Abstract
On viral infection, infected cells can become the target of host immune responses or can go through a programmed cell death process, called apoptosis, as a defense mechanism to limit the ability of the virus to replicate. To prevent this, viruses have evolved elaborate mechanisms to subvert the apoptotic process. Here, we report the identification of a novel antiapoptotic K7 protein of Kaposi's sarcoma-associated herpesvirus (KSHV) which expresses during lytic replication. The KSHV K7 gene encodes a small mitochondrial membrane protein, and its expression efficiently inhibits apoptosis induced by a variety of apoptogenic agents. The yeast two-hybrid screen has demonstrated that K7 targets cellular calcium-modulating cyclophilin ligand (CAML), a protein that regulates the intracellular Ca2+ concentration. Similar to CAML, K7 expression significantly enhances the kinetics and amplitudes of the increase in intracellular Ca2+ concentration on apoptotic stimulus. Mutational analysis showed that K7 interaction with CAML is required for its function in the inhibition of apoptosis. This indicates that K7 targets cellular CAML to increase the cytosolic Ca2+ response, which consequently protects cells from mitochondrial damage and apoptosis. This is a novel viral antiapoptosis strategy where the KSHV mitochondrial K7 protein targets a cellular Ca2+-modulating protein to confer resistance to apoptosis, which allows completion of the viral lytic replication and, eventually, maintenance of persistent infection in infected host.
Apoptosis (programmed cell death) is a mechanism by which the immune system has enlisted every cell of the body to participate in its own death. Whether triggered through signaling by a cell surface receptor such as Fas or through signaling from intracellular molecules, apoptosis proceeds through a cascade of kinases, caspases, phosphatases, and DNases, resulting in the breakdown of mitochondrial membrane potential, DNA fidelity, and cell membrane integrity (30, 31, 68). On viral infection, the host immune system uses apoptosis as an initial defense mechanism to limit the ability of the virus to replicate. To ensure continued virion production, circumvention of the apoptosis response by viruses is necessary for completion of the viral life cycle to maximize the production of viral progeny and thereby prolong the infection (70). In fact, a variety of viruses have evolved elaborate mechanisms to subvert host apoptotic process. One of the best-characterized viral apoptosis inhibitors is the cowpox virus-encoded cytokine response modifier A (CrmA). CrmA inhibits both Fas- and tumor necrosis factor (TNF)-induced apoptosis via interaction with caspase 8 (76). In a similar manner, baculovirus p35 also protects infected cells from apoptosis (15, 76). In addition, adenovirus encodes a set of proteins (E3-10.4K, E3-14.5K or RID complex) that induce internalization and lysosomal degradation of cell surface Fas and other tumor necrosis factor receptor (TNFR)/nerve growth factor family molecules (44, 69). Thus, viruses have developed various mechanisms to inhibit apoptosis-mediated cell killing to escape host immune attack and to persist in infected host.
Kaposi's sarcoma-associated herpesvirus (KSHV) or human herpesvirus 8 is thought to be an etiologic agent of Kaposi's sarcoma (KS) (9, 13, 45, 62). It is also associated with primary effusion lymphoma and an immunoblast variant of Castleman's disease, which are of B-cell origin (12, 63). Genomic sequencing indicates that KSHV is a gamma-2 herpesvirus, closely related to herpesvirus saimiri (27, 47, 57), rhesus monkey rhadinovirus (1), and murine γherpesvirus 68 (71). KSHV has been shown to hijack numerous cellular proteins which, when expressed in the context of virus, can contribute to an abnormal cell growth control (18, 40-42, 46). K13 of KSHV, also termed vFLIP, is a homologue of the cellular FLICE (Fas-associated death domain-like interleukin 1 beta-converting enzyme)-inhibitory protein (c-FLIP) (8, 46). In vivo, the level of vFLIP expression is increased in late-stage KS skin lesions compared with early stage lesions. KSHV vFLIP dramatically blocks anti-Fas receptor antibody-mediated apoptosis, and its expression in tumor cells enhances tumor progression (29). In addition, KSHV orf16 possesses 15 to 20% homology to the cellular Bcl-2 gene product, designated vBcl-2 (4, 59). This homology is largely restricted to the Bcl-2 homology domain 1 (BH1) and BH2 heterodimerization and death repressor domains (4, 14, 59). vBcl-2, which is most probably expressed as a lytic protein during the viral life cycle, inhibits Bax toxicity (5, 65). In fact, vBcl-2 of γHV68 is necessary for the establishment of viral persist and latent infection in the mouse (56). In addition to a putative role in inhibiting the innate apoptotic response, a recent report has posited a possible role for vBcl-2 in blocking apoptosis induced by the KSHV v-cyclin protein (50).
DNA damage and activation of death receptors have long been recognized as initial triggers of apoptosis that induce mitochondrial membrane permeabilization (MMP) and the direct activation of the caspase cascade (20, 24, 39, 60). MMP differentially affects the outer membrane, which becomes protein permeable, and the inner membrane, which continues to retain matrix proteins yet can dissipate the mitochondrial transmembrane potential (ΨTm). This MMP-initiated stimulus triggers the activation of catabolic hydrolases, mainly caspases and nucleases (37, 66). Such hydrolases are activated secondarily to the release of proteins that are normally strictly confined to the mitochondrial intermembrane space, in particular, cytochrome c and apoptosis-inducing factor (67). Cytochrome c stimulates the cytosolic assembly of the apoptosome, the caspase activation complex, whereas apoptosis-inducing factor activates a DNase located in the nucleus (43). Then these activated downstream molecules eventually finish up the final process of apoptosis.
Accumulating evidence indicates that other organelles, including the endoplasmic reticulum (ER), lysosomes, and the Golgi, are also major points of integration of proapoptotic signaling (25, 54, 64). Specifically, the ER senses local stress through chaperones, Ca2+-binding proteins, and Ca2+ release channels, which transmit the ER Ca2+ response to mitochondria (22, 52). Ca2+ in the ER lumen is either free or bound to luminal proteins. Oscillations in the ER Ca2+ concentration participate in the regulation of normal cell function at multiple levels, while perturbation of steady-state Ca2+ levels in the ER as well as acute Ca2+ release can be strongly apoptogenic (19, 51). In fact, several cancer therapeutic agents including thapsigargin (TG) induce rapid ER Ca2+ release and trigger MMP, leading to apoptosis (19). Thus, exceeding normal spatial and temporal boundaries of intracellular Ca2+ concentration ultimately results in deleterious effect on cell growth through apoptosis.
A newly described consensus sequence for mitochondrial targeting signals consists of a hydrophobic region flanked by positively charged residues at both sides (23). Recent studies have further demonstrated that the N-terminal helical structure of the mitochondrial targeting sequence is necessary but not sufficient for efficient mitochondrial import, whereas hydrophobic residues play an essential role in in vivo mitochondrial targeting (21, 49). Since mitochondria play a central role in apoptosis, viruses have also established mechanisms to modulate the mitochondrial component of the apoptotic pathway (32). Here, we report that KSHV K7 localizes to the mitochondria through its amino-terminal leucine-rich hydrophobic region and that its expression modulates intracellular Ca2+ concentration and inhibits apoptosis on stress stimulation. The yeast two-hybrid screen has isolated the cellular calcium-modulating cyclophilin ligand (CAML) as a target for K7 action. K7 interaction of CAML alters the kinetics and amplitudes of intracellular Ca2+ responses on stress stimulation, thereby inhibiting apoptosis. These results indicate that the KSHV mitochondrial K7 protein plays an important role in the viral life cycle by inhibiting apoptosis on viral infection, which allows completion of the viral lytic replication and, eventually, maintenance of persistent infection in the infected host.
MATERIALS AND METHODS
Cell culture and transfection.
293T cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. BJAB and BCBL-1 cells were grown in RPMI 1640 supplemented with 10% fetal calf serum. BCBL-1 cells were induced with phorbol-12-O-tetradecanoylphorbol-13-acetate (20 ng/ml). Fugene 6 (Roche, Indianapolis, Ind.) or calcium phosphate (Clontech, Palo Alto, Calif.) was used for transient expression of K7 in 293T cells, and electroporation at 220V and 975 μF was used for transient expression of K7 in BJAB cells.
Cloning of KSHV K7 DNA from BCBL-1 cells.
A DNA fragment corresponding to the KSHV K7 coding sequence was amplified from BCBL-1 genomic DNA by PCR. PCR-amplified DNA was cloned into pEF-His vector, pTracer-EF vector, or pFRT/TO vector (Invitrogen, San Diego, Calif.). To generate EGFP-K7 fusion proteins, K7 alleles were cut from pFRT/TO derivatives and ligated to pEGFPC2 digested with EcoRI and Smal. KSHV K7 DNA was completely sequenced to verify 100% agreement with the original sequence by using an ABI PRISM 377 automatic DNA sequencer. Mutations in the K7 gene were generated by PCR using oligonucleotide-directed mutagenesis. After each K7 mutant was completely sequenced to verify the presence of the mutation and the absence of any other changes, it was subcloned into the green fluorescent protein (GFP) expression vector pTracer-EF or pEGFP-C1.
Yeast strains and library screen.
Yeast transformation with library cDNA was performed by the method of Schiestp and Gietz (61) as follows. Yeast strain AH109 bearing the Gal4-K7 fusion gene plasmid was grown overnight in yeast extract-peptone-deglucose agar (YPDA) medium to a density of approximately 107 cells/ml and then diluted in 100 ml of warmed YPDA medium to a density of 2 × 106 cells/ml and grown to the exponential stage. The cells were harvested and washed in water, resuspended in 1 ml of water, transferred to a sterile microcentrifuge tube, and pelleted. The pellet was resuspended in 0.5 ml of 10 mM Tris-HCl (pH 7.5)-1 mM EDTA-0.1 M Li acetate. A 50-μl volume of the resulting suspension was mixed with 1 μg of transforming DNA and 50 μg of single-stranded salmon sperm DNA, after which 0.3 ml of a solution of 40% polyethylene glycol- 4000 in Tris-EDTA-Liacetate was added and mixed thoroughly; the mixture was then incubated at 30°C with agitation for 30 min. After a heat pulse at 42°C for 15 min, the cells were pelleted in a microcentrifuge and the pellets were resuspended in 1 ml of Tris-EDTA and plated in selective medium. Library screening and recovery of plasmids were performed as specified by the manufacturer (Clontech.). For assessing the interaction of K7 with CAML, cells of the yeast strain AH109 were transformed with the indicated bait construct and selected on SD/His− Trp− Leu− glucose plates. Several colonies from each bait-interactor combination were picked and transferred to SD/Ade− His− Trp− Leu− X-α-galactose (X-α-Gal) plates containing 2% glucose.
Immunoblotting and mitochondrial fractionation.
Cells were harvested and lysed with lysis buffer (0.15 M NaCl, 0.25% NP-40, 0.25% Triton X-100, 50 mM Tris [pH 7.5]) containing 0.1 mM Na2VO3, 1 mM NaF, and protease inhibitor cocktail (Roche, Mannheim, Germany). For immunoblot analyse polypeptides from whole-cell lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membrane filters. Immunoblot detection was performed with a 1:5,000 dilution of anti-V5 antibody (Invitrogen, Carlsbad, Calif.), a 1:1000 dilution of anti-CAML antibody, or a 1:100 dilution of anti-COX 4 antibody (11). The protein was visualized with chemical luminescent detection reagent (Pierce, Rockford, Ill.) and detected with a Fuji phosphorimager. For mitochondrion fractionation, the protocol provided by the ApoAlert kit vendor (Clontech) was followed. Briefly, 293T cells expressing K7 were washed and lysed with a Dounce homogenizer. Nuclei were removed from the whole-cell lysate by centrifugation at low speed. The cytoplasmic portion was subjected to high-speed centrifugation and separated into mitochondrial and cytoplasmic fractions.
Immunofluorescence tests.
Cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with cold acetone for 15 min, blocked with 10% goat serum in phosphate-buffered saline (PBS) for 30 min, and reacted with 1:1,00 diluted primary antibody in PBS for 30 min at room temperature. After incubation, the cells were washed extensively with PBS, incubated with 1:100-diluted fluorescein isothiocyanate-conjugated secondary antibody (Molecular Probes, Eugene, Oreg.) in PBS for 30 min at room temperature, and washed three times with PBS. Mitochondrial staining was performed with 250 nM MitoTracker (Molecular Probes) for 20 min followed by washing with PBS at room temperature for 5 min. Confocal microscopy was performed using a Leica TCS SP laser-scanning microscope (Leica Microsystems, Exton, Pa.) fitted with a 100× Leica objective (PL APO; 1.4NA) and using the Leica image software. Images were collected at 512- by 512-pixel resolution. The stained cells were optically sectioned in the z axis, and the images in the different channels (photomultiplier tubes) were collected simultaneously. The step size in the z axis varied from 0.2 to 0.5 μm to obtain 16 slices per imaged file. The images were transferred to a Macintosh G4 computer (Apple Computer, Cupertino, Calif.), and NIH Image v1.61 software was used to render the images.
Measurement of mitochondrial membrane potential.
At 24 h after electroporation with pTracer-EF vector or its derivatives, 0.5 million cells were treated with apoptosis-inducing agents for the indicated time and washed with RPMI containing 10% fetal calf serum. Changes in mitochondrial membrane potential were measured using 0.1 μM teramethylrhodamine methyl ester (TMRM; Molecular Probes), since the accumulation of this dye in the mitochondria correlates directly with the magnitude of the negative mitochondrial membrane potential. After additional washing, FACS analysis was performed with a FACS Scan instrument (Becton Dickinson, Mountain View, Calif.) and CellQuest software.
Calcium mobilization analysis.
At 24 h after electroporation, cells (2 × 106) were loaded with 1 μg of indo-1 in 1 ml of complete RPMI for 45 min at 37°C as described previously (42). Baseline calcium levels were established for 1 min prior to the addition of TG. The cells were stimulated with 12.5 nM TG. Baseline absolute intracellular calcium levels were determined by using ionophore and EGTA. Data were collected and analyzed on a FACS Vantage instrument (Becton-Dickinson) with FlowJo Software.
RESULTS
Identification of K7 protein.
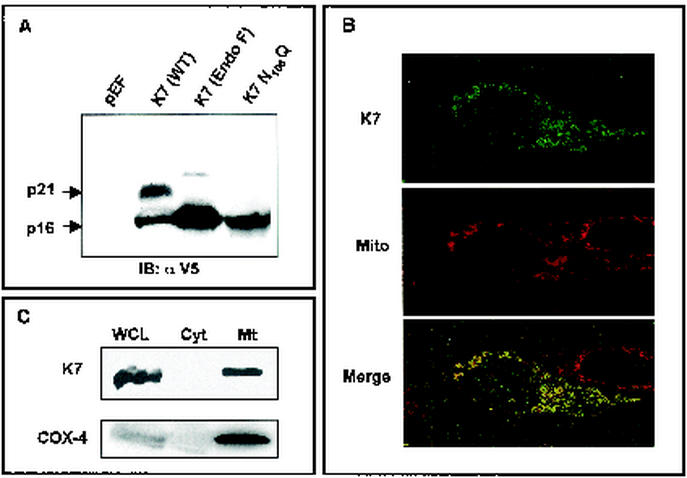
Initial DNA sequence analysis of the KSHV genome predicts that the K7 gene, which expresses during viral lytic replication, contains 126 amino acids with a considerably leucine-rich hydrophobic region flanked by positively charged amino acids at both sides (57). To identify the K7 protein, 293T cells were transfected with an expression vector containing full-length K7 with the V5 epitope tag at the carboxyl terminus. At 48 h posttransfection, 293T cells were lysed with 1% NP-40 detergent and used for an immunoblot assay with anti-V5 monoclonal antibody. Anti-V5 antibody readily reacted with both 16- and 21-kDa proteins (Fig 1A, lane 2). DNA sequence analysis predicts that K7 has a molecular mass of 14.4 kDa and that it contains two potential modifications: myristylation at amino acid 2 and N glycosylation at amino acid 108. Either or both modifications might contribute to the slow migration of K7 protein in SDS-PAGE. To test this, the glycine 2 and asparagine 108 were replaced with alanine and glutamine, respectively, to remove the potential myristylation and glycosylation. While the replacement of glycine 2 with alanine (G2A) did not affect K7 migration in SDS-PAGE (data not shown), the replacement of asparagine 108 with glutamine (N108Q) eliminated the 21-kDa protein, leaving the 16-kDa protein unaffected (Fig. 1A, lane 3). In addition, lysates of 293T cells transfected with K7 expression vector were treated with N-glycosidase F, which cleaves N-linked oligosaccharides. On N-glycosidase F treatment, the 21-kDa protein disappeared and the 16-kDa protein became the major band (Fig. 1A, lane 2). These results indicate that the glycosylation at asparagine 108 is responsible for the slow migration of K7 in SDS-PAGE and that if present, myristolyation does not contribute to the K7 migration rate.
FIG. 1.
Identification of K7 and its localization. (A) Identification and glycosylation of K7. At 48 h after transfection with pEF or pEF-K7, 293T cells were lysed by lysis buffer. Whole-cell lysates (WCL) were used for the immunoblot (IB) assay with anti-V5 antibody. Lanes (from left to right): WCL of 293T cells transfected with pEF; WCL of 293T cells transfected with pEF-K7-V5; WCL of 293T cells transfected with pEF-K7-V5 with N-glycosidase F treatment for 12 h; WCL of 293T cells transfected with pEF-K7 N108Q. Arrows indicate the 16- and 21-kDa K7 proteins. (B) Mitochondrial localization of K7 protein. After staining with Mitotracker (red), 293T cells expressing K7 protein were fixed, permeabilized, and reacted with anti-V5 antibody and Alexa 488-conjugated anti-mouse secondary antibody (green). Immunofluorescence was examined using a Leica confocal immunofluorescence microscope, and a single representative optical section is presented. The yellow areas in the merged panel indicate colocalization of the red and green labels. (C) Subcellular fractionation of K7. 293T cells expressing K7 were used for subcellular fractionation as described in Materials and Methods. Whole-cell lysates (WCL), cytosolic fraction (Cyt), and mitochondrial fraction (Mt) were used for an immunoblot assay with anti-V5 and anti-COX-4 antibodies.
K7 localization at the mitochondrial compartment.
A newly described consensus for directing proteins to mitochondria consists of a hydrophobic region flanked by an N-terminal positively charged residue and a C-terminal short positively charged tail (23). Detailed inspection reveals that the internal hydrophobic region of K7 protein is flanked by the N-terminal hydrophilic region (amino acids 1 to 21) and by the C-terminal positively charged tail (amino acids 74 to 103), suggesting the potential mitochondrial localization of the K7 protein. To examine the subcellular localization of K7, we used indirect immunofluorescence and confocal microscopy. COS-1 cells were transfected with expression vector containing V5-tagged K7. At 24 h after transfection, fixed cells were reacted with anti-V5 antibody, followed by an Alexa 488-conjugated anti-mouse secondary antibody (green). In addition, Mitotracker Red was used for the specific staining of mitochondrial compartment (red). Confocal microscopy analysis revealed that most, but not all, K7 had a punctate, cytoplasmic distribution reminiscent of mitochondrial localization. In fact, when the fluorescent signals of K7 and Mitotracker Red were superimposed, a yellow image was uniformly obtained (Fig. 1B). In addition, the subcellular fractionation further confirmed the mitochondrial localization of K7 (Fig. 1C). Mitochondrial resident COX-4 protein was used as a positive control in the subcellular fractionation (Fig. 1C). These results provide evidence that K7 localizes predominantly to the mitochondrial compartment.
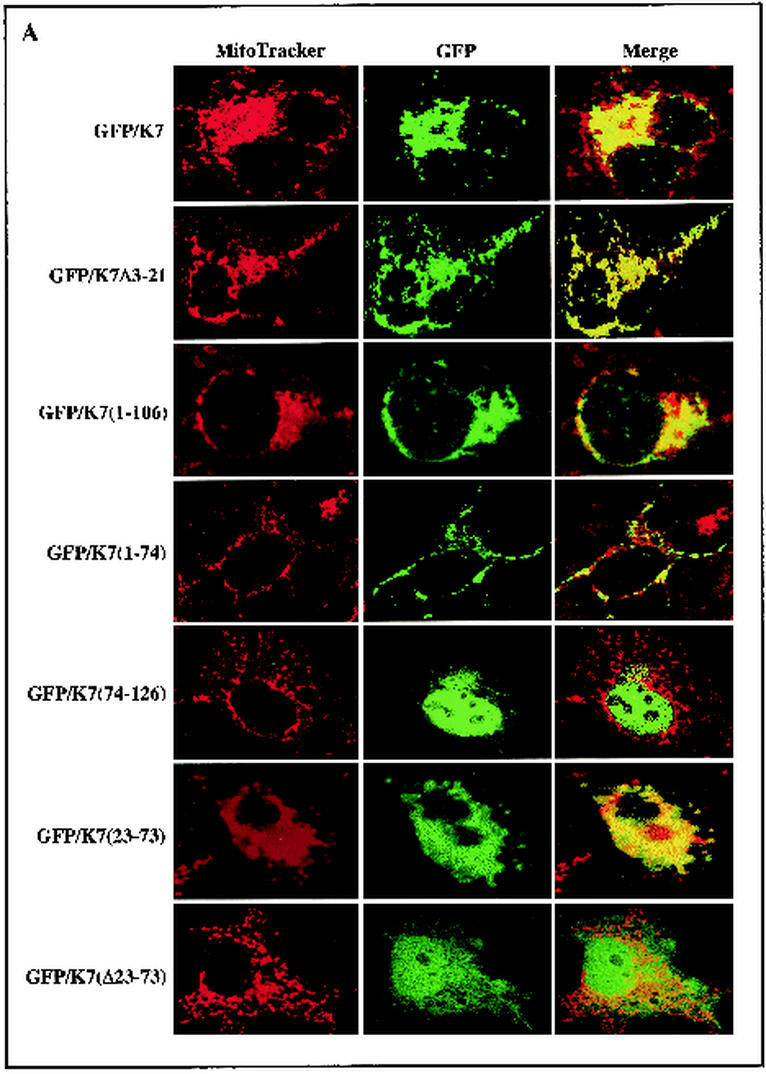
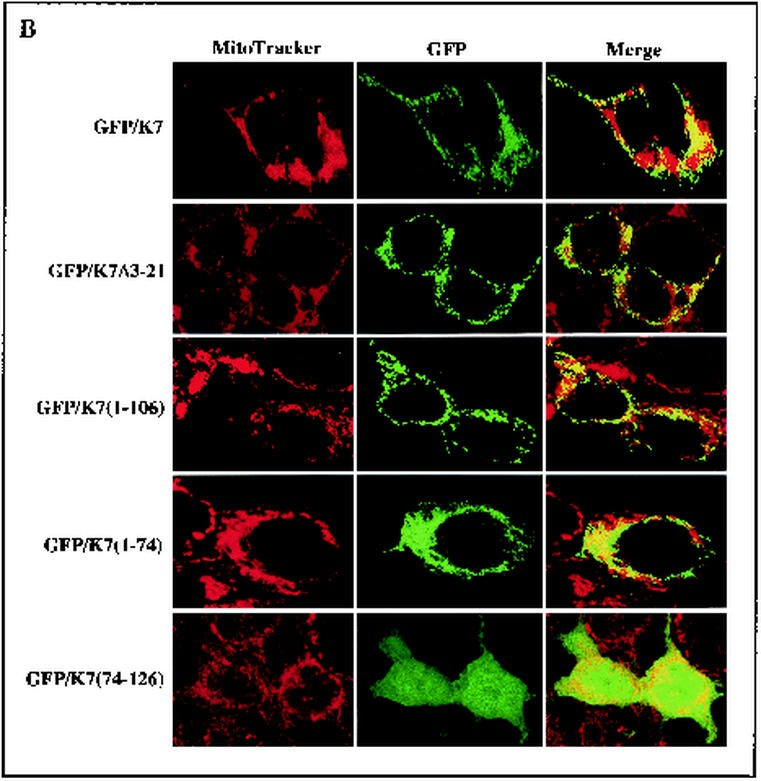
To determine the specific region required for the mitochondrial localization of K7, deletion mutants of K7 were constructed. K7(Δ3-21) contains the deletion of the N-terminal hydrophilic region, K7(1-106) contains the deletion of the C-terminal 20 amino acids, K7(1-74) contains the deletion of the C-terminal positively charged sequence, K7(75-126) contains the deletion of the N-terminal hydrophilic region and internal hydrophobic region, K7(Δ23-73) contains the deletion of the internal leucine-rich hydrophobic region, and K7(23-73) contains only the internal leucine-rich hydrophobic region. Wild-type and K7 mutant proteins were fused into the GFP vector to generate GFP/K7, GFP/K7(Δ3-21), GFP/K7(1-106), GFP/K7(1-74), GFP/K7(75-126), GFP/K7(Δ23-73), and GFP/K7(23-73) constructs, respectively. To examine the subcellular localization of K7 mutants, COS-1 epithelial cells and ECV endothelial cells were transfected with the vector containing GFP/K7 or GFP/K7 mutants. At 48 h posttransfection, cells were stained with Mitotracker Red to visualize the mitochondrial compartment. Confocal microscopy analysis revealed that as seen with GFP/K7, GFP/K7(Δ3-21), GFP/K7(1-106), GFP/K7(1-74), GFP/K7(1-106), and GFP/K7(23-73) were detected as a punctate, cytoplasmic distribution (Fig. 2). Multiple merged images of GFP/K7, GFP/K7(Δ3-21), GFP/K7(1-74), and GFP/K7(23-73) with MitoTracker Red staining indicated mitochondrial localization of these proteins (Fig. 2). In striking contrast, GFP/K7(75-126) and GFP/K7(Δ23-73) were distributed diffusely throughout the cells including the nucleus (Fig. 2). These results demonstrate that the internal leucine-rich hydrophobic region of K7 is necessary and likely sufficient for mitochondrial localization.
FIG.2.
The N-terminal hydrophobic sequence of K7 is sufficient for mitochondrial localization. COS-1 epithelial cells (A) and ECV endothelial cells (B) were transfected with GFP-K7 expression vector. After Mitotracker staining (red), transfected cells were fixed with paraformaldehyde. Immunofluorescence was examined using a Leica confocal immunofluorescence microscope, and a single representative optical section of each GFP-K7 mutant is presented.
K7 prevents the loss of mitochondrial membrane potential (ΨTm) on apoptosis.
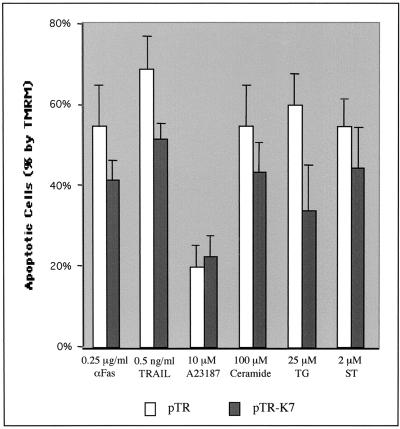
Events such as the loss of the mitochondrial membrane potential, the release of cytochrome c, and, in some cases, the production of reactive oxygen species have been linked with the onset of apoptosis induced by a variety of stimuli including FasL, TRAIL, TNF-α, staurosporin, ceramide, A23187, and TG (3, 25, 73). The loss of the mitochondrial potential has been identified as a common early event in apoptosis. Because of the mitochondrial localization of K7, we investigated whether K7 expression could affect the loss of the mitochondrial ΨTm induced by apoptotic signals. To test this, we have generated a pTracer-GFP/K7 vector where K7 and GFP are expressed from elongation factor 1 and cytomegalovirus early promoters, respectively. After electroporation with pTracer-GFP and pTracer-GFP/K7 vectors, BJAB cells were treated with apoptosis-inducing agents for various times and monitored by TMRM staining for 15 min. TMRM was used to detect mitochondrial membrane potential changes because it displays decreased fluorescence intensity as the mitochondrial membrane potential diminishes but emits a signal in the orange spectral range and therefore does not interfere with GFP fluorescence. Cells were analyzed for TMRM intensity after gating of GFP-positive cells by flow cytometry. K7 expression broadly prevented the loss of mitochondrial ΨTm on treatment with proapoptotic agents, in particular TG, which increases the cytosolic Ca2+ concentration by inhibiting the sarcoplasmic/endoplasmic Ca2+-ATPase (SERCA) (Fig. 3). In contrast, K7 expression showed no effect on apoptosis induced by the Ca2+ ionophore A23187, which increases cytosolic Ca2+ concentrations through the plasma membrane Ca2+ channel (Fig. 3). These results indicate that K7 expression in BJAB cells protects mitochondria from undergoing a permeabilization induced by proapoptotic agents, specifically TG.
FIG. 3.
Inhibition of apoptosis by K7. At 48 h after electroporation with pTracer-GFP (pTR) or pTracer-GFP/K7 (pTR-K7), BJAB cells were exposed to various apoptogenic agents. The treatment with different apoptogenic reagents is as follows: anti-Fas and TRAIL, 16 h; staurosporin (ST), 2 h; C2 ceramide, A23187, and TG, 4 h. After incubation with these agents, the cells were stained with the mitochondrial membrane-specific dye TMRM for 15 min to assess the mitochondrial membrane potential (ΨTM). After being washed with PBS twice, the cells were analyzed for TMRM intensity by gating of GFP-positive cells. The y axis indicates the reduction of the percentage of TMRM-positive cells after stimulation, indicated as apoptotic cells (%). Data represent an average of triplicate measurements, and error bars indicate standard deviations.
Identification of CAML that interacts with K7.
To delineate the molecular mechanism of K7-mediated anti-apoptosis, we used yeast two-hybrid screening to identify the cellular targets. A DNA fragment containing full-length K7 was fused in frame to the Gal4 DNA-binding domain to use as bait in the yeast two-hybrid screen with a cDNA library of activated human peripheral blood mononuclear cells. Transformants were plated on selective Leu-, Trp-, His-, and Ade-deficient plates. Leu, His, Trp, and Ade prototrophs were transferred to plates containing X-α-Gal, and colonies yielding a dark blue color were recovered and further analyzed.
Of the 107 transformants that were tested for growth in the absence of leucine, histidine, adenosine, and tryptophan and for color development in the presence of X-α-Gal, 20 colonies from K7 screening showed moderate growth and were positive for α-galactosidase expression. DNA sequence analysis showed that 6 of 20 K7-interacting clones contained a complete coding sequence of cellular CAML with different sizes of 5′ untranslated sequence. Transformation of the CAML cDNA clone with pGBK-T7 bait vector or pGBK-T7-lamin C neither supported growth on selective leucine-, tryptophan-, adenosine-, and histidine-deficient plates nor yielded color development on X-α-Gal-containing plates, suggesting that CAML is an authentic K7-binding protein (data not shown).
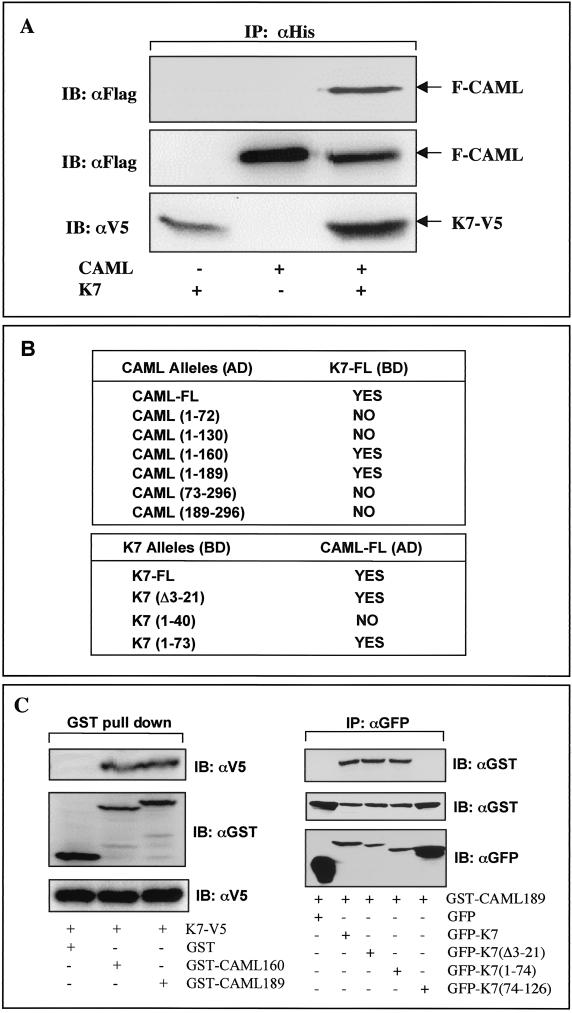
CAML contains 296 amino acids with three predicted transmembrane sequences in the carboxyl-terminal region and with the amino-terminal region projecting into the cytoplasm (11). CAML expression elevates the cytosolic Ca2+ concentration, leading to the activation of cellular NF-AT transcription activity (72). Since K7 efficiently inhibited apoptosis induced by the Ca2+-ATPase inhibitor TG, we hypothesized that K7 interacted with CAML in vivo and that this interaction might alter the intracellular Ca2+ concentration, thereby inhibiting apoptosis. To demonstrate complex formation between CAML and K7, 293T cells were transfected with expression vectors containing CAML or K7. To detect these proteins, the amino terminus of CAML and the carboxyl terminus of K7 were tagged with Flag and V5/His epitopes, respectively. Lysates of transfected cells were used for coimmunoprecipitation with anti-His antibody, followed by immunoblot with anti-Flag antibody. This showed that K7 specifically interacted with CAML in cells expressing both proteins whereas this interaction was not detected in cells expressing CAML or K7 individually (Fig. 4A).
FIG. 4.
Interaction between K7 and CAML. (A) Interaction between K7 and CAML in 293T cells. 293T cells were cotransfected with an expression vector containing V5/His-tagged K7 and/or Flag-tagged CAML. (Top) At 48 h posttransfection, whole-cell lysates were used for immunoprecipitation (IP) with an anti-His antibody, followed by immunoblotting (IB) with an anti-Flag antibody. (Middle and bottom) Whole-cell lysates were also used for immunoblotting with anti-Flag and anti-V5 antibodies to examine the expression of K7 and CAML. Transfection with K7 and CAML expression vector is indicated at the bottom of figure. Arrows indicate V5/His-tagged K7 and Flag-tagged CAML protein. (B) Mapping of regions required for the K7-CAML interaction in the yeast two-hybrid screening. Full-length CAML and its deletion mutants were fused to the Gal4 activation domain (AD) vector, and full-length K7 and its deletion mutants were fused to the Gal4 DNA binding domain (BD) vector. The BD vector containing full-length K7 together with AD vector carrying CAML or its mutants were cotransformed into the AH109 yeast strain. Conversely, the AD vector containing full-length CAML together with the BD vector carrying K7 or its mutants were cotransformed into the AH109 yeast strain. After transformation, AH109 yeast cells were examined for growth on selective Leu-, Trp-, His-, and Ade-deficient plates and for color development on X-α-Gal-containing plate. “Yes” indicates positive results from both assays, and “No” indicates negative results from both assays. None of the transformants showed single positivity in either assay. (C). Mapping of interacting regions of K7 and CAML in 293T cells. (Left) 293T cells were transfected with the GST-CAML mammalian expression vector together with K7 expression vector as indicated at the bottom. At 48 h posttransfection, whole-cell lysates were subjected to precipitation with glutathione-Sepharose beads followed by immunoblotting with an anti-V5 antibody (top). Whole-cell lysates were used for immunoblotting to show the equivalent level of expression of K7 and GST-CAML (middle and bottom). (Right) 293T cells were transfected with the GST-CAML mammalian expression vector together with the GFP-K7 expression vector as indicated at the bottom. At 48 h posttransfection, whole-cell lysates were subjected to immunoprecipitation with an anti-GFP antibody, followed by immunoblotting with an anti-GST antibody (top). Whole-cell lysates were used for immunoblotting to show the equivalent level of expression of GFP-K7 and GFP-K7 mutants (middle and bottom).
To further delineate an interaction of K7 with cellular CAML, we used the yeast two-hybrid system to map the regions required for this interaction. Several deletion mutant forms of K7 and CAML were generated and cloned into the Gal4 DNA binding domain (DB) vector and Gal4 activation domain (AD) vector, respectively. After transformation, the AH109 yeast strain containing both plasmids was examined for growth on selective Leu-, Trp-, His-, and Ade-deficient plates and for color development on X-α-Gal-containing plate. The yeast two-hybrid screening demonstrate that the N-terminal 1 to 160 amino acids of CAML and the N-terminal 22 to 74 amino acids of K7 are responsible for an efficient interaction between K7 and CAML (Fig. 4B). Because of its self-activation, K7(75-126) could not be tested in the yeast two-hybrid assay. In vivo binding assays further confirmed that the N-terminal 1 to 160 amino acids of CAML and the N-terminal 22 to 74 amino acids of K7 were required for their interaction (Fig. 4C). In addition, this result supports a specific interaction between K7 and CAML.
K7 interaction with CAML is necessary for preventing the loss of mitochondrial potential on apoptotic stimulation.
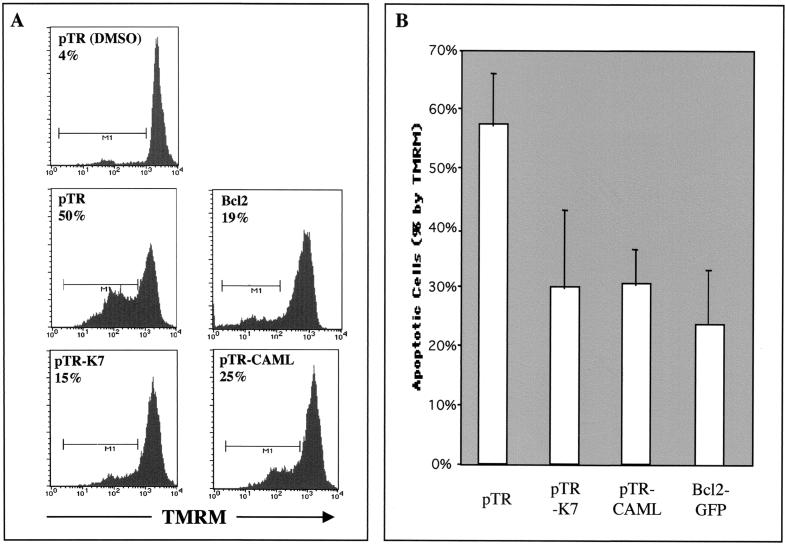
CAML expression increases the cytosolic Ca2+ concentration on TG treatment (72). To investigate the role of the K7-CAML interaction, we first compared the level of TG-induced apoptosis in the presence of K7 and CAML expression. At 24 h after electroporation with pTracer-GFP, pTracer-GFP/K7, or pTracer-GFP/CAML, BJAB cells were treated with 25 μM TG for 4 h and stained with TMRM for 15 min. The cells were analyzed for TMRM intensity after gating of GFP-positive cells by flow cytometry. As a positive control, expression vector containing Bcl-2-GFP was included. The experiment showed that K7 and CAML exhibited similar levels of antiapoptotic activity induced by TG treatment whereas Bcl-2 had an increased level of antiapoptotic activity (Fig. 5). However, no significant additive effect of K7 and CAML coexpression on antiapoptosis was observed, suggesting that endogenous CAML expression may be sufficient for K7 activity (data not shown).
FIG. 5.
Inhibition of apoptosis by K7 and CAML expression. At 24 h after electroporation with pTracer-GFP (pTR), pTracer-GFP/K7 (pTR-K7), pTracer-GFP/CAML (pTR-CAML), or pcDNA-GFP-Bcl-2 (Bcl-2), BJAB cells were treated with dimethyl sulfoxide (DMSO) (top) only or 25 μM TG (other panels) for 4 h. After TG treatment, these cells were stained with the mitochondrial membrane-specific dye TMRM for 15 min to assess the mitochondrial membrane potential (ΨTM). After being washed with PBS twice, the cells were analyzed for TMRM intensity by gating of GFP-positive cells. (A) Histogram of TMRM staining. Representative results of one of three separate experiments are shown. The x axis represents TMRM intensity. Control BJAB cells without TG treatment displayed intense staining with the dye, indicating normal mitochondrial function. The M1 gate indicates the cell population showing a reduction of TMRM fluorescent signal and is presented as the percentage of total GFP-positive cells. (B) Average percent difference in TMRM-positive cells following TG treatment. The reduction of the percentage of TMRM-positive cells after stimulation, indicated as apoptotic cells (%), is represented. Data represent an average of triplicate measurements, and error bars indicate standard deviation.
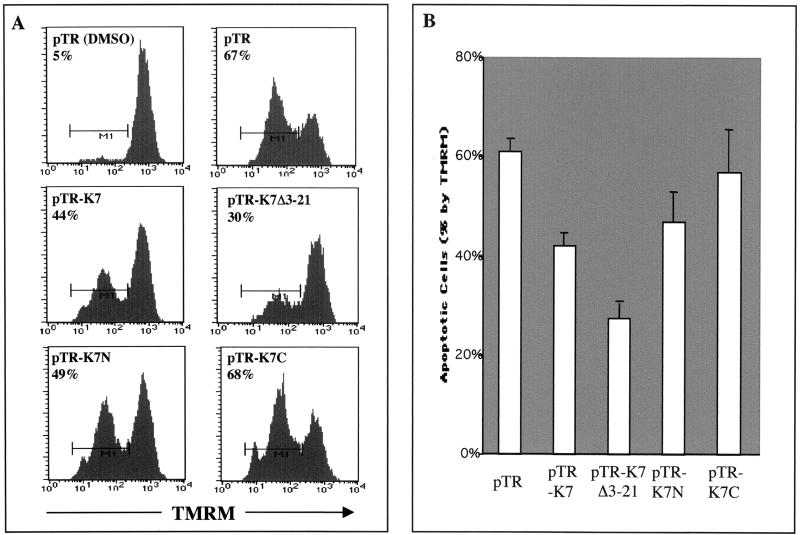
To further define a role for the K7-CAML interaction, we tested whether this interaction was necessary for K7 activity to prevent the loss of mitochondrial membrane potential on TG treatment. At 24 h after electroporation with pTracer-GFP, pTracer-GFP/wt K7, or pTracer-GFP/K7 mutants, BJAB cells were treated with 25 μM TG, stained with TMRM, and then analyzed for TMRM intensity by gating of GFP-positive cells with flow cytometry. As shown in Fig. 6, expression of K7(Δ3-21) and K7(1-74), which both were able to interact with CAML, suppressed the loss of mitochondrial membrane potential on TG treatment. Interestingly, repeated experiments showed that the K7(Δ3-21) mutant exhibited slightly higher level of antiapoptotic activity than did wild-type K7 (Fig. 6). In contrast, the K7(75-126) mutant, which no longer interacted with CAML, only poorly suppressed the TG-induced mitochondrial membrane potential loss under the same conditions (Fig. 6). These results suggest that K7 interaction with CAML is necessary for preventing the loss of the mitochondrial membrane potential on apoptotic stimulation.
FIG. 6.
K7-CAML interaction is necessary for efficient inhibition of apoptosis. BJAB cells were electroporated with pTracer-GFP (pTR), pTracer-GFP/K7 (pTR-K7), pTracer-GFP/K7 Δ3-21 (pTR-K7 Δ3-21), pTracer-GFP-K7 1-74 (pTR-K7N), or pTracer-GFP-K7 74-126 (pTR-K7C). At 48 h postelectroporation, BJAB cells were treated with dimethyl sulfoxide (DMSO) only or 25 μM TG for 4 h and stained with the mitochondrial membrane-specific dye TMRM for 15 min. These cells were analyzed for TMRM intensity by gating of GFP-positive cells. (A) Histogram of TMRM staining. Representative results of one of three separate experiments are shown. The x axis represents TMRM intensity. The M1 gate indicates the cell population showing a reduction of TMRM fluorescent signal and is presented as the percentage of total GFP-positive cells. (B) Average percent difference in TMRM-positive cells following TG treatment. The reduction of the percentage of TMRM-positive cells after stimulation, indicated as apoptotic cells (%), is represented. Data represent an average of triplicate measurements, and error bars indicate standard deviation.
K7 and CAML expression alters the kinetics and amplitudes of intracellular Ca2+ concentration on apoptotic stimulation.
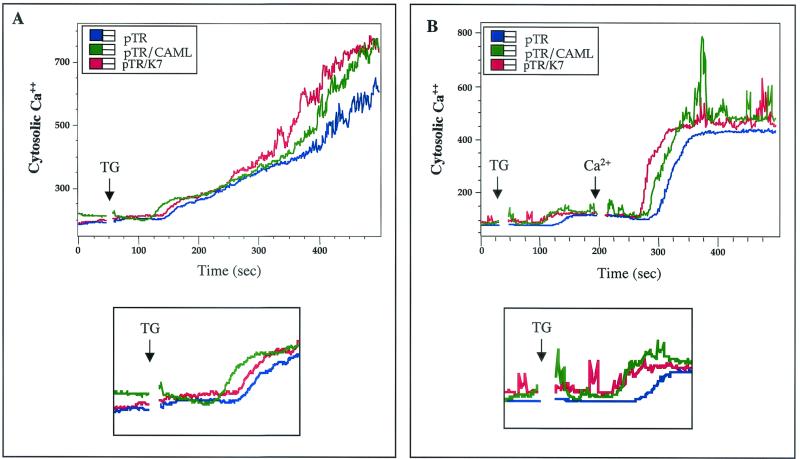
A high concentration of Ca2+ is maintained in the ER lumen by a member of the SERCA family that pumps Ca2+ into the ER, counterbalancing a leak of Ca2+ ions through the ER membrane into the cytoplasm. In this context, TG-induced SERCA inhibition allows Ca2+ to flow from the ER lumen to the cytoplasm, producing a transient elevation of the cytosolic Ca2+ concentration followed by additional elevation of the cytosolic Ca2+ concentration in response to capacitative Ca2+ entry (CCE) (48, 53). Since higher cytoplasmic Ca2+ concentrations could facilitate the refilling of deplete intracellular calcium stores, especially the ER, where calcium signals initiate, early upregulation of the cytoplasmic Ca2+ concentration protects cells against apoptosis (22). To investigate the mechanism of K7-mediated inhibition of apoptosis, we examined the effect of K7 on the cytosolic Ca2+ concentration after TG treatment. BJAB cells were electroporated with pTracer-GFP or pTracer-GFP/wt K7. In addition, pTracer-GFP/CAML was included as a control. At 24 h after electroporation, the cells were loaded with the Ca2+ indicator Indo-1 dye and treated with 12.5 nM TG. The cytosolic Ca2+ concentration in GFP-positive cells was monitored by flow cytometry. On TG treatment, BJAB cells expressing GFP induced rapid Ca2+ flow from the ER to the cytoplasm, producing a transient elevation of the cytosolic Ca2+ concentration followed by a sustained elevation of the cytosolic Ca2+ concentration as a consequence of CCE (Fig. 7A). As previously shown (34), CAML expression in BJAB cells enhanced the transient elevation of the cytosolic Ca2+ concentration on TG stimulation (Fig. 7A, bottom panel). Either K7 or CAML expression in BJAB cells induced faster kinetics and slightly higher amplitude of the transient elevation of the cytosolic Ca2+ concentration than did GFP expression alone (Fig. 7A, bottom panel). Furthermore, CCE-induced elevation of the cytosolic Ca2+ concentration was also significantly higher in BJAB cells expressing K7 or CAML than in those expressing GFP alone (Fig. 7A).
FIG. 7.
K7 and CAML expression alters the kinetics and amplitudes of intracellular Ca2+ concentration on apoptotic stimulation. (A) Increase in cytosolic calcium concentration in the presence of extracellular calcium. BJAB cells electroporated with pTracer-GFP (pTR), pTracer-GFP/K7 (pTR/K7), or pTracer-GFP/CAML (pTR/CAML) were gated for GFP-positive population and treated with 12.5 nM TG. (B) Increase in cytosolic calcium concentration in the absence of extracellular calcium and after addition of extracellular calcium. BJAB cells were extensively washed with Ca2+-free medium before TG treatment. After 200 ms of 12.5 nM TG treatment, 1 mM Ca2+ was added to the cells. The intracellular calcium concentration was monitored over time by noting changes in the ratio of violet to blue (405 to 485 nm) fluorescence of cells loaded with the calcium-sensitive dye indo-1 and analyzed by flow cytometry. Data are presented as a histogram of the number of cells with a particular fluorescence ratio (y axis) against time (x axis). Arrowheads indicate the addition of TG or Ca2+. The breaks in the graphs indicate the time intervals during the addition of TG or Ca2+. The bottom panel of each figure shows a higher magnification of the transient elevation of intracellular Ca2+ concentration immediately after TG treatment. Data were similar in three independent experiments.
To further demonstrate the effect of K7 on cytosolic Ca2+ homeostasis, we examined ER Ca2+ release in the absence of extracellular Ca2+. Transfected cells were extensively washed with Ca2+-free medium before being treated with TG. Under these conditions, the kinetics of Ca2+ release from the ER to the cytoplasm in BJAB cells expressing either K7 or CAML was significantly faster and stronger than in those expressing GFP only (Fig. 7B). While the addition of extracellular Ca2+ induced a CCE-mediated elevation of cytosolic Ca2+ flow in transfected cells, cells expressing K7 or CAML showed faster kinetics and higher amplitude of the CCE-mediated cytosolic Ca2+ flow than did cells expressing GFP alone (Fig. 7B). Thus, these results demonstrate that similar to cellular CAML expression, K7 expression alters the kinetics and amplitude of the cytosolic Ca2+ concentration in response to apoptotic stimuli.
DISCUSSION
Here, we report the identification of a mitochondrial protein of KSHV, K7. KSHV K7 efficiently inhibits apoptosis induced by a variety of apoptotic agents. The yeast two-hybrid screen has demonstrated that K7 targets CAML and that this interaction is necessary for K7 activity in the inhibition of apoptosis. These results indicate that K7 interacts with a cellular CAML and that this interaction activates CAML to increase the cytosolic Ca2+ concentration by enhancing ER Ca2+ release and activating the CCE pathway. In consequence, K7 expression protects cells from mitochondrial damage and apoptosis. Thus, this is a novel viral antiapoptosis strategy where KSHV mitochondrial K7 protein targets a cellular Ca2+-modulating protein to inhibit apoptosis, which allows completion of the viral lytic replication and, eventually, maintenance of persistent infection in the infected host.
A newly described consensus sequence for directing proteins to mitochondria consists of a hydrophobic region flanked by positively charged residues at both sides (23). This sequence has been identified in the vesicle-associated membrane protein 1B (VAMP-1B) (36), monoamine oxidases A and B, Bcl-2, Bcl-XL, Nip3, Nix, myxomavirus M11L (23), Epstein-Barr virus BHRF-1 (38), and KSHV vBcl-2 (14). A recent report (75) has predicted that the first 30 amino acids of K7 at the amino terminus contains a plant-form mitochondrial targeting sequence. However, our mutational analysis has found that the internal hydrophobic region of K7 protein with surrounding positive amino acid sequences is necessary and sufficient for mitochondrial localization. This indicates that the region of K7 from amino acids 23 to 74 probably has sufficient information for mitochondrial targeting and importing. In addition, K7 does not appear to be proteolytically processed in a similar manner to cellular mitochondrial proteins that undergo proteolytic processing during import into mitochondria. Detail mechanism of the internal hydrophobic region of K7 for mitochondrial targeting is under active investigation.
We have demonstrated that K7 expression prevents mitochondria from undergoing a permeability transition after initiation of an apoptotic signal by a variety of stresses, in particular, TG. In addition, an interaction of K7 with cellular CAML appears to be essential for this activity. CAML contains multiple transmembrane sequence at the carboxyl-terminal region and has the amino-terminal region projecting into the cytoplasm. CAML participates in the activation of transcription factors in lymphocytes. Specifically, it stimulates calcineurin, thus causing the subsequent activation of NF-AT (11, 35). The stimulation of calcineurin arises due to the elevated cytosolic Ca2+ levels. While the way in which CAML increases cytosolic Ca2+ levels is not entirely clear, it seems to do so by activating the CCE pathway, which is normally triggered after hormone-induced release of ER Ca2+ stores (34). Consistent with this, CAML expression induces increases in cytosolic Ca2+ levels that are acutely dependent on the extracellular Ca2+ concentration (11, 35). The yeast two-hybrid screening and in vivo coimmunoprecipitation demonstrate that K7 specifically interacts with CAML and that the internal hydrophobic region of K7 is necessary for CAML interaction. This suggests the potential overlapping role of the internal hydrophobic sequence of K7, as with mitochondrial targeting and CAML interaction. It would be of interest to determine whether these activities of the internal hydrophobic region of K7 are functionally or genetically separable. However, it is also conceivable that the two activities are interconnected in such a way that mitochondrial localization of K7 is necessary for binding to CAML or vice versa. In addition, CAML is present primarily in the ER whereas K7 is extensively localized in the mitochondria. While the different localization may not be simply reconcilable for their interaction, there are numerous evidences that mitochondrial compartment is located in close proximity to the ER. Recent findings have demonstrated numerous contacts between the ER and mitochondria (55). Furthermore, this close apposition between the ER and mitochondria may explain why, on opening of the Ca2+-gated channel of the ER, the mitochondrial surface is exposed to a higher concentration of Ca2+ than is the bulk cytosol (2, 17, 74). Therefore, it is possible that CAML and K7 interact at the contacting regions between the ER and mitochondria and that this interaction activates CAML to open the ER Ca2+ channel in response to apoptotic stimulation. On the other hand, since a portion of K7 is also present in the ER, the K7-CAML interaction may occur in the ER. Further study is needed to provide evidence for a detailed mechanism of the K7-CAML interaction. Nevertheless, these results suggest that KSHV has evolved a novel antiapoptotic strategy by harboring K7 that targets cellular Ca2+-modulating protein to protect cells from mitochondrial damage and apoptosis.
Wang et al. (75) have recently demonstrated that K7 interacts with Bcl-2 and active caspase 3 and that this interaction plays an important role in inhibition of apoptosis. While the results in their report are mostly in agreement with ours, there were slight differences; for example, the domain responsible for K7-mediated antiapoptotic activity was found to be the carboxyl-terminal region (amino acids 71 to 126). In contrast, we demonstrated that the amino-terminal region (amino acids 1 to 74) of K7 was sufficient to protect cells against TG-induced apoptosis. Since the conditions used by our group are very different from those reported by Wang et al. (75), our results are not necessarily in disagreement. In fact, Wang et al. (75) examined annexin -V exposure, which is the phenotype of the late stage of apoptosis, as a measurement for apoptosis, whereas we measured the mitochondrial membrane potential loss, which occurs an early step of apoptosis. In addition, while Wang et al. convincingly demonstrated an interaction of K7 with Bcl-2 and active caspase 3, we did not find Bcl-2 and caspase genes from our yeast two-hybrid screening with K7 as bait. This difference is probably due to the different selectivity of each assay in identifying protein-protein interactions. Since K7 probably targets multiple cellular proteins to inhibit apoptosis signal transduction, it is also conceivable that the separate regions of K7 may function at different stages of the apoptosis signal transduction pathway. Both studies suggest that KSHV K7 targets multiple cellular proteins to ensure comprehensive protection from apoptosis, which is the first line of host defense mechanism on viral infection.
The complexity and versatility of calcium fluxes within mammalian cells are achieved by virtue of the temporal and spatial distribution as well as the amplitude of intracellular calcium concentrations. Notably, early upregulation of cytoplasmic calcium concentration ([Ca2+]i) protects cells against apoptosis (22). However, sustained high [Ca2+]i contributes to programmed cell death. On the other hand, a higher [Ca2+]i could facilitate the refilling of depleted intracellular calcium stores, especially the ER, where calcium signals initiate (22). For example, the cellular antiapoptotic Bcl-2 protein, which forms an ion channel in isolated lipid bilayers, alters the state of filling of intracellular Ca2+ stores as well as the kinetics and amplitudes of cellular Ca2+ responses (26, 33). Similar to Bcl- 2, KSHV K7 alters the kinetics and amplitudes of cellular Ca2+ responses on apoptogenic stress, which regulates the organelle Ca2+ concentration to prevent mitochondrial damage. Further study should be directed toward examining the effect of K7 on the Ca2+ concentration in the ER and mitochondria on stimulation, using specific dyes and Ca2+-binding proteins. This would allow us to understand the K7-mediated regulatory mechanism of intracellular calcium concentration.
The universality of calcium as an intracellular messenger depends on its enormous versatility (6, 58). Cellular calcium signal pathways are often in communication with many components that can be mixed and matched to create a wide range of spatial and temporal signals. This versatility is exploited to control processes as diverse as fertilization, proliferation, cell cycle, transcription, signal transduction, and apoptosis (6, 7, 16). It is therefore, not surprising that a number of viruses encode regulatory proteins that alter cellular Ca2+ homeostasis to assist various aspects of viral replication (28). For example, the hepatitis B virus X (HBx) protein has been shown to activate Pyk2 or Ca2+ signaling mediated by mitochondrial Ca2+ channels, which is necessary for HBV replication (10). Thus, the KSHV lytic K7 protein may not only protect cells from mitochondrial damage and apoptosis but also provide a favorable cellular environment for viral replication, which allows for completion of the viral life cycle and maximizes the production of viral progeny for maintenance of persistent infection. A detailed study of the molecular mechanisms of antiapoptosis and calcium modulation by KSHV K7 will not only lead to a better understanding of viral persistence and disease progression but also provide a novel means of investigating cellular apoptosis regulatory systems.
Acknowledgments
We thank B. R. Oh for providing TRAIL protein, B. R. Means for critical reading, and M. Connole for flow cytomtery analysis.
This work was partly supported by U.S. Public Health Service grants CA82057, CA91819, and RR00168 and ACS grant RPG001102. J. Jung is a Leukemia & Lymphoma Society Scholar.
REFERENCES
- 1.Alexander, L., L. Denekamp, A. Knapp, M. R. Auerbach, B. Damania, and R. C. Desrosiers. 2000. The primary sequence of rhesus monkey rhadinovirus isolate 26-95: sequence similarities to Kaposi's sarcoma-associated herpesvirus and rhesus monkey rhadinovirus isolate 17577. J. Virol. 74:3388-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnaudeau, S., W. L. Kelley, J. V. Walsh, Jr., and N. Demaurex. 2001. Mitochondria recycle Ca2+ to the endoplasmic reticulum and prevent the depletion of neighboring endoplasmic reticulum regions. J. Biol. Chem. 276:29430-29439. [DOI] [PubMed] [Google Scholar]
- 3.Ashkenazi, A., and V. M. Dixit. 1998. Death receptors: signaling and modulation. Science 281:1305-1308. [DOI] [PubMed] [Google Scholar]
- 4.Bellows, D. S., B. N. Chau, P. Lee, Y. Lazebnik, W. H. Burns, and J. M. Hardwick. 2000. Antiapoptotic herpesvirus Bcl-2 homologs escape caspase-mediated conversion to proapoptotic proteins. J. Virol. 74:5024-5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bellows, D. S., M. Howell, C. Pearson, S. A. Hazlewood, and J. M. Hardwick. 2002. Epstein-Barr virus BALF1 is a BCL-2-like antagonist of the herpesvirus antiapoptotic BCL-2 proteins. J. Virol. 76:2469-2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berridge, M. J., M. D. Bootman, and P. Lipp. 1998. Calcium—a life and death signal. Nature 395:645-648. [DOI] [PubMed] [Google Scholar]
- 7.Berridge, M. J., P. Lipp, and M. D. Bootman. 2000. The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell. Biol. 1:11-21. [DOI] [PubMed] [Google Scholar]
- 8.Boshoff, C., Y. Endo, P. D. Collins, Y. Takeuchi, J. D. Reeves, V. L. Schweickart, M. A. Siani, T. Sasaki, T. J. Williams, P. W. Gray, P. S. Moore, Y. Chang, and R. A. Weiss. 1997. Angiogenic and HIV-inhibitory functions of KSHV-encoded chemokines. Science 278:290-294. [DOI] [PubMed] [Google Scholar]
- 9.Boshoff, C., T. F. Schulz, M. M. Kennedy, A. K. Graham, C. Fisher, A. Thomas, J. O. McGee, R. A. Weiss, and J. J. O'Leary. 1995. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat. Med. 1:1274-1278. [DOI] [PubMed] [Google Scholar]
- 10.Bouchard, M. J., L. H. Wang, and R. J. Schneider. 2001. Calcium signaling by HBx protein in hepatitis B virus DNA replication. Science 294:2376-2378. [DOI] [PubMed] [Google Scholar]
- 11.Bram, R. J., and G. R. Crabtree. 1994. Calcium signalling in T cells stimulated by a cyclophilin B-binding protein. Nature 371:355-358. [DOI] [PubMed] [Google Scholar]
- 12.Cesarman, E., Y. Chang, P. S. Moore, J. W. Said, and D. M. Knowles. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N. Engl. J. Med. 332:1186-1191. [DOI] [PubMed] [Google Scholar]
- 13.Chang, Y., E. Cesarman, M. S. Pessin, F. Lee, J. Culpepper, D. M. Knowles, and P. S. Moore. 1994. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science 266:1865-1869. [DOI] [PubMed] [Google Scholar]
- 14.Cheng, E. H., J. Nicholas, D. S. Bellows, G. S. Hayward, H. G. Guo, M. S. Reitz, and J. M. Hardwick. 1997. A Bcl-2 homolog encoded by Kaposi sarcoma-associated virus, human herpesvirus 8, inhibits apoptosis but does not heterodimerize with Bax or Bak. Proc. Natl. Acad. Sci. USA 94:690-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clem, R. J., and L. K. Miller. 1994. Control of programmed cell death by the baculovirus genes p35 and iap. Mol. Cell. Biol. 14:5212-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crabtree, G. R., and E. N. Olson. 2002. NFAT signaling: choreographing the social lives of cells. Cell 109(Suppl.):S67-S79. [DOI] [PubMed] [Google Scholar]
- 17.Csordas, G., and G. Hajnoczky. 2001. Sorting of calcium signals at the junctions of endoplasmic reticulum and mitochondria. Cell Calcium 29:249-262. [DOI] [PubMed] [Google Scholar]
- 18.Damania, B., J. K. Choi, and J. U. Jung. 2000. Signaling activities of gammaherpesvirus membrane proteins. J. Virol. 74:1593-1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Distelhorst, C. W., and G. Dubyak. 1998. Role of calcium in glucocorticosteroid-induced apoptosis of thymocytes and lymphoma cells: resurrection of old theories by new findings. Blood 91:731-734. [PubMed] [Google Scholar]
- 20.Du, C., M. Fang, Y. Li, L. Li, and X. Wang. 2000. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 102:33-42. [DOI] [PubMed] [Google Scholar]
- 21.Duby, G., M. Oufattole, and M. Boutry. 2001. Hydrophobic residues within the predicted N-terminal amphiphilic alpha-helix of a plant mitochondrial targeting presequence play a major role in in vivo import. Plant J. 27:539-549. [DOI] [PubMed] [Google Scholar]
- 22.Ermak, G., and K. J. Davies. 2002. Calcium and oxidative stress: from cell signaling to cell death. Mol. Immunol. 38:713-721. [DOI] [PubMed] [Google Scholar]
- 23.Everett, H., M. Barry, S. F. Lee, X. Sun, K. Graham, J. Stone, R. C. Bleackley, and G. McFadden. 2000. M11L: a novel mitochondria-localized protein of myxoma virus that blocks apoptosis of infected leukocytes. J. Exp. Med. 191:1487-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferri, K. F., E. Jacotot, J. Blanco, J. A. Este, and G. Kroemer. 2000. Mitochondrial control of cell death induced by HIV-1-encoded proteins. Ann. N. Y. Acad. Sci. 926:149-164. [DOI] [PubMed] [Google Scholar]
- 25.Ferri, K. F., and G. Kroemer. 2001. Organelle-specific initiation of cell death pathways. Nat. Cell Biol. 3:E255-E263. [DOI] [PubMed] [Google Scholar]
- 26.Foyouzi-Youssefi, R., S. Arnaudeau, C. Borner, W. L. Kelley, J. Tschopp, D. P. Lew, N. Demaurex, and K. H. Krause. 2000. Bcl-2 decreases the free Ca2+ concentration within the endoplasmic reticulum. Proc. Natl. Acad. Sci. USA 97:5723-5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ganem, D. 1997. KSHV and Kaposi's sarcoma: the end of the beginning? Cell 91:157-160. [DOI] [PubMed] [Google Scholar]
- 28.Ganem, D. 2001. Virology. The X files-one step closer to closure. Science 294:2299-2300. [DOI] [PubMed] [Google Scholar]
- 29.Glykofrydes, D., H. Niphuis, E. M. Kuhn, B. Rosenwirth, J. L. Heeney, J. Bruder, G. Niedobitek, I. Muller-Fleckenstein, B. Fleckenstein, and A. Ensser. 2000. Herpesvirus saimiri vFLIP provides an antiapoptotic function but is not essential for viral replication, transformation, or pathogenicity. J. Virol. 74:11919-11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goyal, L. 2001. Cell death inhibition: keeping caspases in check. Cell 104:805-808. [DOI] [PubMed] [Google Scholar]
- 31.Green, D. R. 2000. Apoptotic pathways: paper wraps stone blunts scissors. Cell 102:1-4. [DOI] [PubMed] [Google Scholar]
- 32.Hardwick, J. M. 1998. Viral interference with apoptosis. Semin. Cell Dev. Biol. 9:339-349. [DOI] [PubMed] [Google Scholar]
- 33.He, H., M. Lam, T. S. McCormick, and C. W. Distelhorst. 1997. Maintenance of calcium homeostasis in the endoplasmic reticulum by Bcl-2. J. Cell Biol. 138:1219-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Holloway, M. P., and R. J. Bram. 1996. A hydrophobic domain of Ca2+-modulating cyclophilin ligand modulates calcium influx signaling in T lymphocytes. J. Biol. Chem. 271:8549-8552. [DOI] [PubMed] [Google Scholar]
- 35.Hunton, D. L., P. A. Lucchesi, Y. Pang, X. Cheng, L. J. Dell'Italia, and R. B. Marchase. 2002. Capacitative calcium entry contributes to nuclear factor of activated T-cells nuclear translocation and hypertrophy in cardiomyocytes. J. Biol. Chem. 277:14266-14273. [DOI] [PubMed] [Google Scholar]
- 36.Isenmann, S., Y. Khew-Goodall, J. Gamble, M. Vadas, and B. W. Wattenberg. 1998. A splice-isoform of vesicle-associated membrane protein-1 (VAMP-1) contains a mitochondrial targeting signal. Mol. Biol. Cell 9:1649-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joza, N., S. A. Susin, E. Daugas, W. L. Stanford, S. K. Cho, C. Y. Li, T. Sasaki, A. J. Elia, H. Y. Cheng, L. Ravagnan, K. F. Ferri, N. Zamzami, A. Wakeham, R. Hakem, H. Yoshida, Y. Y. Kong, T. W. Mak, J. C. Zuniga-Pflucker, G. Kroemer, and J. M. Penninger. 2001. Essential role of the mitochondrial apoptosis-inducing factor in programmed cell death. Nature 410:549-554. [DOI] [PubMed] [Google Scholar]
- 38.Khanim, F., C. Dawson, C. A. Meseda, J. Dawson, M. Mackett, and L. S. Young. 1997. BHRF1, a viral homologue of the Bcl-2 oncogene, is conserved at both the sequence and functional level in different Epstein-Barr virus isolates. J. Gen. Virol. 78:2987-2999. [DOI] [PubMed] [Google Scholar]
- 39.Korsmeyer, S. J., M. C. Wei, M. Saito, S. Weiler, K. J. Oh, and P. H. Schlesinger. 2000. Pro-apoptotic cascade activates BID, which oligomerizes BAK or BAX into pores that result in the release of cytochrome c. Cell Death Differ. 7:1166-1173. [DOI] [PubMed] [Google Scholar]
- 40.Lagunoff, M., R. Majeti, A. Weiss, and D. Ganem. 1999. Deregulated signal transduction by the K1 gene product of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA 96:5704-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee, B. S., X. Alvarez, S. Ishido, A. A. Lackner, and J. U. Jung. 2000. Inhibition of intracellular transport of B cell antigen receptor complexes by Kaposi's sarcoma-associated herpesvirus K1. J. Exp. Med. 192:11-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee, H., R. Veazey, K. Williams, M. Li, J. Guo, F. Neipel, B. Fleckenstein, A. Lackner, R. C. Desrosiers, and J. U. Jung. 1998. Deregulation of cell growth by the K1 gene of Kaposi's sarcoma-associated herpesvirus. Nat. Med. 4:435-440. [DOI] [PubMed] [Google Scholar]
- 43.Li, P., D. Nijhawan, I. Budihardjo, S. M. Srinivasula, M. Ahmad, E. S. Alnemri, and X. Wang. 1997. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 91:479-489. [DOI] [PubMed] [Google Scholar]
- 44.Li, Y., J. Kang, and M. S. Horwitz. 1997. Interaction of an adenovirus 14.7- kilodalton protein inhibitor of tumor necrosis factor alpha cytolysis with a new member of the GTPase superfamily of signal transducers. J. Virol. 71:1576-1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mesri, E. A., E. Cesarman, L. Arvanitakis, S. Rafii, M. A. Moore, D. N. Posnett, D. M. Knowles, and A. S. Asch. 1996. Human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus is a new transmissible virus that infects B cells. J. Exp. Med. 183:2385-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moore, P. S., C. Boshoff, R. A. Weiss, and Y. Chang. 1996. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science 274:1739-1744. [DOI] [PubMed] [Google Scholar]
- 47.Moore, P. S., and Y. Chang. 1995. Kaposi's sarcoma findings. Science 270:15. [DOI] [PubMed] [Google Scholar]
- 48.Mori, Y., M. Wakamori, T. Miyakawa, M. Hermosura, Y. Hara, M. Nishida, K. Hirose, A. Mizushima, M. Kurosaki, E. Mori, K. Gotoh, T. Okada, A. Fleig, R. Penner, M. Iino, and T. Kurosaki. 2002. Transient receptor potential 1 regulates capacitative Ca2+ entry and Ca2+ release from endoplasmic reticulum in B lymphocytes. J. Exp. Med. 195:673-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Neve, E. P., and M. Ingelman-Sundberg. 2001. Identification and characterization of a mitochondrial targeting signal in rat cytochrome P450 2E1 (CYP2E1). J. Biol. Chem. 276:11317-11322. [DOI] [PubMed] [Google Scholar]
- 50.Ojala, P. M., K. Yamamoto, E. Castanos-Velez, P. Biberfeld, S. J. Korsmeyer, and T. P. Makela. 2000. The apoptotic v-cyclin-CDK6 complex phosphorylates and inactivates Bcl-2. Nat. Cell Biol. 2:819-825. [DOI] [PubMed] [Google Scholar]
- 51.Pinton, P., D. Ferrari, E. Rapizzi, F. D. Di Virgilio, T. Pozzan, and R. Rizzuto. 2001. The Ca2+ concentration of the endoplasmic reticulum is a key determinant of ceramide-induced apoptosis: significance for the molecular mechanism of Bcl-2 action. EMBO J. 20:2690-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pozzan, T., P. Magalhaes, and R. Rizzuto. 2000. The comeback of mitochondria to calcium signalling. Cell Calcium 28:279-283. [DOI] [PubMed] [Google Scholar]
- 53.Putney, J. W., Jr., L. M. Broad, F. J. Braun, J. P. Lievremont, and G. S. Bird. 2001. Mechanisms of capacitative calcium entry. J. Cell Sci. 114:2223-2229. [DOI] [PubMed] [Google Scholar]
- 54.Rizzuto, R., P. Bernardi, and T. Pozzan. 2000. Mitochondria as all-round players of the calcium game. J. Physiol. 529:37-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rizzuto, R., P. Pinton, W. Carrington, F. S. Fay, K. E. Fogarty, L. M. Lifshitz, R. A. Tuft, and T. Pozzan. 1998. Close contacts with the endoplasmic reticulum as determinants of mitochondrial Ca2+ responses. Science 280:1763-1766. [DOI] [PubMed] [Google Scholar]
- 56.Roy, D. J., B. C. Ebrahimi, B. M. Dutia, A. A. Nash, and J. P. Stewart. 2000. Murine gammaherpesvirus M11 gene product inhibits apoptosis and is expressed during virus persistence. Arch. Virol. 145:2411-2420. [DOI] [PubMed] [Google Scholar]
- 57.Russo, J. J., R. A. Bohenzky, M. C. Chien, J. Chen, M. Yan, D. Maddalena, J. P. Parry, D. Peruzzi, I. S. Edelman, Y. Chang, and P. S. Moore. 1996. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 93:14862-14867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santella, L., and E. Carafoli. 1997. Calcium signaling in the cell nucleus. FASEB J. 11:1091-1109. [PubMed] [Google Scholar]
- 59.Sarid, R., T. Sato, R. A. Bohenzky, J. J. Russo, and Y. Chang. 1997. Kaposi's sarcoma-associated herpesvirus encodes a functional bcl-2 homologue. Nat. Med. 3:293-298. [DOI] [PubMed] [Google Scholar]
- 60.Scaffidi, C., S. Fulda, A. Srinivasan, C. Friesen, F. Li, K. J. Tomaselli, K. M. Debatin, P. H. Krammer, and M. E. Peter. 1998. Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 17:1675-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schiestl, R. H., and R. D. Gietz. 1989. High efficiency transformation of intact yeast cells using single stranded nucleic acids as a carrier. Curr. Genet. 16:339-346. [DOI] [PubMed] [Google Scholar]
- 62.Simpson, G. R., T. F. Schulz, D. Whitby, P. M. Cook, C. Boshoff, L. Rainbow, M. R. Howard, S. J. Gao, R. A. Bohenzky, P. Simmonds, C. Lee, A. de Ruiter, A. Hatzakis, R. S. Tedder, I. V. Weller, R. A. Weiss, and P. S. Moore. 1996. Prevalence of Kaposi's sarcoma-associated herpesvirus infection measured by antibodies to recombinant capsid protein and latent immunofluorescence antigen. Lancet 348:1133-1138. [DOI] [PubMed] [Google Scholar]
- 63.Soulier, J., L. Grollet, E. Oksenhendler, P. Cacoub, D. Cazals-Hatem, P. Babinet, M. F. d'Agay, J. P. Clauvel, M. Raphael, L. Degos, et al. 1995. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 86:1276-1280. [PubMed] [Google Scholar]
- 64.Stout, A. K., H. M. Raphael, B. I. Kanterewicz, E. Klann, and I. J. Reynolds. 1998. Glutamate-induced neuron death requires mitochondrial calcium uptake. Nat. Neurosci. 1:366-373. [DOI] [PubMed] [Google Scholar]
- 65.Sun, R., S. F. Lin, K. Staskus, L. Gradoville, E. Grogan, A. Haase, and G. Miller. 1999. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 73:2232-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Susin, S. A., E. Daugas, L. Ravagnan, K. Samejima, N. Zamzami, M. Loeffler, P. Costantini, K. F. Ferri, T. Irinopoulou, M. C. Prevost, G. Brothers, T. W. Mak, J. Penninger, W. C. Earnshaw, and G. Kroemer. 2000. Two distinct pathways leading to nuclear apoptosis. J. Exp. Med. 192:571-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Susin, S. A., H. K. Lorenzo, N. Zamzami, I. Marzo, B. E. Snow, G. M. Brothers, J. Mangion, E. Jacotot, P. Costantini, M. Loeffler, N. Larochette, D. R. Goodlett, R. Aebersold, D. P. Siderovski, J. M. Penninger, and G. Kroemer. 1999. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 397:441-446. [DOI] [PubMed] [Google Scholar]
- 68.Susin, S. A., N. Zamzami, and G. Kroemer. 1998. Mitochondria as regulators of apoptosis: doubt no more. Biochim. Biophys. Acta 1366:151-165. [DOI] [PubMed] [Google Scholar]
- 69.Tollefson, A. E., T. W. Hermiston, D. L. Lichtenstein, C. F. Colle, R. A. Tripp, T. Dimitrov, K. Toth, C. E. Wells, P. C. Doherty, and W. S. Wold. 1998. Forced degradation of Fas inhibits apoptosis in adenovirus-infected cells. Nature 392:726-730. [DOI] [PubMed] [Google Scholar]
- 70.Tortorella, D., B. E. Gewurz, M. H. Furman, D. J. Schust, and H. L. Ploegh. 2000. Viral subversion of the immune system. Annu. Rev. Immunol. 18:861-926. [DOI] [PubMed] [Google Scholar]
- 71.Virgin, H. W., IV, P. Latreille, P. Wamsley, K. Hallsworth, K. E. Weck, A. J. Dal Canto, and S. H. Speck. 1997. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol. 71:5894-5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.von Bulow, G. U., and R. J. Bram. 1997. NF-AT activation induced by a CAML-interacting member of the tumor necrosis factor receptor superfamily. Science 278:138-141. [DOI] [PubMed] [Google Scholar]
- 73.Wallach, D., E. E. Varfolomeev, N. L. Malinin, Y. V. Goltsev, A. V. Kovalenko, and M. P. Boldin. 1999. Tumor necrosis factor receptor and Fas signaling mechanisms. Annu. Rev. Immunol. 17:331-367. [DOI] [PubMed] [Google Scholar]
- 74.Wang, H. J., G. Guay, L. Pogan, R. Sauve, and I. R. Nabi. 2000. Calcium regulates the association between mitochondria and a smooth subdomain of the endoplasmic reticulum. J. Cell Biol. 150:1489-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang, H. W., T. V. Sharp, A. Koumi, G. Koentges, and C. Boshoff. 2002. Characterization of an anti-apoptotic glycoprotein encoded by Kaposi's sarcoma-associated herpesvirus which resembles a spliced variant of human survivin. EMBO J. 21:2602-2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou, Q., and G. S. Salvesen. 2000. Viral caspase inhibitors CrmA and p35. Methods Enzymol. 322:143-154. [DOI] [PubMed] [Google Scholar]