Abstract
In vitro studies show that human immunodeficiency virus type 1 (HIV-1) does not replicate in freshly isolated monocytes unless monocytes differentiate to monocyte-derived macrophages. Similarly, HIV-1 may replicate in macrophages in vivo, whereas it is unclear whether blood monocytes are permissive to productive infection with HIV-1. We investigated HIV-1 replication in CD14+ monocytes and resting and activated CD4+ T cells by measuring the levels of cell-associated viral DNA and mRNA and the genetic evolution of HIV-1 in seven acutely infected patients whose plasma viremia had been <100 copies/ml for 803 to 1,544 days during highly active antiretroviral therapy (HAART). HIV-1 DNA was detected in CD14+ monocytes as well as in activated and resting CD4+ T cells throughout the course of study. While significant variation in the decay slopes of HIV-1 DNA was seen among individual patients, viral decay in CD14+ monocytes was on average slower than that in activated and resting CD4+ T cells. Measurements of HIV-1 sequence evolution and the concentrations of unspliced and multiply spliced mRNA provided evidence of ongoing HIV-1 replication, more pronounced in CD14+ monocytes than in resting CD4+ T cells. Phylogenetic analyses of HIV-1 sequences indicated that after prolonged HAART, viral populations related or identical to those found only in CD14+ monocytes were seen in plasma from three of the seven patients. In the other four patients, HIV-1 sequences in plasma and the three cell populations were identical. CD14+ monocytes appear to be one of the potential in vivo sources of HIV-1 in patients receiving HAART.
Highly active antiretroviral therapy (HAART) has generally been successful in reducing human immunodeficiency virus type 1 (HIV-1) RNA in plasma to “undetectable” levels (<50 copies/ml), with dramatic improvements in the clinical course of HIV-1 infection (3, 19, 20, 34, 44). However, low levels of viral replication persist in persons on prolonged HAART (6, 13, 17, 19a, 26a, 32, 38a, 47, 58, 61). While resting CD4+ T cells are a potential reservoir for virus persistence (9, 10, 15, 16, 57), recent studies suggest the existence of other sources of emergent HIV-1 upon discontinuing HAART (8, 60).
Blood monocytes, derived from mononuclear phagocyte precursor cells in bone marrow, may circulate in peripheral blood for 1 to 3 days before entering tissues and differentiating to tissue-specific macrophages (29). CD14 is expressed exclusively on the mononuclear phagocyte lineage, at high levels on the surfaces of most blood monocytes (48) and at lower but detectable levels in macrophages in tissues such as lung (49). However, CD14 is absent in macrophages from small intestine, T cells, B cells, and natural killer (NK) cells (48). Tissue macrophages may be productively infected with HIV-1 and simian immunodeficiency virus-HIV-1 chimeras and act as viral reservoirs (18, 24, 25, 27, 33). In vitro studies suggest that HIV-1 does not replicate in freshly isolated peripheral blood monocytes unless monocytes differentiate to monocyte-derived macrophages (11, 30, 31, 39, 50). Although HIV-1 can be detected in blood monocytes (18, 23, 26, 27), it is unknown whether the virus is produced or is maintained latently in monocytes in vivo (28, 45). Most recently, infectious HIV-1 has been isolated from monocyte-derived macrophages of patients on prolonged HAART (51), indicating that monocytes harbor replication-competent HIV-1 and confirming that HIV-1 can be produced after monocytes differentiate to macrophages (11, 26, 30, 31, 50). Whether HIV-1 replicates in undifferentiated blood monocytes remains unclear. In the present study, we investigated HIV-1 replication in purified CD14+ monocytes without the aid of in vitro adherence, activation, and differentiation (11, 26, 30, 31, 50). Moreover, we compared the replication characteristics of HIV-1 in CD14+ monocytes with those in activated and resting CD4+ T cells. Finally, we determined the contribution of these three cell populations to HIV-1 persistence in patients on prolonged HAART.
(This study was presented in part at the 4th International Workshop on HIV, Cells of Macrophage Lineage, and other Reservoirs, Donnini, Florence, Italy, 1 to 4 December 1999, and the 1st International Workshop on Acute HIV-1 Infection 2000, Arlington, Va., 16 to 17 October 2000.)
MATERIALS AND METHODS
Study patients.
We studied seven homosexual men who initiated HAART containing indinavir (2,400 mg/day), zidovudine (600 mg/day), and lamivudine (300 mg/day) between 18 and 121 days after the onset of symptoms of acute HIV infection (Table 1) (3, 44). All patients were highly compliant with therapy. Six of them had maintained undetectable levels of plasma viremia (<50 HIV-1 RNA copies/ml of plasma, determined by a Roche ultrasensitive reverse transcriptase PCR [RT-PCR] assay) for 803 to 1,544 days with 1 or 2 transient episodes of plasma viremia (<100 copies/ml), while patient 7 had consistently maintained plasma HIV-1 RNA levels of fewer than 50 copies/ml. Leukapheresis from each patient included a time point prior to or on the day of the initiation of therapy (sample I) and two or three time points at 346 to 1,630 days into therapy (samples II, III, and IV).
TABLE 1.
Characteristics of HIV-1-infected patients receiving HAART
| Patient and sample | Time (days)a of:
|
No. of CD4+ cells/mm3 | No. of HIV-1 RNA copies/ml of plasmab | ||
|---|---|---|---|---|---|
| Sampling | HAART initiation | Seroconversion | |||
| 1 | 90 | 62 | |||
| I | 64 | 452 | 38,900 | ||
| II | 729 | 860 | <50 (20) | ||
| III | 954 | 986 | <50 (12) | ||
| IV | 1,458 | 1,002 | <50 (31) | ||
| 2 | 121 | 41 | |||
| I | 41 | 702 | 492,000 | ||
| II | 1,051 | 842 | <50 (20) | ||
| III | 1,235 | 815 | <50 (8) | ||
| IV | 1,751 | 788 | <50 (7) | ||
| 3 | 79 | 58 | |||
| I | 78 | 777 | 22,200 | ||
| II | 425 | 1,423 | <50 (25) | ||
| III | 739 | 1,518 | <50 (14) | ||
| IV | 1,153 | 1,265 | <50 (6) | ||
| 4 | 20 | 30 | |||
| I | 10 | 381 | 3,500,000 | ||
| II | 423 | 928 | <50 (30) | ||
| III | 1,251 | 841 | <50 (12) | ||
| 5 | 18 | 18 | |||
| I | 18 | 755 | 6,000,000 | ||
| II | 578 | 1,061 | <50 (14) | ||
| III | 941 | 1,001 | <50 (17) | ||
| 6 | 80 | 60 | |||
| I | 35 | 435 | 67,862 | ||
| II | 820 | 718 | <50 (18) | ||
| III | 1,017 | 848 | <50 (10) | ||
| IV | 1,202 | 803 | <50 (11) | ||
| 7 | 21 | 20 | |||
| I | 20 | 680 | 12,500 | ||
| II | 485 | 729 | <50 (4) | ||
| III | 725 | 801 | <50 (4) | ||
| IV | 1,205 | 832 | <50 (5) | ||
Isolation of CD14+ monocytes and resting and activated CD4+ T lymphocytes.
CD14+ monocytes, resting CD4+ T lymphocytes, and activated CD4+ T lymphocytes were purified from peripheral blood mononuclear cells (PBMC) by negative selection using magnetic bead depletion followed by fluorescent-activated cell sorting (FACS) (7, 10, 15, 16). Specifically, monocytes were selected using monoclonal antibodies (MAbs) against CD3, CD8, CD19, and CD16 expressed on the surfaces of T cells, CD8+ T cells, B cells, and NK cells, respectively. The resulting cells were further positively sorted by FACS of monocytes with antibodies to CD14. Resting CD4+ T cells (HLA-DR−, CD25−, CD38−, and CD69−) were purified from PBMC by negative selection and an additional 6-day incubation to remove residual activated cells as described previously (7, 10). To purify activated CD4+ T cells, negative selection was used to deplete CD8+ T cells, B cells, NK cells, and monocytes/macrophages. The resulting cells were then selected by FACS using antibodies to activated cell surface markers HLA-DR, CD25, CD38, and CD69. The purity of isolated cells was analyzed by flow cytometry.
Quantification of HIV-1 DNA.
Genomic DNA was isolated from purified cells using the QIAmp tissue kit (Qiagen) according to the manufacture’s protocol. HIV-1 DNA copies were quantified by real-time PCR (TagMan) (3, 5). The results shown in Fig. 2 and Table 2 are the mean values of three independent measurements for each sample. The detection limits for this assay were 5 copies per 1 to 5 μ g of total DNA per PCR, as described previously (3, 5).
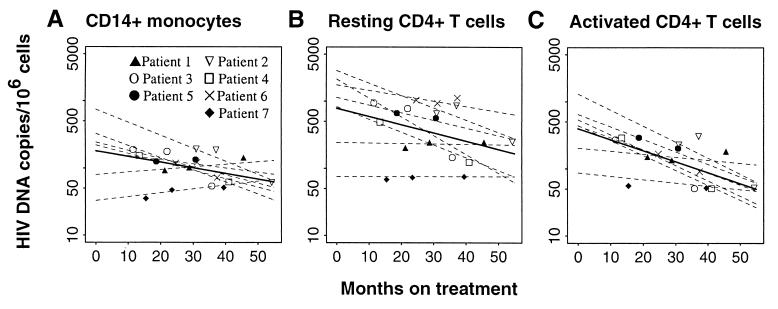
FIG. 2.
Decay rates of HIV-1 DNA in peripheral blood CD14+ monocytes (A), resting CD4+ T cells (B), and activated CD4+ T cells (C) in seven acutely infected patients taking HAART who had undetectable levels of plasma virus. Patients 1 to 6 had maintained undetectable levels of plasma viremia with one or two episodes of low-level plasma viremia (<100 copies/ml), while patient 7 had consistently maintained plasma HIV-1 RNA at levels lower than 50 copies/ml. Average decay slopes were estimated using linear random-effects regression (12) for one phase decay. The bold line in each panel represents the average slope and intercept of the decay in that cell compartment, and the dashed lines with different symbols represent predicted individual intercepts and decay slopes. The data are summarized in Table 2.
TABLE 2.
Decay of HIV-1 infected CD14+ monocytes and resting and activated CD4+ T cells
| Cell type | Decay slope (per mo)
|
t1/2 (mo)
|
||
|---|---|---|---|---|
| Mean | 95% CI | Mean | 95% CI | |
| CD14+ monocytes | −0.0073 | −0.0168 to 0.0022 | 41.3 | 17.9 to ∞ |
| Resting CD4+ T cells | −0.0128 | −0.0240 to −0.0015 | 23.6 | 12.5 to 196.6 |
| Activated CD4+ T cells | −0.0152 | −0.0253 to −0.0050 | 19.8 | 11.9 to 59.8 |
Quantification of cell-associated unspliced (US) and multiply spliced (MS) viral mRNA and virion RNA in plasma.
Cell-associated RNA was isolated with the QIAmp RNA kit (Qiagen), digested with DNase I, and reverse transcribed as described previously (5) with antisense primer DM104 (residues 1674 to 1645 of HIV-1 HXB2 sequence in GenBank, 5′-AGTCTCTAAAGGGTTCCTTTGGTCCTTGTC-3′) for US gag or primer DT1R (8556 to 8525, 5′-GCAATCAAGAGTAAGTCGATCAAGCGGTGGTA-3′) for MS tat. cDNA diluted in 10-fold series in triplicate was used for nested PCR with the following primers: DM102 (HIV-1 gag, residues 1395 to 1427, 5′-GAGACCATCAATGAGGAAGCTGCAGAATGGGAT-3′) and DM104 for a first round of PCR and SK38 and SK39 (41) for a second round of PCR. For tat, DT1F (residues 5780 to 7807, 5′-TGGGTGTCGACATAGCAGAATAGGCATT-3′) and DT1R were used for the first round of amplification, and DT2F (residues 5831 to 5859, 5′-GGAGGCCAGTAGATCATAGACTAGAGCCCT-3′) and DT2R (residues 8451 to 8432, 5′-TCTCTGTCTCTCTCTCCACCTTCTTCTTC-3′) were used for second-round reactions. PCR products were separated on polyacrylamide gels after liquid hybridization with 32P-labeled probes (SK39 for gag; DMTP1, residues 6025 to 6059, 5′-GGGCTGGAGGTGGGTTGCTTTGATAGAGAATCTTG-3′, for tat), blotted, and autoradiographed. Conditions of PCR and liquid hybridization procedure have been described previously (5, 63). Controls without RT were negative, which confirmed the absence of viral DNA. Each PCR amplification contained primers for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (14) as an internal control for the amount of amplifiable cDNA. The amounts of gag and tat HIV-1 mRNA were calculated following limiting dilution with the computer program QUALITY (40). Viral RNA was isolated from plasma (63) containing fewer than 50 HIV-1 RNA copies/ml and then HIV-1 gag was quantified as described above. Utilization of nested PCR plus 32P liquid hybridization was sensitive enough to detect one copy per μ g of cDNA per reaction and specific for the detection of HIV-1 (data not shown). The averages of two independent measurements for HIV-1 mRNA and virion RNA for each sample are shown in Table 1 and Fig. 3.
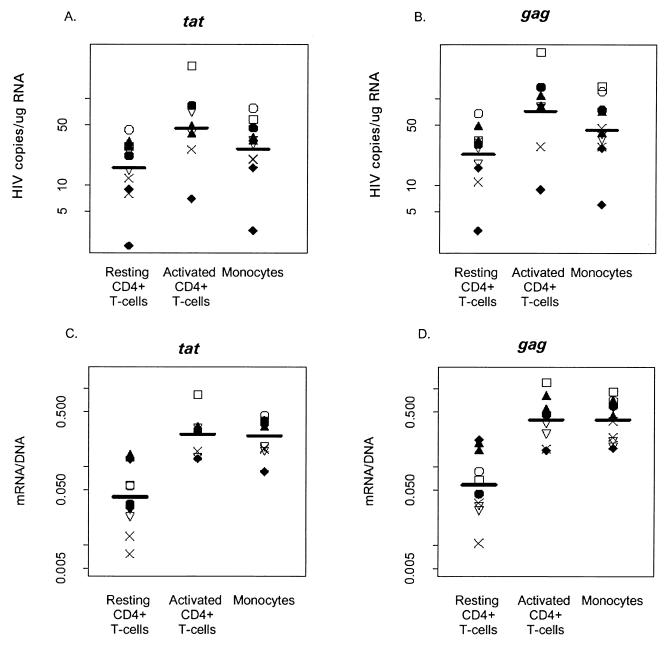
FIG. 3.
Comparison of levels of HIV-1 mRNA in CD14+ monocytes and activated CD4+ T cells and resting CD4+ T cells in seven patients following 1 to 3 years of HAART. (A) Levels of MS tat mRNA in the three cell types. Pairwise comparisons of the mean level of the log of tat mRNA using general estimating equations (12) detected significant differences between all three cell types (P < 0.001 for resting compared to activated cells, P < 0.001 for resting cells compared to monocytes, P = 0.003 for activated cells compared to monocytes. (B) Corresponding levels of MS gag mRNA. Again, significant differences were found between all three cell types (P < 0.001 for resting compared to activated cells, P < 0.001 for resting cells compared to monocytes, P = 0.020 for activated cells compared to monocytes). (C) Ratio of HIV-1 tat mRNA to DNA. (D) Ratio of HIV-1 gag mRNA to DNA. Pairwise comparisons of the log of means ratios of both tat and gag mRNA and DNA suggest significant differences between resting CD4+ T cells and both monocytes and activated CD4+ T cells (for both gag and tat, P < 0.001 for resting cells compared to activated cells and monocytes; for tat, P = 0.984 for activated cells compared to monocytes; for gag, P < 0.828 for activated cells compared to monocytes).
PCR, sequencing, and sequence analyses.
Cellular DNA and cDNA which had been reverse transcribed from plasma viral RNA with primer PE2 were used to amplify HIV-1 env gp120 sequences using a nested PCR with the outer primers PE0 and PE2 and the inner primers PE1 and P2 (63). Multiple independent PCR products generated from target sources containing 20 to 200 copies of HIV-1 DNA or cDNA of each sample (purified cells or plasma) were cloned and sequenced (63). Twelve to seventeen clone sequences were aligned by using Clustal W (52). Likelihood ratio tests were implemented through MODELTEST (38) and used to derive a maximum-likelihood model (PAUP 4.0; Sinauer Associates, Inc., Sunderland, Mass.) of evolution that statistically fit the data while making the fewest assumptions about the evolution of the sequences themselves. Parameters derived from the best-fit model were applied to the data sets to obtain maximum-likelihood distances in PAUP*. The distances were used to construct neighbor-joining (42) trees from which we calculated the most recent common ancestor (MRCA) for the entire ingroup of each patient. We then calculated the distances to the MRCA for each sequence and divided these distances into two groups, those from MRCA to sample I and those from MRCA to sample II. A t test was performed to compare the mean distances between the MRCA and sequences from samples I and II.
Statistical analyses.
Estimates of decay slopes of proviral DNA were obtained with linear random effects regression models (12) of log-transformed data beginning at the time the patient started HAART. Cell-associated HIV-1 DNA levels prior to the initiation of treatment were used as the DNA levels at treatment start for individuals who did not have these measurements on the day of treatment initiation. The associated coefficient of time covariant provided an estimate of the mean decay slope, while individual decay slopes were estimated using empirical Bayes methods (12). All comparisons of means (DNA, mRNA, and ratios) and decay slopes (DNA) were made with generalized estimating equations (12) to account for correlations which could arise for different cell types sampled from the same individual and repeated sampling of individuals over time.
Nucleotide sequence accession numbers.
The nucleotide sequences reported in this paper have been submitted to GenBank and were given accession numbers AF405731 to AF406313.
RESULTS
Decay rate of HIV-1 DNA in CD14+ monocytes and activated and resting CD4+ T cells.
We used negative selection and FACS to isolate monocytes (CD14+), activated CD4+ T cells (CD69+, CD25+, CD38+, and HLA-DR+), and resting CD4+ T cells (CD69−, CD25−, CD38−, and HLA-DR−). As shown in Fig. 1, these cells were highly pure and free from contamination with the other two cell populations. HIV-1 DNA was detected in all three cell types throughout the study (Fig. 2 and Table 2). The mean numbers of copies of HIV-1 DNA before the initiation of treatment were 272 ± 26.2 per 106 CD14+ monocytes, 1,247 ± 115.1 per 106 activated CD4+ T cells, and 1,286 ± 102.7 per 106 resting CD4+ T cells (P < 0.01 for the comparison of monocytes versus activated or resting CD4+ T cells). HIV-1 DNA levels decreased over the course of therapy in all three cell compartments. Previous studies have shown an initial fast decay of HIV-1 in a variety of sample types (CD4+ T cells, resting CD4+ T cells, bulk PBMC, and/or blood plasma) that was followed by a much slower and varying decay after the initiation of HAART (4, 17, 22, 35, 36, 55). We were not able to define a similar decay in the three cell populations during early treatment because of the inability to access large volumes of PBMC via leukapheresis at such frequent time points. We compared the decay of HIV-1 DNA in these three cell compartments between samples I and II and found that the viral decay was significantly slower in CD14+ monocytes (mean decay rate of −0.0195 [range, −0.0260 to −0.0130] log10 per month) and resting CD4+ T cells (−0.0191 [−0.0290 to −0.00092]) than in activated CD4+ T cells (−0.0380 [−0.0490 to −0.0271]) during the first 2 years of treatment. We then focused on the decay of HIV-1 DNA in CD14+ monocytes and activated and resting CD4+ T cells after patients had attained less than 50 RNA copies per ml of plasma during HAART. We estimated the decay rate in each cell compartment by using only HIV-1 DNA copies of samples II to IV (Table 1) from each patient. As shown in Fig. 2 and Table 2, there was a significant variation in the viral decay rate in all three cell compartments and among individual patients, ranging from −0.0193 log10 HIV-1 DNA copies per month (half-life [t1/2], 15.6 months) to +0.0058 (t1/2, infinite) for CD14+ monocytes (Fig. 2A), −0.0281/month (t1/2, 10.7 months) to 0.0001 (t1/2, infinity) for resting CD4+ T cells (Fig. 2B), and −0.0239 (t1/2, 12.6 months) to −0.0045/month (t1/2, 67 months) for activated CD4+ T cells (Fig. 2C). The mean decay rate of HIV-1 DNA in CD14+ monocytes (−0.0073; 95% confidence interval [CI], −0.0168 to +0.0022) was significantly lower than that estimated in the activated CD4+ T cells (−0.0152; CI, −0.0253 to −0.0050) (P = 0.0002). No significant differences were seen in the rate of viral decay between CD14+ monocytes and resting CD4+ T cells (−0.0128; CI, −0.0240 to −0.0015) (P = 0.668) (Table 2). The corresponding estimated t1/2 of HIV-1 DNA were 41.3 months in CD14+ monocytes, 23.6 months in resting CD4+ T cells, and 19.8 months in activated CD4+ T cells (Table 2). We could not determine the impact of intermittent episodes of plasma viremia on viral decay (38a), because all patients except one (patient 7) had at least one documented episode of low-level viremia (<100 HIV RNA copies/ml).
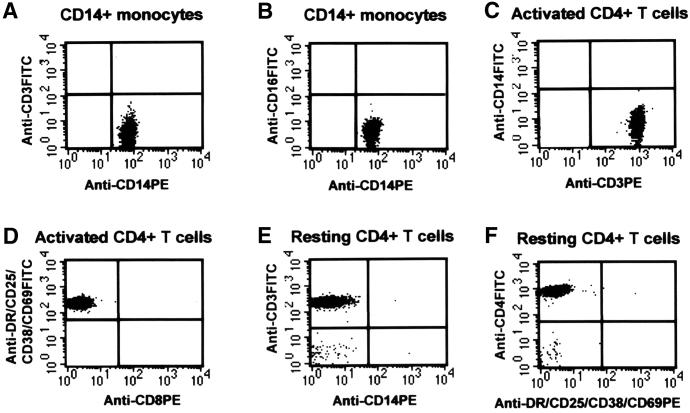
FIG. 1.
Representative two-color fluorescence analysis of enriched CD14+ monocytes (A and B) and activated (C and D) and resting (E and F) CD4+ T cells. Purified cells were stained either with unconjugated MAb (negative control) or with fluorescein isothiocyanate (FITC)- or phycoerythrin (PE)-conjugated MAb. The purity of CD14+ monocytes obtained by using a combination of negative selection and FACS (see Materials and Methods) was 98.31 to 99.99% (mean ± standard deviation, 99.37% ± 0.58%). There was no contamination of T cells (A) or B cells (B). Activated CD4+ T cells, also purified by both negative selection and FACS for the activation markers HLA-DR, CD25, CD38, and CD69, showed a purity of 98.02 to 99.95% (99.21% ± 0.63%) without CD14+ monocytes (C) and resting CD4+ T cells (D). Resting CD4+ T cells were isolated by negative selection with a 6-day incubation as described previously (7, 10, 15). These cells had undetectable (below the negative control) contamination with CD14+ monocytes (E) and activated CD4+ T cells (F). The unstained cells in the left bottom corner of panels E and F are red blood cells.
HIV-1 transcription activity in CD14+ monocytes and activated and resting CD4+ T cells.
The apparent half-lives of HIV-1-infected cells from our patients (Table 2) were much longer than the estimated mean intermitotic life spans (21, 22, 35, 36, 54–56) of monocytes/macrophages (41.3 months versus 14 days) and activated (19.8 months versus 2 days) and resting memory (23.6 versus 6 months) CD4+ T cells, suggesting that these reservoirs may be renewed as a result of continued viral replication. We then examined HIV-1 transcription activity by assessing the levels of cell-associated MS (tat) and US (gag) viral mRNA in samples II and/or III. HIV-1 mRNA in samples II and III, taken after plasma virus had been undetectable for 301 to 1,028 days (Table 1), would indicate ongoing HIV-1 transcription in vivo (17, 32, 43). We detected both MS and US HIV-1 mRNA in all three cell populations (Fig. 3). The mean concentrations of gag and tat mRNA showed significant differences between the three cell populations (Fig. 3A and B). We also estimated HIV-1 transcriptional activity by measuring the ratio between viral DNA and RNA in these three cell populations. The mean mRNA/DNA ratios of tat and gag for CD14+ monocytes were similar to that for activated CD4+ T cells and were significantly higher than that for resting CD4+ T cells (Fig. 3C and D), indicating higher levels of viral transcription in CD14+ monocytes and activated CD4+ T cells than in resting CD4+ T cells.
HIV-1 sequence evolution in CD14+ monocytes and activated and resting CD4+ T cells.
While the above data indicate ongoing viral transcriptional activity, the production of infectious virus could still be blocked at assembly by the protease inhibitor included in HAART. We therefore evaluated HIV-1 sequence evolution in all three cell populations, since mutational changes accumulate as a result of completed rounds of viral replication in vivo. As shown in Table 3, the genetic distances from the deduced MRCA to sequences in sample II were longer than those for sample I in most patients (Table 3), suggesting sequence evolution that varied by patient (8, 60, 61) and by cell population. Four of seven patients had minor sequence evolution over the course of follow-up; three others (patients 1, 6, and 7) exhibited significant sequence evolution. When HIV-1 sequences in CD14+ monocytes from all seven patients were analyzed together, we found a significant difference between the mean genetic distances from MRCA to sample I (mean, 0.43) and from MRCA to sample II (0.68) (P = 0.02). For activated and resting CD4+ T cells, the mean distances from MRCA to sample II (0.70 for activated CD4+ T cells and 0.54 for resting CD4+ T cells) tended to be longer than those from MRCA to sample I (0.51 for activated and 0.50 for resting CD4+ T cells). However, the sequence evolution in the two CD4+-T-cell populations was not statistically significant (P = 0.08 for activated CD4+ T cells; P = 0.45 for resting CD4+ T cells).
TABLE 3.
Analysis of HIV-1 genetic distances in patients receiving HAARTa
| Patient | Cell type | Mean genetic distance (%) from MRCA to:
|
P value | Continued evolution | |
|---|---|---|---|---|---|
| Sample I | Sample II | ||||
| 1 | Monocytes | 0.51 | 1.24 | 0.0309 | Yes |
| Activated T cells | 0.91 | 2.01 | 0.0074 | Yes | |
| Resting T cells | 0.44 | 0.63 | 0.4465 | No | |
| 2 | Monocytes | 0.75 | 0.84 | 0.4841 | No |
| Activated T cells | 0.75 | 0.75 | 0.5035 | No | |
| Resting T cells | 0.78 | 0.82 | 0.2138 | No | |
| 3 | Monocytes | 0.21 | 0.25 | 0.3547 | No |
| Activated T cells | 0.33 | 0.26 | 0.3465 | No | |
| Resting T cells | 0.24 | 0.25 | 0.8285 | No | |
| 4 | Monocytes | ND | ND | ND | ND |
| Activated T cells | ND | ND | ND | ND | |
| Resting T cells | 0.60 | 0.57 | 0.5089 | No | |
| 5 | Monocytes | 0.28 | 0.28 | 0.9897 | No |
| Activated T cells | 0.20 | 0.25 | 0.4048 | No | |
| Resting T cells | 0.20 | 0.21 | 0.6969 | No | |
| 6 | Monocytes | 0.64 | 0.68 | 0.3782 | No |
| Activated T cells | 0.64 | 0.66 | 0.6914 | No | |
| Resting T cells | 0.63 | 0.71 | 0.0059 | Yes | |
| 7 | Monocytes | 0.20 | 0.77 | ≤0.0001 | Yes |
| Activated T cells | 0.25 | 0.28 | 0.4215 | No | |
| Resting T cells | 0.44 | 0.63 | 0.4465 | No | |
ND, not done.
Comparison of HIV-1 sequences in plasma and purified cells: evidence for production of HIV-1 virions from provirus in CD14+ monocytes.
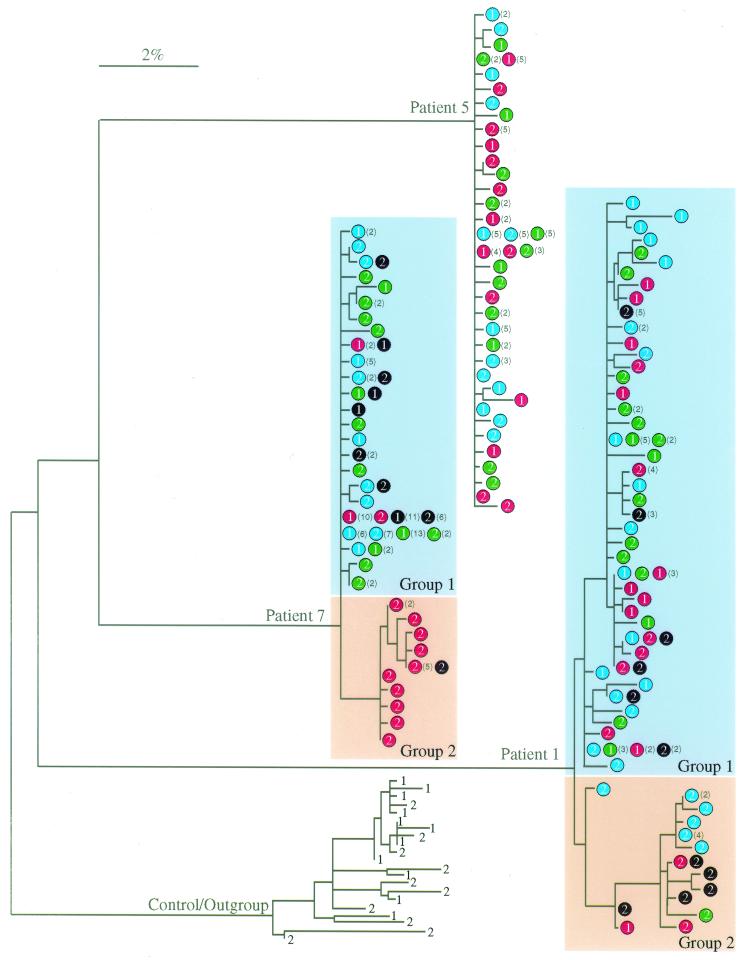
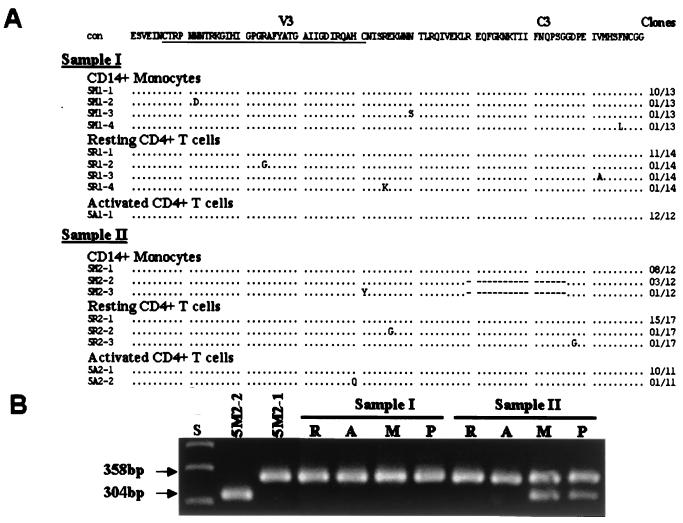
We were able to detect low levels of plasma viremia (4 to 30 HIV-1 RNA copies/ml of plasma) at the time corresponding to sample II or III (Table 1), indicating ongoing production of HIV-1 in vivo after prolonged HAART (13). We then compared HIV-1 env sequences from plasma with proviral sequences obtained from purified cell populations. In patients 2, 3, 4, and 6, HIV-1 sequences in plasma and in all three cell types were homogenous and indistinguishable (data not shown) within each individual. However, there were two distinct but associated groups of sequences in patient 1 (Fig. 4). Group 1 encompassed the major variant populations found in all cell compartments. Group 2 sequences were largely observed in sample II (639 days posttherapy) and were closely related to a variant that was derived from CD14+ monocytes of sample I (26 days before therapy) (Fig. 4). Sequences similar to this monocyte-associated lineage were not detected in activated or resting CD4+ T cells of sample I by sequencing of additional clones or by screening of PCR products with a quantitative homoduplex tracking assay capable of detecting minor variants (63). However, both groups of sequences were observed in blood plasma at a time point corresponding to sample II, suggesting that both groups of proviruses were able to produce virus. In patient 7, HIV-1 sequences were homogenous before therapy (sample I) and remained so in resting and activated CD4+ T cells after 464 days of therapy (sample II), whereas new variants with significant evolution (Table 3) were seen in CD14+ monocytes of sample II as well as in plasma 2 months after sample II (group 2 of patient 7 in Fig. 4). In patient 5, two groups of variants with and without a 54-bp deletion in HIV-1 region C3 (clones 5 M2-2 and 5 M2-1, respectively, in Fig. 5) were identified in CD14+ monocytes of sample II (560 days posttherapy). We then performed additional PCR studies to assess the representation of virus with the 54-bp deletion in all three cell populations and plasma. The sequence populations without the 54-bp deletion (5 M2-1-like, 358 bp) (Fig. 5B) were detected in all cells and plasma, whereas the sequences with the 54-bp deletion (5 M2-2-like, 304 bp) (Fig. 5B) were detected only in CD14+ monocytes and plasma at day 560 posttherapy (sample II). These results are consistent with the production of HIV-1 from CD14+ monocytes under suppressive HAART.
FIG. 4.
Phylogenetic tree analysis of HIV-1 envelope sequences in CD14+ monocytes (red circles), activated (blue circles) and resting CD4+ T cells (green circles), and plasma (black circles) of patients receiving HAART. The number within each circle is the sample number (i.e., 1 and 2 indicate samples I and II). Numbers in parentheses are the numbers of sequences that were identical in the sample to the left. The outgroup is a patient (46) who was not on therapy but whose samples were obtained over a time frame similar to that of the ingroup. Numbers on these sequences are relative sampling times. The bar indicates genetic distance.
FIG. 5.
Comparison of HIV-1 V3-C3 env gp120 sequences in CD14+ monocytes, activated and resting CD4+ T cells, and blood plasma from patient 5. (A) Alignment of deduced amino acid sequences in CD14+ monocytes and activated and resting CD4+ T cells. The consensus sequence (con) is at the top, with the V3 loop underlined. Dots indicate amino acids identical to the consensus sequence; dashes indicate sequence deletions. The numbers of clones with identical sequences are shown on the right. (B) PCR analysis of HIV-1 V3-C3 sequences in CD14+ monocytes (M), activated (A) and resting (R) CD4+ T cells, and blood plasma (P). 5 M2-2 is a minor variant population with the 54-bp deletion; 5 M2-1 is the major population of sequences present in all cell populations and plasma samples. S, size markers.
DISCUSSION
Previous in vitro studies showed that HIV-1 replication in freshly isolated blood monocytes and resting CD4+ T cells was blocked prior to the completion of reverse transcription and integration (50, 59). However, a recent study showed that treating, but not activating, resting CD4+ T cells with the cytokines interleukin 2 (IL-2), IL-4, IL-7, and IL-15 was able to overcome this block, resulting in HIV-1 replication in resting CD4+ T cells (53). Thus, it is likely that replication of HIV-1 in vivo occurs in resting T cells that are exposed to cytokines at sites of infection or in tissues (53, 62). Whether cytokines similarly render monocytes susceptible to HIV-1 infection in vivo is unknown. Since the original presentation of these results, Sonza et al. have shown that HIV-1 can be isolated when patient’s monocytes are differentiated into monocyte-derived macrophages (51). Our studies were conducted on freshly isolated patient CD14+ monocytes without adherence-induced differentiation, which is known to alter the susceptibility of mononuclear phagocytes to HIV-1 replication (11, 26, 30, 31, 39, 50, 51). Our findings indicate that HIV-1 replicates in CD14+ monocytes in vivo, even in patients receiving HAART. Our studies also confirm that HIV-1 replication can occur in activated and to lesser extent in resting CD4+ T cells in patients undergoing suppressive HAART (62).
A recent study concluded that in five of eight patients who discontinued HAART, a rebound HIV-1 in plasma could have resulted from the activation of virus in resting CD4+ T cells (60). Another study showed that HIV-1 reservoirs other than resting CD4+ T cells could prompt the emergence of plasma virus (8). In three of our seven patients, the viral populations that were closely related or identical to those detected either initially (patients 1 and 7) (Fig. 4) or only (patient 5) (Fig. 5) in CD14+ monocytes were seen in plasma after prolonged HAART, indicating that CD14+ monocytes may serve as a potentially important source of HIV-1 in patients taking HAART. This hypothesis is supported by our findings of higher levels of HIV-1 transcripts and sequence evolution in CD14+ monocytes than in resting CD4+ T cells, which suggests a higher level of HIV-1 replication in CD14+ monocytes than in resting CD4+ T cells. The differences in both HIV-1 DNA and RNA between the three cell populations, the consistency between the DNA and RNA data, and the independently performed sequence analyses confirm our findings and verify the lack of “contamination” within the laboratory as an explanation of our results. While we demonstrate CD14+ monocytes as a potential reservoir of HIV-1 replication in patients on HAART, it is possible that other reservoirs, especially in tissues (18, 24, 25, 27, 33, 59), may contribute to the persistence of HIV-1. In four other patients, HIV-1 sequence populations in plasma were identical to those isolated from resting CD4+ T cells (8, 59) which were also identical to viral sequences from CD14+ monocytes and activated CD4+ T cells, suggesting that not only resting CD4+ T cells (60) but also CD14+ monocytes and activated CD4+ T cells are potential viral sources in these patients.
Although the apparent long half-life of HIV-1-infected CD14+ monocytes appears to be similar to that in resting CD4+ T cells, HIV-1 could turn over at a higher rate in CD14+ monocytes than in resting CD4+ T cells in the presence of HAART. Given the fact that monocytes may circulate in peripheral blood for only a few days before differentiating to macrophages in tissues (29), the persistence of HIV-1 in blood monocytes itself suggests ongoing virus replication and/or recent infection in monocytes. Our findings of more evident HIV-1 replication in CD14+ monocytes suggest that the HIV-1 pool in monocytes could be renewed, as a result of viral replication, more frequently in CD14+ monocytes than in resting CD4+ T cells. However, the viral pool in CD14+ monocytes, as well as in resting CD4+ T cells, could also be renewed by virus produced from activated CD4+ T cells. The source of HIV-1 in blood monocytes and the role they play in the overall pool of HIV-1 replication remain to be defined. It is possible that infected monocytes produce relatively small amounts of virus but are a major carrier of virus into tissue sites where tissue macrophages may produce virus. Hence, blood monocytes may serve as an indirect source of HIV-1. One potential explanation for pronounced viral replication in CD14+ monocytes is that antiretroviral drugs may not block viral replication in monocytes/macrophages as efficiently as in CD4+ T cells (1, 2, 24, 37). As reported elsewhere (26), we have not found evidence for the evolution of drug resistance in patients taking HAART (T. Zhu et al., unpublished data). The establishment of HIV-1 infection in CD14+ monocytes during primary infection and the ongoing viral replication in CD14+ monocytes as well as in CD4+ T cells and other tissues constitute the major problem of HIV-1 eradication. Therapies with greater potency against viral production in monocytes may provide more complete suppression of HIV-1.
Acknowledgments
We are grateful to K. Diem, M. Berrey, T. Shea, L. Stensland, H. Liu, and E. Peterson for assistance.
We acknowledge financial support from NIH grants (AI41535, AI45206, AI35605, AI45402, and AI 49109).
REFERENCES
- 1.Aquaro, S., C. F. Perno, E. Balestra, J. Balzarini, A. Cenci, A. Bertoli, M. Francesconi, S. Panti, F. Serra, N. Villani, and R. Calio. 1997. Inhibition of replication of HIV in primary monocyte/macrophages by different antiviral drugs and comparative efficacy in lymphocytes. J. Leukoc. Biol. 62:138–143. [DOI] [PubMed] [Google Scholar]
- 2.Aquaro, S., R. Calio, E. Balestra, P. Bagnarelli, A. Cenci, A. Bertoli, B. Tavazzi, D. DiPierro, M. Francesconi, D. Abdelahad, and C. F. Perno. 1998. Clinical implications of HIV dynamics and drug resistance in macrophages. J. Biol. Regul. Homeost. Agents 12(Suppl. 1–2):23–27. [PubMed] [Google Scholar]
- 3.Berrey, M. M., T. Schacker, A. C. Collier, T. Shea, S. J. Brodie, D. Mayers, R. Coombs, J. Krieger, T. W. Chun, A. S. Fauci, S. G. Self, and L. Corey. 2001. Treatment of primary human immunodeficiency virus type 1 infection with potent antiretroviral therapy reduces frequency of rapid progression to AIDS. J. Infect. Dis. 183:1466–1475. [DOI] [PubMed] [Google Scholar]
- 4.Blankson, J. N., D. Finzi, T. C. Pierson, B. P. Sabundayo, K. Chadwick, J. B. Margolick, T. C. Quinn, and R. F. Siliciano. 2000. Biphasic decay of latently infected CD4+ T cells in acute human immunodeficiency virus type 1 infection. J. Infect. Dis. 182:1636–1642. [DOI] [PubMed] [Google Scholar]
- 5.Brodie, S. J., B. K. Patterson, D. A. Lewinsohn, K. Diem, D. Spach, P. D. Greenberg, S. R. Riddell, and L. Corey. 2000. HIV-specific cytotoxic T lymphocytes traffic to lymph nodes and localize at sites of HIV replication and cell death. J. Clin. Investig. 105:1407–1417. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Cavert, W., D. W. Notermans, K. Staskus, S. W. Wietgrefe, M. Zupancic, K. Gebhard, K. Henry, Z. Q. Zhang, R. Mills, H. McDade, C. M. Schuwirth, J. Goudsmit, S. A. Danner, and A. T. Haase. 1997. Kinetics of response in lymphoid tissues to antiretroviral therapy of HIV-1 infection. Science 276:960–964. [DOI] [PubMed] [Google Scholar]
- 7.Chun, T. W., L. Carruth, D. Finzi, X. Shen, J. A. DiGiuseppe, H. Taylor, M. Hermankova, K. Chadwick, J. Margolick, T. C. Quinn, Y. H. Kuo, R. Brookmeyer, M. A. Zeiger, P. Barditch-Crovo, and R. F. Siliciano. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183–188. [DOI] [PubMed] [Google Scholar]
- 8.Chun, T. W., R. T. Davey, Jr., M. Ostrowski, J. S. Justement, D. Engel, J. I. Mullins, and A. S. Fauci. 2000. Relationship between pre-existing viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat. Med. 6:757–761. [DOI] [PubMed] [Google Scholar]
- 9.Chun, T. W., D. Engel, M. M. Berrey, T. Shea, L. Corey, and A. S. Fauci. 1998. Early establishment of a pool of latently infected, resting CD4(+) T cells during primary HIV-1 infection. Proc. Natl. Acad. Sci. USA 95:8869–8873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chun, T. W., L. Stuyver, S. B. Mizell, L. A. Ehler, J. A. Mican, M. Baseler, A. L. Lloyd, M. A. Nowak, and A. S. Fauci. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94:13193–13197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collman, R., N. F. Hassan, R. Walker, B. Godfrey, J. Cutilli, J. C. Hastings, H. Friedman, S. D. Douglas, and N. Nathanson. 1989. Infection of monocyte-derived macrophages with human immunodeficiency virus type 1 (HIV-1): monocyte-tropic and lymphocyte-tropic strains of HIV-1 show distinctive patterns of replication in a panel of cell types. J. Exp. Med. 170:1149–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diggle, P. J., K. Y. Liang, and S. L. Zeger. 1994. Analysis of longitudinal data. Oxford University Press, Oxford, United Kingdom.
- 13.Dornadula, G., H. Zhang, B. VanUitert, J. Stern, Jr., L. Livornese, M. J. Ingerman, J. Witek, R. J. Kedanis, J. Natkin, J. DeSimone, and R. J. Pomerantz. 1999. Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA 282:1627–1632. [DOI] [PubMed] [Google Scholar]
- 14.Ercolani, L., B. Florence, M. Denaro, and M. Alexander. 1988. Isolation and complete sequence of a functional human glyceraldehyde-3-phosphate dehydrogenase gene. J. Biol. Chem. 263:15335–15341. [PubMed] [Google Scholar]
- 15.Finzi, D., J. Blankson, J. D. Siliciano, J. B. Margolick, K. Chadwick, T. Pierson, K. Smith, J. Lisziewicz, F. Lori, C. Flexner, T. C. Quinn, R. E. Chaisson, E. Rosenberg, B. Walker, S. Gange, J. Gallant, and R. F. Siliciano. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512–517. [DOI] [PubMed] [Google Scholar]
- 16.Finzi, D., M. Hermankova, T. Pierson, L. M. Carruth, C. Buck, R. E. Chaisson, T. C. Quinn, K. Chadwick, J. Margolick, R. Brookmeyer, J. Gallant, M. Markowitz, D. D. Ho, D. D. Richman, and R. F. Siliciano. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. [DOI] [PubMed] [Google Scholar]
- 17.Furtado, M. R., D. S. Callaway, J. P. Phair, K. J. Kunstman, J. L. Stanton, C. A. Macken, A. S. Perelson, and S. Wolinsky. 1999. Persistence of HIV-1 transcription in peripheral-blood mononuclear cells in patients receiving potent antiretroviral therapy. N. Engl. J. Med. 340:1614–1622. [DOI] [PubMed] [Google Scholar]
- 18.Gartner, S., P. Markovits, D. M. Markovitz, M. H. Kaplan, R. C. Gallo, and M. Popovic. 1986. The role of mononuclear phagocytes in HTLV-III/LAV infection. Science 233:215–219. [DOI] [PubMed] [Google Scholar]
- 19.Gulick, R. M., J. W. Mellors, D. Havlir, J. J. Eron, C. Gonzalez, D. McMahon, D. D. Richman, F. T. J. L. Valentine, A. Meibohm, E. A. Emini, and J. A. Chodakewitz. 1997. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N. Engl. J. Med. 337:734–739. [DOI] [PubMed] [Google Scholar]
- 19a.Gunthard, H. F., S. D. Frost, A. J. Leigh-Brown, C. C. Ignacio, K. Kee, A. S. Perelson, C. A. Spina, D. V. Havlir, M. Hezareh, D. J. Looney, D. D. Richman, and J. K. Wong. 1999. Evolution of envelope sequences of human immunodeficiency virus type 1 in cellular reservoirs in the setting of potent antiviral therapy. J. Virol. 73:9404–9412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hammer, S. M., K. E. Squires, M. D. Hughes, J. M. Grimes, L. M. Demeter, J. S. Currier, Jr., J. J. Eron, J. E. Feinberg, Jr., H. H. Balfour, L. R. Deyton, J. A. Chodakewitz, and M. A. Fischl. 1997. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N. Engl. J. Med. 337:725–733. [DOI] [PubMed] [Google Scholar]
- 21.Hellerstein, M., M. B. Hanley, D. Cesar, S. Siler, C. Papageorgopoulos, E. Wieder, D. Schmidt, R. Hoh, R. Neese, D. Macallan, S. Deeks, and J. M. McCune. 1999. Directly measured kinetics of circulating T lymphocytes in normal and HIV-1-infected humans. Nat. Med. 5:83–89. [DOI] [PubMed] [Google Scholar]
- 22.Ho, D. D., A. U. Neumann, A. S. Perelson, W. Chen, J. M. Leonard, and M. Markowitz. 1995. Rapid turnover of plasma virions and CD4 lymphocytes in HIV-1 infection. Nature 373:123–126. [DOI] [PubMed] [Google Scholar]
- 23.Ho, D. D., T. R. Rota, and M. S. Hirsch. 1986. Infection of monocyte/macrophages by human T lymphotropic virus type III. J. Clin. Investig. 77:1712–1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Igarashi, T., C. R. Brown, Y. Endo, A. Buckler-White, R. Plishka, N. Bischofberger, V. Hirsch, and M. A. Martin. 2001. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): implications for HIV-1 infections of humans. Proc. Natl. Acad. Sci. USA 98:658–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koenig, S., H. E. Gendelman, J. M. Orenstein, M. C. Dal Canto, G. H. Pezeshkpour, M. Yungbluth, F. Janotta, A. Aksamit, M. A. Martin, and A. S. Fauci. 1986. Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science 233:1089–1093. [DOI] [PubMed] [Google Scholar]
- 26.Lambotte, O., Y. Taoufik, M. G. de Goer, C. Wallon, C. Goujard, and J. F. Delfraissy. 2000. Detection of infectious HIV in circulating monocytes from patients on prolonged highly active antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 23:114–119. [DOI] [PubMed] [Google Scholar]
- 26a.Lewin, S. R., M. Vesanen, L. Kostrikis, A. Hurley, M. Duran, L. Zhang, L., D. D. Ho, and M. Markowitz. 1999. Use of real-time PCR and molecular beacons to detect virus replication in human immunodeficiency virus type 1-infected individuals on prolonged effective antiretroviral therapy. J. Virol. 73:6099–6103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McElrath, M. J., J. E. Pruett, and Z. A. Cohn. 1989. Mononuclear phagocytes of blood and bone marrow: comparative roles as viral reservoirs in human immunodeficiency virus type 1 infection. Proc. Natl. Acad. Sci. USA 86:675–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McElrath, M. J., R. M. Steinman, and Z. A. Cohn. 1991. Latent HIV-1 infection in enriched populations of blood monocytes and T cells from seropositive patients. J. Clin. Investig. 87:7–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meuret, G., J. Bammert, and G. Hoffmann. 1974. Kinetics of human monocytopoiesis. Blood 44:801–816. [PubMed] [Google Scholar]
- 30.Naif, H. M., S. Li, M. Alali, J. Chang, C. Mayne, J. Sullivan, and A. L. Cunningham. 1999. Definition of the stage of host cell genetic restriction of replication of human immunodeficiency virus type 1 in monocytes and monocyte-derived macrophages by using twins. J. Virol. 73:4866–4881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Naif, H. M., S. Li, M. Alali, A. Sloane, L. Wu, M. Kelly, G. Lynch, A. Lloyd, and A. L. Cunningham. 1998. CCR5 expression correlates with susceptibility of maturing monocytes to human immunodeficiency virus type 1 infection. J. Virol. 72:830–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Natarajan, V., M. Bosche, J. A. Metcalf, D. J. Ward, H. C. Lane, and J. A. Kovacs. 1999. HIV-1 replication in patients with undetectable plasma virus receiving HAART. Lancet 353:119–120. [DOI] [PubMed] [Google Scholar]
- 33.Orenstein, J. M., C. Fox, and S. M. Wahl. 1997. Macrophages as a source of HIV during opportunistic infections. Science 276:1857–1861. [DOI] [PubMed] [Google Scholar]
- 34.Palella, F. J., Jr., K. M. Delaney, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, and S. D. Holmberg. 1998. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. N. Engl. J. Med. 338:853–860. [DOI] [PubMed] [Google Scholar]
- 35.Perelson, A. S., P. Essunger, Y. Cao, M. Vesanen, A. Hurley, K. Saksela, M. Markowitz, and D. D. Ho. 1997. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 387:188–191. [DOI] [PubMed] [Google Scholar]
- 36.Perelson, A. S., A. U. Neumann, M. Markowitz, J. M. Leonard, and D. D. Ho. 1996. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science 271:1582–1586. [DOI] [PubMed] [Google Scholar]
- 37.Perno, C. F., F. M. Newcomb, D. A. Davis, S. Aquaro, R. W. Humphrey, R. Calio, and R. Yarchoan. 1998. Relative potency of protease inhibitors in monocytes/macrophages acutely and chronically infected with human immunodeficiency virus. J. Infect. Dis. 178:413–422. [DOI] [PubMed] [Google Scholar]
- 38.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817–818. [DOI] [PubMed] [Google Scholar]
- 38a.Ramratnam, B., J. E. Mittler, L. Zhang, D. Boden, A. Hurley, F. Fang, C. A. Macken, A. S. Perelson, M. Markowitz, and D. D. Ho. 2000. The decay of the latent reservoir of replication-competent HIV-1 is inversely correlated with the extent of residual viral replication during prolonged anti-retroviral therapy. Nat. Med. 6:82–85. [DOI] [PubMed] [Google Scholar]
- 39.Rich, E. A., I. S. Chen, J. A. Zack, M. L. Leonard, and W. A. O’Brien. 1992. Increased susceptibility of differentiated mononuclear phagocytes to productive infection with human immunodeficiency virus-1 (HIV-1). J. Clin. Investig. 89:176–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rodrigo, A. G., P. C. Goracke, K. Rowhanian, and J. I. Mullins. 1997. Quantitation of target molecules from polymerase chain reaction-based limiting dilution assays. AIDS Res. Hum. Retrovir. 13:737–742. [DOI] [PubMed] [Google Scholar]
- 41.Saiki, R. K., D. H. Gelfand, S. Stoffel, S. J. Scharf, R. Higuchi, G. T. Horn, K. B. Mullis, and H. A. Erlich. 1988. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239:487–491. [DOI] [PubMed] [Google Scholar]
- 42.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406–425. [DOI] [PubMed] [Google Scholar]
- 43.Saksela, K., C. E. Stevens, P. Rubinstein, and D. Baltimore. 1995. HIV-1 messenger RNA in peripheral blood mononuclear cells as an early marker of risk for progression to AIDS. Ann. Intern. Med. 123:641–648. [DOI] [PubMed] [Google Scholar]
- 44.Schacker, T. W., J. P. Hughes, T. Shea, R. W. Coombs, and L. Corey. 1998. Biological and virologic characteristics of primary HIV infection. Ann. Intern. Med. 128:613–620. [DOI] [PubMed] [Google Scholar]
- 45.Schrier, R. D., J. A. McCutchan, and C. A. Wiley. 1993. Mechanisms of immune activation of human immunodeficiency virus in monocytes/macrophages. J. Virol. 67:5713–5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shankarappa, R., J. B. Margolick, S. J. Gange, A. G. Rodrigo, D. Upchurch, H. Farzadegan, P. Gupta, C. R. Rinaldo, G. H. Learn, X. He, X. L. Huang, and J. I. Mullins. 1999. Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J. Virol. 73:10489–10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharkey, M. E., I. Teo, T. Greenough, N. Sharova, K. Luzuriaga, J. L. Sullivan, R. P. Bucy, L. G. Kostrikis, A. Haase, C. Veryard, R. E. Davaro, S. H. Cheeseman, J. S. Daly, C. Bova, R. T. Ellison III, B. Mady, K. K. Lai, G. Moyle, M. Nelson, B. Gazzard, S. Shaunak, and M. Stevenson. 2000. Persistence of episomal HIV-1 infection intermediates in patients on highly active anti-retroviral therapy. Nat. Med. 6:76–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith, P. D., E. N. Janoff, M. Mosteller-Barnum, M. Merger, J. M. Orenstein, J. F. Kearney, and M. F. Graham. 1997. Isolation and purification of CD14-negative mucosal macrophages from normal human small intestine. J. Immunol. Methods 202:1–11. [DOI] [PubMed] [Google Scholar]
- 49.Smith, P. D., A. F. Suffredini, J. B. Allen, L. M. Wahl, J. E. Parrillo, and S. M. Wahl. 1994. Endotoxin administration to humans primes alveolar macrophages for increased production of inflammatory mediators. J. Clin. Immunol. 14:141–148. [DOI] [PubMed] [Google Scholar]
- 50.Sonza, S., A. Maerz, N. Deacon, J. Meanger, J. Mills, and S. Crowe. 1996. Human immunodeficiency virus type 1 replication is blocked prior to reverse transcription and integration in freshly isolated peripheral blood monocytes. J. Virol. 70:3863–3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sonza, S., H. P. Mutimer, R. Oelrichs, D. Jardine, K. Harvey, A. Dunne, D. F. Purcell, C. Birch, and S. M. Crowe. 2001. Monocytes harbor replication-competent, non-latent HIV-1 in patients on highly active antiretroviral therapy. AIDS 15:17–22. [DOI] [PubMed] [Google Scholar]
- 52.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Unutmaz, D., V. N. KewalRamani, S. Marmon, and D. R. Littman. 1999. Cytokine signals are sufficient for HIV-1 infection of resting human T lymphocytes. J. Exp. Med. 189:1735–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.van Furth, R. 1989. Origin and turnover of monocytes and macrophages. Curr. Top. Pathol. 79:125–150. [PubMed] [Google Scholar]
- 55.Wei, X., S. K. Ghosh, M. E. Taylor, V. A. Johnson, E. A. Emini, P. Deutsch, J. D. Lifson, S. Bonhoeffer, M. A. Nowak, B. H. Hahn, M. S. Saag, and G. M. Shaw. 1995. Viral dynamics in human immunodeficiency virus type 1 infection. Nature 373:117–122. [DOI] [PubMed] [Google Scholar]
- 56.Whitelaw, D. M. 1972. Observations on human monocyte kinetics after pulse labeling. Cell Tissue Kinet. 5:311–317. [DOI] [PubMed] [Google Scholar]
- 57.Wong, J. K., M. Hezareh, H. F. Gunthard, D. V. Havlir, C. C. Ignacio, C. A. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291–1295. [DOI] [PubMed] [Google Scholar]
- 58.Yerly, S., T. V. Perneger, S. Vora, B. Hirschel, and L. Perrin. 2000. Decay of cell-associated HIV-1 DNA correlates with residual replication in patients treated during acute HIV-1 infection. AIDS 14:2805–2812. [DOI] [PubMed] [Google Scholar]
- 59.Zack, J. A., S. J. Arrigo, S. R. Weitsman, A. S. Go, A. Haislip, and I. S. Y. Chen. 1990. HIV-1 entry into quiescent primary lymphocytes: molecular analysis reveals a labile, latent viral structure. Cell 61:213–222. [DOI] [PubMed] [Google Scholar]
- 60.Zhang, L., C. Chung, B. S. Hu, T. He, Y. Guo, A. J. Kim, E. Skulsky, X. Jin, A. Hurley, B. Ramratnam, M. Markowitz, and D. D. Ho. 2000. Genetic characterization of rebounding HIV-1 after cessation of highly active antiretroviral therapy. J. Clin. Investig. 106:839–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, L., B. Ramratnam, K. Tenner-Racz, Y. He, M. Vesanen, S. Lewin, A. Talal, P. Racz, A. S. Perelson, B. T. Korber, M. Markowitz, and D. D. Ho. 1999. Quantifying residual HIV-1 replication in patients receiving combination antiretroviral therapy. N. Engl. J. Med. 340:1605–1613. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, Z., T. Schuler, M. Zupancic, S. Wietgrefe, K. A. Staskus, K. A. Reimann, T. A. Reinhart, M. Rogan, W. Cavert, C. J. Miller, R. S. Veazey, D. Notermans, S. Little, S. A. Danner, D. D. Richman, D. Havlir, J. Wong, H. L. Jordan, T. W. Schacker, P. Racz, K. Tenner-Racz, N. L. Letvin, S. Wolinsky, and A. T. Haase. 1999. Sexual transmission and propagation of SIV and HIV in resting and activated CD4+ T cells. Science 286:1353–1357. [DOI] [PubMed] [Google Scholar]
- 63.Zhu, T., N. Wang, A. Carr, D. S. Nam, R. Moor-Jankowski, D. A. Cooper, and D. D. Ho. 1996. Genetic characterization of human immunodeficiency virus type 1 in blood and genital secretions: evidence for viral compartmentalization and selection during sexual transmission. J. Virol. 70:3098–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]