Abstract
The human serum human immunodeficiency virus type 1 (HIV-1)-neutralizing serum 2 (HNS2) neutralizes many primary isolates of different clades of HIV-1, and virus expressing envelope from the same donor, clone R2, is neutralized cross-reactively by HIV-immune human sera. The basis for this cross-reactivity was investigated. It was found that a rare mutation in the proximal limb of variable region 3 (V3), 313-4 PM, caused virus pseudotyped with the R2 envelope to be highly sensitive to neutralization by monoclonal antibodies (MAbs) directed against conformation-sensitive epitopes at the tip of the V3 loop, such as 19b, and moderately sensitive to MAbs against CD4 binding site (CD4bs) and CD4-induced (CD4i) epitopes, soluble CD4 (sCD4), and HNS2. In addition, introduction of this sequence by mutagenesis caused enhanced sensitivity to neutralization by 19b, anti-CD4i MAb, and HNS2 in three other primary HIV-1 envelopes and by anti-CD4bs MAb and sCD4 in one of the three. The 313-4 PM sequence also conferred increased infectivity for CD4+ CCR5+ cells and the ability to infect CCR5+ cells upon all of these four and two of these four HIV-1 envelopes, respectively. Neutralization of R2 by HNS2 was substantially inhibited by the cyclized R2 V3 35-mer synthetic peptide. Similarly, the peptide also had some lesser efficacy in blocking neutralization of R2 by other sera or of neutralization of other primary viruses by HNS2. Together, these results indicate that the unusual V3 mutation in the R2 clone accounts for its uncommon neutralization sensitivity phenotype and its capacity to mediate CD4-independent infection, both of which could relate to immunogenicity and the neutralizing activity of HNS2. This is also the first primary HIV-1 isolate envelope glycoprotein found to be competent for CD4-independent infection.
The capacity to induce broadly cross-reactive neutralizing antibodies against epidemiologically important viral strains is a characteristic of all successful viral vaccines (42). The failure of experimental vaccines intended for prevention of human immunodeficiency virus type 1 (HIV-1) infection to induce such responses is a major concern (29). The objective of inducing broadly cross-reactive neutralizing antibodies against HIV-1 is problematic because of the high sequence variability of the viral envelope proteins and the general resistance of primary isolates to neutralization. With regard to this sequence variability at neutralization epitopes, attempts to identify antigenic groupings of HIV-1 strains based on neutralization by antibodies have achieved surprisingly limited success. There is evidence that clade B and E strains can be distinguished based on sensitivity to neutralizing antibodies, but other groupings based on neutralization responses have not been identified (34). The general resistance of primary isolates to neutralization has been referred to as a global neutralization resistance phenotype, since those strains resist neutralization by human sera, monoclonal antibodies (MAbs) directed at multiple epitopes, and soluble CD4 (sCD4) (38, 40). Increased understanding of the nature of neutralizing antibody responses capable of broadly cross-reactive neutralization of primary isolates should promote efforts to develop an effective vaccine.
The HIV-1-neutralizing serum 2 (HNS2) (2) was prepared from an HIV-1-infected participant in a cohort study conducted at the National Institutes of Health for use as a reference reagent for laboratories conducting neutralizing antibody tests (1, 19). This serum contains antibodies capable of neutralizing a wide variety of primary HIV-1 isolates (10, 11). Previously, we reported the cloning and characterization of envelope genes from peripheral blood mononuclear cells collected from the donor of HNS2 approximately 1 to 2 years before the plasma used for preparation of HNS2 (43). The envelope proteins encoded by these genes were similar to each other in general neutralization sensitivity when expressed on pseudotyped viruses. One of the envelopes, termed R2, was tested more extensively. It cross-reacted in neutralization assays with all tested sera from people infected with clade B strains of HIV-1 and was neutralized by the majority of tested sera from people infected with clade A, C, and F and from some patients infected with clade D or E strains. Although virus pseudotyped with the R2 envelope was only moderately sensitive to neutralization, it was substantially more cross-reactive than the other clade B strains to which it was compared, including laboratory strains and primary isolates. This unusual neutralizing cross-reactivity of R2 may indicate that the envelope expresses epitopes that induced cross-reactive neutralizing antibodies in the donor. The R2 gene sequence is similar in many respects to other clade B HIV-1 envelope gene sequences, including the number and location of potential N-linked glycosylation sites. The V3 region sequence was unique, however, compared to other sequences in the Human Retroviruses and AIDS Database. The predicted amino acid sequence of R2, with the uncommon amino acids underlined, is as follows: CSRPNNNTRKSIPMGPGRAFYTTGQIIGDIRQAHC (amino acids [aa] 301 to 335). Even though other clones from the swarm of clones infecting the same donor may have induced antibodies against neutralization epitopes not represented on R2, the neutralizing cross-reactivity of R2 is of substantial interest. The studies described in this report were conducted in an attempt to elucidate the basis for the neutralizing cross-reactivity of HNS2 and clone R2.
MATERIALS AND METHODS
HIV envelope expression and genome plasmids.
The cloning of the R2 envelope gene into the pSV7d vector has been described previously (43) (GenBank accession no. AF128126). Similar production of envelope clones from patients in the Multicenter AIDS Cohort Study (MACS) and of the T-cell line-adapted (TCLA) MN strain has also been described (37–39, 44, 52; GenBank accession no. AF130388, AF130391, AF130396, and AF130401). Clones of the primary MN strain (MN-P) were produced in a similar manner. Reverse transcription-PCR was used to synthesize envelope genes from RNA extracted from a suspension of the primary virus. John Sullivan, University of Massachusetts, kindly provided the primary virus. One representative clone was selected for use in these studies (GenBank accession no. AF443202). PNL4-3.luc.E-R- was obtained from N. Landau (38). Mutagenesis procedures were carried out using Pfu polymerase (Quick Change Mutagenesis Kit; Stratagene) following the manufacturer’s instructions. The reactions were performed in an automated thermal cycler (Perkin-Elmer model 2400). Mutations were verified by nucleotide sequence analysis, performed using the dideoxy cycle sequencing technique and AmpliTaq FS DNA polymerase, according to the manufacturer’s directions (Applied Biosystems). After the sequencing reaction, the DNA was purified using Centriflex gel filtration cartridges (Advanced Genetic Technologies, Corp., Gaithersburg, Md.). Sequencing gels were run and analyzed using an Applied Biosystems Prism (model 377) DNA sequencer.
Sera and MAbs.
The HNS2 (AIDS Research and Reference Reagent Program [ARRRP] catalog no. 1983) has been described previously (49). Sera were obtained from participants in the MACS, a longitudinal study of the natural history of HIV-1 infection in homosexual men (24, 44). The sera used in this study were from the same MACS participants from whom envelope clones were obtained previously, donors, 3, 4, 6, 8, 9, and 10 (44, 52). The sera were obtained after 3 to 4 years of follow-up of HIV-1 infection diagnosed at entry into the study. A number of MAbs were obtained through the ARRRP (catalog numbers are shown in parentheses): 257-D IV (1510), 268-D IV (1511), IIIB-V3-13 (1727), IIIB-V3-21 (1725), R/V3-50-1 (1289), F105 (857), immunoglobulin G1b12 (IgG1b12) (2640), 4.8D (1756), 17b (4091), 2F5 (1475), and 2G12 (1476); these were provided by J. Laman, D. R. Burton, M. R. Posner, J. E. Robinson, and S. Zolla-Pazner (3–5, 8, 10, 11, 15, 16, 18–22, 25, 28, 30, 35, 36, 41, 48). Other MAbs were provided directly by collaborators in this study, specifically, 418-D, 391/95D and 694/98-D (21), 838-D and 1027-D (20), 19b (3), and 15e (2).
Synthetic peptides.
R2 strain V3 peptides were synthesized using 9-fluorenylmethoxy carbonyl chemistry with N,N-dicyclohexylcarbodiimide-N-methylpyrrolidone/1-hydroxybenzotriazole esters and an automated ABI synthesizer, model 433 (51). The sequences of these peptides were KSIPMGPGRAFYTTGQI (aa 310 to 326) and CSRPNNNTRKSIPMGPGRAFYTTGQIIGDIRQAHC (aa 301 to 335). The mutant R2 (313-4PM/HI, 325Q/D [PM-to-HI mutation at positions 313 and Q-to-D mutation at position 325]) V3 peptide was prepared similarly. Strain 93TH966.8 V3 peptide (sequence: CTRPSNNTRTSTTIGPGQVFYRTGDITGNIRKAYC) was synthesized using the same methods. The peptides were purified using C18, acetonitrile-in-water gradient chromatography with a Waters high-performance liquid chromatograph. Sequences of the purified peptides were verified using an ABI automated sequencer. In general, the peptides were then lyophilized and stored at 4 to 8°C. In one case the peptide was cyclized before lyophilization, as described below. Preparation of a linear MN strain V3 peptide has been described previously (6). Cyclized MN strain 35-mer peptide was obtained from the ARRRP (catalog no. 1841; provided by P. Catasti [7]).
Cyclization of peptides was accomplished by one of two methods. Two different preparations of the R2 V3 35-mer were synthesized. One of these was oxidized before lyophilization. The pH of the fraction of the C18 column eluate containing the peptide was adjusted to 8.0, air was slowly bubbled through it for 12 h, the pH was readjusted to 7.4, and the peptide was lyophilized. This peptide was essentially insoluble in water. To prepare it for use, it was first dissolved in dimethyl sulfoxide (DMSO) and then diluted with medium at 37°C. Under these conditions the peptide appeared to be mostly soluble, although some particulate matter could be seen adhering to the wall of the vial. The other R2 V3 35-mer peptide preparation synthesized and the R2 (313-4PM/HI, 325Q/D) and 93TH966.8 V3 35-mer peptides were cyclized after lyophilization. The lyophilized, noncyclized R2 V3 35-mer was also essentially insoluble in water, while all other peptides tested were soluble in water to at least 10 mg/ml. To obtain cyclized peptides, solutions were prepared from the previously lyophilized R2 and R2 (313-4PM/HI, 325Q/D) V3 35-mers in DMSO (10 mg/ml at 37°C). These solutions were diluted 1:10 in water. The 93TH966.8 peptide was dissolved in water without presolution in DMSO. The pH of each was adjusted to 8.5 with ammonium hydroxide. These solutions were aerated by bubbling air through the solutions for periods of ≥1 h. Following aeration, the pH was adjusted to 7.4 using HCl. A proportion of the R2 V3 35-mer remained insoluble as white particles throughout the postlyophilization cyclization procedure, while all other peptides prepared in this manner remained fully soluble throughout. To obtain an approximate quantitation of the amount of R2 V3 35-mer in solution, the turbidity of the suspension was determined at 480 nm using a spectrophotometer. The spectrophotometer was blanked with a solution of 10% DMSO in water, and a standard curve was produced using slurries of known amounts of the 35-mer peptide suspended in water. The amount of precipitate estimated by turbidity was subtracted from the amount of peptide added at the beginning of the preparation procedure to estimate the amount remaining in solution. The solubility of the R2 V3 35-mer in 10% DMSO-in-water solution at 37°C, pH 7.4, cyclized after or before lyophilization, was estimated to be ≥950, 300 to 850, or ≥950 μg/ml, respectively. Peptides were sterilized by passage through 0.22-μm-pore-size filters prior to use.
The R2 V3 35-mer peptide dissolved in 10% DMSO, which had been aerated for 1 h after lyophilization, was analyzed by high-performance liquid chromatography, using a C18 column and a 5 to 80% acetonitrile-in-water gradient. Four peaks were seen (results not shown). The principal molecular species observed eluted earlier than the unoxidized peptide, consistent with it being monomeric. The other three peaks eluted at progressively higher acetonitrile concentrations, consistent with the inclusion of multimeric forms. Similar profiles were seen on analysis of peptide after aeration but before pH adjustment, as well as after the full process of aeration, pH adjustment, and clarification. Aeration of the peptide solution for 12 h, compared to 1 h, caused no change in the elution profile of the oxidized peptide, indicating that oxidation had been essentially complete within 1 h. The same profile was observed when the peptide was oxidized before lyophilization was analyzed. Thus, the principal molecular species generated during aeration was probably cyclized monomer.
Pseudotyped virus preparation, infectivity, and neutralization assays.
Pseudotyped viruses were prepared by transfection of 293T cells with pNL4-3.luc.E-R and envelope-expressing plasmids, as previously described (38, 44). Infectivity and neutralization assays were carried out using HOS CD4+ CCR5+, HOS CD4+ CXCR4+, HOS CCR5+, or HOS CXCR4+ cells (37–39, 43, 44, 52). Infectivity was determined on the basis of luminescence measured 3 days after infection. Neutralization assays were carried out, as described previously, by preincubation of serial serum or MAb dilutions with pseudotyped viruses prior to cell infection (38, 43, 44). The 90% neutralization endpoints were determined for assays involving human serum, and 50% neutralization endpoints were determined for assays using MAbs. A modified Reed-Muench approach was used in which the endpoint was considered to be the highest dilution at which ≥50% of wells had luminescence <90% or <50% of the nonneutralized control (44). Alternatively, the mean luminescence readings for triplicate wells were determined, and the endpoint was considered to be the last dilution at which the mean results from the test samples were less than 10 or 50% of the nonneutralized control mean. The endpoints obtained using the two methods of calculation (modified Reed-Muench versus comparison of means) were identical in almost every case, and this differences were never more than twofold. Differences in calculated endpoints did not significantly alter the results of comparisons made. The results presented in this report are those obtained using the first method. The blocking effects of synthetic V3 peptides on antibody-mediated neutralization were tested by preincubation of sera and peptides for 1 h prior to use of the sera in serial dilutions in neutralization assays.
Statistical analyses.
Statistical analyses were conducted using Microsoft Excel. Averages, variance, and standard deviations (SDs) were determined using standard functions. The 50% inhibitory doses (ID50s) of sera, MAb, and sCD4 were determined using regression analyses, as follows. Best-fit trend lines were determined for each data set by selecting the method for determining a line equation that resulted in the strongest correlation coefficient (R2 value) over the range of test values under consideration. The resulting line equations were used to determine the calculated values associated with 50% inhibitory effects.
RESULTS
Epitope-specific neutralization of virus pseudotyped with R2 strain envelope.
The sensitivity of the R2 envelope to neutralization by antibodies directed against various epitopes was assessed using MAbs and sCD4. The results of those assays are shown in Table 1 as 50% inhibitory concentrations (concentrations of MAb or sCD4 that inhibited infectivity by 50% or more) (11). The results are shown in comparison to results obtained in similar assays using viruses pseudotyped with envelopes from the MN-TCLA and MN-P strains. Only the 19b antibody inhibited infection of the R2 strain with ≥90% efficiency (not shown). The only anti-V3 antibodies that neutralized R2 with ≥50% efficiency were 19b and 694/98-D. Neutralization of R2 was also observed with two of the three anti-CD4bs MAbs (15e and IgG1b12), with sCD4, with both of the anti-CD4i MAbs tested, and with 2G12 and 2F5. The 2G12 antibody reacts with an undefined epitope on gp120, and the 2F5 antibody reacts with a conserved epitope in gp41. In comparison, the MN-TCLA strain was neutralized by a number of anti-V3 MAbs and by the 19b and 694/98D MAbs at concentrations substantially lower than those required for neutralization of R2. The MN-TCLA strain was also neutralized at concentrations of anti-CD4bs and anti-CD4i MAbs and sCD4 that were substantially less than the concentrations that neutralized R2. The MN strain has a mutation in the 2F5 epitope and is relatively resistant to neutralization by that MAb; it is known to be resistant to neutralization by 2G12 (37–39). The MN-P strain was tested for neutralization by representative antibodies directed against the different envelope regions. It was generally more resistant than R2 to neutralization by these reagents, except for IgG1B12, to which the two strains had similar sensitivities. Overall, these neutralization results indicate that multiple epitopes on R2 are functional targets for neutralization by MAb and sCD4 and that the neutralization sensitivity profile of R2 is intermediate between the highly sensitive MN-TCLA strain and the typically resistant MN-P strain.
TABLE 1.
Neutralization of clone R2-, MN-TCLA-, and MN-P-pseudotyped viruses by MAbs and sCD4a
| Envelope region | Antibody and/or ligand | Epitope | IC50 (μg/ml) for strain
|
||
|---|---|---|---|---|---|
| R2 | MN-TCLA | MN-P | |||
| V3 | 257D-IV | RIHIG | >0.5 | 0.01 | —b |
| 268D-IV | HIGPGR | >0.5 | 0.05 | — | |
| IIIB/V3-13 | IRIQRGPGR | >10 | — | — | |
| IIIB/V3-21 | INCTRPN | >10 | — | — | |
| R/V3-50-1 | RIHIG | >10 | ≤0.01 | — | |
| 19b | XIXXXXGXXFYXT | 1.7 | 1.0 | 10.0 | |
| 391/95D | KRIHIGPGRAFY | >10 | 0.01 | — | |
| 418-D | HIGPGRA | >10 | — | — | |
| 694/98-D | GRAF | 2.5 | 0.01 | >20 | |
| 447-52D | GPGR | >10 | <0.05 | — | |
| 838-D | KISITK | >10 | — | — | |
| 1027-D | KISITKGP | >10 | — | — | |
| CD4bs | F105 | Conformational | >10 | >100 | — |
| 15e | Conformational | 3.7 | 1.2 | >20 | |
| IgG1B12 | Conformational | 6.6 | 0.02 | 1.3 | |
| sCD4 | CD4bs | 0.28 | 0.11 | 5.2 | |
| CD4i | 4.8D | Conformational | 8.6 | 1.0 | >20 |
| 17b | Conformational | 9.4 | 5.0 | — | |
| gp41 | 2F5 | ELDKWA | 7.5 | 5.0 | — |
| Other | 2G12 | Unknown | 7.5 | >50 | — |
Role of V3 sequence in sensitivity to neutralization by MAb.
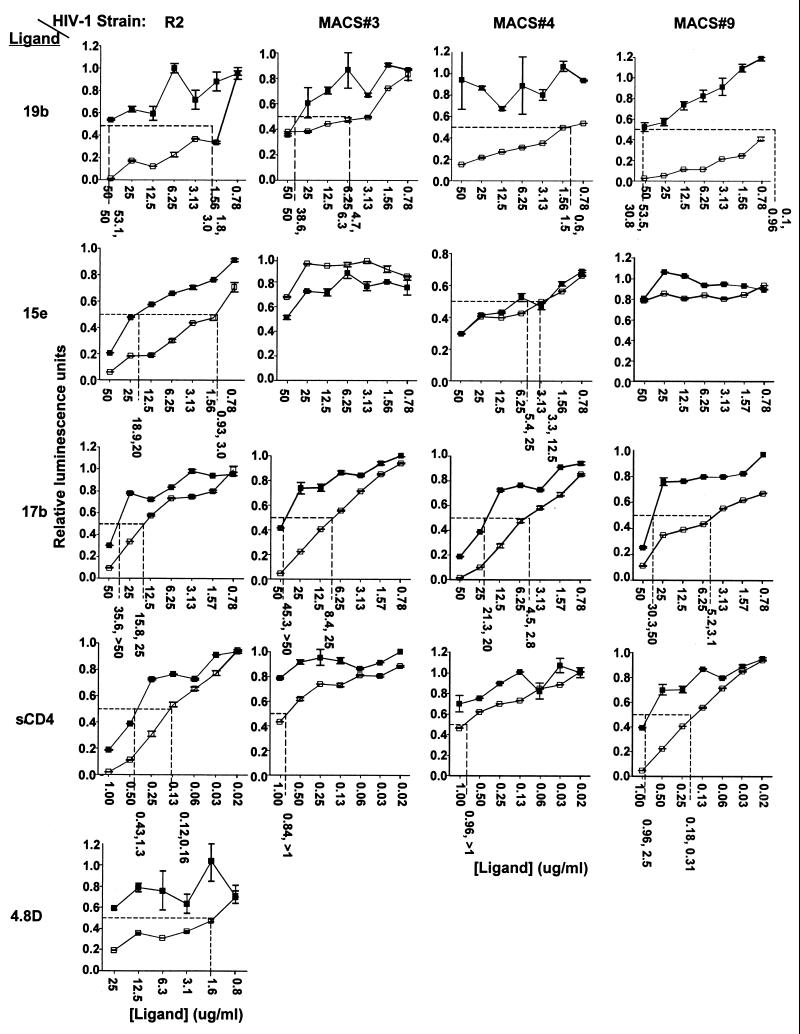
The R2 strain has a very unusual mutation, 313-4 PM, that lies within the discontinuous cluster of amino acids near the apex of the V3 loop that comprise the epitope recognized by the MAb 19b (35, 38). To test whether this mutation contributes to the sensitivity to neutralization of R2 by this MAb, the effect of substitution of amino acids at these positions was evaluated. The consensus sequence residues, HI, were introduced into the R2 clone, and the PM sequence was introduced into three other primary clade B envelope genes, replacing HI or SI sequences (43). In each case the PM sequence was associated with significantly enhanced sensitivity to neutralization by the 19b MAb, as shown in Fig. 1 at left. This difference averaged 21.5-fold for the R2 strain and 8.1, >50, and >50-fold for MACS strains 3, 4, and 9, respectively.
FIG. 1.
Neutralization of viruses pseudotyped with the R2 or MACS strain 3, 4, or 9 envelopes with V3 residue 313-4 PM (□) or HI/SI (▪) sequences. The standard error for each data point is shown as error bars. A best-fit trend line was calculated for each series of data points using Microsoft Excel. The line equations were used to derive the 50% neutralizing concentration of each MAb or sCD4. The dashed lines in each graph indicate the 50% luminescence and neutralizing concentrations for each data series. The numbers shown below each vertical dashed line correspond to the IC50s obtained in the experiment shown and the same concentrations obtained in a similar experiment.
To test whether the effect of the PM sequence was limited to effects on neutralization by anti-V3 MAb, the sensitivity of the four pairs of mutants to neutralization at other sites was evaluated, as is also shown in Fig. 1. The sensitivity of R2 to neutralization by the MAbs 15e, 17b, and 4.8D and sCD4 was reduced significantly by substitution of the 313-4 PM sequence with HI. Introduction of the PM mutation into the other three strains did not detectably alter sensitivity to the 15e MAb (anti-CD4bs), and sensitivity to neutralization by sCD4 was only enhanced consistently in the case of MACS strain 9. The sensitivity of all three viruses to neutralization by the MAb 17b (anti-CD4i) was enhanced by the PM mutation. Thus, there was a more consistent effect on neutralization by MAb directed against epitopes associated with coreceptor interaction than against those associated with CD4 binding.
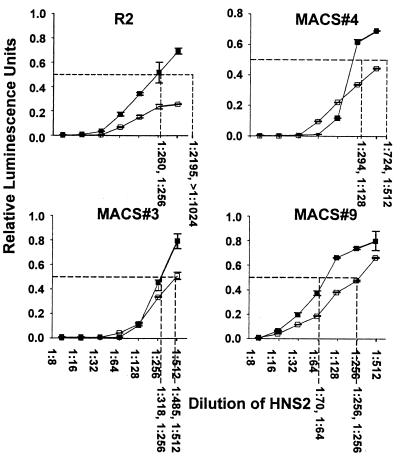
Effect of V3 loop mutation on neutralization by HNS2.
In view of the effects of the 313-4 PM mutation on sensitivity to neutralization by MAb and sCD4, it was of interest to determine if this mutation would also affect neutralization by human serum. Comparative neutralization of the four pairs of mutant clones by HNS2 is shown in Fig. 2. The neutralization of the R2 strain was consistently more than fourfold greater than that of R2 (313-4PM/HI). The neutralization of the other three strains was enhanced significantly in each case by introduction of the 313-4 PM sequence, although the magnitude of the effect in each case was less than in the R2 strain.
FIG. 2.
Neutralization by HNS2 of viruses pseudotyped with R2 or MACS strain 3, 4, or 9 envelope, each with the PM (□) or HI/SI (▪) sequence at residues 313 and 314. Error bars indicate the SD calculated for each data point. The 50% neutralization endpoints were calculated using methods similar to those described in the Fig. 1 legend. Values obtained from duplicate experiments are shown below each vertical dashed line.
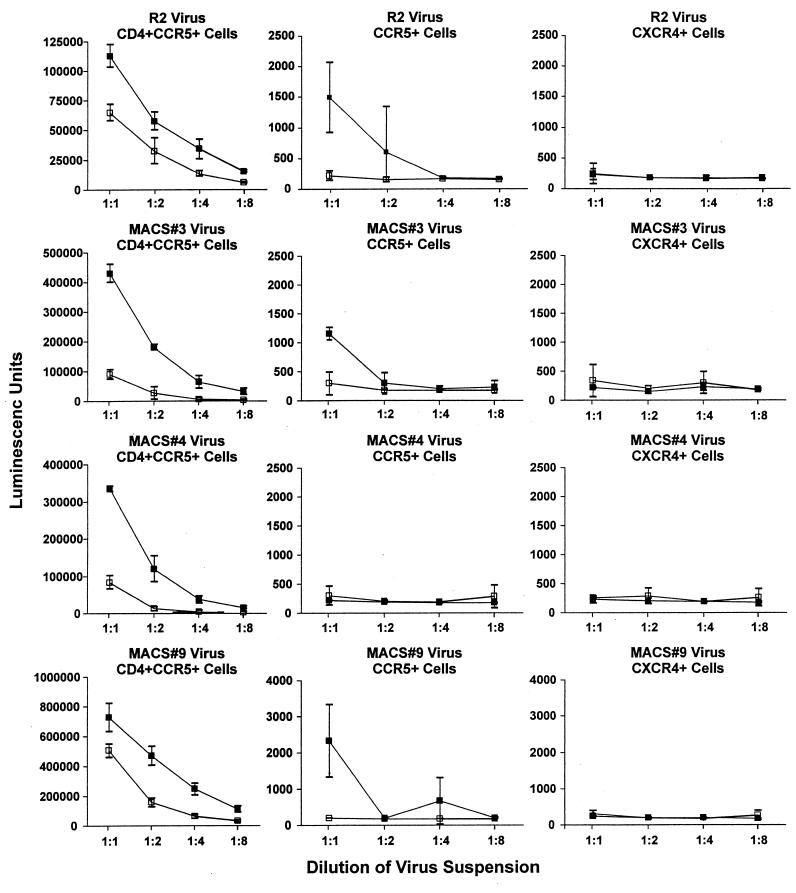
Effect of the 313-4 PM V3 mutation on infectivity.
During the course of preparation and characterization of the four pairs of mutant genes it was noted unexpectedly that the 313-4 PM mutation resulted in significantly enhanced infectivity of each clone. Since the mutation also caused sensitivity to neutralization by anti-CD4i MAb in the absence of CD4, the possibility that the mutation also resulted in CD4 independence of infectivity was evaluated. Results of comparative infectivity assays of the four pairs of clones are shown in Fig. 3. Infectivity of the R2 strain for HOS CD4+CCR5+ cells was reduced by approximately half by the HI mutation in the experiment shown and on average in five other experiments. Introduction of the 313-4 PM sequence enhanced infectivity of the MACS strains 3, 4, and 9 about eight-, eight-, and twofold, respectively, in repeated experiments. Infectivity of R2 and MACS strain 9 for HOS CCR5+ cells that did not express CD4 was increased significantly in each of three experiments by the presence of the 313-4 PM mutation. There was some evidence of infectivity of the MACS strain 3 (313-4HI/PM) clone for HOS CCR5+ cells in one experiment (the experiment shown), but not in two other experiments. Neither the MACS strain 4 clone nor its 313-4 PM mutant infected HOS CCR5+ cells in any of three experiments. As expected, none of the clones infected HOS CXCR4+ cells in any experiment.
FIG. 3.
Comparative infectivity of viruses pseudotyped with R2 and MACS strain 3, 4, and 9 envelopes, each with the PM (▪) or HI/SI (□) sequence at residues 313 and 314. Results are shown as means and SDs (error bars) for each data point.
V3 peptide blocking of HNS2 antibody-mediated neutralization of clone R2-pseudotyped virus.
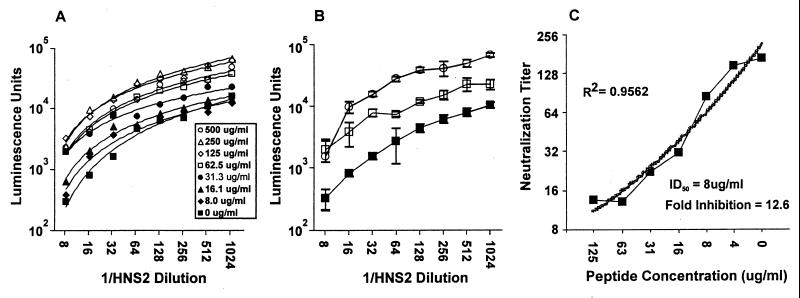
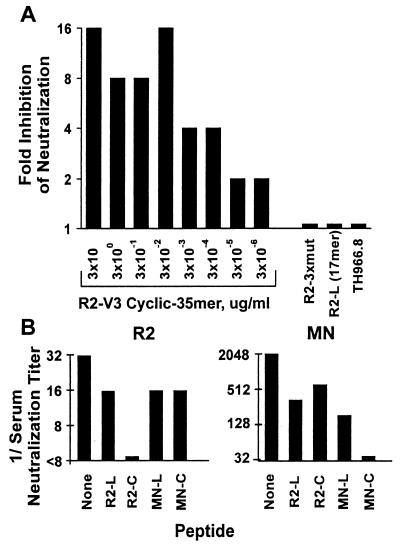
Peptide blocking studies were carried out to evaluate the possibility that anti-V3 antibodies might contribute to the cross-reactivity of HNS2 (6, 46). In preliminary studies it was found that addition of peptide to nonimmune serum did not cause the serum to enhance infectivity of virus pseudotyped with R2 envelope (not shown). The blocking effect of cyclized R2 V3 35-mer peptide on neutralization of R2 pseudovirus by HNS2 is shown in Fig. 4. Figure 4A illustrates the concentration-dependent inhibitory effects of increasing peptide concentrations on neutralization. Best-fit regression lines were essentially parallel, with maximum inhibitory effect being achieved at a peptide concentration of 250 μg/ml. The variance of individual data points is illustrated in Fig. 4B. Neutralization endpoints were calculated using the trend lines determined from the regression analyses shown in Fig. 4A. The relationship between neutralization titer and peptide concentration obtained from this analysis is shown in Fig. 4C. A highly significant correlation between peptide concentration and neutralization titer was obtained by regression analysis. There was 12.6-fold inhibition of neutralization at a peptide concentration of 250 μg/ml, and the IC50 of the peptide calculated by regression analysis was 8.0 μg/ml. Similar results were obtained in two identical experiments.
FIG. 4.
Inhibition of HNS2 neutralization of virus pseudotyped with R2 envelope by synthetic cyclized R2 V3 35-mer peptide. (A) The results shown indicate the average luminescence units obtained from triplicate determinations. The data points correspond to peptide concentrations, as indicated in the box insert. The lines shown are best-fit trend lines for each peptide concentration, which were calculated using Microsoft Excel. (B) The results shown indicate the means and SDs of luminescence units obtained in the absence (▪) and presence of peptide, 0.06 (□) and 0.5 (○) μg/ml, respectively. (C) The results shown were obtained when the trend lines shown in panel A were used to calculate 90% serum neutralizing concentrations corresponding to each concentration of peptide. The results are presented to show the relationship between peptide concentration and residual serum neutralizing activity. The best-fit trend line shown (heavy line) and the regression coefficient obtained (R2) were calculated in Microsoft Excel. The ID50 and fold inhibition at maximum peptide concentration were determined using the line equation for the best-fit line.
A number of peptides were compared to the cyclized R2 V3 peptide for capacity to inhibit neutralization of virus pseudotyped with the R2 strain envelope. In three repeat experiments, in which each peptide was tested at 20 μg/ml, there was no significant inhibition by the linear R2 17-mer peptide, the cyclized or linear MN peptides, or the R2 (313-4PM/HI, 325Q/D) peptide, while the cyclized R2 V3 35-mer peptide inhibited neutralization four- to eightfold in each experiment. Results of one of these experiments are shown in Fig. 5A. None of the R2-3x mutant (313-4PM, 325Q), 17-mer linear R2, or unrelated TH966.8 V3 peptides inhibited neutralization of R2 by HNS2 in any of these experiments. Reciprocal inhibition of neutralization of viruses pseudotyped with the R2 or MN envelopes by R2 and MN V3 peptides was tested. The MN peptides and pseudotyped virus were used in these comparisons, since that is the only peptide-virus combination for which substantial inhibition of neutralization has been reported. Results of one of these three experiments, all of which yielded similar results, are shown in Fig. 5B. Neutralization of the MN-TCLA strain was inhibited four- to eightfold by MN, cyclized R2 35-mer, and linear R2 17-mer V3 peptides. Thus, it appeared that the blocking effects of the cyclized R2 peptide depended on the specific sequence of the peptide used. The relative specificities of effects of the R2 and MN peptides in the reciprocal experiments suggest that the effect of the R2 peptide on neutralization of R2 by HNS2 is probably related to the specific antigenic structure of R2 envelope. The specificity of the inhibitory effect of the cyclized R2 V3 peptide was further evaluated by testing its capacity to inhibit neutralization by the MAbs 19b and 2F5. Neutralization of R2 by 19b was inhibited 16-fold, while neutralization by 2F5 was not inhibited at a peptide concentration of 20 μg/ml in repeated experiments (data not shown).
FIG. 5.
Specificity of V3 peptide blocking. (A) Inhibition of neutralization of virus pseudotyped with R2 envelope by R2-V3 cyclized 35-mer at various concentrations and absence of inhibition by R2-3xmut (313-4HI, 325D), R2-L(17-mer), and TH966.8 V3 peptides, each tested at 20 μg/ml. Fold inhibition of neutralization equal to one indicates no inhibition. Fold inhibition was determined by comparing the highest serum dilutions at which 50% neutralization was observed. (B) The neutralization titers of HNS2 against viruses pseudotyped with the R2 (left side) and MN-TCLA (right side) envelopes in the presence of various V3 peptides or no V3 peptide are shown. The peptides R2-L (17-mer), R2-C (cyclized R2-V3 35-mer), MN-L (MN linear), and MN-C (MN cyclized) were each used at 20 μg/ml. Serum neutralization titers were the highest dilutions at which there was ≥50% inhibition of neutralization.
Cyclized R2 V3 peptide inhibition of HNS2 neutralization of viruses pseudotyped with envelopes from heterologous donors.
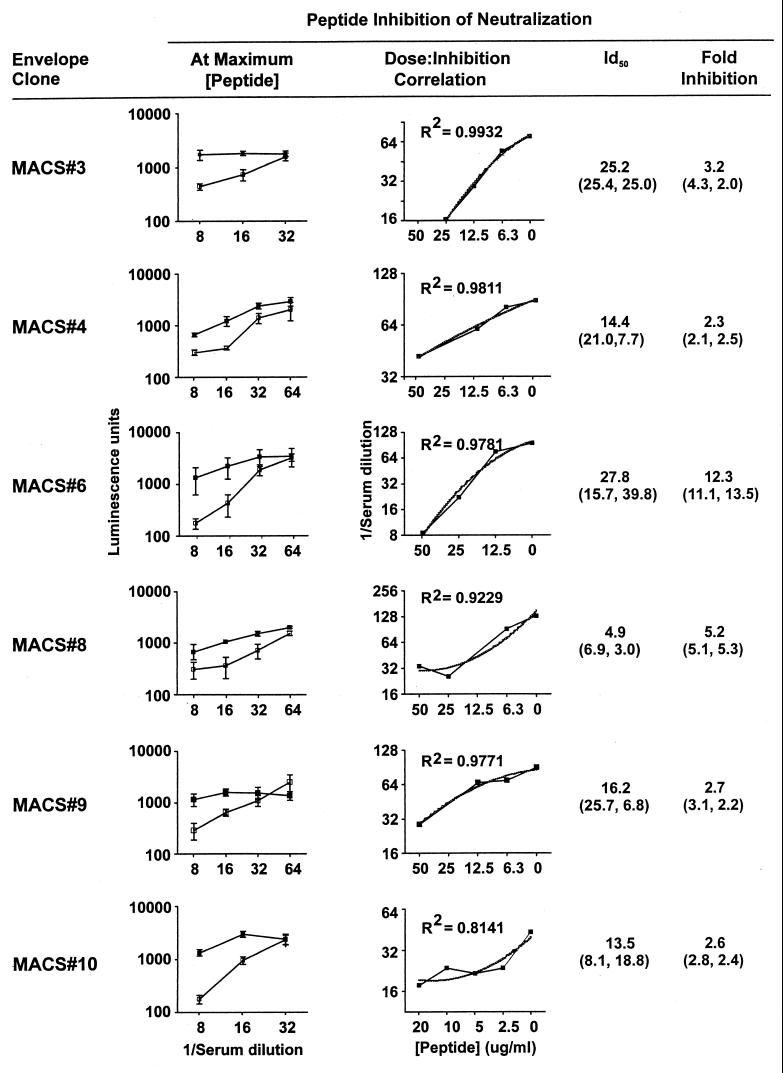
Inhibition by cyclized R2 V3 35-mer peptide of HNS2 neutralization of viruses pseudotyped with heterologous MACS donor envelopes was evaluated to determine whether anti-V3 antibody contributed to the neutralizing cross-reactivity of HNS2. Results of a representative experiment are shown in Fig. 6. The effects of various concentrations of R2 V3 35-mer were compared to those of a similar solution containing no V3 peptide. This solution was used as a negative control, in part because of previous reports demonstrating no effects of various V3 peptides on neutralization of common strains of HIV-1 and our results demonstrating no effects of other peptides on neutralization of R2 pseudotyped virus. More important in the choice of a negative control, however, was that the hypothesis being tested was that the R2 peptide would have similar effects on neutralization of R2 and other strains. Since other peptides did not affect R2 neutralization, their effects on viruses pseudotyped with other envelopes would not have had direct relevance to the hypothesis being tested. Peptide concentrations in twofold increments up to 50 μg/ml were tested. In each case, regression analyses yielded a family of essentially parallel curves (not shown) similar to those shown in Fig. 4A. The means and SDs of the luminescence results obtained in neutralization tests in the presence of 0 and 50 μg/ml of peptide are compared in Fig. 6, left, demonstrating that significant inhibitory effects were seen in each case. The correlations obtained by regression analyses between neutralization titers and peptide concentrations are shown in Fig. 6, center. The IC50s of peptide ranged from 4.9 to 27.8 μg/ml, and the fold inhibition observed ranged from 2.3 to 12.3. The results were consistent with the hypothesis that anti-V3 antibodies contribute to the neutralization of these strains by HNS2.
FIG. 6.
Inhibition of HNS2 neutralization of virus pseudotyped with MACS donor envelopes by synthetic cyclized R2 V3 35-mer peptide. (Left) The means and SDs (error bars) of luminescence results obtained in the absence (□) and presence (▪) of peptide, 50 or 20 μg/ml, are shown. Results were determined in triplicate. Variation is shown as SD. (Center) Serum neutralization endpoints, calculated as shown in Fig. 4, at the various peptide concentrations tested. Regression coefficients and trend lines are shown. (Right) The ID50s and fold inhibition at maximum peptide concentrations are shown in parentheses for the experiments shown and for duplicate experiments; the numbers preceding the parentheses are the averages of the parenthetical results.
Cyclized R2 V3 peptide inhibition of R2 neutralization by sera from MACS donors.
Inhibition of heterologous serum neutralization of pseudotyped R2 virus by cyclized R2 V3 35-mer peptide was evaluated to determine if cross-reactivity of these sera with R2 was mediated by anti-V3 antibodies. The comparative neutralization titers were determined for sera from the same six donors from the MACS described above against virus pseudotyped with the R2 envelope in the presence and absence of cyclized R2 V3 35-mer peptide. These sera have been described previously and have been shown to neutralize some heterologous primary HIV-1 clade B envelope viruses, but to a lesser extent than HNS2 (52). The neutralizing activity of each of the sera was consistently inhibited by the peptide. At 25 μg of peptide/ml the range of inhibition of neutralization titers ranged from 1-fold (no inhibition) to 4-fold (mean, 2.2-fold), while at 50 μg/ml the inhibition ranged from 2- to 4-fold (mean, 3.7-fold; results not shown). The peptide concentration required to achieve 50% inhibition of neutralization by these heterologous sera was about 5- to 10-fold higher than that required for 50% inhibition of homologous neutralization of R2 pseudotyped virus.
DISCUSSION
Here we report on a series of studies that were conducted to elucidate the basis for the neutralizing cross-reactivity of antibodies in the reference neutralizing human serum HNS2 and the primary HIV-1 envelope clone R2, which had been prepared previously from the donor of serum HNS2. Since the R2 envelope and HNS2 both display broad neutralizing cross-reactivity, we hypothesized that R2 expresses epitopes that are neutralization functional and immunogenic and that are responsible for at least some of the cross-reactivity of the HNS2 serum. Virus pseudotyped with the R2 envelope, in comparison to neutralization-sensitive and -resistant viruses, was moderately sensitive to neutralization by sCD4 and by MAb against conserved, cross-reactive epitopes near the CD4 and coreceptor binding sites and on gp41 and was highly sensitive to neutralization by MAbs 19b and 694/98-D, directed against conformation-sensitive epitopes at the apex of the V3 loop. Since the discontinuous sequence of amino acids recognized by the 19b MAb spans a region that includes a rare V3 sequence in the R2 clone, we tested whether this mutation might account for the sensitivity of the clone to this MAb. Substitution of consensus sequence amino acids for the two unusual residues (313-4PM/HI) resulted in not only substantially reduced sensitivity to neutralization by 19b but also reduced sensitivity to neutralization by anti-CD4bs and anti-CD4i MAbs and sCD4. Introduction of the reverse mutation into three other primary virus envelopes had the reverse effect on sensitivity to neutralization by 19b and anti-CD4i MAb in all three cases and on sensitivity to sCD4 in the third case. Finally, the 313-4 PM mutation caused increased infectivity of all four primary virus envelopes for cells expressing CD4 and CCR5 and caused R2 and one of the three heterologous envelopes to be capable of CD4-independent infection.
The possibility that this neutralization sensitivity or high infectivity phenotype of the R2 envelope was related to the immune response of the donor of R2 and HNS2 was supported by results of experiments evaluating the blocking effects of cyclized R2 V3 peptide on neutralization. Our experiments with the cyclized R2 V3 35-mer peptide were aimed at evaluating its ability to competitively inhibit the neutralizing activity of HNS2. Here we have shown that a significant portion of the neutralizing activity of that serum is blocked by the synthetic R2 V3 35-mer peptide. The magnitude of the anti-V3 response found in this donor was significantly greater than in the other donors tested, and the antibody response to this epitope appeared to contribute to the neutralizing cross-reactivity of HNS2. The lack of a direct effect of the cyclized R2 V3 35-mer peptide, and of mixtures of neutralizing sera with various other control peptides, on infectivity or neutralization of R2-pseudotyped virus supports the interpretation that the inhibitory effect of R2 V3 peptide probably indicates the presence of anti-V3 neutralizing antibodies. Overall, we interpret our results as indicating the possibility that the moderate global neutralization sensitivity, partially CD4-independent infectivity phenotype associated with the 313-4 PM mutation in the R2 clone may have enhanced the immunogenicity of conserved neutralization epitopes that are functional on primary isolates of HIV-1.
The MAbs IgG1b12, 2G12, and 2F5 neutralize some primary virus isolates (40). There have also been conflicting reports regarding neutralization of primary isolates by various anti-V3 MAbs (8, 10, 11, 35). In general, other MAbs have not displayed significant neutralizing cross-reactivity among primary virus isolates, even though a number bind to virions with significant cross-reactivity (50). Further, differential sensitivity to neutralization by sCD4 also distinguishes primary and laboratory strains of HIV-1 (9). TCLA laboratory strains of HIV-1 are generally sensitive to neutralization by sCD4 at concentrations of around 0.1 μg/ml, while primary isolates are commonly neutralized only by concentrations approaching 10 μg/ml or higher. We found the R2 envelope to be highly sensitive to neutralization by the 19b and 694/98-D antibodies, moderately sensitive to the IgG1b12, 2G12, and 2F5 antibodies, and highly sensitive to sCD4 in comparison to the MN-P strain (Table 1), three other primary virus isolate envelopes (Fig. 1), and other primary virus isolates specified in published reports, but less sensitive than the highly sensitive MN-TCLA strain. The R2 envelope’s sensitivity to neutralization by the antibodies 15e (CD4bs epitope), 4.8D, and 17b (both with CD4i epitopes) also identified it as being moderately sensitive to neutralization.
The nucleotide and predicted amino acid sequences of the R2 gene are characteristic of clade B envelopes, and the protein is predicted to be normally glycosylated (43). However, the V3 amino acid sequence of clone R2 is unique compared to other V3 sequences available to date. The complete amino acid sequence of the V3 loop (residues 301 to 335) is CSRPNNNTRKSIPMGPGRAFYTTGQIIGDIRQAHC. The PM sequence at residues 313 and 314 appears in 1.5% of the clade B V3 sequences listed in The Human Retroviruses and AIDS 1999 Compendium (http://phage.lanl.gov:80/HTML/98compendium.html), and the combination of the 313-4 PM and the Q at residue 325 appears in only 0.2%. Other substitutions that distinguish this clone’s V3 region from the clade B consensus sequence include serine for threonine at residue 302 and arginine for glutamine at residue 318. The 313-4 PM residues precede the GPGRAF sequence at the apex of the V3 loop, which is highly conserved in North American clade B strains. A double turn is a reported feature at the apex of the V3 loop, involving the proline normally present at the position corresponding to residue 316 of the R2 V3 loop (17). This double turn is involved in the formation of the epitope recognized by 19b, which includes the residues I310 and G315 that flank the 313-4 PM mutation in R2. Proline residues often have significant consequences regarding local secondary structure (31, 33). Variations in residues flanking the tip of the V3 loop are known to affect conformation (53, 54). The synthetic R2 V3 35-mer peptide was much less soluble in aqueous solution than other peptides, consistent with the possibility that the conformation of the peptide was significantly altered by the 313-4PM/HI and/or 325Q/D mutation. With these considerations in mind, it was reasonably likely that the 313-4 PM sequence could have significant consequences with respect to conformation of the neutralization epitope at the apex of V3, even though the PM sequence is not required for binding by the 19b MAb (17).
The global neutralization resistance phenotype that is characteristic of primary isolates of HIV-1 is associated with enhanced infectivity (37–39) and is further evidenced by comparison of the infectivity of the MN-P and MN-TCLA clones described in this report. The MN-P clone is approximately 250- to 500-fold more infectious (results not shown) and about 250- to 500-fold more resistant to neutralization by HNS2 than the MN-TCLA clone. Studies aimed at addressing the basis for this relationship are in progress, but it seems likely, based on published data, that the neutralization resistance and enhanced infectivity are both dependent upon the efficiency of steps leading to fusion after receptor and coreceptor engagement by the HIV-1 envelope (37–39). In relation to the global neutralization resistance-high infectivity phenotype, the combination of high infectivity and neutralization sensitivity resulting from the 313-4 PM mutation in R2 is paradoxical. We suggest that the effect of the 313-4 PM sequence on infectivity of R2 may be related to competence for coreceptor interaction. The findings of enhanced sensitivity to neutralization by MAbs directed against epitopes involved in coreceptor interaction and of sensitivity to neutralization at these epitopes in the absence of CD4 are consistent with such a possibility. The association of neutralization sensitivity with increased infectivity found with the R2 envelope may be essential in maintaining the neutralization sensitivity phenotype if it is the basis for enhanced capacity to induce cross-reactive neutralizing antibodies.
It has been hypothesized that neutralization of primary isolates of HIV-1 may depend on the presence of antibodies against epitopes that are not normally exposed on the surface of the virus until after interaction of the envelope with CD4 (27). Efforts to enhance the immunogenicity of HIV-1 envelopes with respect to capacity to induce primary virus-neutralizing antibodies have included removal of variable loop sequences and mutation of predicted N-linked glycosylation sites, based on the hypothesis that these structures may cloak important neutralization epitopes (32, 45). Alternative approaches to enhance exposure of neutralization epitopes have included the selection of CD4-independent envelopes and envelope-CD4 complexes (23, 27). A mutation that confers a CD4-independent infectivity phenotype has been shown to cause enhanced sensitivity to neutralization and exposure of a CD4-induced epitope, consistent with the possibility that this phenotype may be relevant to immunogenicity of epitopes involved in coreceptor interaction (13). An intriguing possibility is that the neutralization sensitivity-infectivity phenotype of the R2 envelope that is dependent on the 313-4 PM sequence may account for the primary virus cross-reactive neutralizing activity of HNS2, based on a similar principle of exposure of specific neutralization epitopes. In addition, our results using R2 V3 35-mer peptide as a blocking reagent are consistent with the possibility of enhanced immunogenicity of the R2 envelope at certain epitopes, because a substantial part of the homologous neutralizing activity of HNS2 and some of its heterologous neutralizing activity appeared to be directed against V3, and the magnitude of the anti-V3 neutralization against the primary viruses tested appeared to be greater in HNS2 than in other sera from HIV-1-infected donors. Others have suggested the possibility that conformation-dependent neutralizing antibodies against V3 may contribute to HIV-1 neutralization (47).
Finally, primary HIV-1 strains with CD4-independent infectivity have not been found previously, although primary strains of HIV-2 and simian immunodeficiency virus are commonly found to be competent for CD4-independent infectivity (12, 14). The CD4 independence of a clone of HIV-1 developed by in vitro selection has been found to depend upon the interactions of mutated residues in the proximal and distal limbs of V3 with other residues in the envelope complex (26). Further, this CD4 independence was also associated with neutralization sensitivity. Thus, our results confirm that the phenomenon represented by this in vitro selection process can occur in nature. An intriguing possibility that merits additional study is that an enhanced capacity for induction of primary virus cross-reactive neutralizing antibodies, which may result from mutations such as those we have studied here, could be associated with the reduced pathogenicity that distinguishes naturally occurring HIV-2 and simian immunodeficiency virus infections from HIV-1 infections. The capacity of a natural infection to induce an immune response that can attenuate the progress of infection is generally an indication of the potential for successful immunization (42).
Acknowledgments
We are grateful to Xi Chen for technical assistance and for the expert assistance and advice of Karen Wolcott of the Biomedical Instrumentation Center, Uniformed Services University of the Health Sciences.
This work was supported by NIH grants RO1-AI37438, AI44339, AI 32424, HL AI/HL 36085, HL 59725, UO1-AI-35042, AI24030, and MO1-RR-00772 (GCRC); USUHS grant RO87EZ; and research funds from the Department of Veterans Affairs supporting the Research Center for AIDS and HIV Infection and a Merit Review Award.
REFERENCES
- 1.Alter, H. J., J. S. Epstein, S. G. Swenson, M. J. VanRaden, J. W. Ward, R. A. Kaslow, J. E. Menitove, H. G. Klein, S. G. Sandler, and M. H. Sayers. 1990. Prevalence of human immunodeficiency virus type 1 p24 antigen in U.S. blood donors—an assessment of the efficacy of testing in donor screening. The HIV-Antigen Study Group. N. Engl. J. Med. 323:1312–1317. [DOI] [PubMed] [Google Scholar]
- 2.Bagley, J., P. J. Dillon, C. Rosen, J. Robinson, J. Sodroski, and W. A. Marasco. 1994. Structural characterization of broadly neutralizing human monoclonal antibodies against the CD4 binding site of HIV-1 gp120. Mol. Immunol. 31:1149–1160. [DOI] [PubMed] [Google Scholar]
- 3.Boots, L. J., P. M. McKenna, B. A. Arnold, P. M. Keller, M. K. Gorny, S. Zolla-Pazner, J. E. Robinson, and A. J. Conley. 1997. Anti-human immunodeficiency virus type 1 human monoclonal antibodies that bind discontinuous epitopes in the viral glycoproteins can identify mimotopes from recombinant phage peptide display libraries. AIDS Res. Hum. Retrovir. 13:1549–1559. [DOI] [PubMed] [Google Scholar]
- 4.Buchacher, A., R. Predl, K. Strutzenberger, W. Steinfellner, A. Trkola, M. Purtscher, G. Gruber, C. Tauer, F. Steindl, and A. Jungbauer. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retrovir. 10:359–369. [DOI] [PubMed] [Google Scholar]
- 5.Burton, D. R., C. F. Barbas, M. A. Persson, S. Koenig, R. M. Chanock, and R. A. Lerner. 1991. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc. Natl. Acad. Sci. USA 88:10134–10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carrow, E. W., L. K. Vujcic, W. L. Glass, K. B. Seamon, S. C. Rastogi, R. M. Hendry, R. Boulos, N. Nzila, and G. V. Quinnan, Jr. 1991. High prevalence of antibodies to the gp120 V3 region principal neutralizing determinant of HIV-1MN in sera from Africa and the Americas. AIDS Res. Hum. Retrovir. 7:831–838. [DOI] [PubMed] [Google Scholar]
- 7.Catasti, P., J. D. Fontenot, E. M. Bradbury, and G. Gupta. 1995. Local and global structural properties of the HIV-MN V3 loop. J. Biol. Chem. 270:2224–2232. [DOI] [PubMed] [Google Scholar]
- 8.Conley, A. J., M. K. Gorny, J. A. Kessler, L. J. Boots, M. Ossorio-Castro, S. Koenig, D. W. Lineberger, E. A. Emini, C. Williams, and S. Zolla-Pazner. 1994. Neutralization of primary human immunodeficiency virus type 1 isolates by the broadly reactive anti-V3 monoclonal antibody, 447-52D. J. Virol. 68:6994–7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daar, E. S., X. L. Li, T. Moudgil, and D. D. Ho. 1990. High concentrations of recombinant soluble CD4 are required to neutralize primary human immunodeficiency virus type 1 isolates. Proc. Natl. Acad. Sci. USA 87:6574–6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Souza, M. P., D. Livnat, J. A. Bradac, and S. H. Bridges. 1997. Evaluation of monoclonal antibodies to human immunodeficiency virus type 1 primary isolates by neutralization assays: performance criteria for selecting candidate antibodies for clinical trials. J. Infect. Dis. 175:1056–1062. [DOI] [PubMed] [Google Scholar]
- 11.D’Souza, M. P., P. Durda, C. V. Hanson, and G. Milman. 1991. Evaluation of monoclonal antibodies to HIV-1 by neutralization and serological assays: an international collaboration. AIDS 5:1061–1070. [DOI] [PubMed] [Google Scholar]
- 12.Edinger, A. L., C. Blanpain, K. J. Kunstman, S. M. Wolinsky, M. Parmentier, and R. W. Doms. 1999. Functional dissection of CCR5 coreceptor function through the use of CD4-independent simian immunodeficiency virus strains. J. Virol. 73:4062–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards, T. G., T. L. Hoffman, F. Baribaud, S. Wyss, C. C. LaBranche, J. Romano, J. Adkinson, M. Sharron, J. A. Hoxie, and R. W. Doms. 2001. Relationships between CD4 independence, neutralization sensitivity, and exposure of a CD4-induced epitope in a human immunodeficiency virus type 1 envelope protein. J. Virol. 75:5230–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Endres, M. J., P. R. Clapham, M. Marsh, M. Ahuja, J. D. Turner, A. McKnight, J. F. Thomas, B. Stoebenau-Haggarty, S. Choe, P. J. Vance, T. N. Wells, C. A. Power, S. S. Sutterwala, R. W. Doms, N. R. Landau, and J. A. Hoxie. 1996. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell 87:745–756. [DOI] [PubMed] [Google Scholar]
- 15.Forthal, D. N., G. Landucci, M. K. Gorny, S. Zolla-Pazner, and W. E. J. Robinson. 1995. Functional activities of 20 human immunodeficiency virus type 1 (HIV-1)-specific human monoclonal antibodies. AIDS Res. Hum. Retrovir. 11:1095–1099. [DOI] [PubMed] [Google Scholar]
- 16.Fouts, T. R., J. M. Binley, A. Trkola, J. E. Robinson, and J. P. Moore. 1997. Neutralization of the human immunodeficiency virus type 1 primary isolate JR-FL by human monoclonal antibodies correlates with antibody binding to the oligomeric form of the envelope glycoprotein complex. J. Virol. 71:2779–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghiara, J. B., D. C. Ferguson, A. C. Satterthwait, H. J. Dyson, and I. A. Wilson. 1997. Structure-based design of a constrained peptide mimic of the HIV-1 V3 loop neutralization site. J. Mol. Biol. 266:31–39. [DOI] [PubMed] [Google Scholar]
- 18.Gorny, M. K., A. J. Conley, S. Karwowska, A. Buchbinder, J. Y. Xu, E. A. Emini, S. Koenig, and S. Zolla-Pazner. 1992. Neutralization of diverse human immunodeficiency virus type 1 variants by an anti-V3 human monoclonal antibody. J. Virol. 66:7538–7542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorny, M. K., J. R. Mascola, Z. R. Israel, T. C. VanCott, C. Williams, P. Balfe, C. Hioe, S. Brodine, S. Burda, and S. Zolla-Pazner. 1998. A human monoclonal antibody specific for the V3 loop of HIV type 1 clade E cross-reacts with other HIV type 1 clades. AIDS Res. Hum. Retrovir. 14:213–221. [DOI] [PubMed] [Google Scholar]
- 20.Gorny, M. K., T. C. VanCott, C. Hioe, Z. R. Israel, N. L. Michael, A. J. Conley, C. Williams, J. A. Kessler, P. Chigurupati, S. Burda, and S. Zolla-Pazner. 1997. Human monoclonal antibodies to the V3 loop of HIV-1 with intra- and interclade cross-reactivity. Rev. Infect. Dis. 159:5114–5122. [PubMed] [Google Scholar]
- 21.Gorny, M. K., J. Y. Xu, S. Karwowska, A. Buchbinder, and S. Zolla-Pazner. 1993. Repertoire of neutralizing human monoclonal antibodies specific for the V3 domain of HIV-1 gp120. Rev. Infect. Dis. 150:635–643. [PubMed] [Google Scholar]
- 22.Hioe, C. E., S. Xu, P. Chigurupati, S. Burda, C. Williams, M. K. Gorny, and S. Zolla-Pazner. 1997. Neutralization of HIV-1 primary isolates by polyclonal and monoclonal human antibodies. Int. Immunol. 9:1281–1290. [DOI] [PubMed] [Google Scholar]
- 23.Hoxie, J. A., C. C. LaBranche, M. J. Endres, J. D. Turner, J. F. Berson, R. W. Doms, and T. J. Matthews. 1998. CD4-independent utilization of the CXCR4 chemokine receptor by HIV-1 and HIV-2. J. Reprod. Immunol. 41:197–211. [DOI] [PubMed] [Google Scholar]
- 24.Kaslow, R. A., D. G. Ostrow, R. Detels, J. P. Phair, B. F. Polk, and C. R. Rinaldo, Jr. 1987. The Multicenter AIDS Cohort Study: rationale, organization, and selected characteristics of the participants. Am. J. Epidemiol. 126:310–318. [DOI] [PubMed] [Google Scholar]
- 25.Laal, S., S. Burda, M. K. Gorny, S. Karwowska, A. Buchbinder, and S. Zolla-Pazner. 1994. Synergistic neutralization of human immunodeficiency virus type 1 by combinations of human monoclonal antibodies. J. Virol. 68:4001–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaBranche, C. C., T. L. Hoffman, J. Romano, B. S. Haggarty, T. G. Edwards, T. J. Matthews, R. W. Doms, and J. A. Hoxie. 1999. Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions required for coreceptor specificity. J. Virol. 73:10310–10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaCasse, R. A., K. E. Follis, M. Trahey, J. D. Scarborough, D. R. Littman, and J. H. Nunberg. 1999. Fusion-competent vaccines: broad neutralization of primary isolates of HIV. Science 283:357–362. [DOI] [PubMed] [Google Scholar]
- 28.Laman, J. D., M. M. Schellekens, Y. H. Abacioglu, G. K. Lewis, M. Tersmette, R. A. Fouchier, J. P. Langedijk, E. Claassen, and W. J. Boersma. 1992. Variant-specific monoclonal and group-specific polyclonal human immunodeficiency virus type 1 neutralizing antibodies raised with synthetic peptides from the gp120 third variable domain. J. Virol. 66:1823–1831. (Erratum, 66:5175.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Letvin, N. L. 1998. Progress in the development of an HIV-1 vaccine. Science 280:1875–1880. [DOI] [PubMed] [Google Scholar]
- 30.Li, A., H. Katinger, M. R. Posner, L. Cavacini, S. Zolla-Pazner, M. K. Gorny, J. Sodroski, T. C. Chou, T. W. Baba, and R. M. Ruprecht. 1998. Synergistic neutralization of simian-human immunodeficiency virus SHIV-vpu+ by triple and quadruple combinations of human monoclonal antibodies and high-titer anti-human immunodeficiency virus type 1 immunoglobulins. J. Virol. 72:3235–3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu, M., and P. S. Kim. 1997. A trimeric structural subdomain of the HIV-1 transmembrane glycoprotein. J. Biomol. Struct. Dyn. 15:46–71. [DOI] [PubMed] [Google Scholar]
- 32.Ly, A., and L. Stamatatos. 2000. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J. Virol. 74:6769–6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacArthur, M. W., and J. M. Thornton. 1991. Influence of proline residues on protein conformation. J. Mol. Biol. 218:397–412. [DOI] [PubMed] [Google Scholar]
- 34.Mascola, J. R., M. K. Louder, S. R. Surman, T. C. VanCott, X. F. Yu, J. Bradac, K. R. Porter, K. E. Nelson, M. Girard, J. G. McNeil, F. E. McCutchan, D. L. Birx, and D. S. Burke. 1996. Human immunodeficiency virus type 1 neutralizing antibody serotyping using serum pools and an infectivity reduction assay. AIDS Res. Hum. Retrovir. 12:1319–1328. [DOI] [PubMed] [Google Scholar]
- 35.Moore, J. P., A. Trkola, B. Korber, L. J. Boots, J. A. Kessler, F. E. McCutchan, J. Mascola, D. D. Ho, J. Robinson, and A. J. Conley. 1995. A human monoclonal antibody to a complex epitope in the V3 region of gp120 of human immunodeficiency virus type 1 has broad reactivity within and outside clade B. J. Virol. 69:122–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nyambi, P. N., M. K. Gorny, L. Bastiani, G. van der Groen, C. Williams, and S. Zolla-Pazner. 1998. Mapping of epitopes exposed on intact human immunodeficiency virus type 1 (HIV-1) virions: a new strategy for studying the immunologic relatedness of HIV-1. J. Virol. 72:9384–9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park, E. J., and G. V. Quinnan, Jr. 1999. Both neutralization resistance and high infectivity phenotypes are caused by mutations of interacting residues in the human immunodeficiency virus type 1 gp41 leucine zipper and the gp120 receptor- and coreceptor-binding domains. J. Virol. 73:5707–5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Park, E. J., L. K. Vujcic, R. Anand, T. S. Theodore, and G. V. Quinnan, Jr. 1998. Mutations in both gp120 and gp41 are responsible for the broad neutralization resistance of variant human immunodeficiency virus type 1 MN to antibodies directed at V3 and non-V3 epitopes. J. Virol. 72:7099–7107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Park, E. J., S. Zolla-Pazner, and G. V. Quinnan, Jr. 2000. Distinct mechanisms mediating enhanced infectivity and envelope conformational change determine global neutralization resistance phenotype of human immunodeficiency virus type 1. J. Virol. 74:4183–4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Parren, P. W. H. I., M. Wang, A. Trkola, J. M. Binley, M. Purtscher, H. Katinger, J. P. Moore, and D. R. Burton. 1998. Antibody neutralization-resistant primary isolates of human immunodeficiency virus type 1. J. Virol. 72:10270–10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Posner, M. R., L. A. Cavacini, C. L. Emes, J. Power, and R. Byrn. 1993. Neutralization of HIV-1 by F105, a human monoclonal antibody to the CD4 binding site of gp120. J. Acquir. Immune Defic. Syndr. 6:7–14. [PubMed] [Google Scholar]
- 42.Quinnan, G. V. 1997. Immunization against viral diseases, p.791–834. In G. Galasso, R. Whitley, and T. C. Merigan (ed.), Antiviral agents and human viral diseases. Raven Press, New York, N.Y.
- 43.Quinnan, G. V., P. F. Zhang, D. W. Fu, M. Dong, and H. J. Alter. 1999. Expression and characterization of HIV type 1 envelope protein associated with a broadly reactive neutralizing antibody response. AIDS Res. Hum. Retrovir. 15:561–570. [DOI] [PubMed] [Google Scholar]
- 44.Quinnan, G. V., P. F. Zhang, D. W. Fu, M. Dong, and J. B. Margolick. 1998. Evolution of neutralizing antibody response against HIV type 1 virions and pseudovirions in multicenter AIDS cohort study participants. AIDS Res. Hum. Retrovir. 14:939–949. [DOI] [PubMed] [Google Scholar]
- 45.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679–684. [DOI] [PubMed] [Google Scholar]
- 46.Spear, G. T., D. M. Takefman, S. Sharpe, M. Ghassemi, and S. Zolla-Pazner. 1994. Antibodies to the HIV-1 V3 loop in serum from infected persons contribute a major proportion of immune effector functions including complement activation, antibody binding, and neutralization. Virology 204:609–615. [DOI] [PubMed] [Google Scholar]
- 47.Steimer, K. S., C. J. Scandella, P. V. Skiles, and N. L. Haigwood. 1991. Neutralization of divergent HIV-1 isolates by conformation-dependent human antibodies to Gp120. Science 254:105–108. [DOI] [PubMed] [Google Scholar]
- 48.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vujcic, L. K., and G. V. Quinnan, Jr. 1995. Preparation and characterization of human HIV type 1 neutralizing reference sera. AIDS Res. Hum. Retrovir. 11:783–787. [DOI] [PubMed] [Google Scholar]
- 50.York, J., K. E. Follis, M. Trahey, P. N. Nyambi, S. Zolla-Pazner, and J. H. Nunberg. 2001. Antibody binding and neutralization of primary and T-cell line-adapted isolates of human immunodeficiency virus type 1. J. Virol. 75:2741–2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng, W., P. O. Regamey, K. Rose, Y. Wang, and E. Bayer. 1997. Use of Fmoc-N-(2-hydroxy-4-methoxybenzyl)amino acids in peptide synthesis. J. Peptide Res. 49:273–279. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, P. F., X. Chen, D. W. Fu, J. B. Margolick, and G. V. Quinnan, Jr. 1999. Primary virus envelope cross-reactivity of the broadening neutralizing antibody response during early chronic human immunodeficiency virus type 1 infection. J. Virol. 73:5225–5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zvi, A., D. J. Feigelson, Y. Hayek, and J. Anglister. 1997. Conformation of the principal neutralizing determinant of human immunodeficiency virus type 1 in complex with an anti-gp120 virus neutralizing antibody studied by two-dimensional nuclear magnetic resonance difference spectroscopy. Biochemistry 36:8619–8627. [DOI] [PubMed] [Google Scholar]
- 54.Zvi, A., I. Kustanovich, Y. Hayek, S. Matsushita, and J. Anglister. 1995. The principal neutralizing determinant of HIV-1 located in V3 of gp120 forms a 12-residue loop by internal hydrophobic interactions. FEBS Lett. 368:267–270. [DOI] [PubMed] [Google Scholar]