Abstract
It is now accepted that an effective vaccine against AIDS must include effective cytotoxic-T-lymphocyte (CTL) responses. The simian immunodeficiency virus (SIV)-infected rhesus macaque is the best available animal model for AIDS, but analysis of macaque CTL responses has hitherto focused mainly on epitopes bound by a single major histocompatibility complex (MHC) class I molecule, Mamu-A*01. The availability of Mamu-A*01-positive macaques for vaccine studies is therefore severely limited. Furthermore, it is becoming clear that different CTL responses are able to control immunodeficiency virus replication with varying success, making it a priority to identify and analyze CTL responses restricted by common MHC class I molecules other than Mamu-A*01. Here we describe two novel epitopes derived from SIV, one from Gag (Gag71-79 GY9), and one from the Nef protein (Nef159-167 YY9). Both epitopes are bound by the common macaque MHC class I molecule, Mamu-A*02. The sequences of these two eptiopes are consistent with the molecule's peptide-binding motif, which we have defined by elution of natural ligands from Mamu-A*02. Strikingly, we found evidence for the selection of escape variant viruses by CTL specific for Nef159-167 YY9 in 6 of 6 Mamu-A*02-positive animals. In contrast, viral sequences encoding the Gag71-79 GY9 epitope remained intact in each animal. This situation is reminiscent of Mamu-A*01-restricted CTL that recognize Tat28-35 SL8, which reproducibly selects for escape variants during acute infection, and Gag181-189 CM9, which does not. Differential selection by CTL may therefore be a paradigm of immunodeficiency virus infection.
As the search for a safe and effective vaccine against AIDS enters its third decade, the pandemic continues, with over 40 million human immunodeficiency virus (HIV)-infected individuals worldwide as of the end of 2001 (36). The need for a prophylactic vaccine, and thus for a clear understanding of effective antiviral immune responses, is increasing in urgency. There is much evidence that cellular immune responses play a major role in controlling HIV and simian immunodeficiency virus (SIV) infection (19), and it is therefore logical that candidate vaccines target cytotoxic-T-lymphocyte (CTL) responses. However, what constitutes an effective cellular immune response is far from clear. Studies in the SIV-infected rhesus macaque have proven to be invaluable for the analysis of immunodeficiency virus pathogenesis, cellular and humoral immune responses, and viral evolution. Many insights into AIDS pathogenesis drawn from the study of SIV-infected rhesus macaques could not have been gleaned from studies of HIV-infected humans, so the development of this model remains a priority.
Most studies of CTL responses against SIV have involved macaques that express the common major histocompatibility complex (MHC) class I molecule, Mamu-A*01. These studies have been fundamental to our understanding of antiviral CTL responses. Crucially, the identification the peptide-binding motif (4) of Mamu-A*01 allowed for the comprehensive screening of the viral genome for possible CTL epitopes, and resulted in the identification of multiple Mamu-A*01-restricted epitopes (2). Knowledge of minimal optimal epitopes has also enabled the synthesis of Mamu-A*01 tetrameric reagents specific for CTL that recognize these epitopes (16). Tetramers that bind CTL specific for the immunodominant epitope Gag181-189 CM9 have been used routinely as a benchmark measurement of immune responses in SIV-infected rhesus macaques and have facilitated analysis of the impact of CTL on viremia (9, 17, 25, 32). However, since investigators have focused on analysis of the Mamu-A*01-restricted CTL for which reagents were available, there is a paucity both of Mamu-A*01-positive animals available for research and of information regarding immune responses restricted by other MHC class I molecules.
Identification of minimal optimal CTL epitopes has also enabled us and others to demonstrate the effects of selective pressure exerted by particular CTL on SIV. Strikingly, we described rapid escape in the Mamu-A*01-restricted CTL epitope Tat28-35 SL8 (3) during the first 4 weeks of infection with the molecularly cloned virus SIVmac239. One unresolved question arising from CTL escape studies, however, is why some high-frequency CTL responses reproducibly select for escape variants and some do not. Most recently, we showed that, whereas the previously characterized Mamu-A*01-restricted CTL recognizing Tat28-35 SL8 and Gag181-189 CM9 were present at similar frequencies during acute infection (21), only Tat28-35 SL8-specific CTL are capable of rapidly selecting for escape variant virus. It was, therefore, of interest to determine whether this pattern of differential escape was recapitulated in epitopes bound by common macaque MHC class I alleles other than Mamu-A*01. Such information would not only shed light on the kinetics of viral evolution within a host but also expand the pool of animals available for vaccine and pathogenesis studies.
In the present work, then, we characterized SIV epitopes bound by the common Indian macaque MHC class I molecule, Mamu-A*02. We found high levels of Mamu-A*02-restricted CTL in vaccinated and naive macaques directed against an epitope in Gag (Gag71-79 GY9) and one in Nef (Nef159-167 YY9). The amino acid sequences of these epitopes were consistent with the Mamu-A*02 peptide binding motif, which we identified by elution of natural ligands from the Mamu-A*02 molecule. Furthermore, we observed that Nef159-167 YY9-specific CTL selected for escape variants of SIVmac239 in six of six Mamu-A*02-positive macaques, while we detected no evidence of escape from Gag71-79 GY9-specific CTL.
MATERIALS AND METHODS
Animals.
Rhesus macaques (Macaca mulatta) were maintained in accordance with the NIH Guide to the Care and Use of Laboratory Animals and with the approval of the University of Wisconsin Research Animal Resource Center review committee.
Peptides.
Overlapping peptides (20-mers, 15-mers, 10-mers, 9-mers, and 8-mers) were synthesized by Chiron (Raleigh, N.C.), the Natural and Medical Science Institute (University of Tübingen, Tübingen, Germany) or by the Biotechnology Center (University of Wisconsin-Madison) based on SIVmac239 protein sequences, with the exception of Pol peptides, which corresponded to the SIVmac251 sequence. Lyophilized peptides were resuspended in phosphate-buffered saline (PBS) with 10% dimethyl sulfoxide (Sigma). Consecutive 20-mer, 15-mer, and 9-mer peptides overlap by 10, 11, or 8 amino acids, respectively. Pools of peptides contained 10 peptides, each at a final concentration of 1 mg/ml.
PBMC.
Peripheral blood mononuclear cells (PBMC) were separated from whole heparin- or EDTA-preserved blood by Ficoll-diatrizoate (Histopaque; Sigma, St. Louis, Mo.) density gradient centrifugation. The PBMC were either used immediately or stored at −180°C in liquid nitrogen. PBMC were cultured in R-15 medium (RPMI 1640 supplemented with 15% fetal calf serum, 2 mM l-glutamine, 25 mM HEPES, 25 μM 2-mercaptoethanol, 50 μg of streptomycin/ml, 50 U of penicillin/ml) supplemented with 20 to 100 IU of recombinant interleukin-2 (IL-2; Proleukin; Chiron Therapeutica, Emeryville, Calif.)/ml.
B-lymphoblastoid cell lines.
Rhesus monkey B-lymphoblastoid cell lines (B-LCL) were generated as described previously (38) by incubating PBMC with supernatants from S594 cells chronically infected with Herpesvirus papio. All B-LCL lines were cultured in R-10 medium (RPMI 1640 supplemented with 10% fetal calf serum, 2 mM l-glutamine, 25 mM HEPES, 25 μM 2-mercaptoethanol, 50 μg of streptomycin/ml, 50 U of penicillin/ml).
Creation of stable MHC class I transfectants.
Stable transfectants expressing rhesus macaque MHC class I molecules were created in the HLA class I-deficient human B-cell line 721.221 as described elsewhere (12).
Peptide-specific T-cell lines.
In order to generate peptide-specific T-cell lines, fresh or thawed PBMC were stimulated in vitro with peptide-pulsed, autologous B-LCL as stimulator cells. Briefly, stimulator cells were generated by incubation of 5 × 106 autologous B-LCL with 0.1 to 0.5 μM peptide at 37°C in a humidified atmosphere with 5% CO2. After 1.5 h, the cells were gamma irradiated (3,000 rads), washed twice with R-15 medium, and added to 5 × 106 fresh autologous PBMC in R-15 medium. At day 0, 2.5 kU of rhIL-7 (R&D Systems, Minneapolis, Minn.)/ml were added to the culture medium. After 2 days, 20 U of recombinant IL-2 (Proleukin)/ml was added, and the cultures were fed afterward every second day with R-15 medium containing 100 U of IL-2/ml. At day 7, CD8β-positive cells were enriched by using the MiniMACS system (Miltenyi Biotec, Inc., Auburn, Calif.). Briefly, live lymphocytes from the 7 day old in vitro-stimulated culture were purified (Ficoll-diatrizoate, Histopaque; Sigma), washed twice with fluorescence-activated cell sorting (FACS) buffer (PBS, 2% fetal bovine serum), and incubated with 6 μl of anti CD8β-phycoerythrin (PE; Immunotech, Westbrook, Maine) for 30 min at 6°C in MACS buffer (FACS buffer, 2 mM EDTA). CD8β-positive cells were then incubated with anti-PE-labeled beads (Miltenyi) and enriched by using MS+ columns (Miltenyi) for MiniMACS according to the manufacturer's protocol. These CD8β-positive cells were again stimulated in vitro by using peptide-pulsed, autologous B-LCL as stimulator cells. After a total of 14 days of in vitro stimulation, the cells were used as effectors in the intracellular cytokine staining (ICS [see below]) assay to test for peptide-specific cells.
DNA/MVA vaccinations and intrarectal SIVmac239 challenge.
Animal 87082 was immunized 10 times with DNA by using the PowderJectXR1 device (PowderJect Vaccines, Inc., Madison, Wis.) at intervals of 4 to 9 weeks, as described elsewhere (14). Briefly, four vaccine plasmids were coadministered, representing all open reading frames (ORFs) of SIV. SIV gag, pol, env, vif, vpr, vpx, tat, and rev were encoded by pSIV17E-Fr gag-pol-env; SIV nef was encoded by pSIVNef-TPA and pSIVNef, and SIV rev was encoded by pSIVrev. The sequence of each ORF was derived from the macrophage-tropic clone SIVmac17E-Fred except for rev, which was derived from SIVmac239 (5, 29, 33). SIVmac17E-Fred shares significant sequence identity with SIVmac239 (5, 33). The construction of these DNA vectors has been described elsewhere (14). Approximately 1 year after the last DNA vaccination, animal 87082 was inoculated twice with recombinant modified vaccinia Ankara (rMVA [7, 10, 20]) vectors within a 13-week interval (14). The animal received 108 infectious units of each rMVA vector, which together encode the gag-pol, env, nef, rev, and tat genes of SIVmacJ5 (31). No rMVA was available that expressed vif, vpr, or vpx. MVA vectors were delivered intradermally and intrarectally (14). No side effects or lesions were found associated with the inoculations. At 9 weeks after the last MVA boost, animal 87082 was challenged intrarectally with the molecularly cloned virus SIVmac239/nef-open (29) by using a dose of 9 ng of p27, or ca. 10 intrarectal 50% monkey infectious doses (23), as described elsewhere (14). All other Mamu-A*02+ animals investigated in this study were naive animals challenged with the same dose and virus, by the same route, as 87082.
ICS of fresh PBMC.
A total of 106 freshly isolated PBMC were incubated with either staphylococcal enterotoxin B (10 μg/ml; Sigma, St. Louis, Mo.) as a positive control, or pools of 10 15- or 20-mer peptides, together with 0.5 μg of anti-CD28 (clone L293) and 0.5 μg of anti-CD49d (clone 9F10; BD Pharmingen, San Diego, Calif.), in a total volume of 200 μl of R-10 medium. For functional avidity assays, minimal optimal peptides were tested in dilutions ranging from 0.005 to 5,000 μg/ml, as reported in detail elsewhere (26). Anti-CD28 and anti-CD49d antibodies were added to provide optimal costimulation (27). After 1.5 h at 37°C, 10 μg/ml of brefeldin A was added and the cells were incubated for another 5 h at 37°C. Brefeldin A inhibits the export of proteins from the endoplasmic reticulum and results, therefore, in the intracellular accumulation of cytokines, which would otherwise be secreted. Cells were washed twice with 1 ml of FACS buffer and then stained with 6 μl of anti-CD8α-PerCP (clone SK1; Becton Dickinson, San Diego, Calif.) and 4 μl of anti-CD4-APC (clone SK3, Becton Dickinson) in 100 μl of FACS buffer for 40 min. After two washes with 1 ml of FACS buffer, the cells were fixed with 2% paraformaldehyde in PBS overnight at 4°C. The cells were then washed once with FACS buffer, treated with permeabilization buffer (0.1% saponin in FACS buffer) for 10 min at room temperature, washed once more with 0.1% saponin buffer, and resuspended in 100 μl of 0.1% saponin buffer. Then, 1 μl of anti-human gamma interferon (IFN-γ)-fluorescein isothiocyanate monoclonal antibody (clone 4S.B3; BD Pharmingen) and either 6 μl of anti-CD69-PE (clone L78; Becton Dickinson) or 1 μl of anti-human tumor necrosis factor alpha (TNF-α)-PE monoclonal antibody (clone MAb11; Pharmingen) was added. After 50 min of incubation at room temperature, the cells were washed twice with 0.1% saponin buffer and then fixed with 2% paraformaldehyde. Samples were stored in the dark at 4°C until analysis. Between 100,000 and 200,000 lymphocyte-gated events were acquired on a FACSCalibur flow cytometer (Becton Dickinson) and analyzed with FlowJo software (Treestar). The background level of IFN-γ staining in PBMC (induced by a control influenza peptide SNEGSYFFG) varied among animals but was typically <0.05% in the CD8+ lymphocytes and <0.02% in the CD4+ lymphocytes. The only samples considered positive were those in which IFN-γ staining was at least twice that of the background or in which there was a distinct population of brightly IFN-γ-positive cells also positive for CD69 or TNF-α.
ICS of T-cell lines for fine mapping, MHC restriction analysis, and testing of mutant peptides.
When T-cell lines were analyzed by ICS, the method described above (for fresh PBMC) was modified so that 105 B-LCL were used instead of anti-CD28 and anti-CD49d. The background level of ICS in T-cell lines (induced by the control peptide SNEGSYFFG) was usually <0.5% and was subtracted from all values. To determine the restricting MHC class I allele, heterologous B-LCL from MHC defined animals were initially pulsed with the epitope peptide, washed after 1.5 h of incubation at 37°C, and used as stimulus for peptide-specific T-cell lines. Afterward, transferents expressing only one of the potentially restricting MHC class I molecules were pulsed with the epitope and tested in ICS as stimuli. In this MHC restriction assay, only samples with ICS at least half of the maximal response induced by autologous peptide-pulsed B-LCL were considered positive. To compare the T-cell recognition of the wild-type sequence and mutant sequences detected in an epitope, peptides containing the corresponding sequence were tested in serial dilutions as stimuli for peptide-specific T-cell lines. The peptide was either present during the incubation time or was pulsed onto autologous B-LCL and washed away before the effectors were added.
Cytotoxicity assay.
Target cells were prepared by incubating B-LCL with peptides at a final concentration of 0.5 μM or by infecting B-LCL with rMVA overnight with a multiplicity of infection of 10. Target cells were labeled with 0.5 mCi of Na251CrO4/ml for 1.5 h at 37°C in a humidified atmosphere with 5% CO2, followed by six washes in R-10 medium. T-cell lines were used as effector cells. The chromium release assay was performed by using standard procedures in U-bottom 96-well microtiter plates. Briefly, autologous peptide-pulsed or rMVA-infected B-LCL target cells were plated at 104 cells per well, and effector cells were added to give a final volume of 200 μl/well. The plates were incubated for 4 h at 37°C. Supernatants were then harvested and measured for gamma emission. Spontaneous release was determined by incubating target cells with medium alone and was <25% of maximal release with detergent (3% Triton X-100) in all assays. Results were expressed as the percent specific chromium release (lysis) calculated by using the following formula: % lysis = [(cpmeffectors − cpmmedia)/cpmtotal − cpmmedia)] × 100, where cpm are the counts per minute.
MHC class I typing of rhesus monkeys for Mamu-A*02 by PCR-SSP.
The Mamu-A*02 allele was detected by sequence-specific DNA amplification (PCR-SSP) concurrent with the molecular typing of seven other MHC class I alleles from the rhesus macaque (W. Rehrauer et al., unpublished data), which have also been shown to bind SIV-derived peptides. The 3′-terminal region of PCR-SSP primers targeted nucleotide polymorphisms unique to these eight Indian rhesus MHC class I alleles. For Mamu-A*02, the following primers amplify a 718-bp product that corresponds to a portion of exons 2 and 3 and the intervening intron B: the 5′ primer, 5′-GGGGCCCTGGCCCTGACT-3′; and the 3′ primer, 5′-CTCGCCCTCCAGGTAGGT-3′. Primers yielding a 260-bp product corresponding to conserved sequences of Mamu-DRB exon 2 were included as an internal control in amplification reactions (15). PCR conditions were optimized for magnesium ion concentration and pH by using a PCR Optimizer Kit (Invitrogen, San Diego, Calif.). Genomic DNA was isolated from 200 μl of EDTA anticoagulated whole blood or buffy coat by using the Qiagen blood kit (Qiagen, Valencia, Calif.) according to manufacturer's guidelines. Approximately 75 ng of genomic DNA was amplified for each sample in a total reaction volume of 25 μl. Final reaction mixtures contained Invitrogen 1× PCR buffer (60 mM Tris HCl [pH 9.5], 2 mM MgCl2, 15 mM ammonium sulfate), 410 mM concentrations of each deoxynucleoside triphosphate [Promega], 0.5 mM concentrations of each PCR-SSP primer, 0.3 mM concentrations of each internal control primer, and 0.961 U of platinum Taq polymerase (Gibco-BRL/Life Technologies, Gaithersburg, Md.). Thermal cycling conditions were as follows: an initial 1-min denaturation at 96°C, followed by 5 cycles of 96°C for 25 s, 70°C for 50 s, and 72°C for 45 s; 21 cycles of 96°C for 25 s, 67°C for 50 s, and 72°C for 45 s; and finally, 4 cycles of 96°C for 25 s, 55°C for 60 s, and 72°C for 120 s. Subsequently, PCR products were electrophoresed on 2% agarose gels (0.5× Tris-borate-EDTA) at a constant voltage and analyzed for the presence of the required internal control product and the Mamu A*02-specific amplicon relative to a 100-bp DNA ladder (Gibco-BRL/Life Technologies).
Amplification of vRNA from plasma.
Cell-free plasma was obtained by centrifugation of EDTA anticoagulated whole blood on a Ficoll density gradient as described above. Viral RNA (vRNA) was extracted from 560 μl of plasma by using the Qiagen QIAmp viral RNA Mini kit according to the manufacturer's instructions for large-volume samples. Amplicons (ca. 1.1 to 1.7 kbp) containing the Mamu-A*02-restricted Nef CTL epitope were generated by using the sense primers SIV 8534-F (5′-GCTGGGATAGTGCAGCAACAGCAAC-3′) or SIV 9029-F (5′-GGTTCTCTTCCCCACCCTC-3′) and the antisense primer SIV 10203-R (5′-ATCAAGAAAGTGGGCGTTCCCGACC-3′). Amplicons (ca. 1.3 kbp) containing the Mamu-A*02-restricted Gag CTL epitope were generated by using the primers SIV 1151-F (5′-AGGAACCAACCACGACGGAG-3′) and SIV 2445-R (5′-AAAGGGATTGGCACTGGTGCGAGG-3′). For each vRNA sample, the reverse transcription-PCR analyses were performed with the Qiagen One-Step RT-PCR kit under the following conditions: 1 cycle of 50° for 1 h; 1 cycle of 95°C for 15 min; 45 cycles of 94°C for 30 s, 59°C for 30 s, and 72°C for 2 min; 1 cycle of 68°C for 20 min; and a 4°C refrigeration.
Mixed-base sequence detection.
The amplified viral cDNA was purified by using the Qiagen PCR purification kit. Approximately 150 ng of purified, amplified cDNA was used as a template for each direct sequencing reaction. Both strands of the CTL epitope-spanning portion (ca. 400 bp) of each amplicon were sequenced by using BigDye chemistry (Applied Biosystems, Foster City, Calif.). For the Mamu-A*02-restricted Nef CTL epitope, the sense oligonucleotide was 9536-F (5′-TCCATGGAGAAACCCAGCTG-3′) and the antisense oligonucleotide was 9899-R (5′-ATCCTCCTGTGCCTCATCTG-3′). For the Mamu-A*02-restricted Gag CTL epitope, the sense oligonucleotide was SIV 1502-F (5′-CTCCATTAGTGCCAACAGGC-3′) and the antisense oligonucleotide was SIV 1826-R (5′-CCTGGCACTACTTCTGCTCC-3′). All sequencing was performed with an ABI 377 automated DNA sequencer (Applied Biosystems).
Factura v.2.2 (Applied Biosystems) was used to process the raw sequence data, identifying all nucleotide sites wherein the ratio of the highest signal peak to the second highest signal peak was less than 2:1, indicating that two viral sequences coexist at a single position. These edited files were then imported into AutoAssembler v2.1 and assembled into contiguous sequences. The contigs were manually edited and a consensus sequence, based on both the sense and the antisense sequences, was generated. This consensus sequence was aligned to SIVmac239 (GenBank accession no. MM33262 [29]) by using the CLUSTALW alignment function in MacVector 7.0 trial version. All mixed-base sites and variant sites described in the present study were verified in two or more independent sequences.
Clone analysis.
For analysis of individual cloned viral cDNA sequences, viral amplicons were obtained as described above. Amplicon DNA was purified from agarose gels by using the QIAquick kit (Qiagen), and ∼150 ng of amplicon DNA was cloned in the pCRII-TOPO TA vector (Invitrogen). Plasmid DNA was isolated from bacterial lysates by using the Qiagen Spin miniprep kit or on the Qiagen BioRobot 3000 by using the QIAPrep 96 Turbo kit according to the manufacturer's instructions. Plasmid DNA (600 ng to 1 μg of each clone) was then used as a template in cycle sequencing of a ∼400-bp region surrounding the epitope, with BigDye chemistry and primers as specified above. Sequence data were edited and analyzed essentially as described above, except that no sites of mixed-base heterogeneity were detected within individual cloned sequences. Sequences derived from both sense and antisense strands were then aligned to the wild-type SIVmac239 nucleotide sequence by using the CLUSTALW routine in MacVector as described above. Conceptual translations of consensus clone sequences were also multiply aligned to a conceptual translation of the wild-type SIVmac239 sequence in this manner.
Statistical analysis of nucleotide variation.
Sequence variation was analyzed statistically as described previously (3, 13, 26). The numbers of nonsynonymous substitutions per nonsynonymous site (dN) and of synonymous substitutions per synonymous site (dS) were estimated as detailed previously (24). The means of the dN and the dS values were calculated for each sequence sample from each animal for (i) all pairwise comparisons between each viral sequence and the wild-type inoculum sequence and (ii) all pairwise comparisons among sequences obtained during chronic infection. These quantities were calculated for the nucleotide sequence encoding the minimal optimal epitope Nef159-167 YY9 (SIVmac239 amino acid sequence YTSGPGIRY) and compared to the values obtained for surrounding regions of the nef ORF. Regions for which dN was greater than dS were considered likely to be under positive selective pressure and therefore are good candidates for CTL escape mutations. If Mamu-A*02-restricted CD8+ T cells were reactive in ICS to synthetic peptides representing the index Nef159-167 YY9 epitope, but not to synthetic peptides derived from variant sequences, the variants were considered to be CTL escape mutants.
Mamu-A*02 peptide motif.
Pooled peptide motifs and individual ligand sequences were derived as previously described (37). Briefly, a construct encoding Mamu-A*02 without the cytoplasmic or transmembrane domain was transfected into the HLA class I-deficient human B-cell line 721.221. Stable transfectants were cultured in hollow fiber bioreactors, and Mamu-A*02-containing supernatant was collected. Approximately 50 mg of molecule was purified on W6/32 affinity columns and acid denatured to elute bound peptide. Peptides were separated from free heavy- and light-chain by passage through a 3-Kd cutoff-stirred cell (Millipore, Bedford, Mass.). One-tenth of the stirred-cell flow through was used for automated Edman sequencing on an ABI 492A pulsed-phase protein sequencer (Perkin-Elmer Applied Biosystems Division, Norwalk, Conn.). A peptide motif was compiled by using resultant data and previously described methods (6). The remaining peptide was fractionated by reversed-phase high-pressure liquid chromatography prior to tandem mass spectrometry (MS/MS sequencing on a Q-STAR quadrupole time-of-flight mass spectrometer (Sciex; Applied Biosystems, Foster City, Calif.). For assays of SIV-derived peptide binding to Mamu-A*02, the molecule was purified from 721.221 transfectant supernatants by affinity chromatography with monoclonal antibody W6/32, as described previously (34). SIV-derived peptides were then tested for their affinities to purified Mamu-A*02 molecules in standard competition assays. Affinities of SIV-derived peptides for Mamu-A*02 are expressed as the concentration of unlabeled SIV peptide required to reduce binding of a labeled reference peptide to 50% of its maximum level; this is the 50% inhibitory concentration.
RESULTS
Mamu-A*02 is expressed at a high frequency in rhesus macaques of Indian descent.
Since Mamu-A*02 was reported to bind an epitope from SIVmac251 Env (39), we designed a PCR-SSP-based method for detection of this allele. We determined that this PCR-SSP reaction was robust and yet specific for the Mamu-A*02 allele (data not shown). The PCR amplicon from eight animals was sequenced and corresponded to the published sequence of Mamu-A*02 (data not shown). We used the assay to determine the frequency of this allele in rhesus macaques of Indian descent (Table 1). Mamu-A*02 was present in approximately one-fifth (19%) of the 992 animals that were typed. This allele is thus almost as common as Mamu-A*01 (15). About one of six Mamu-A*02-positive animals in our colony was also positive for Mamu-A*01 (Table 1).
TABLE 1.
Frequency of Mamu-A∗02 in different Indian rhesus macaque colonies
| Colonya | No. of animals positive/total no. of animals (%)
|
||
|---|---|---|---|
| Mamu-A∗02+ | Mamu-A∗01+ | Mamu-A∗02+ and Mamu-A∗01+ | |
| WRPRC | 137/561 (24.4) | 267/1,096 (24.4) | 23/136 (16.9) |
| Yerkes | 36/337 (10.7) | 192/513 (37.4) | 8/36 (22.2) |
| Various NIH/NCI- funded projects | 18/94 (19.1) | 96/245 (39.2) | 4/18 (22.2) |
| Total | 191/992 (19.3) | 555/1,854 (29.9) | 35/190 (18.4) |
NIH/NCI, National Institutes of Health and National Cancer Institute.
Identification of SIV-derived epitopes recognized by CD8-positive lymphocytes in fresh PBMC from a Mamu-A*02-positive animal.
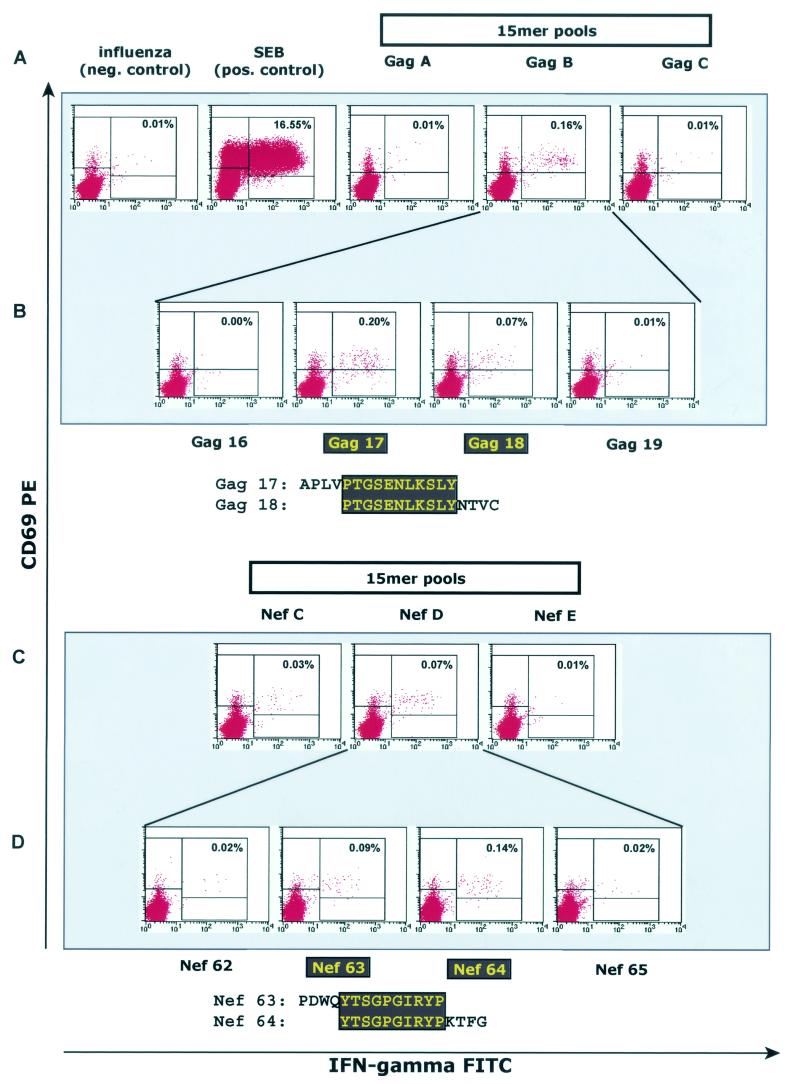
The Mamu-A*02-positive animal 87082 had been immunized against SIVmac Gag, Pol, Env, Nef, Tat, and Rev proteins as described above. At 1 week after the rMVA boost, a time point at which levels of vaccine-induced CTL are expected to be high, fresh PBMC were isolated and tested in the ICS assay for the presence of SIV-specific CD8+ lymphocytes. Pools of peptides (10 overlapping peptides per pool) spanning the SIV proteins Gag, Pol, Env, Nef, Tat, and Rev were used as stimuli in these assays (14). We identified three peptide pools that induced IFN-γ production in the CD8+ lymphocyte population in this animal (Gag pool B, Nef pool C, and Nef pool D; Fig. 1A and C). If a pool elicited an IFN-γ response, its 10 constituent peptides were tested individually with fresh PBMC isolated at week 2 post-rMVA. The individual 15-mer peptides Gag 17 and 18 and Nefs 63 and 64 were recognized by CD8+ lymphocytes from animal 87082 (Fig. 1B and D). The peptide recognized in Nef pool C was also identified (results not shown).
FIG. 1.
Vaccinated animal 87082 makes CD8-positive T-cell responses to regions of viral Gag and Nef proteins. At 1 week after the boost with recombinant MVA expressing SIV proteins, PBMC from animal 87082 were isolated and tested in ICS against pools of 15mer peptides (A and C), as described in Materials and Methods. (B and D) If a peptide pool sensitized cells to produce IFN-γ and CD69 (see Gag pool B and Nef pool D), its constituent peptides were then tested with PBMC from the second week after immunization. The 15mer peptides Gag 17 and 18 (B) and Nef 63 and 64 (D) were found to stimulate CD8-positive lymphocytes. The sequences of the regions where these peptides overlap are boxed in dark gray.
Fine mapping of the CTL epitopes contained in peptide Gag 17 and Nef 64 by using in vitro-stimulated T-cell lines.
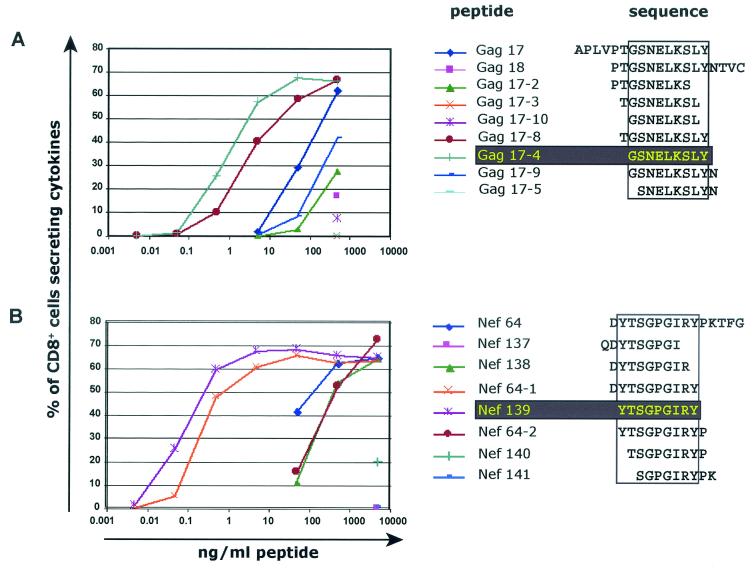
The optimal length of peptides bound by MHC class I molecules is between 8 and 10 amino acids (11, 18, 28). To fine-map the newly identified epitopes, T-cell lines were generated by using the 15mer peptides Gag 17 and Nef 64. Then, overlapping 9mers were tested by ICS to define further the region of the epitope. Thereafter, dilutions of peptides of various lengths were tested by ICS to determine which peptide elicited optimal stimulation of the T-cell line. The 9mer peptide Gag 17-4 (GSENLKSLY; Gag71-79 GY9) effectively stimulated Gag 17-specific T cells at lower concentrations than all other peptides (Fig. 2A). Similarly, the 9mer peptide Nef 139 (YTSGPGIRY; Nef159-167 YY9) stimulated the Nef 64-specific T cells optimally (Fig. 2B). The fine specificity of Nef159-167 YY9-specific T cells was also confirmed with a T-cell line from a naive animal infected with SIV (animal 95084; data not shown). We therefore conclude that Gag71-79 GY9 and Nef159-167 YY9 represent the minimal optimal CTL epitopes recognized in vivo. The amino acid sequences of each epitope are conserved among SIVmac17E-Fred, SIVmac239, SIVmac251, and SIVmac32H.
FIG. 2.
Definition of minimal optimal CD8-positive T-cell epitopes. T-cell lines specific for Gag 17 and Nef 64 were generated and tested in ICS against a range of concentrations of overlapping peptides in order to determine which peptide results in maximal stimulation of the T-cell line. The two 9mer peptides, Gag 17-4 (GSENLKSLY; Gag71-79 GY9) and Nef 139 (YTSGPGIRY; Nef159-167 YY9), optimally stimulated the Gag 17-specific (A) or the Nef 64-specific T cells (B).
Identification of the restricting MHC class I molecule by using heterologous B-LCL from MHC class I-defined animals and MHC class I transferents.
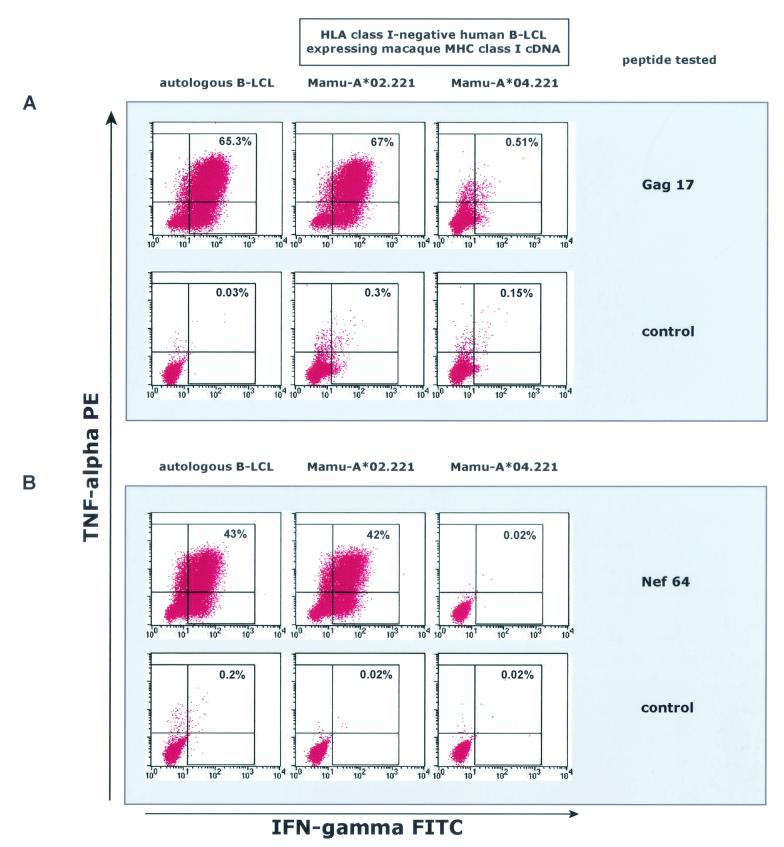
After definition of these two new CTL epitopes, we determined the restricting MHC class I molecules. A family of MHC-defined animals were screened to determine whether the restricting MHC class I molecule was expressed by any of these MHC-typed animals. B-LCL from these animals were pulsed with the appropriate epitope-containing peptide and were used in the ICS assay, together with the peptide-specific T-cell lines from animal 87082. B-LCL from two of these MHC-defined animals (animals 83098 and 90135) were able to present both the Gag 17 and the Nef 64 epitopes to the corresponding T-cell lines from animal 87082; these were the only two animals in the family that expressed Mamu-A*02 (data not shown). To confirm that Mamu-A*02 was the restricting allele for these two new CD8 epitopes, a 721.221-derived cell line expressing Mamu-A*02 was pulsed with the appropriate peptide and used in the ICS assay for the peptide-specific T-cell lines from animal 87082 (Fig. 3). Both epitopes were indeed restricted by Mamu-A*02 (Fig. 3)
FIG. 3.
Mamu-A*02 restricts SIV-derived CTL epitopes. Cells of the MHC class I-negative human B-lymphoblastoid cell line 721.221, transfected with Mamu-A*02 (Mamu-A*02.221) or Mamu-A*04 (Mamu-A*04.221), were pulsed with the appropriate peptide and used in the ICS assay, together with peptide-specific T-cell lines from animal 87082. Both T-cell lines specific for Gag71-79 GY9 (A) and for Nef159-167 YY9 (B) were stimulated by Mamu-A*02-transferents pulsed with the relevant peptide but not by Mamu-A*04 transferents.
Mamu-A*02 binds peptides with a threonine or serine at position 2 and a hydrophobic position 9 anchor.
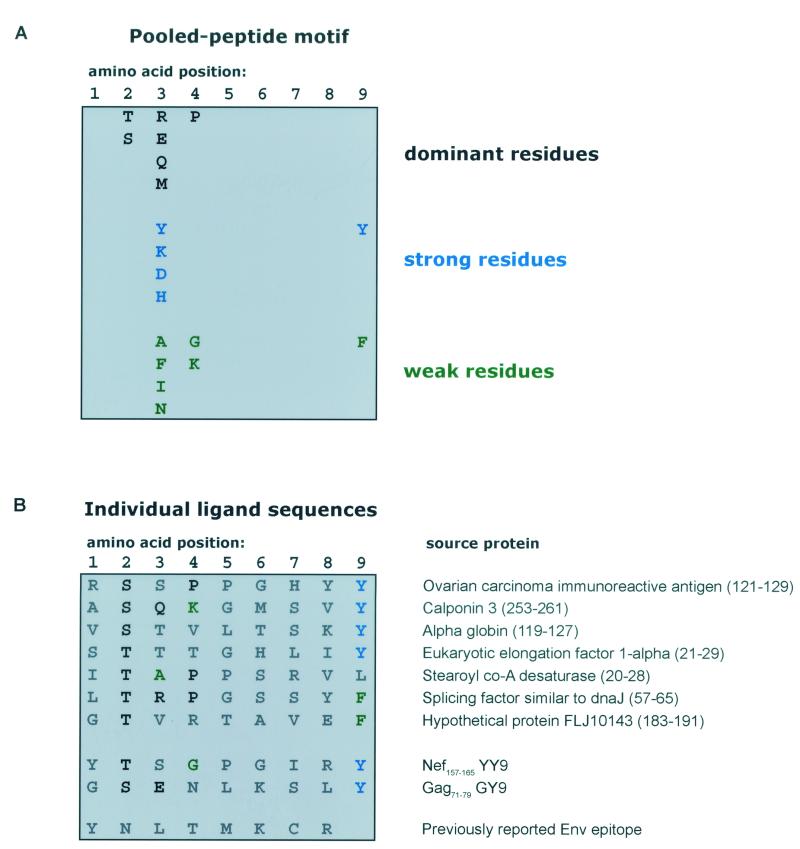
We next determined that the Nef- and Gag-derived epitopes were similar to peptides naturally bound by Mamu-A*02. Endogenously loaded peptides were eluted from Mamu-A*02 molecules and subjected to pooled Edman sequencing. Compilation of a motif from raw data (Fig. 4A) revealed a strong preference for the amino acids threonine (T) or serine (S) at position 2 (P2) of the peptides bound by this molecule. The C termini of bound peptides were composed of a hydrophobic tyrosine (Y) or, to a lesser extent, phenylalanine (F). In order to characterize further the peptides bound by this molecule, individual ligands were selected for MS/MS sequencing. Seven ligands were sequenced and are reported in Fig. 4B. Peptide sequences obtained by MS/MS reiterate the importance of the P2 and P9 anchors for peptide binding; each ligand possesses either a T or S at its second position, and each terminates with a hydrophobic residue. Comparison of endogenously loaded peptides with the Mamu-A*02-restricted SIV epitopes (Fig. 4B) indeed reveals a similar sequence of amino acids, particularly at the P2 and P9 positions.
FIG. 4.
Pooled motif and individual ligand sequences for Mamu-A*02. (A) Peptides eluted from Mamu-A*02 were pooled and subjected to Edman sequencing. The resultant data were analyzed as previously described (6) to generate a peptide motif based on the fold increase in picomoles over the prior round of sequencing. Dominant residues (in boldface) exhibited a ≥3.5-fold increase over the prior round; strong residues (in blue) exhibited a 2.5- to 3.5-fold increase; weak residues (in green) exhibited a 2.0- to 2.5-fold increase. (B) Individual ligand sequences were derived from MS/MS sequencing of selected ions. The two new Gag and Nef SIV epitopes and the previously described Env epitope (39) are listed below the naturally loaded ligands.
T cells specific for Gag 17 and Nef 64 have cytotoxic activity.
To investigate whether the T cells that recognize the two Mamu-A*02-restricted epitopes possess cytolytic activity, the peptide-specific T-cell lines were also tested in standard chromium-release CTL assays. Peptide-pulsed or rMVA-infected, autologous B-LCL, as well as peptide-pulsed Mamu-A*02-transfected 721.221 cells, were specifically lysed by the peptide-specific T-cell lines (data not shown).
Nef159-167 YY9-specific CTL select for escape mutations, but Gag71-79 GY9-specific CTL do not.
Having defined the minimal optimal Mamu-A*02-restricted epitopes, we sought to determine whether CTL recognizing these epitopes selected for escape in SIVmac239. We therefore amplified regions of the gag and nef genes from vRNA in plasma isolated from 6 Mamu-A*02-positive animals during the acute (4 to 16 weeks postinfection) and chronic (30 to 67 weeks postinfection) phases. Regions of each amplicon (∼400 bp) surrounding the epitope were directly sequenced. In each acute-phase sample, we observed at least one site of nonsynonymous mixed-base heterogeneity in the region encoding the Nef159-167 YY9 epitope, whereas Gag71-79 GY9 epitope sequences remained intact (Fig. 5). In the chronic phase of infection, direct sequencing showed at least one complete amino acid replacement in the Nef159-167 YY9 epitope of each animal sequenced, whereas no substitutions were detected in the Gag71-79 GY9 epitope (data not shown). We confirmed the presence of CTL against both Mamu-A*02-restricted epitopes by generating T-cell lines from four of these animals (animals 87082, 97086, 95084, and 96020) by using thawed PBMC (not shown).
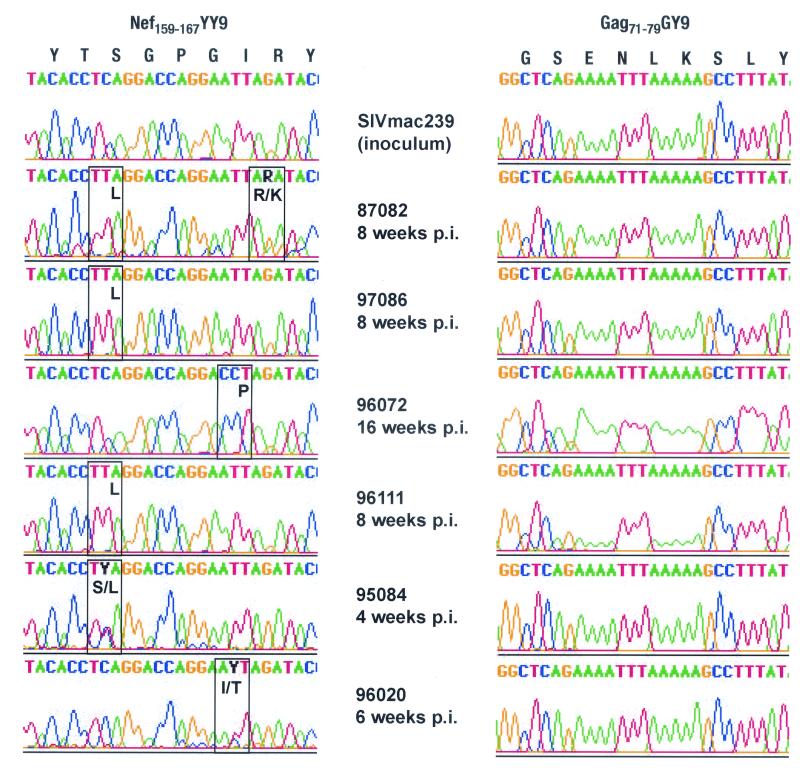
FIG. 5.
Amino acid variation accumulates in the Mamu-A*02-restricted Nef159-167 YY9 epitope during acute infection with SIVmac239. Virus isolated from Mamu-A*02-positive monkeys 4 to 16 weeks after infection with SIVmac239 was analyzed by direct sequencing of viral amplicons spanning the SIV genes nef and gag. The Mamu-A*02-restricted Nef159-167 YY9 epitope accumulated amino acid variation in all animals analyzed, whereas the second Mamu-A*02-restricted epitope Gag71-79 GY9 did not. Sites of mixed-base heterogeneity are shown in black; codons containing these sites are boxed, with the encoded amino acid(s) indicated in black.
We next examined the evolution of the Nef159-167 YY9 epitope more thoroughly by sequencing individually cloned nef amplicons. At least 10 independently cloned amplicons were prepared from chronic-phase vRNA for each Mamu-A*02-positive animal. Cloned amplicons each contained at least one variant codon in the Nef159-167 YY9 sequence, whereas relatively few nonsynonymous substitutions were detected in surrounding regions (Fig. 6). Clone sequences were also subjected to dN/dS analysis, according to the method of Nei and Gojobori (24). dN exceeded dS within the Nef159-167 YY9 epitope sequence (P < 0.005; Table 2), whereas dS was greater than dN outside the epitope region (P < 0.05). These results provide strong evidence that substitutions within the epitope were the result of positive selection by CTL.
FIG. 6.
Sequencing of individually cloned viral cDNA amplicons. At least 10 individual nef amplicons derived from chronic-phase vRNA of each animal were cloned and sequenced as described in the text. No wild-type sequences were detected within the Nef159-167 YY9 epitope, whereas there was relatively little variation outside the epitope sequence (see Table 2). The sequence of the clonal SIVmac239 inoculum is indicated at the top; the epitope is indicated by a yellow box.
TABLE 2.
Comparison of dN and dS rates within and outside the Nef159-167 YY9 epitopea
| Region | Mean rate ± SE
|
|
|---|---|---|
| dN | dS | |
| Epitope | 0.0804 ± 0.0133b | 0.0039 ± 0.0025 |
| Remainder | 0.0053 ± 0.0016c | 0.0090 ± 0.0024 |
dN and dS rates with respect to the wild-type SIVmac239 sequence are shown for chronic-phase viral cDNA clones (see Fig. 6). dN and dS were calculated for the epitope coding region and the remainder of the amplicon (≈400 bp) for each animal. P values refer to differences between dN and dS in the epitope or remainder.
P < 0.0005.
P < 0.05.
CTL recognition is diminished by Nef159-167 YY9 peptide variants.
We next synthesized peptides encoded by Nef159-167 YY9 escape variants and tested them in Mamu-A*02 binding assays and in serial dilutions with Nef159-167 YY9-specific CTL. The only peptide variant that significantly reduced binding to MHC class I had a serine-for-tyrosine substitution at position 9 (YTSGPGIRS [variant residue is boldfaced]; Table 3), and resulted in a 500-fold reduction in binding capacity. This is consistent with the binding motif we defined for Mamu-A*02, since this variant is the only peptide tested in which an anchor position substitution introduces a residue not tolerated by the molecule. Interestingly, we also found that a previously described epitope derived from SIV Env (YNLTMKR [39]) and reported to be bound by Mamu-A*02, failed to bind appreciably to this MHC class I molecule. Again, this finding is consistent with the Mamu-A*02 binding motif, since this epitope lacks the preferred residues at anchor positions 2 and 9. We were also unable to culture CTL that recognized this peptide from any of our Mamu-A*02-positive animals infected with SIVmac239 (not shown).
TABLE 3.
Binding capacity to Mamu-A∗02 of reported SIV-derived, Mamu-A∗02-restricted epitopes and variants
| Sequence | Protein | Source or reference | IC50 (nM) |
|---|---|---|---|
| GSENLKSLY | Gag | This study | 4.9 |
| YTSGPGIRY (index) | Nef | 30; this study | 2.7 |
| YTLGPGIRY | Nef | This study | 2.2 |
| YTSGPGIRF | Nef | This study | 2.0 |
| YTLGPGIRF | Nef | This study | 2.2 |
| YTFGPGIRY | Nef | This study | 4.6 |
| YTSGPGIRS | Nef | This study | 1,077 |
| YNLTMKCR | Env | 39 | 858 |
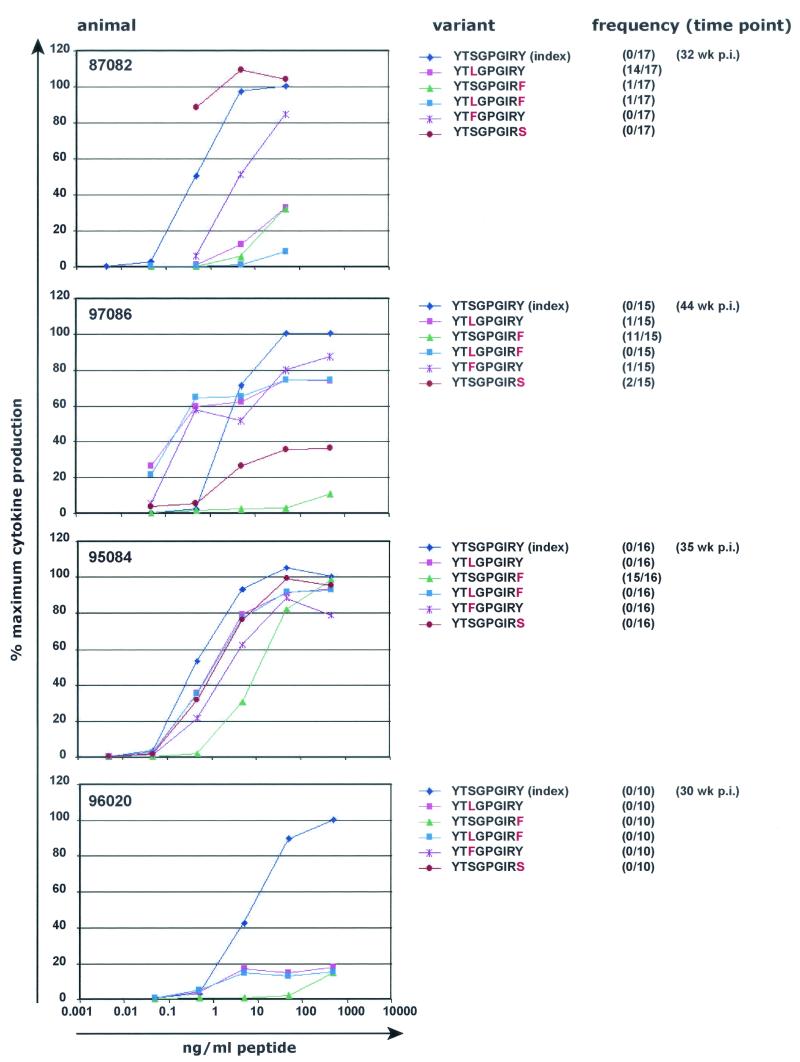
On the other hand, nearly all Nef159-167 YY9 variants diminished recognition by CTL raised against the wild-type peptide, although the degree of reduction varied among animals (Fig. 7 ). One variant detected in animal 97086, YTSGPGIRS, was poorly recognized in this animal but was recognized nearly as well as, or even better than, the wild-type peptide in animals 95084 and 87082, although this peptide binds very poorly to Mamu-A*02 in vitro (see Table 3). Interestingly, this variant sequence was not detected in any other animals. The most frequent variant detected in animal 95084 (YTSGPGIRF, 15 of 16 clones) demonstrated the greatest reduction in recognition by CTL from this animal (Fig. 7). All other variant peptides demonstrated only slight reduction in recognition by CTL from animal 95084, even when excess peptide was washed from the B-LCL before the were used in the assay (results not shown). All tested variant peptides were poorly recognized by CTL from animal 96020 (Fig. 7).
FIG. 7.
Mutant peptides reduce recognition by Nef159-167 YY9-specific CTL. T-cell lines specific for Nef159-167 YY9 were generated from four Mamu-A*02-positive, SIV-infected animals by using thawed PBMC from weeks 4 (animal 87082), 6 (animal 95084), 8 (animal 96020), or 10 (animal 97086) postchallenge. These T-cell lines were then tested in ICS against a range of concentrations of the index peptide and variant peptides representing some of the mutant epitope sequences detected in these animals (see Fig. 6).
CTL specific for Nef159-167 YY9 show high “functional avidity,” whereas Gag71-79 GY9-specific CTL do not.
In a recent cross-sectional study of acute SIVmac239 infection in 21 macaques, we found that viruses from 19 of 21 animals showed evidence of escape from CTL responses within the first 4 weeks of infection, leading us to hypothesize that acute-phase CTL escape is a hallmark of SIV infection (26). In that study, we showed that CTL responses that rapidly selected for escape mutant viruses were capable of recognizing lower concentrations of peptide than those CTL that did not select for acute-phase escape. CTL that are sensitized by low levels of peptide have been termed high “functional avidity” CTL (1, 8, 35); it is thought that such CTL may be more effective at eliminating virus-infected cells than CTL that require more peptide for activation (low “functional avidity”). We determined that Nef159-167 YY9-specific CTL were of high “functional avidity” and capable of 50% maximal IFN-γ production in the ICS assay when stimulated with peptide at a concentration of 4.8 μg/ml (26). Since high-avidity, Nef159-167 YY9-specific CTL selected for escape variant viruses but Gag71-79 GY9-specific CTL did not, we sought to measure the functional avidity of CTL recognizing the latter epitope. Fresh PBMC from animal 87082 obtained at 2 and 3 weeks postinfection yielded 50% maximal IFN-γ production when stimulated with 12.2 μg of peptide/ml and are thus of lower functional avidity than CTL that recognize Nef159-167 YY9 (data not shown).
DISCUSSION
We have identified two SIV-derived epitopes, Nef159-167 YY9 and Gag71-79 GY9, restricted by a common rhesus macaque MHC class I molecule, Mamu-A*02. We developed a robust, sequence-specific DNA amplification technique allowing for rapid typing of macaques for the Mamu-A*02 allele and found it to be common in captive-bred Indian rhesus macaques, expressed by 19% of 992 animals screened. We also defined the peptide binding motif of the Mamu-A*02 molecule by sequencing of eluted natural ligands; the amino acid sequences of both epitopes fit this motif exactly. Knowledge of the motif will make it possible to screen proteins from SIV and other pathogens to identify more potential epitopes restricted by Mamu-A*02. This method has proven successful for Mamu-A*01 and Mamu-B*17 and has resulted in the identification of 14 Mamu-A*01-restricted and 16 Mamu-B*17-restricted epitopes (2, 22).
We were also interested in determining whether CTL that recognized either of these two new epitopes were capable of exerting selective pressure on SIV. Direct sequencing of viral amplicons showed that the Nef159-167 YY9 epitope sequence accumulated nucleotide substitutions beginning in the acute phase of infection and that variant codons appeared fixed in viruses sequenced during chronic infection. Meanwhile, we detected no nucleotide substitutions in the Gag71-79 GY9 epitope, even in chronic infection. Moreover, we found the rate of nonsynonymous substitutions within the Nef159-167 YY9 epitope to be significantly elevated in comparison to surrounding sequences, indicating that the variation we detected was the result of positive selection. While most Nef159-167 YY9 variant peptides tested bound Mamu-A*02 as well as the index peptide, one variant, YTSGPGIRS, effectively prevented binding. Still, T-cell lines generated by using wild-type Nef159-167 YY9 peptide showed a diminished capacity to recognize most variant peptides. We therefore conclude that Nef159-167 YY9-specific CTL selected for escape variant viruses, which altered either epitope binding to the cognate MHC class I molecule or recognition by T-cell receptors. CTL that recognized the novel epitope Gag71-79 GY9, however, did not select for variant viruses, even late in infection. Surprisingly, we also found that a previously described Mamu-A*02-restricted epitope (39) was not bound effectively by the Mamu-A*02 molecule. The sequence of this peptide did not fit the Mamu-A*02 binding motif, and we could not detect CTL directed against this epitope in any Mamu-A*02-positive infected macaque.
The Nef159-167 YY9 epitope was recently described in macaques chronically infected with SIVmac251 or SHIV by other researchers (30). Because significant numbers of Nef159-167 YY9-specific CTL were detected in chronic-phase PBMC from two animals infected with SHIV-89.6 or SHIV-89.6P, the authors of that study concluded that these CTL did not select for escape variant viruses. In striking contrast, we detected no wild-type sequences in the region encoding Nef159-167 YY9 in cloned viral amplicons obtained during chronic infection from five animals. Indeed, the results reported above and elsewhere (26) suggest that Nef159-167 YY9-specific CTL reproducibly select for escape variants during acute SIVmac239 infection of Mamu-A*02-positive animals.
Interestingly, the relationship between Nef159-167 YY9-specific CTL responses, which rapidly select for escape, and Gag71-79 GY9-specific responses, which do not, recapitulates the relationship between the immunodominant Mamu-A*01-restricted CTL responses directed against Tat28-35 SL8 (rapid escape) and Gag181-189 CM9 (slow escape). These phenomena suggest that, whereas CTL directed against different epitopes may be present at similar levels, they exert differential selective pressure on the virus. This may be explained in part by a difference in “functional avidity” of CTL for their cognate epitopes, as we have recently argued elsewhere (26). With the identification of epitopes bound by another high-frequency macaque MHC class I molecule, one “high avidity” and one “low avidity,” we are presented with the opportunity to better define the mechanisms by which CTL select for escape variant viruses. In any case, these data, along with those we have reported elsewhere (26), suggest a model wherein acute-phase CTL responses facilitate control, but not clearance, of primary viremia. Escape from these effective CTL responses would then enable the virus to persist in the host, in the presence of other CTL responses that are less effective at controlling viral replication. Vaccine or immunotherapy regimens that preferentially stimulate CTL of “high functional avidity” may therefore induce immune responses that are particularly effective at elimination of virus-infected cells.
Knowledge of two high-frequency CTL responses restricted by the high-frequency Mamu-A*02 molecule will also help alleviate the pressure to use Mamu-A*01-positive animals for vaccine challenge experiments. An understanding of CTL responses restricted by MHC class I molecules other than Mamu-A*01 will enable us to elucidate interactions between SIV and the macaque immune system with greater breadth, thus increasing the utility of this model for HIV pathogenesis and immunity.
Acknowledgments
T.U.V. and T.C.F. contributed equally to this work.
We thank Elizabeth Meek and Millicent A. Shultz for their excellent technical assistance in typing animals by PCR-SSP, Peicheng Jing for generating T-cell lines and for performing CTL assays, and Donald Carter for assistance in performing ICS.
This work was supported by NIH grants RR1537, AI46366, AI45461, and RR00167. David I. Watkins is a recipient of an Elizabeth Glaser Scientist award.
REFERENCES
- 1.Alexander-Miller, M. A., G. R. Leggatt, and J. A. Berzofsky. 1996. Selective expansion of high- or low-avidity cytotoxic T lymphocytes and efficacy for adoptive immunotherapy. Proc. Natl. Acad. Sci. USA 93:4102-4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen, T. M., B. R. Mothe, J. Sidney, P. Jing, J. L. Dzuris, M. E. Liebl, T. U. Vogel, D. H. O'Connor, X. Wang, M. C. Wussow, J. A. Thomson, J. D. Altman, D. I. Watkins, and A. Sette. 2001. CD8+ lymphocytes from simian immunodeficiency virus-infected rhesus macaques recognize 14 different epitopes bound by the major histocompatibility complex class I molecule Mamu-A*01: implications for vaccine design and testing. J. Virol. 75:738-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, T. M., D. H. O'Connor, P. Jing, J. L. Dzuris, B. R. Mothe, T. U. Vogel, E. Dunphy, M. E. Liebl, C. Emerson, N. Wilson, K. J. Kunstman, X. Wang, D. B. Allison, A. L. Hughes, R. C. Desrosiers, J. D. Altman, S. M. Wolinsky, A. Sette, and D. I. Watkins. 2000. Tat-specific cytotoxic T lymphocytes select for SIV escape variants during resolution of primary viraemia. Nature 407:386-390. [DOI] [PubMed] [Google Scholar]
- 4.Allen, T. M., J. Sidney, M. F. del Guercio, R. L. Glickman, G. L. Lensmeyer, D. A. Wiebe, R. DeMars, C. D. Pauza, R. P. Johnson, A. Sette, and D. I. Watkins. 1998. Characterization of the peptide binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from simian immunodeficiency virus. J. Immunol. 160:6062-6071. [PubMed] [Google Scholar]
- 5.Anderson, M. G., D. Hauer, D. P. Sharma, S. V. Joag, O. Narayan, M. C. Zink, and J. E. Clements. 1993. Analysis of envelope changes acquired by SIVmac239 during neuroadaption in rhesus macaques. Virology 195:616-626. [DOI] [PubMed] [Google Scholar]
- 6.Barber, L. D., B. Gillece-Castro, L. Percival, X. Li, C. Clayberger, and P. Parham. 1995. Overlap in the repertoires of peptides bound in vivo by a group of related class I HLA-B allotypes. Curr. Biol. 5:179-190. [DOI] [PubMed] [Google Scholar]
- 7.Carroll, M. W., and B. Moss. 1997. Host range and cytopathogenicity of the highly attenuated MVA strain of vaccinia virus: propagation and generation of recombinant viruses in a nonhuman mammalian cell line. Virology 238:198-211. [DOI] [PubMed] [Google Scholar]
- 8.Derby, M., M. Alexander-Miller, R. Tse, and J. Berzofsky. 2001. High-avidity CTL exploit two complementary mechanisms to provide better protection against viral infection than low-avidity CTL. J. Immunol. 166:1690-1697. [DOI] [PubMed] [Google Scholar]
- 9.Donahoe, S. M., W. J. Moretto, R. V. Samuel, K. J. Metzner, P. A. Marx, T. Hanke, R. I. Connor, and D. F. Nixon. 2000. Direct measurement of CD8+ T-cell responses in macaques infected with simian immunodeficiency virus. Virology 272:347-356. [DOI] [PubMed] [Google Scholar]
- 10.Drexler, I., K. Heller, B. Wahren, V. Erfle, and G. Sutter. 1998. Highly attenuated modified vaccinia virus Ankara replicates in baby hamster kidney cells, a potential host for virus propagation, but not in various human transformed and primary cells. J. Gen. Virol. 79:347-352. [DOI] [PubMed] [Google Scholar]
- 11.Dzuris, J. L., J. Sidney, E. Appella, R. W. Chesnut, D. I. Watkins, and A. Sette. 2000. Conserved MHC class I peptide binding motif between humans and rhesus macaques. J. Immunol. 164:283-291. [DOI] [PubMed] [Google Scholar]
- 12.Evans, D. T., P. Jing, T. M. Allen, D. H. O'Connor, H. Horton, J. E. Venham, M. Piekarczyk, J. Dzuris, M. Dykhuzen, J. Mitchen, R. A. Rudersdorf, C. D. Pauza, A. Sette, R. E. Bontrop, R. DeMars, and D. I. Watkins. 2000. Definition of five new simian immunodeficiency virus cytotoxic T-lymphocyte epitopes and their restricting major histocompatibility complex class I molecules: evidence for an influence on disease progression. J. Virol. 74:7400-7410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans, D. T., D. H. O'Connor, P. Jing, J. L. Dzuris, J. Sidney, J. da Silva, T. M. Allen, H. Horton, J. E. Venham, R. A. Rudersdorf, T. Vogel, C. D. Pauza, R. E. Bontrop, R. DeMars, A. Sette, A. L. Hughes, and D. I. Watkins. 1999. Virus-specific cytotoxic T-lymphocyte responses select for amino acid variation in simian immunodeficiency virus Env and Nef. Nat. Med. 5:1270-1276. [DOI] [PubMed] [Google Scholar]
- 14.Horton, H., T. U. Vogel, D. K. Carter, K. Vielhuber, D. H. Fuller, T. Shipley, J. T. Fuller, K. J. Kunstman, G. Sutter, D. C. Montefiori, V. Erfle, R. C. Desrosiers, N. Wilson, L. J. Picker, S. M. Wolinsky, C. Wang, D. B. Allison, and D. I. Watkins. 2002. Immunization of rhesus macaques with a DNA prime/modified vaccinia virus Ankara boost regimen induces broad simian immunodeficiency virus (SIV)-specific T-cell responses and reduces initial viral replication but does not prevent disease progression following challenge with pathogenic SIVmac239. J. Virol. 76:7187-7202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knapp, L. A., E. Lehmann, M. S. Piekarczyk, J. A. Urvater, and D. I. Watkins. 1997. A high frequency of Mamu-A*01 in the rhesus macaque detected by polymerase chain reaction with sequence-specific primers and direct sequencing. Tissue Antigens 50:657-661. [DOI] [PubMed] [Google Scholar]
- 16.Kuroda, M. J., J. E. Schmitz, D. H. Barouch, A. Craiu, T. M. Allen, A. Sette, D. I. Watkins, M. A. Forman, and N. L. Letvin. 1998. Analysis of Gag-specific cytotoxic T lymphocytes in simian immunodeficiency virus-infected rhesus monkeys by cell staining with a tetrameric major histocompatibility complex class I-peptide complex. J. Exp. Med. 187:1373-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuroda, M. J., J. E. Schmitz, W. A. Charini, C. E. Nickerson, M. A. Lifton, C. I. Lord, M. A. Forman, and N. L. Letvin. 1999. Emergence of CTL coincides with clearance of virus during primary simian immunodeficiency virus infection in rhesus monkeys. J. Immunol. 162:5127-5133. [PubMed] [Google Scholar]
- 18.Madden, D. R. 1995. The three-dimensional structure of peptide-MHC complexes. Annu. Rev. Immunol. 13:587-622. [DOI] [PubMed] [Google Scholar]
- 19.McMichael, A. J., and S. L. Rowland-Jones. 2001. Cellular immune responses to HIV. Nature 410:980-987. [DOI] [PubMed] [Google Scholar]
- 20.Meyer, H., G. Sutter, and A. Mayr. 1991. Mapping of deletions in the genome of the highly attenuated vaccinia virus MVA and their influence on virulence. J. Gen. Virol. 72:1031-1038. [DOI] [PubMed] [Google Scholar]
- 21.Mothe, B. R., H. Horton, D. K. Carter, T. M. Allen, M. E. Liebl, P. Skinner, T. U. Vogel, S. Fuenger, K. Vielhuber, W. Rehrauer, N. Wilson, G. Franchini, J. D. Altman, A. Haase, L. J. Picker, D. B. Allison, and D. I. Watkins. 2002. Dominance of CD8 responses specific for epitopes bound by a single major histocompatibility complex class I molecule during the acute phase of viral infection. J. Virol. 76:875-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mothe, B. R., J. Sidney, J. L. Dzuris, M. E. Liebl, S. Fuenger, D. I. Watkins, and A. Sette. 2002. Characterization of the Peptide-binding specificity of Mamu-B*17 and identification of Mamu-B*17-restricted epitopes derived from simian immunodeficiency virus proteins. J. Immunol. 169:210-219. [DOI] [PubMed] [Google Scholar]
- 23.Murphy, C. G., W. T. Lucas, R. E. Means, S. Czajak, C. L. Hale, J. D. Lifson, A. Kaur, R. P. Johnson, D. M. Knipe, and R. C. Desrosiers. 2000. Vaccine protection against simian immunodeficiency virus by recombinant strains of herpes simplex virus. J. Virol. 74:7745-7754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 25.Nixon, D. F., S. M. Donahoe, W. M. Kakimoto, R. V. Samuel, K. J. Metzner, A. Gettie, T. Hanke, P. A. Marx, and R. I. Connor. 2000. Simian immunodeficiency virus-specific cytotoxic T lymphocytes and protection against challenge in rhesus macaques immunized with a live attenuated simian immunodeficiency virus vaccine. Virology 266:203-210. [DOI] [PubMed] [Google Scholar]
- 26.O'Connor, D. H., T. M. Allen, T. U. Vogel, P. Jing, I. P. DeSouza, E. Dodds, E. J. Dunphy, C. Melsaether, B. Mothe, H. Yamamoto, H. Horton, N. Wilson, A. L. Hughes, and D. I. Watkins. 2002. Acute phase cytotoxic T lymphocyte escape is a hallmark of simian immunodeficiency virus infection. Nat. Med. 8:493-499. [DOI] [PubMed] [Google Scholar]
- 27.Picker, L. J., M. K. Singh, Z. Zdraveski, J. R. Treer, S. L. Waldrop, P. R. Bergstresser, and V. C. Maino. 1995. Direct demonstration of cytokine synthesis heterogeneity among human memory/effector T cells by flow cytometry. Blood 86:1408-1419. [PubMed] [Google Scholar]
- 28.Rammensee, H. G., T. Friede, and S. Stevanoviic. 1995. MHC ligands and peptide motifs: first listing. Immunogenetics 41:178-228. [DOI] [PubMed] [Google Scholar]
- 29.Regier, D. A., and R. C. Desrosiers. 1990. The complete nucleotide sequence of a pathogenic molecular clone of simian immunodeficiency virus. AIDS Res. Hum. Retrovir. 6:1221-1231. [DOI] [PubMed] [Google Scholar]
- 30.Robinson, S., W. A. Charini, M. H. Newberg, M. J. Kuroda, C. I. Lord, and N. L. Letvin. 2001. A commonly recognized simian immunodeficiency virus Nef epitope presented to cytotoxic T lymphocytes of Indian-origin rhesus monkeys by the prevalent major histocompatibility complex class I allele Mamu-A*02. J. Virol. 75:10179-10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rud, E. W., M. Cranage, J. Yon, J. Quirk, L. Ogilvie, N. Cook, S. Webster, M. Dennis, and B. E. Clarke. 1994. Molecular and biological characterization of simian immunodeficiency virus macaque strain 32H proviral clones containing nef size variants. J. Gen. Virol. 75:529-543. [DOI] [PubMed] [Google Scholar]
- 32.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 33.Sharma, D. P., M. C. Zink, M. Anderson, R. Adams, J. E. Clements, S. V. Joag, and O. Narayan. 1992. Derivation of neurotropic simian immunodeficiency virus from exclusively lymphocytetropic parental virus: pathogenesis of infection in macaques. J. Virol. 66:3550-3556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sidney, J., S. Southwood, D. L. Mann, M. A. Fernandez-Vina, M. J. Newman, and A. Sette. 2001. Majority of peptides binding HLA-A*0201 with high affinity crossreact with other A2-supertype molecules. Hum. Immunol. 62:1200-1216. [DOI] [PubMed] [Google Scholar]
- 35.Slifka, M. K., and J. L. Whitton. 2001. Functional avidity maturation of CD8+ T cells without selection of higher affinity TCR. Nat. Immunol. 2:711-717. [DOI] [PubMed] [Google Scholar]
- 36.UNAIDS, JUNPoHA, and World Health Organization. 2001. AIDS epidemic update. World Health Organization, Geneva, Switzerland.
- 37.Urvater, J. A., N. Otting, J. H. Loehrke, R. Rudersdorf, I. I. Slukvin, M. S. Piekarczyk, T. G. Golos, A. L. Hughes, R. E. Bontrop, and D. I. Watkins. 2000. Mamu-I: a novel primate MHC class I B-related locus with unusually low variability. J. Immunol. 164:1386-1398. [DOI] [PubMed] [Google Scholar]
- 38.Vogel, T., S. Norley, B. Beer, and R. Kurth. 1995. Rapid screening for Mamu-A1-positive rhesus macaques using a SIVmac Gag peptide-specific cytotoxic T-lymphocyte assay. Immunology 84:482-487. [PMC free article] [PubMed] [Google Scholar]
- 39.Watanabe, N., S. N. McAdam, J. E. Boyson, M. S. Piekarczyk, Y. Yasutomi, D. I. Watkins, and N. L. Letvin. 1994. A simian immunodeficiency virus envelope V3 cytotoxic T-lymphocyte epitope in rhesus monkeys and its restricting major histocompatibility complex class I molecule Mamu-A*02. J. Virol. 68:6690-6696. [DOI] [PMC free article] [PubMed] [Google Scholar]