Abstract
Efficient expression of the human immunodeficiency virus type 1 (HIV-1) structural gene products Gag, Pol, and Env involves the regulation by viral Rev and Rev-responsive elements (RRE). Removal of multiple inhibitory sequences (INS) in the coding regions of these structural genes or modification of the codon usage patterns of HIV-1 genes to those used by highly expressed human genes has been found to significantly increase HIV-1 structural protein expression in the absence of Rev and RRE. In this study, we show that efficient and stable expression of the HIV-1 structural gene products Gag and Env could be achieved by transfection with a noncytopathic Sindbis virus expression vector by using HIV-1 sequences from primary isolates without any sequence modification. Stable expression of these Gag and Env proteins was observed for more than 12 months. The fact that the Sindbis virus expression vector replicates its RNA only in the cytoplasm of the transfected cells and the fact that the lack of expression of HIV-1 Gag by the DNA vector containing unmodified HIV-1 gag sequences was associated with a lack of detectable cytoplasmic gag RNA suggest that a major blockage in the expression of HIV-1 structural proteins in the absence of Rev/RRE is caused by inefficient accumulation of mRNA in the cytoplasm. Efficient long-term expression of structural proteins of diverse HIV-1 strains by the noncytopathic Sindbis virus expression system may be a useful tool for functional study of HIV-1 gene products and vaccine research.
Expression of the human immunodeficiency virus type 1 (HIV-1) structural proteins Gag, Pol, and Env is regulated by the viral protein Rev, the Rev-responsive elements (RRE), and their interaction with host proteins (11). Gag is expressed in the form of a 55-kDa precursor, pr55Gag, which is posttranslationally cleaved into p17MA, p24CA, p7NC, and p6Gag by the viral protease during virus assembly and maturation (11, 22). The expression of retroviral Gag proteins, which constitute the principal structural components of the virus, is sufficient to drive viral particle assembly and budding (11, 22). The Pol proteins of HIV-1 (protease, reverse transcriptase, and integrase) are synthesized in the form of a fusion protein that includes part of the Gag protein (MA, CA, and NC, but not p6Gag). The Gag-Pol fusion protein is the result of a −1 ribosomal frameshift event between the p7NC and p6Gag domains of Gag, which occurs with a frequency of 5 to 10% of the total Gag proteins synthesized (11, 22). Full-length Env gp160 is synthesized from the singly spliced mRNA in the endoplasmic reticulum, cleaved into its noncovalently associated subunits, gp120 and gp41, in the Golgi, and subsequently transported to the cell surface (11, 15).
Expression of the HIV-1 Gag, Pol, and Env proteins by DNA vectors has been restricted by the presence of multiple inhibitory sequences (INS) in the structural genes encoding the Gag, Pol, and Env proteins of HIV-1. This situation makes the expression of the structural HIV-1 proteins dependent on the viral regulatory protein Rev, which is responsible for the nuclear export and efficient expression of unspliced HIV-1 mRNAs (8, 10, 16, 17). Rev binds specifically to an RNA site within HIV-1 mRNA named RRE. In the absence of functional Rev/RRE, mRNAs containing INS are either retained in the nucleus or degraded rapidly; therefore, little protein can be expressed from these mRNAs. Modification of HIV-1 sequences, presumably removing the inhibitory sequences, can result in significantly enhanced HIV-1 protein expression in the absence of Rev/RRE (4, 6, 14, 19, 21, 24).
Transient expression of HIV-1 or SIV structural proteins in the absence of Rev/RRE has also been achieved by using several recombinant viral vectors including recombinant vaccinia virus (3, 13), alphaviruses (5, 23), poliovirus (7), and vesicular stomatitis virus (20). However, strategies for long-term expression of HIV-1 structural proteins are limited. Recently it was reported that a noncytopathic Sindbis virus-derived expression vector could be used for the long-term expression of foreign genes (2, 12). In the present study, we have demonstrated that this noncytopathic Sindbis virus expression vector could induce efficient and stable expression of the HIV-1 structural proteins Gag and Env by using unmodified sequences from primary HIV-1 isolates. These data suggest that this noncytopathic Sindbis virus expression system can be a rapid and efficient means for achieving long-term expression of HIV proteins of diverse origin for use in functional studies and vaccine research.
MATERIALS AND METHODS
DNA constructs.
The plasmid pcDNA3.1(−) and pSinRep5 vector system were purchased from Invitrogen (San Diego, Calif.). The noncytopathic Sindbis vector, pSinRep19, was a gift from Charles Rice (Department of Molecular Microbiology, Washington University School of Medicine). pGag and pGagins have been described previously (19). The gag coding regions of CRF08 and CRF01 (18) were amplified by PCR with the primers Gag-for (5′GCTAGAAGGTCTAGAAATGG GTGCGAGAGCG3′, with the XbaI site underlined) and Gag-rev (5′AGTTGCCCCCGAATCCTTATTGTGACGAGG3′, with the EcoRI site underlined). The PCR products were digested with XbaI and EcoRI and cloned into pcDNA3.1(−) downstream from the human cytomegalovirus(CMV) immediate-early promoter (Pcmv) sequence, by use of XbaI and EcoRI sites, to generate pgagCRF08 and pgagCRF01. To generate pSinRep5-gag, pSinRep5-gagins, pSinRep5-gagCRF08, and pSinRep5-gagCRF01, XbaI-PmeI fragments from plasmids pGag, pGagins, pGagCRF08, and pGagCRF01 were cloned into pSinRep5 downstream from subgenomic promoter and upstream from polyadenylation signal, using the XbaI and PmlI sites. To generate pSinRep19-gag, pSinRep19-gagins, pSinRep19-gagCRF08, pSinRep19-gagCRF01, XbaI-PmeI fragments from plasmids pGag, pGagins, pGagCRF08, and pGagCRF01 were cloned into pSinRep19 downstream from the subgenomic promoter and upstream from the polyadenylation signal, by using XbaI and PmlI sites.
The HIV-1 env coding region from CRF08 (18) was amplified by PCR using the primers Envc-for (5′GGCTCTAGAATGAGAGTGAGGGGGACAC3′, with that XbaI site underlined) and Envc-rev (5′GGCCACGTGTTACTGTTATTGCAAAGCTGC3′, with the PmlI site underlined). The PCR products was digested with XbaI and PmlI and cloned into pSinRep19 using XbaI and PmlI sites to generate pSinRep19-envCRF08. All plasmids used in this study were constructed by standard molecular biology techniques and verified by restriction enzyme analysis and DNA sequencing.
In vitro transcription.
Plasmid DNAs were purified with the QIAGEN MAXiprep kit (Valencia, Calif.). For pSinRep5 and pSinRep19-derived vectors, purified plasmid DNAs were linearized with either NotI or PacI for runoff transcription. An invitroScript CAP kit (Invitrogen) was used to synthesize RNA transcripts. Transcribed RNAs were purified by phenol-chloroform extractions, concentrated by ethanol precipitation, and resuspended in RNase-free Tris-EDTA buffer.
Transfection and establishment of cell lines.
COS-7 cells were maintained in Dulbecco's modified Eagle's medium with 10% fetal bovine serum and antibiotics and passaged upon confluence. BHK cells were maintained in alpha minimal essential medium supplemented with 10% fetal bovine serum and antibiotics, and passaged upon confluence. RNA and DNA were transfected into cells by DMRIE-C (Life Technologies, Gaithersburg, Md.) according to the manufacturer's recommendations. To establish stable cell lines expressing HIV-1 structural proteins, pSinRep-19-derived constructs were transfected into BHK cells. Cells were allowed to rest for about 12 to 18 h after transfection, and the medium was changed to standard growth medium with 5 μg of puromycin/ml (Sigma) for 1 to 2 weeks. The established BHK cell lines were maintained in standard growth medium with 5 μg of puromycin/ml.
Immunoblotting.
At 24 h after RNA transfection or 72 h after DNA transfection, the transfected cells were collected, and cell lysates and virus-like particles (VLP) were prepared as previously described (9, 19). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out by standard methods. Proteins were transferred to nitrocellulose (Schleicher & Schuell) as described previously (9, 19). The blots were stained with an HIV-1-positive human serum or a polyclonal sheep anti-HIV-1 Env serum in a phosphate-buffered saline solution with 3% nonfat dried milk. Secondary antibodies were alkaline phosphatase-conjugated antihuman or antigoat (Jackson Immunoresearch, Inc., West Grove, Pa.), and staining was carried out with BCIP (5-bromo-4-chloro-3-indolylphosphate) and nitroblue tetrazolium solutions prepared from chemicals obtained from Sigma.
RNA isolation and Northern blotting.
Cytoplasmic and nucleic RNAs were isolated from transfected cells by using the RNeasy Mini Kit (QIAGEN) according to the manufacturer's recommendations. The RNA samples were resolved on a 1% agarose gel containing formaldehyde and transferred to nitrocellulose membranes. To detect gag and gagins mRNA, the blots were blocked and probed with a 32P-labeled random-primed HIV-1 gag fragment (nucleotides 1231 to 1550 of the gag coding region). To detect gag and gagins RNA expressed by pSinRep5-gag and pSinRep5-gagins (see Fig. 2), the blots were blocked and probed with a 32P-labeled random-primed pSinRep5 vector fragment [310 to 500 bp before poly(A)]. Blots were washed and exposed to X-ray film.
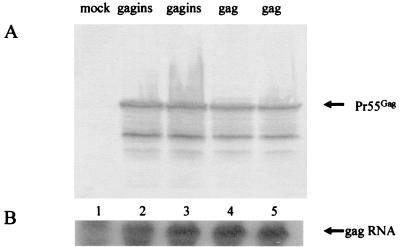
FIG. 2.
Gag expression and RNA levels in pSin-Rep5GAG and pSin-Rep5GAGINS-transfected COS-7 cells. (A) Immunoblotting of pSin-Rep5GAG-(gag) and pSin-Rep5GAGINS (gagins)-transfected cells. Cell lysates were separated by SDS-PAGE, transferred to nitrocellulose filters, and reacted with HIV-1-positive human serum. The positions of the Gag precursor molecules are indicated by arrows. (B) Northern blot of RNA from pSin-Rep5GAG (gag)- and pSin-Rep5GAGINS (gagins)-transfected cells. Lane 1, sample from mock-transfected cell; lanes 2 and 3, samples from pSin-Rep5GAGINS-transfected cells; lanes 4 and 5, samples from pSin-Rep5GAG-transfected cells.
RESULTS
Inhibitory sequences in the coding region of HIV-1 gag reduced the accumulation of gag-specific mRNA in the cytoplasm.
Expression of HIV-1 Gag could be significantly enhanced by removing inhibitory sequences in the gag coding region. The constructs pGAG and pGAGINS have been described previously (19). The coding region for GAGINS contains nucleotide modifications that remove ins sequences without changing the amino acid sequences in HXB2 Gag. After transfection of COS-7 cells, Gag was detected in the cell lysates of pGAGINS DNA-transfected cells but not in wild-type gag gene encoding DNA pGAG or in control DNA-transfected cells (Fig. 1A). Transfection efficiency was comparable in all cells, as monitored by cotransfection with a β-galactosidase (β-Gal) expression DNA vector and detection of β-Gal activity in the cell lysates (data not shown).
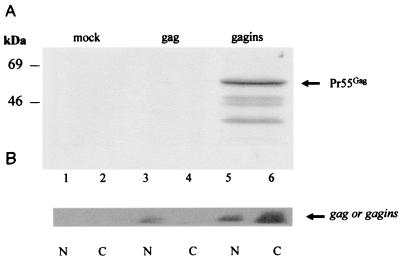
FIG. 1.
Gag expression and mRNA distribution in pGAG-and pGAGINS-transfected COS-7 cells. (A) Immunoblotting of pGAG (gag)- and pGAGINS (gagins)-transfected cells. Cell lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, transferred to nitrocellulose filters, and reacted with an HIV-1-positive human serum. The positions of the Gag precursor molecules are indicated by arrows. (B) Northern blot of mRNA from pGAG (gag)- and pGAGINS (gagins)-transfected cells. Lanes 1 and 2, samples from mock-transfected cells; lanes 3 and 4, samples from pGAG-transfected cells; lanes 5 and 6, samples from pGAGINS-transfected cells. N, nuclear fraction; C, cytoplasmic fraction.
To examine whether gag-specific mRNA was influenced by INS mutation, total RNA was extracted from either the cytoplasm or nuclei of transfected COS-7 cells. The amount of gag-specific mRNA was detected by Northern blotting using a radio-labeled RNA probe specific for gag. Although gag-specific mRNA was detected in the nuclei of pGAG- and pGAGINS-transfected COS-7 cells, cytoplasmic gag-specific mRNA was detected only in the pGAGINS-transfected COS-7 cells (Fig. 1B). Therefore, it seemed that the lack of expression of Gag protein by pGAG-transfected COS-7 cells was associated with reduced accumulation of gag-specific mRNA in the cytoplasm.
Wild-type and INS mutant Gag proteins were expressed to comparable extents in a Sindbis expression system, which replicates in the cytoplasm.
To address the question of whether selective targeting of wild-type gag mRNA to the cytoplasm could enhance Gag protein expression, we cloned both wild-type gag and INS mutant gag genes into a Sindbis expression system. The Sindbis expression system contains only nonstructural viral genes and self-replicates in the cytoplasm of transfected cells. Therefore, the mRNA encoded by the Sindbis expression system will not go through a nucleus-to-cytoplasm transport process.
Efficient Gag expression was detected in cells transfected with the Sindbis RNA vector (pSinRep-5) containing wild-type gag or gagins sequences (Fig. 2). This result was in sharp contrast to that obtained for the DNA vectors containing these two gag sequences (Fig. 1). There was little variation between two separate clones of Sindbis vectors containing wild-type gag or gagins sequences (Fig. 2). RNAs containing wild-type gag or gagins sequences were detected in the transfected cells (Fig. 2). When normalized for the amount of cytoplasmic gag RNA, comparable quantities of Gag molecules were detected in the Sindbis vector-containing wild-type gag and gagins sequences (Fig. 2). As expected, no Gag was detected in COS-7 cells transfected with a control Sindbis vector containing a β-Gal gene (Fig. 2).
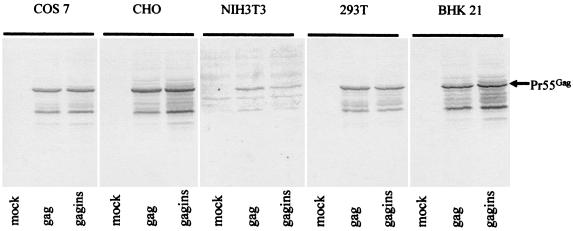
Expression of Gag by the Sindbis vectors containing wild-type gag or gagins sequences in human (293T), monkey (COS-7), mouse (NIH 3T3), and hamster (BHK21) cells was further compared. No significant differences between cells transfected with the Sindbis vector containing wild-type gag sequence and cells transfected with that containing the gagins sequence were seen (Fig. 3). The overall expression of Gag by the Sindbis vectors containing wild-type gag or gagins sequences in mouse NIH 3T3 cells was less than that seen in COS-7, CHO, 293T, or BHK21 cells (Fig. 3). This difference may be related to the transfection of the NIH 3T3 cells being less efficient than that of other cells. The transfection efficiency was measured by cotransfection with a β-Gal expression DNA vector and the β-Gal activity was lower in NIH 3T3 cells than in the other cell lines (data not shown). However, we cannot rule out the possibility that the replication of the Sindbis expression system is less efficient in NIH 3T3 cells than in other cell lines.
FIG. 3.
Immunoblot of pSin-Rep5GAG (gag)-and pSin-Rep5GAGINS (gagins)-transfected cells. Cell lysates were prepared from 106 cells 24 h after transfection, separated by SDS-PAGE, transferred to nitrocellulose filters, and reacted with an HIV-1-positive human serum.
Efficient expression of primary HIV-1 gag and env genes by the Sindbis expression system.
We next addressed the question of whether efficient expression of Gag using unmodified sequences from primary HIV-1 isolates could be achieved by using the Sindbis expression vector. To answer this question, we cloned gag genes from primary HIV-1 isolates representing CRF01 (subtype E) and CRF08 (B/C recombinants) from China (18) into the DNA expression vector pcDNA3.1 and the Sindbis RNA expression vector.
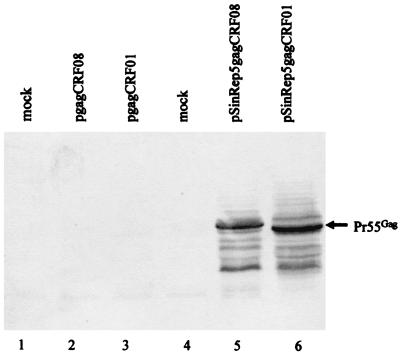
After transfection of COS-7 cells, Gag molecules were detected in cells transfected with the Sindbis vectors containing CRF01 or CRF08 gag sequences (Fig. 4, lanes 5 and 6). In contrast, no Gag was detected in cells transfected with pcDNA3.1 containing CRF01 and CRF08 gag sequences (Fig. 4, lanes 2 and 3). The DNA transfection was successful, as evidenced by the expression of β-Gal molecules from the cotransfected control DNA vector (data not shown). These results suggest that inhibitory sequences are present in the gag sequences of diverse strains, including primary HIV-1 isolates from different subtypes.
FIG. 4.
Comparison of Gag expression by DNA vectors and pSinRep5 vectors. Cell lysates were prepared from 106 cells 3 days after transfection, separated by SDS-PAGE, transferred to nitrocellulose filters, and reacted with HIV-1-positive human serum. Lanes 2 and 3, samples from cells transfected with the DNA vectors pgagCRF08 and pgagCRF01-transfected cells, respectively; lanes 5 and 6, samples from cells transfected with Sindbis vectors pSINRep5-gagCRF08 and pSINRep5-gagCRF01, respectively.
Long-term and stable expression of primary HIV-1 gag and env genes by a novel noncytopathic Sindbis expression system.
Because of the stringent requirement for Rev/RRE for the expression of HIV-1 structural gene products, strategies for efficient and stable expression of HIV-1 structural proteins are scarce. As demonstrated above, efficient expression of HIV-1 Gag proteins from unmodified gag sequences would be achieved by using the Sindbis virus vector. However, because of the cytopathic effect induced by the Sindbis virus vector, only transient expression of Gag proteins was obtained. Recently, a noncytopathic Sindbis virus vector which allows long-term expression of target gene products has been developed (2). We first evaluated the expression of unmodified gag and gagins gene expression in this noncytopathic Sindbis virus vector.
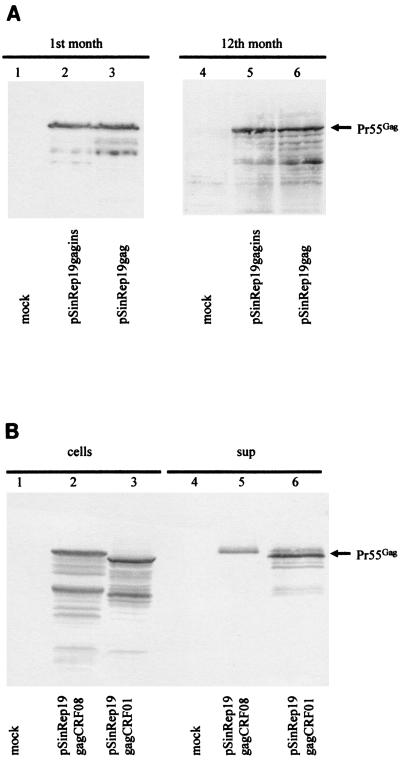
BHK cells were transfected with the Sindbis Rep-19-containing gag or gagins sequences and selected by resistance to puromycin. One month after transfection, comparable Gag molecules were detected in cells transduced with the Sindbis Rep-19-containing gag and gagins sequences (Fig. 5A, lanes 2 and 3). The expression of Gag molecules was rather stable in these cells. One year after transfection, efficient expression of Gag molecules could still be detected in the BHK cells transduced with the Sindbis Rep-19-containing gag or gagins sequences (Fig. 5A, lanes 5 and 6).
FIG. 5.
Long-term expression of Gag from diverse HIV-1 strains. (A) Gag expression by the noncytopathic Sindbis virus vector containing either the gag INS mutant sequence (pSinRep19-gagins) or the unmodified gag sequence (pSinRep19-gag). Cell lysates were prepared from 106 BHK-21 cells 1 month or 12 months after transfection and analyzed by immunoblotting by using HIV-1-positive human serum. (B) Long-term expression of primary HIV-1 isolates by the noncytopathic Sindbis virus vector containing either the CRF08 gag sequence (pSinRep19-gagCRF08) or the CRF01 gag sequence (pSinRep19-gagCRF01). Cell lysates (lanes 1 to 3) were prepared from 106 BHK-21 cells at 12 months after transfection and analyzed by immunoblotting with HIV-1-positive human serum. VLP from the supernatants of these cells (lanes 4 to 6) were prepared and analyzed by immunoblotting as described in Materials and Methods by use of an HIV-1-positive human serum.
Next, we addressed the question of whether efficient and stable expression could be achieved with the Sindbis Rep-19 vector by using unmodified gag sequences of CRF01 and CRF08. BHK cells transfected with Rep-19-containing gag sequences of CRF01 or CRF08 were established, and efficient expression of Gag molecules was detected in these cells 1 month after transfection (data not shown). One year after transfection, Gag proteins corresponding to CRF08 (Fig. 5B, lane 2) and CRF01 (Fig. 5B, lane 3) were still detected in transfected BHK cells. The Gag expression levels in these cells 1 year after transfection were not significantly different from those of cells at 1 month after transfection (data not shown), suggesting that expressions of CRF01 and CRF08 Gag by the Rep-19 vector were stable. Also, pelletable Gag could be detected in the supernatants of transduced BHK cells (Fig. 5B, lanes 5 and 6), suggesting that VLP may be formed from these cells.
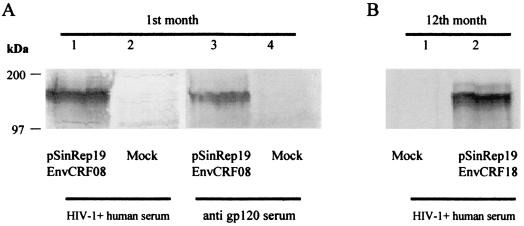
Finally, we tested the ability of the Sindbis Rep-19 vector to express HIV-1 Env proteins, using unmodified env sequences from the CRF08 isolate. As was the case for gag, expression of the CRF08 Env protein was not detected when the control DNA vector pcDNA3.1 was used (data not shown). In contrast, efficient expression of CRF08 Env proteins from transfected BHK cells was detected 1 month after transfection (Fig. 6A, lane 1). As expected, no Env proteins were detected in mock-transfected BHK cells (Fig. 6A, lane 2). The specificity of the Env proteins was confirmed by reactivity with a polyclonal sheep anti-gp120 antiserum (Fig. 6A, lane 3). Furthermore, efficient and stable expression of CRF08 Env proteins was achieved in these cells 1 year after transfection (Fig. 6B, lane 2).
FIG. 6.
Long-term expression of HIV-1 Env protein from CRF08 by the noncytopathic Sindbis virus vector. Cell lysates were prepared from 106 BHK21 cells at 1 month (A) or 12 months (B) after transfection and analyzed by immunoblotting with an HIV-1-positive human serum or a polyclonal sheep anti-gp120 antibody.
DISCUSSION
Expression of HIV-1 structural genes depends on the regulation of Rev/RRE. In the absence of Rev, DNA vectors containing gag, pol, or env genes cannot be expressed efficiently (11). The reason for the restricted expression of HIV-1 structural genes in the absence of Rev is not totally clear. Removal of inhibitory sequences from these genes results in efficient Rev-independent expression of Gag and Pol (19, 21). One explanation for this phenomenon is that the inhibitory sequences prevent efficient transport of these mRNAs from the nucleus to the cytoplasm. Another possibility is that the inhibitory sequences result in rapid degradation of the mRNA in the nucleus and/or cytoplasm (1). It has also been suggested that the codons used in the HIV-1 structural genes do not match those that are used in the highly expressed human genes. Use of codon-optimized HIV-1 sequences has resulted in significantly enhanced gene expression (4, 24). In the present study, we have demonstrated that efficient expression of HIV-1 structural proteins with unmodified HIV-1 sequences can be achieved by using the Sindbis expression system but not with a DNA expression vector. Unlike DNA vectors, which require nucleus-to-cytoplasm transport of the transcribed mRNA, the Sindbis expression system replicates its RNA only in the cytoplasm. Our results are consistent with the idea that the major blockage in the expression of HIV-1 structural genes is at the level of cytoplasmic accumulation of mRNA.
Because of the stringent requirement for Rev/RRE, achieving efficient expression of HIV-1 structural proteins, especially long-term expression, has been difficult. In the absence of sequence modification, expression of HIV-1 structural proteins by DNA expression vectors is usually undetectable (4, 14, 19, 21, 24). In contrast to the results obtained with DNA vectors, efficient expression of HIV-1 structural proteins by use of unmodified sequences has been achieved with several recombinant viral vectors, including vaccinia virus (3, 13), alphaviruses (5, 23), vesicular stomatitis virus (20), and poliovirus (7). However, this expression is usually transient, due to cytopathogenicity of the recombinant viruses. The results presented in this study suggest that the noncytopathic Sindbis vector pSinRep-19 could offer a useful system for achieving long-term expression of HIV-1 structural proteins from diverse HIV-1 strains including primary HIV-1 isolates.
Transduction with DNA vectors containing selection markers can result in the establishment of cell lines expressing the gene of interest. Previously, we had established mouse p815 cell lines expressing HIV-1 Gag by using pcDNA3.1 with a modified HIV-1 gag sequence (19). However, we observed that the expression of Gag in this cell line decreased significantly over time (B. Liu and X.-F. Yu, unpublished results). We have also tried to stably express HIV-1 in BHK cells by using pGAGINS. Although stable cells resistant to selection with G418 were obtained, the expression of HIV-1 Gag by the DNA vector pGAGINS was extremely low (Liu and Yu, unpublished). This observation was in sharp contrast to the results that we obtained with the noncytopathic Sindbis virus vector (Fig. 5). These results suggest that the noncytopathic Sindbis virus vector may have advantages for long-term protein expression in certain cell types over the traditional DNA expression vectors with a CMV promoter.
Efficient long-term expression of HIV-1 structural proteins with unmodified viral sequences from primary HIV-1 isolates may have several advantages. HIV-1 structural proteins expressed in these cells may assemble into VLP. Retroviral Gag protein alone can drive the formation of VLP (11, 22). Coexpression of Pol and Env proteins with Gag may allow incorporation of Pol and Env into VLP and these VLP could be part of HIV vaccine strategies for the induction of anti-HIV immune responses. Since viral RNA and other critical viral regulatory proteins are not part of the system, these VLP will be safer than whole killed viral particles. Efficient long-term expression of HIV-1 Env proteins from diverse HIV-1 strains obtained in the field could offer an alternative means of producing modified Env proteins, as opposed to a transient expression system such as a vaccinia virus or baculovirus vector. Efficient and stable expression of HIV-1 structural proteins may also enable functional studies of these proteins from primary HIV-1 isolates.
Acknowledgments
We are grateful to Charles Rice of Rockefeller University for the noncytopathic Sindbis virus system and George Pavlakis of National Cancer Institute for the gagins sequence. We thank Xianghui Yu for technical assistance. The antiserum against HIV-1 gp120 (catalogue no. 288) reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
Support also came from National Institutes of Health grants (AI-46324 and AI-48280 to X.-F.Y.).
REFERENCES
- 1.Afonina, E., M. Neumann, and G. N. Pavlakis. 1997. Preferential binding of poly(A)-binding protein 1 to an inhibitory RNA element in the human immunodeficiency virus type 1 gag mRNA. J. Biol. Chem. 272:2307-2311. [DOI] [PubMed] [Google Scholar]
- 2.Agapov, E. V., I. Frolov, B. D. Lindenbach, B. M. Pragai, S. Schlesinger, and C. M. Rice. 1998. Noncytopathic Sindbis virus RNA vectors for heterologous gene expression. Proc. Natl. Acad. Sci. USA 95:12989-12994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 4.Andre, S., B. Seed, J. Eberle, W. Schraut, A. Bultmann, and J. Haas. 1998. Increased immune response elicited by DNA vaccination with a synthetic gp120 sequence with optimized codon usage. J. Virol. 72:1497-1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caley, I. J., M. R. Betts, D. M. Irlbeck, N. L. Davis, R. Swanstrom, J. A. Frelinger, and R. E. Johnston. 1997. Humoral, mucosal, and cellular immunity in response to a human immunodeficiency virus type 1 immunogen expressed by a Venezuelan equine encephalitis virus vaccine vector. J. Virol. 71:3031-3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casimiro, D. R., A. Tang, H. C. Perry, R. S. Long, M. Chen, G. J. Heidecker, M. E. Davies, D. C. Freed, N. V. Persaud, S. Dubey, J. G. Smith, D. Havlir, D. Richman, M. A. Chastain, A. J. Simon, T. M. Fu, E. A. Emini, and J. W. Shiver. 2002. Vaccine-induced immune responses in rodents and nonhuman primates by use of a humanized human immunodeficiency virus type 1 pol gene. J. Virol. 76:185-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crotty, S., C. J. Miller, B. L. Lohman, M. R. Neagu, L. Compton, D. Lu, F. X. Lu, L. Fritts, J. D. Lifson, and R. Andino. 2001. Protection against simian immunodeficiency virus vaginal challenge by using Sabin poliovirus vectors. J. Virol. 75:7435-7452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullen, B. R. 1992. Mechanism of action of regulatory proteins encoded by complex retroviruses. Microbiol. Rev. 56:375-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dettenhofer, M., and X. F. Yu. 1999. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J. Virol. 73:1460-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felber, B. K., M. Hadzopoulou-Cladaras, C. Cladaras, T. Copeland, and G. N. Pavlakis. 1989. rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. USA 86:1495-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freed, E. O., and M. A. Martin. 2001. HIVs and their replication, p. 1971-2041. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 12.Frolov, I., E. Agapov, T. A. Hoffman, Jr., B. M. Pragai, M. Lippa, S. Schlesinger, and C. M. Rice. 1999. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J. Virol. 73:3854-3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanke, T., R. V. Samuel, T. J. Blanchard, V. C. Neumann, T. M. Allen, J. E. Boyson, S. A. Sharpe, N. Cook, G. L. Smith, D. I. Watkins, M. P. Cranage, and A. J. McMichael. 1999. Effective induction of simian immunodeficiency virus-specific cytotoxic T lymphocytes in macaques by using a multiepitope gene and DNA prime-modified vaccinia virus Ankara boost vaccination regimen. J. Virol. 73:7524-7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang, Y., W. P. Kong, and G. J. Nabel. 2001. Human immunodeficiency virus type 1-specific immunity after genetic immunization is enhanced by modification of Gag and Pol expression. J. Virol. 75:4947-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter, E. 1997. Viral entry and receptors, p. 71-120. In H. E. Varmus et al. (ed.), Retroviruses, chapter 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 16.Parslow, T. C. 1993. Human retroviruses: post-transcriptional regulation of human retroviral gene expression. IRL Press at Oxford University, Oxford, United Kingdom.
- 17.Pavlakis, G. N. 1996. The molecular biology of human immunodeficiency virus type 1, 4th ed. Lippincott-Raven, Philadelphia, Pa.
- 18.Piyasirisilp, S., F. E. McCutchan, J. K. Carr, E. Sanders-Buell, W. Liu, J. Chen, R. Wagner, H. Wolf, Y. Shao, S. Lai, C. Beyrer, and X. F. Yu. 2000. A recent outbreak of human immunodeficiency virus type 1 infection in southern China was initiated by two highly homogeneous, geographically separated strains, circulating recombinant form AE and a novel BC recombinant. J. Virol. 74:11286-11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu, J. T., R. Song, M. Dettenhofer, C. Tian, T. August, B. K. Felber, G. N. Pavlakis, and X. F. Yu. 1999. Evaluation of novel human immunodeficiency virus type 1 Gag DNA vaccines for protein expression in mammalian cells and induction of immune responses. J. Virol. 73:9145-9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rose, N. F., P. A. Marx, A. Luckay, D. F. Nixon, W. J. Moretto, S. M. Donahoe, D. Montefiori, A. Roberts, L. Buonocore, and J. K. Rose. 2001. An effective AIDS vaccine based on live attenuated vesicular stomatitis virus recombinants. Cell 106:539-549. [DOI] [PubMed] [Google Scholar]
- 21.Schneider, R., M. Campbell, G. Nasioulas, B. K. Felber, and G. N. Pavlakis. 1997. Inactivation of the human immunodeficiency virus type 1 inhibitory elements allows Rev-independent expression of Gag and Gag/protease and particle formation. J. Virol. 71:4892-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swanstrom, R., and J. W. Wills. 1997. Synthesis, assembly, and processing of viral proteins, p. 263-334. In H. E. Varmus et al. (ed.), Retroviruses, chapter 7. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 23.Vajdy, M., J. Gardner, J. Neidleman, L. Cuadra, C. Greer, S. Perri, D. O'Hagan, and J. M. Polo. 2001. Human immunodeficiency virus type 1 Gag-specific vaginal immunity and protection after local immunizations with Sindbis virus-based replicon particles. J. Infect. Dis. 184:1613-1616. [DOI] [PubMed] [Google Scholar]
- 24.zur Megede, J., M. C. Chen, B. Doe, M. Schaefer, C. E. Greer, M. Selby, G. R. Otten, and S. W. Barnett. 2000. Increased expression and immunogenicity of sequence-modified human immunodeficiency virus type 1 gag gene. J. Virol. 74:2628-2635. [DOI] [PMC free article] [PubMed] [Google Scholar]