Abstract
The spread of most strains of vaccinia virus in cell monolayers occurs predominantly via extracellular enveloped virions that adhere to the tips of actin-containing microvilli and to a lesser extent via diffusion of released virions. The mechanism by which virions adhere to the cell surface is unknown, although several viral proteins may be involved. The present investigation was initiated with the following premise: spontaneous mutations that increase virus release will be naturally selected by propagating a virus unable to spread by means of actin tails. Starting with an A36R deletion mutant that forms small, round plaques, five independent virus clones with enhanced spread due to the formation of comet or satellite plaques were isolated. The viral membrane glycoprotein genes of the isolates were sequenced; four had mutations causing C-terminal truncations of the A33R protein, and one had a serine replacing proline 189 of the B5R protein. The comet-forming phenotype was specifically reproduced or reversed by homologous recombination using DNA containing the mutated or natural sequence, respectively. Considerably more extracellular enveloped virus was released into the medium by the second-site mutants than by the parental A36R deletion mutant, explaining their selection in tissue culture as well as their comet-forming phenotype. The data suggest that the B5R protein and the C-terminal region of the A33R protein are involved in adherence of cell-associated enveloped virions to cells. In spite of their selective advantage in cultured cells, the second-site mutants were not detectably more virulent than the A36R deletion mutant when administered to mice by the intranasal route.
Vaccinia virus, a member of the poxvirus family, replicates in the cytoplasm of infected cells (20). There are several infectious forms of vaccinia virus: (i) intracellular mature virions (IMV), which assemble in factory regions (6, 19); (ii) intracellular enveloped virions (IEV), which result from the wrapping of IMV with trans-Golgi or endosomal cisternae (11, 30, 34); (iii) cell-associated enveloped virions (CEV), which form after the outer IEV membrane fuses with the plasma membrane and remain attached to the cell surface and the tips of thick microvilli (3, 12, 33); and (iv) extracellular enveloped virions (EEV), presumably derived by the release of CEV into the medium (1, 23). Recent studies indicated that the IEV are transported to the cell periphery via microtubules and that actin tails form near the base of the CEV and propel them at the tips of microvilli (10, 14, 21, 25, 38, 39). The CEV and EEV mediate cell-to-cell and longer-range spread, respectively (3).
The EEV membrane contains at least four glycoproteins, which are products of the A33R (27), A34R (17), A56R (32), and B5R (7, 15) genes and one nonglycosylated protein encoded by the F13L gene (13). Two additional proteins, A36R (22, 37) and F12L (36), found on the IEV are absent from extracellular virions. Mutations of A56R cause syncytia (31), whereas deletion of any of the other IEV or EEV proteins results in a small-plaque phenotype. The small-plaque phenotypes of B5R and F13L deletion mutants result from a defect in wrapping of IMV (2, 8, 41), and those of A33R, A34R and A36R deletion mutants and an A36R point mutant result at least partly from a failure to form actin tails (26, 28, 29, 38, 42, 43).
The relative amounts of CEV and EEV produced during infection vary with different virus strains. The WR and IHD strains, although both derived from the New York City Board of Health strain, make small and large amounts of EEV, respectively, resulting in different plaque phenotypes (24). In liquid culture medium, the WR and New York City Board of Health strains form large, round plaques, whereas IHD forms numerous satellite plaques that take on a comet shape. The plaque phenotype of IHD can be largely explained by a single amino acid mutation in the A34R open reading frame (ORF), which regulates the dissociation of virions from the cell surface (4). Deletions of segments of the extracellular domain of the B5R protein and abnormal expression of the A33R protein can also produce comet-shaped plaques (16, 26).
The present study was initiated after we noted that the small plaques, which formed when an A36R deletion mutant virus stock was titered under a liquid overlay, were heterogeneous. Although the majority of plaques were round, a small number looked like comets. Because the A36R deletion mutant spreads very slowly, we suspected that the comet plaques originated from a virus with a spontaneous second-site mutation that had a selective growth advantage because of enhanced EEV release. We tested this hypothesis by serially passing virus from independent cultures of a plaque-purified A36R deletion mutant. Comet-forming viruses with either C-terminal truncations in the A33R ORF or a point mutation in the B5R ORF were selected after only a few passages.
MATERIALS AND METHODS
Cells and viruses.
BS-C-1 and RK13 cell monolayers were grown in minimum essential medium with Earl’s salts (EMEM; Quality Biologicals, Gaithersburg, Md.). The WR strain of vaccinia virus and the virus mutants lacking A33R or A36R genes (22, 26) are referred to as vΔA33R and vΔA36R, respectively.
Plaque assay.
Following adsorption of the virus to monolayers of BS-C-1 cells for 1 h at 37°C, EMEM containing 2% fetal bovine serum was added. Two days later the medium was removed and crystal violet (0.1% in 20% ethanol) was added for 30 min. The cells were then washed and dried. When a semisolid overlay was used, 0.6% SeaKem ME agarose (BioWhittaker Molecular Applications, Rockland, Maine) and 5% fetal bovine serum were included in the medium. After 2 days, the overlaid cells were fixed with formaldehyde (20% in saline), the semisolid overlay was removed, and the cells were stained with crystal violet.
One-step growth.
BS-C-1 cells were infected at a multiplicity of 5 PFU per cell for 1 h and were then washed twice with fresh medium to remove unadsorbed virus. At time intervals thereafter, the medium was collected and clarified by low-speed centrifugation to sediment detached cells and debris. The adherent cells were scraped into 1 ml of fresh medium and combined with the cell pellet from the clarified medium. The cells were frozen and thawed three times and sonicated for 30 s. Virus titers in cell lysates and media were determined by plaque assays with an agar overlay.
Transfection.
Approximately 2 μg of PCR product in 100 μl of Opti-MEM (Invitrogen, Carlsbad, Calif.) was mixed with 8 μl of Lipofectamine 2000 (Gibco BRL, Gaithersburg, Md.) in 100 μl of Opti-MEM and kept at room temperature for 15 min. Then 800 μl of Opti-MEM was added. BS-C-1 cells were infected at a multiplicity of 1 PFU per 20 cells. After 2 h, the cells were washed with Opti-MEM and the DNA plus Lipofectamine mixture was added to the cell monolayer. Three hours later, EMEM containing 2% fetal bovine serum was added. The cultures were harvested after 2 days at 37°C, and the virus was analyzed by plaque assay.
PCR and sequencing of viral DNA.
DNA from virus-infected cells was prepared for PCR by treatment with protease and purified using the QIA DNA blood minikit (Qiagen, Hilden, Germany), using the procedure suggested by the manufacturer. Primers 5′ to 3′ GGTCGTTAGTAGGGAGGAGAACAAAG and GCGCAAGCACTAGGCATCAGTTC were used to amplify the A33R ORF; primers 5′ to 3′ CCGATGTCTTGATTACCTGGCT and CTGTTATAGTCAACACACCCATTGGAG were used for amplification of the A34R ORF, and primers 5′ to 3′ ATTGATGTTTTTAACGCTACAATC and GTACATCTCATTGTCATTTACAAC were used to amplify the B5R ORF. The Pfu Turbo DNA polymerase (Stratagene, La Jolla, Calif.) was added; the extension reaction time for A33R and A34R ORFs was 45 s and for the B5R ORF was 80 s. The PCR products were purified using the Wizard PCR Preps DNA purification resin (Promega, Madison, Wis.). The BigDye terminator cycle sequencing v2 Ready reaction kit (Applied Biosystems, Foster City, Calif.) was applied, and the DNA was sequenced in the 3100 New Genetic Analyzer (Applied Biosystems).
Metabolic labeling and immunoprecipitation.
Confluent BS-C-1 cells (in 35-mm-diameter dishes) were infected at a multiplicity of 5 PFU per cell. At 5 h after infection, the medium (1 ml) was replaced with methionine- and cysteine-free Eagle minimum essential medium (Sigma, St. Louis, Mo.) supplemented with 2% dialyzed fetal bovine serum (Invitrogen) to which 70 μCi of 35S-protein labeling mix (Easy Tag; Perkin-Elmer Life Sciences, Boston, Mass.) had been added. The cells were harvested at 20 h after infection, washed with phosphate-buffered saline, and suspended in 200 μl of lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, and 1% NP-40) containing protease inhibitors (Roche Diagnostics, Mannheim, Germany). After 30 min at 4°C, the extract was centrifuged at 20,000 × g for 30 min and 200 μl of lysis buffer and 50 μl of protein A-agarose (Roche Diagnostics) were added to the supernatant. The mixture was rotated for 2 h at 4oC and centrifuged at 1,000 × g for 1 min. The supernatant was incubated for 20 h at 4°C with 2 μl of serum from a rabbit that had been immunized with a synthetic peptide, comprised of amino acids 84 to 100 of the A33R protein. Protein A-agarose was then added, and the incubation was continued for 2 h at 4°C. The complex was washed several times with phosphate-buffered saline and dissolved in 50 μl of loading buffer. Following electrophoresis in 4 to 20% polyacrylamide (Invitrogen), the gels were dried and exposed to Kodak BioMax MR film. The molecular masses of viral proteins were estimated by comparison with Full-Range-Rainbow protein markers (Amersham, Little Chalfont, Buckinghamshire, England).
CsCl gradient analysis of virus.
Monolayers of RK13 cells were infected at a multiplicity of 5 PFU per cell. Five hours later, the medium (10 ml) was replaced with methionine- and cysteine-free Eagle minimum essential medium (Sigma) supplemented with 2% dialyzed fetal bovine serum (Invitrogen) and 900 μCi of a 35S-protein labeling mix (Easy Tag; Perkin-Elmer Life Sciences). At 40 h after infection, the medium was harvested and cells and large debris were removed by low-speed centrifugation. Cells were scraped and collected by low-speed centrifugation, resuspended in swelling buffer (10 mM Tris, pH 9.0), and disrupted by Dounce homogenization. The cytoplasm was then collected, after removal of the nuclei by low-speed centrifugation. Virus from the cytoplasm or the culture medium was centrifuged through 36% sucrose cushion in an SW41 rotor at 32,000 revolutions/min for 12 min at 4°C. The virus was then resuspended in buffer and layered on the top of a CsCl step gradient, as previously described (8). Fractions were collected from the bottom of the centrifuge tube, and the amount of radioactivity was determined by scintillation counting.
Virulence of purified virus for mice.
BS-C-1 cells were harvested 2 days after infection by low-speed centrifugation. The cells were suspended in swelling buffer (10 mM Tris pH 9.0) and were Dounce homogenized. The cytoplasm was separated from nuclei by low-speed centrifugation and layered on 36% sucrose in the above buffer. After centrifugation at 13,500 revolutions/min in an SW28.1 rotor for 80 min at 4°C, the virus was suspended in a small volume of the buffer and its infectivity was determined by plaque assay. The virulence of vaccinia virus for mice was then determined essentially as described earlier (18, 35, 40). Groups (n = 4) of 5-week-old female BALB/c mice were anesthetized and inoculated intranasally with 5 × 106 PFU of purified virus in 20 μl of 10 mM Tris, pH 9.0. Mice were weighed daily, and those that lost more than 30% of their initial body weight were terminated.
RESULTS
Isolation of comet-forming virus derived from the A36R deletion mutant vΔA36R.
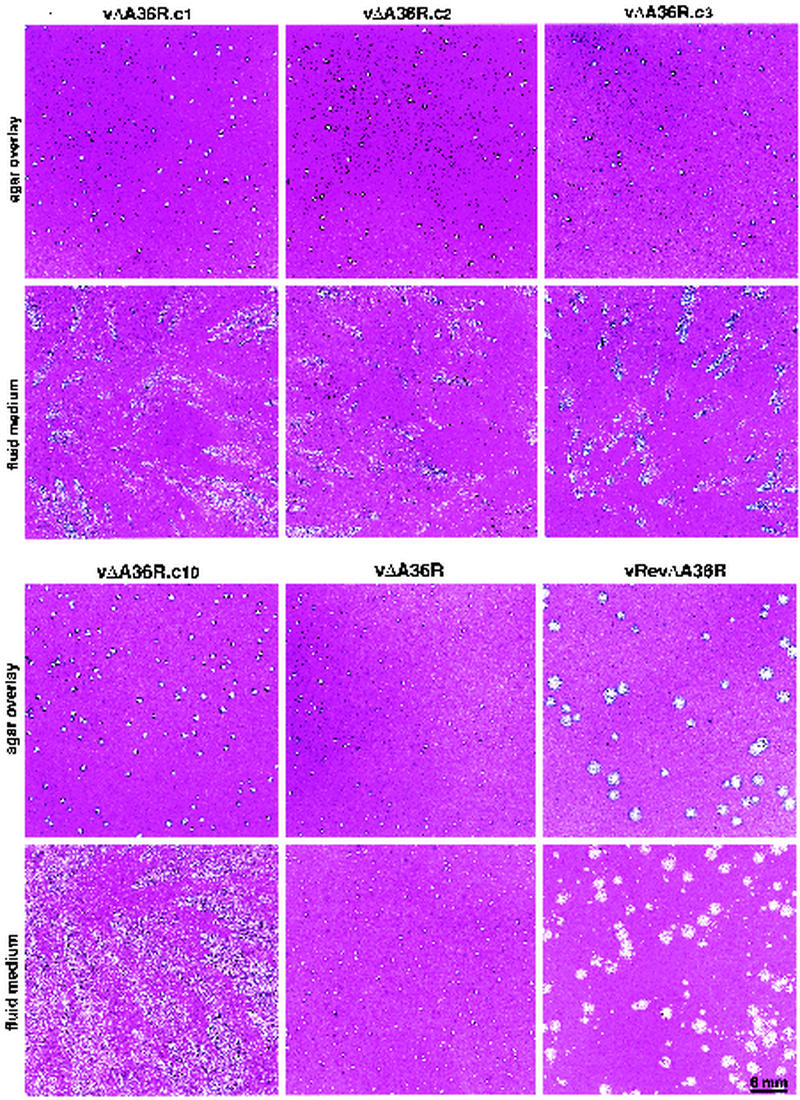
After propagating vΔA36R, obtained from Parkinson and Smith (22), we noted that a few of the plaques formed comets under a liquid overlay. The comet-forming virus could have resulted from a second-site mutation or a contamination in our laboratory. To discriminate between these possibilities, we plaque purified vΔA36R and then serially propagated it in separate BS-C-1 monolayers. After four passages, numerous comet-forming plaques were identified in most cultures. The plaques formed by comet-forming viruses were slightly larger under agar than those of the parental vΔA36R, facilitating their purification. We named the comet-forming mutants as follows: vΔA36R.c1, vΔA36R.c2, vΔA36R.c3, vΔA36R.c5, and vΔA36R.c10. The appearances of these plaques, except for vΔA36R.c5, which was subsequently shown to have the same mutation as vΔA36R.c2, are shown in Fig. 1. Under a liquid overlay, the comet-forming plaques were distinct from those of the parental vΔA36R or a wild-type revertant (vRevΔA36R) obtained by recombination with the A36R gene (22). Under agar, the isolated virus plaques were much smaller than the wild-type revertant virus. These data suggested that the comet-forming isolates resulted from second-site mutations.
FIG. 1.
Plaque morphology of virus clones. BS-C-1 monolayers were infected with vΔA36R.c1, vΔA36R.c2, vΔA36R.c3, vΔA36R.c10, vΔA36R, or vRevΔA36R. Following adsorption, fluid or agar-containing overlays were added. After 2 days of incubation at 37°C, the cultures were fixed and stained with crystal violet.
Occurrence of second-site mutations in the A33R and B5R ORFs.
We suspected that the comet-forming phenotypes were due to spontaneous mutations in one or more of the EEV glycoprotein genes. The A34R gene seemed a likely candidate, as a point mutation in this ORF was largely responsible for the comet-forming phenotype of the IHD strain of vaccinia virus. Surprisingly, the A34R sequences of vΔA36R.c1, vΔA36R.c2, vΔA36R.c3, vΔA36R.c5, and vΔA36R.c10 were identical to each other and to that of vΔA36R and the WR strain of vaccinia virus. Consequently, we sequenced the A33R ORFs of the comet-forming and parental viruses. Mutations truncating the A33R ORF, relative to that of vΔA36R and WR, were found in vΔA36R.c1, vΔA36R.c2, vA36R.c5, and vΔA36R.c10 (Fig. 2). The mutations in vΔA36R.c2 and vA36R.c5 were identical, and therefore the latter was not subjected to further analysis. In each case the mutations were between codons 116 and 143. A nucleotide substitution produced a stop codon in the A33R ORF of vΔA36R.c2 and vΔA36R.c10, whereas in vΔA36R.c1 there was a duplication of 4 nucleotides that altered the reading frame, resulting in a stop codon downstream. The other comet-forming mutant vΔA36R.c3, however, had no mutation in the A33R ORF.
FIG. 2.
Predicted amino acid sequences of the C-terminal regions of the A33R ORFs of vΔA36R, vΔA36R.c1, vΔA36R.c2, and vΔA36R.c10. The C-terminal sequence of vΔA36R is identical to that of the WR strain of vaccinia virus and also to that of vΔA36R.c3. Amino acid numbers are shown. The amino acids of vΔA36R.c1 formed by a frameshift mutation are underlined.
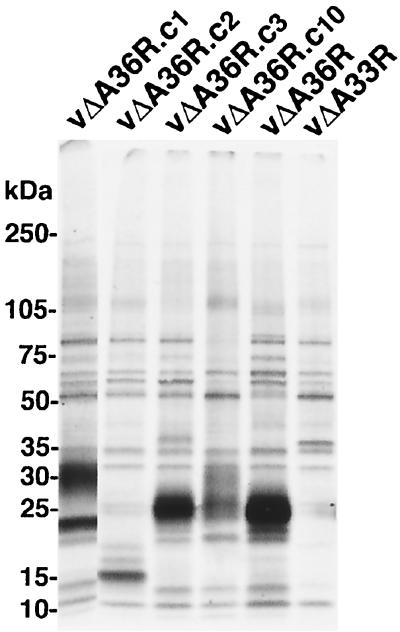
Expression of the A33R ORFs of the mutant viruses was investigated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) of metabolically labeled proteins synthesized in infected cells and immunoprecipitated with antibodies prepared in a rabbit against an A33R synthetic peptide comprised of amino acids 84 to 100 (Fig. 3). The estimated masses of the A33R polypeptides were 27 kDa for vΔA36R and vΔA36R.c3, 23 kDa for vΔA36R.c1, and 16 kDa for vΔA36R.c2. The mobilities of these A33R polypeptides were consistent with the sequencing results, which had indicated an intact A33R ORF for vΔA36R and vΔA36R.c3 and truncated ones for vΔA36R.c1 and vΔA36R.c2. A truncated protein was also predicted for vΔA36R.c10; however, a diffuse band of about 25 kDa was detected. An additional diffuse band of about 30 kDa was detected in immunoprecipitates from lysates of cells infected with vΔA36R.c1 and vΔA36R.c10, perhaps representing a protein that stably interacts with the mutated A33R protein.
FIG. 3.
SDS-PAGE analysis of the A33R proteins made by mutant viruses. Lysates from infected BS-C-1 cells that had been metabolically labeled with 35S-amino acids were incubated with antibody against a synthetic peptide representing amino acids 84 to 100 of A33R. The bound proteins were analyzed by SDS-PAGE and autoradiography. The position and masses of marker proteins are indicated.
We also sequenced the B5R ORFs of each of the viruses. Except for vΔA36R.c3, the sequences were identical to that of wild-type vaccinia virus. The mutation in vΔA36R.c3 was predicted to cause the nonconservative substitution of serine for proline 189. SDS-PAGE analysis and Western blotting indicated that the B5R protein expressed by vΔA36R.c3 and the other viruses all had the same mobility (data not shown). In summary, these data suggested that a second-site mutation in either the A33R or the B5R gene provided a selective advantage for the A36R deletion mutant and was associated with the comet-forming phenotype.
Transfer and reversal of the comet-forming phenotype by recombination.
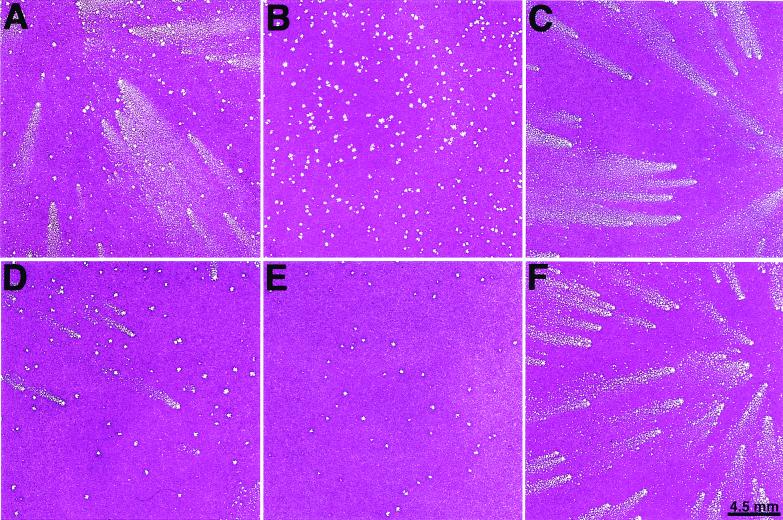
The finding of independent mutations in ORFs encoding membrane glycoproteins, however, provided only circumstantial evidence that these were the cause of the comet-forming phenotype. We needed to exclude contributions from mutations in other genes of vaccinia virus. We chose vΔA36R.c1 as an example to directly prove that truncations of the A33R ORF induce the comet-forming phenotype. The PCR products of the A33R ORF from vΔA36R.c1 or the WR strain were separately transfected into cells infected with vΔA36R. The transfected cells were harvested, and the plaques of the progeny were examined in liquid medium. While 3% of the progeny produced in cells that were transfected with DNA encoding the A33R ORF of vΔA36R.c1 produced comets (Fig. 4A), no comet-forming progeny was detected when the A33R ORF of WR was used for transfection (Fig. 4B). To prove that the comet-forming viruses were derived by recombination and not mutation, three comet-forming viruses were plaque purified under agar. The plaque phenotype of one of these is shown in Fig. 4C. In each case, the DNA sequence of the A33R ORF was identical to that of vΔA36R.c1. As a further proof of the origin of the comet-forming phenotype, we reversed it by transfection of vΔA36R.c1-infected cells with the PCR product of the A33R ORF of the WR strain of vaccinia virus. We noted the presence of non-comet-forming plaques under liquid overlay, which appeared slightly smaller than vΔA36R.c1 under agar. Virus, from six such plaques, was isolated, and each sample produced exclusively small, round plaques that were identical to those of vΔA36R and had an A33R sequence identical to that of the WR strain of vaccinia virus.
FIG. 4.
Marker transfer of the comet-forming phenotype. BS-C-1 cells were infected with vΔA36R and were then transfected with the PCR product of the A33R ORF of vΔA36R.c1 (A) or that of vaccinia virus strain WR (B) or with the PCR product of the B5R ORF of vΔA36R.c3 (D) or with that of WR (E). The cells were harvested after incubation for 2 days, and the diluted lysates were analyzed by plaque assay in BS-C-1 cells with fluid medium. Plaque-purified recombinant viruses, obtained by marker transfer of the A33R (C) and B5R (F) ORFs of vΔA36R.c1 and vΔA36R.c3, respectively, were assayed on BS-C-1 cell monolayers with fluid medium.
The ability of a PCR copy of the B5R gene of vΔA36R.c3 to transfer the comet-forming plaque characteristic to vΔA36R was also determined by transfection. Approximately 11% of the plaques produced by the progeny had a comet-like appearance (Fig. 4D), whereas none was detected when B5R DNA of the WR strain was used for transfection (Fig. 4E). Six of the comet-forming isolates were plaque purified, and the plaque phenotype of one of them is shown in Fig. 4F. The nucleotide sequence of the B5R ORF of each isolate was identical to that of vΔA36R.c3. We then succeeded in reversing the comet-forming phenotype by transfection of vΔA36R.c3-infected cells with the PCR product of the B5R ORF of the WR strain of vaccinia virus. Small plaques without comets were detected, and virus from six were plaque purified three times. The plaques formed by these isolates were identical to those of vΔA36R, and the sequences of the B5R ORFs were identical to that of the WR strain of vaccinia virus.
Formation of IMV and EEV.
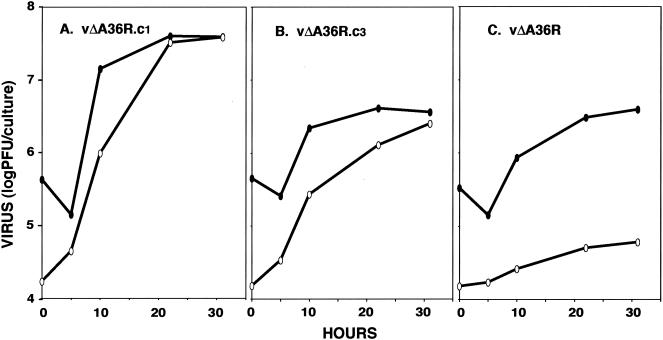
Comets result from enhanced release and spread of extracellular virus. We measured the amounts of cell-associated and released virus produced during single-step growth of vΔA36R.c1, vΔA36R.c3, and the parent vΔA36R in BS-C-1 cells. While 1.5% of the total virus was present in the medium of cells infected with vΔA36R, this percentage was much higher in cells infected with vΔA36R.c1 or vΔA36R.c3, reaching 50.3 or 49.4%, respectively (Fig. 5).
FIG. 5.
One-step growth curves. BS-C-1 cells were infected with vΔA36R.c1, vΔA36R.c3, or vΔA36R at a multiplicity of 5 PFU per cell. At the indicated times, virus titers in the fluid media (white ovals) and the cells (black ovals) were determined by plaque assay.
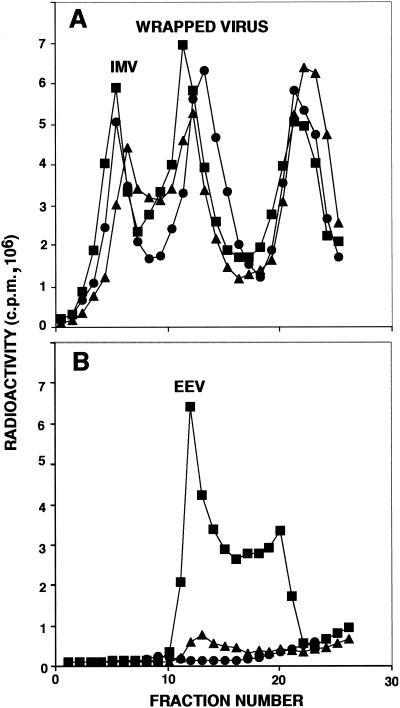
Additional experiments were carried out to confirm that the increased infectivity in the medium represented EEV. RK13 cells were infected with vΔA36R.c1, vΔA36R.c3, or vΔA36R and were metabolically labeled with methionine and cysteine. The labeled viruses formed during 40 h were analyzed in CsCl gradients. The levels of IMV (refractive index of 1.361) and of wrapped viruses (IEV and CEV; refractive index of 1.357) from the cytoplasmic fractions of the cells were similar for the three viruses (Fig. 6A). However, the amount of released virus (EEV) in the culture medium (refractive index of 1.358) was higher for vΔA36R.c1 and for vΔA36R.c3 than for vA36R (Fig. 6B). The higher yield of EEV obtained with vΔA36R.c1 than with vΔA36R.c3 was similar to the difference in infectious virus recovered (Fig. 5). In this experiment, the percentage of the total infectivity that was released into the culture medium of RK13 cells infected with vΔA36R.c1, vΔA36R.c3, or vΔA36R was 60, 34, or 2%, respectively, similar to values obtained with BS-C-1 cells.
FIG. 6.
Analysis of labeled virus in CsCl gradients. RK13 cells were infected with vΔA36R (•), vΔA36R.c1 (▪), or vΔA36R.c3 (▴) and labeled with a mixture of [35S]methionine and [35S]cysteine. Virus particles in the cells (A) and in the culture media (B) were concentrated by sedimentation through a sucrose cushion and were applied to CsCl gradients and centrifuged. Fractions were collected from the bottom of each tube, and radioactivity was determined by scintillation counting.
Virulence for mice.
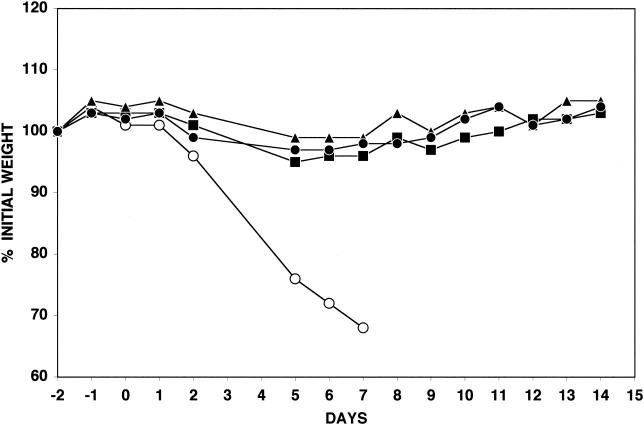
Previous studies showed that vΔA36R was highly attenuated, compared with the WR parental strain of vaccinia virus, when inoculated into mice by the intranasal route (22). We considered that the enhanced ability of the comet-forming viruses to spread in cell monolayers might correlate with increased virulence. Mice were infected intranasally with 5 × 106 PFU of vΔA36R.c1, vΔA36R.c3, vΔA36R, or the WR strain of vaccinia virus, and each mouse was weighed daily. While mice infected with the WR strain of vaccinia virus lost more than 30% of their initial weights within the 1st week following inoculation, those infected with vΔA36R.c1, vΔA36R.c3, or vΔA36R lost little weight during this period of time and fully recovered thereafter (Fig. 7). No significant difference in the weight losses caused by vΔA36R, vΔA36R.c1, and vΔA36R.c3 was observed. All surviving mice had significant neutralizing antibodies, as determined on the 14th day following inoculation (data not shown).
FIG. 7.
Virulence for mice. Purified preparations containing 5 × 106 PFU of vΔA36R (•), vΔA36R.c1 (▪), vΔA36R.c3 (▴), or WR (○) were inoculated intranasally into mice. The weights of the four mice in each group were determined daily, and those that lost more than 30% were sacrificed. The percentages of initial average weight are plotted against days before and after infection.
DISCUSSION
Orthopoxviruses have evolved a unique mechanism for direct cell-to-cell spread, which involves the adherence of extracellular virions called CEV to the tips of long, motile, actin-containing microvilli (3, 5, 12, 33). A minor amount of virus is released from the plasma membrane, allowing the infection to spread through the medium, but the latter mechanism seems less important than direct cell-to-cell spread, as mutations that interfere with the formation of virus-tipped microvilli have a small-plaque phenotype and reduced virulence (26, 28, 29, 42, 43). The IHD strain of vaccinia virus is exceptional because it releases a relatively large amount of extracellular virus that forms numerous satellite plaques in the form of comets (24). IHD was derived from the non-comet-forming New York City Board of Health strain, apparently via a spontaneous mutation in the A34R outer membrane protein (4). Most spontaneous mutations that increase virus release, however, are probably deleterious, as comet-forming viruses do not usually predominate over wild-type virus. In this study, we increased the selective advantage of virus release by starting with a mutant that could not form actin tails.
The A36R protein is a component of the IEV required for actin tail formation but is not present in the outer membrane of extracellular virus (9, 22, 37, 43). Because deletion of the A36R gene restricts the spread of virus in cell monolayers, it provided an ideal genetic background for the isolation of second-site mutants exhibiting enhanced virus spread. After several sequential passages of the deletion mutant, comet-type plaques were noted. The proportion of comet-forming plaques increased with passage number, indicating that they had an advantage over the parental virus. The comet-forming viruses characterized in the present study had mutations in either the A33R or B5R ORF. Four mutants were truncated in the C-terminal domain of the A33R ORF; the truncation was either 35 (vΔA36R.c1 and vΔA36R.c10) or 64 (vΔA36R.c2 and vΔA36R.c5) amino acids. The fifth comet-forming virus, vΔA36R.c3, had proline 189 of the B5R ORF replaced by serine. It was possible to construct the comet-forming viruses by transfecting vA36R-infected cells with either the truncated A33R ORF derived from vΔA36R.c1 or with the mutated B5R ORF from vΔA36R.c3, proving that single mutations were responsible for the comet phenotype. Furthermore, non-comet-forming revertants could be isolated by transfecting vΔA36R.c1-infected cells with the wild-type A33R ORF or vΔA36R.c3 -infected cells with the wild-type B5R ORF.
The A33R and B5R mutants exhibited enhanced release of virus from BS-C-1- or RK13-infected cells. Whereas only 2% of the infectious virus produced by the A36R deletion mutant was found in the medium, approximately half of the infectious virus produced by vΔA36R.c1 or vΔA36R.c3 was released. The additional virus released into the medium by vΔA36R.c1 and vΔA36R.c3 was shown to be EEV by CsCl gradient analysis. Because the second-site mutants still did not make actin tails, as demonstrated by fluorescence microscopy, and because there were no observable differences in morphogenesis, as seen by electron microscopy (data not shown), we attributed their selection solely to increased virus release. Although the second-site mutants were able to outgrow the parent A36R deletion mutant in tissue culture, this was not correlated with higher pathogenicity for mice. Either enhanced spread does not occur in vivo, or the released virus is more susceptible to the immune system than is the CEV.
In conclusion, the present studies provided insights into the modes of orthopoxvirus spread in tissue culture. While direct cell-to-cell spread via virus-tipped microvilli is the dominant mechanism, enhanced virus release provides an alternative when that is prevented. It seems likely that the two mechanisms are antagonistic, since the CEV must adhere tightly to the microvilli. The mechanism of adherence is unknown, but the present and previous studies suggest that the A33R, A34R, and B5R proteins are involved. Whether these proteins interact with cellular proteins on the cell surface or with viral proteins, perhaps derived from the fused outer IEV membrane, remains to be determined. Our finding that three independent A33R mutants have C-terminal truncations may suggest that the deleted region is involved in protein-protein interactions.
Acknowledgments
We thank members of the Laboratory of Viral Diseases, especially Brian Ward for help with the fluorescent microscopy, Andrea Weisberg for electron microscopy, and Norman Cooper for tissue culture cells.
REFERENCES
- 1.Appleyard, G., A. J. Hapel, and E. A. Boulter. 1971. An antigenic difference between intracellular and extracellular rabbitpox virus. J. Gen. Virol. 13:9-17. [DOI] [PubMed] [Google Scholar]
- 2.Blasco, R., and B. Moss. 1991. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-dalton outer envelope protein. J. Virol. 65:5910-5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blasco, R., and B. Moss. 1992. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J. Virol. 66:4170-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasco, R., J. R. Sisler, and B. Moss. 1993. Dissociation of progeny vaccinia virus from the cell membrane is regulated by a viral envelope glycoprotein: effect of a point mutation in the lectin homology domain of the A34R gene. J. Virol. 67:3319-3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cudmore, S., P. Cossart, G. Griffiths, and M. Way. 1995. Actin-based motility of vaccinia virus. Nature 378:636-638. [DOI] [PubMed] [Google Scholar]
- 6.Dales, S., and L. Siminovitch. 1961. The development of vaccinia virus in Earle's L strain cells as examined by electron microscopy. J. Biophys. Biochem. Cytol. 10:475-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engelstad, M., S. T. Howard, and G. L. Smith. 1992. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology 188:801-810. [DOI] [PubMed] [Google Scholar]
- 8.Engelstad, M., and G. L. Smith. 1993. The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology 194:627-637. [DOI] [PubMed] [Google Scholar]
- 9.Frischknecht, F., V. Moreau, S. Rottger, S. Gonfloni, I. Reckmann, G. Superti-Furga, and M. Way. 1999. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature 401:926-929. [DOI] [PubMed] [Google Scholar]
- 10.Geada, M. M., I. Galindo, M. M. Lorenzo, B. Perdiguero, and R. Blasco. 2001. Movements of vaccinia virus intracellular enveloped virions with GFP tagged to the F13L envelope protein. J. Gen. Virol. 82:2747-2760. [DOI] [PubMed] [Google Scholar]
- 11.Hiller, G., and K. Weber. 1985. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J. Virol. 55:651-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hiller, G., K. Weber, L. Schneider, C. Parajsz, and C. Jungwirth. 1979. Interaction of assembled progeny pox viruses with the cellular cytoskeleton. Virology 98:142-153. [DOI] [PubMed] [Google Scholar]
- 13.Hirt, P., G. Hiller, and R. Wittek. 1986. Localization and fine structure of a vaccinia virus gene encoding an envelope antigen. J. Virol. 58:757-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hollinshead, M., G. Rodger, H. Van Eijl, M. Law, R. Hollinshead, D. J. Vaux, and G. L. Smith. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J. Cell Biol. 154:389-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isaacs, S. N., E. J. Wolffe, L. G. Payne, and B. Moss. 1992. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J. Virol. 66:7217-7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mathew, E. C., C. M. Sanderson, R. Hollinshead, and G. L. Smith. 2001. A mutational analysis of the vaccinia virus B5R protein. J. Gen. Virol. 82:1199-1213. [DOI] [PubMed] [Google Scholar]
- 17.McIntosh, A. A., and G. L. Smith. 1996. Vaccinia virus glycoprotein A34R is required for infectivity of extracellular enveloped virus. J. Virol. 70:272-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore, J. B., and G. L. Smith. 1992. Steroid hormone synthesis by a vaccinia enzyme—a new type of virus virulence factor. EMBO J. 11:1973-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morgan, C., S. Ellison, H. Rose, and D. Moore. 1954. Structure and development of viruses observed in the electron microscope. II. Vaccinia and fowl pox viruses. J. Exp. Med. 100:301-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moss, B. 2001. Poxviridae: the viruses and their replication, p. 2849-2883. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed., vol. 2. Lippincott, Williams & Wilkins, Philadelphia, Pa.
- 21.Moss, B., and B. M. Ward. 2001. High-speed mass transit for poxviruses on microtubules. Nat. Cell Biol. 3:E245-E246. [DOI] [PubMed] [Google Scholar]
- 22.Parkinson, J. E., and G. L. Smith. 1994. Vaccinia virus gene A36R encodes a M(r) 43-50 K protein on the surface of extracellular enveloped virus. Virology 204:376-390. [DOI] [PubMed] [Google Scholar]
- 23.Payne, L. 1978. Polypeptide composition of extracellular enveloped vaccinia virus. J. Virol. 27:28-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Payne, L. G. 1980. Significance of extracellular virus in the in vitro and in vivo dissemination of vaccinia virus. J. Gen. Virol. 50:89-100. [DOI] [PubMed] [Google Scholar]
- 25.Rietdorf, J., A. Ploubidou, I. Reckmann, A. Holmström, F. Frischknecht, M. Zettl, T. Zimmermann, and M. Way. 2001. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat. Cell Biol. 3:992-1000. [DOI] [PubMed] [Google Scholar]
- 26.Roper, R., E. J. Wolffe, A. Weisberg, and B. Moss. 1998. The envelope protein encoded by the A33R gene is required for formation of actin-containing microvilli and efficient cell-to-cell spread of vaccinia virus. J. Virol. 72:4192-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roper, R. L., L. G. Payne, and B. Moss. 1996. Extracellular vaccinia virus envelope glycoprotein encoded by the A33R gene. J. Virol. 70:3753-3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Röttger, S., F. Frischknecht, I. Reckmann, G. L. Smith, and M. Way. 1999. Interactions between vaccinia virus IEV membrane proteins and their roles in IEV assembly and actin tail formation. J. Virol. 73:2863-2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanderson, C. M., F. Frischknecht, M. Way, M. Hollinshead, and G. L. Smith. 1998. Roles of vaccinia virus EEV-specific proteins in intracellular actin tail formation and low pH-induced cell-cell fusion. J. Gen. Virol. 79:1415-1425. [DOI] [PubMed] [Google Scholar]
- 30.Schmelz, M., B. Sodeik, M. Ericsson, E. J. Wolffe, H. Shida, G. Hiller, and G. Griffiths. 1994. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J. Virol. 68:130-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seki, M., M. Oie, Y. Ichihashi, and H. Shida. 1990. Hemadsorption and fusion inhibition activities of hemagglutinin analyzed by vaccinia virus mutants. Virology 175:372-384. [DOI] [PubMed] [Google Scholar]
- 32.Shida, H. 1986. Nucleotide sequence of the vaccinia virus hemagglutinin gene. Virology 150:451-462. [DOI] [PubMed] [Google Scholar]
- 33.Stokes, G. V. 1976. High-voltage electron microscope study of the release of vaccinia virus from whole cells. J. Virol. 18:636-643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tooze, J., M. Hollinshead, B. Reis, K. Radsak, and H. Kern. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163-178. [PubMed] [Google Scholar]
- 35.Turner, G. S. 1967. Respiratory infection of mice with vaccinia virus. J. Gen. Virol. 1:399-402. [DOI] [PubMed] [Google Scholar]
- 36.van Eijl, H., M. Hollinshead, G. Rodger, W. H. Zhang, and G. L. Smith. 2002. The vaccinia virus F12L protein is associated with intracellular enveloped virus particles and is required for their egress to the cell surface. J. Gen. Virol. 83:195-207. [DOI] [PubMed] [Google Scholar]
- 37.van Eijl, H., M. Hollinshead, and G. L. Smith. 2000. The vaccinia virus A36R protein is a type Ib membrane protein present on intracellular but not extracellular enveloped virus particles. Virology 271:26-36. [DOI] [PubMed] [Google Scholar]
- 38.Ward, B. M., and B. Moss. 2001. Vaccinia virus intracellular movement is associated with microtubules and independent of actin tails. J. Virol. 75:11651-11663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ward, B. M., and B. Moss. 2001. Visualization of intracellular movement of vaccinia virus virions containing a green fluorescent protein-B5R membrane protein chimera. J. Virol. 75:4802-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williamson, J. D., R. W. Reith, L. J. Jeffrey, J. R. Arrand, and M. Mackett. 1990. Biological characterization of recombinant vaccinia viruses in mice infected by the respiratory route. J. Gen. Virol. 71:2761-2767. [DOI] [PubMed] [Google Scholar]
- 41.Wolffe, E. J., S. N. Isaacs, and B. Moss. 1993. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J. Virol. 67:4732-4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolffe, E. J., E. Katz, A. Weisberg, and B. Moss. 1997. The A34R glycoprotein gene is required for induction of specialized actin-containing microvilli and efficient cell-to-cell transmission of vaccinia virus. J. Virol. 71:3904-3915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wolffe, E. J., A. S. Weisberg, and B. Moss. 1998. Role for the vaccinia virus A36R outer envelope protein in the formation of virus-tipped actin-containing microvilli and cell-to-cell virus spread. Virology 244:20-26. [DOI] [PubMed] [Google Scholar]