs screened. The latter result suggests that deletion of the region from −169 to −369 bp upstream of Wp1 is unfavorable for LCL formation independent of the number of copies of the BamHI W repeat present in the viral genome.
Acknowledgments
This research was supported by NIH research grant R01-CA43143 to S.H.S.
We thank David Leib and Skip Virgin and members of their laboratories for advice on this research.
REFERENCES
- 1.Adhya, S., and M. Gottesman. 1982. Promoter occlusion: transcription through a promoter may inhibit its activity. Cell 29:939-944. [DOI] [PubMed] [Google Scholar]
- 2.Altiok, E., J. Minarovits, L. F. Hu, B. Contreras-Brodin, G. Klein, and I. Ernberg. 1992. Host-cell-phenotype-dependent control of the BCR2/BWR1 promoter complex regulates the expression of Epstein-Barr virus nuclear antigens 2-6. Proc. Natl. Acad. Sci. USA 89:905-909. (Erratum, 89:6225). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bell, A., J. Skinner, H. Kirby, and A. Rickinson. 1998. Characterisation of regulatory sequences at the Epstein-Barr virus BamHI W promoter. Virology 252:149-161. [DOI] [PubMed] [Google Scholar]
- 4.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 5.Hammerschmidt, W., and B. Sugden. 1989. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature 340:393-397. [DOI] [PubMed] [Google Scholar]
- 6.Kempkes, B., D. Spitkovsky, P. Jansen-Durr, J. W. Ellwart, E. Kremmer, H. J. Delecluse, C. Rottenberger, G. W. Bornkamm, and W. Hammerschmidt. 1995. B-cell proliferation and induction of early G1-regulating proteins by Epstein-Barr virus mutants conditional for EBNA2. EMBO J. 14:88-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirby, H., A. Rickinson, and A. Bell. 2000. The activity of the Epstein-Barr virus BamHI W promoter in B cells is dependent on the binding of CREB/ATF factors. J. Gen. Virol. 81:1057-1066. [DOI] [PubMed] [Google Scholar]
- 8.Puglielli, M. T., N. Desai, and S. H. Speck. 1997. Regulation of EBNA gene transcription in lymphoblastoid cell lines: characterization of sequences downstream of BCR2 (Cp). J. Virol. 71:120-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Puglielli, M. T., M. Woisetschlaeger, and S. H. Speck. 1996. oriP is essential for EBNA gene promoter activity in Epstein-Barr virus-immortalized lymphoblastoid cell lines. J. Virol. 70:5758-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ricksten, A., A. Olsson, T. Andersson, and L. Rymo. 1988. The 5′ flanking region of the gene for the Epstein-Barr virus-encoded nuclear antigen 2 contains a cell type specific cis-acting regulatory element that activates transcription in transfected B-cells. Nucleic Acids Res. 16:8391-8410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thorley-Lawson, D. A. 2001. Epstein-Barr virus: exploiting the immune system. Nat. Rev. Immunol. 1:75-82. [DOI] [PubMed] [Google Scholar]
- 12.Tierney, R., H. Kirby, J. Nagra, A. Rickinson, and A. Bell. 2000. The Epstein-Barr virus promoter initiating B-cell transformation is activated by RFX proteins and the B-cell-specific activator protein BSAP/Pax5. J. Virol. 74:10458-10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tomkinson, B., and E. Kieff. 1992. Use of second-site homologous recombination to demonstrate that Epstein-Barr virus nuclear protein 3B is not important for lymphocyte infection or growth transformation in vitro. J. Virol. 66:2893-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walls, D., and M. Perricaudet. 1991. The identification of new transcription elements active in Epstein-Barr virus latent gene expression. C. R. Acad. Sci. Ser. III 312:85-90. [PubMed] [Google Scholar]
- 15.Woisetschlaeger, M., X. W. Jin, C. N. Yandava, L. A. Furmanski, J. L. Strominger, and S. H. Speck. 1991. Role for the Epstein-Barr virus nuclear antigen 2 in viral promoter switching during initial stages of infection. Proc. Natl. Acad. Sci. USA 88:3942-3946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woisetschlaeger, M., J. L. Strominger, and S. H. Speck. 1989. Mutually exclusive use of viral promoters in Epstein-Barr virus latently infected lymphocytes. Proc. Natl. Acad. Sci. USA 86:6498-6502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woisetschlaeger, M., C. N. Yandava, L. A. Furmanski, J. L. Strominger, and S. H. Speck. 1990. Promoter switching in Epstein-Barr virus during the initial stages of infection of B lymphocytes. Proc. Natl. Acad. Sci. USA 87:1725-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yandava, C. N., and S. H. Speck. 1992. Characterization of the deletion and rearrangement in the BamHI C region of the X50-7 Epstein-Barr virus genome, a mutant viral strain which exhibits constitutive BamHI W promoter activity. J. Virol. 66:5646-5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoo, L., and S. H. Speck. 2000. Determining the role of the Epstein-Barr virus Cp EBNA2-dependent enhancer during the establishment of latency by using mutant and wild-type viruses recovered from cottontop marmoset lymphoblastoid cell lines. J. Virol. 74:11115-11120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoo, L. I., M. Mooney, M. T. Puglielli, and S. H. Speck. 1997. B-cell lines immortalized with an Epstein-Barr virus mutant lacking the Cp EBNA2 enhancer are biased toward utilization of the oriP-proximal EBNA gene promoter Wp1. J. Virol. 71:9134-9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoo, L. I., and S. H. Speck. 2000. Regulation of EBNA gene expression. EBV Report 7:175-185. [Google Scholar]